Abstract
Purpose
To investigate the molecular composition of subretinal fluid (SRF) in central serous chorioretinopathy (CSCR) and rhegmatogenous retinal detachment (RRD) using proteomics and metabolomics.
Methods
SRF was obtained from one patient with severe nonresolving bullous CSCR requiring surgical subretinal fibrin removal, and two patients with long-standing RRD. Proteins were trypsin-digested, labeled with Tandem-Mass-Tag and fractionated according to their isoelectric point for identification and quantification by tandem mass spectrometry. Independently, metabolites were extracted on cold methanol/ethanol, and identified by untargeted ultra-high performance liquid chromatography and high-resolution mass spectrometry. Bioinformatics analyses were conducted.
Results
In total, 291 proteins and 651 metabolites were identified in SRF samples. Compared with RRD, 128 proteins (77 downregulated; 51 upregulated) and 76 metabolites (43 downregulated; 33 upregulated) differed in the SRF from CSCR. Protein and metabolites notably deregulated in CSCR were related to glycolysis/gluconeogenesis, inflammation (including serum amyloid P component, versican), alternative complement pathway (complement factor H and complement factor H–related protein), cellular adhesion, biliary acid metabolism (farnesoid X receptor/retinoid X receptor), and gluco- and mineralocorticoid systems (aldosterone, angiotensin, and corticosteroid-binding globulin).
Conclusions
Proteomics and metabolomics can be performed on SRF. A unique SRF sample from CSCR exhibited a distinct molecular profile compared with RRD.
Translational Relevance
This first comparative multiomics analysis of SRF improved the understanding of CSCR and RRD pathophysiology. It identified pathways potentially involved in the better photoreceptor preservation in CSCR, suggesting neuroprotective targets that will require additional confirmation.
Keywords: subretinal space, retinal pigment epithelium, retinal detachment, retinal metabolism, vitreoretinal surgery
Introduction
Central serous chorioretinopathy (CSCR) is characterized by serous detachments of the neurosensory retina, frequently affecting the macula, focal pigment epithelial detachments, increased choroidal thickness and choroidal vascular hyperpermeability. Severe CSCR can present as bullous exudative retinal detachment with persistence of subretinal material,1 that may exceptionally require surgery.2 During the procedure, subretinal fluid (SRF) can be collected, but to the best of our knowledge, the composition of SRF from a CSCR patient has never been analyzed. Mechanisms of SRF accumulation in CSCR are still uncertain. Several theories have been proposed to explain fluid entry from the choroid toward the subretinal space: dilated and hyperpermeable choroidal vessels favoring trans- or interretinal pigment epithelium (RPE) entry flow,3 changes in RPE cell polarity altering hydroionic pumping direction,4 uni- or multifocal rupture of the RPE barrier,5 or active reverse flow by unknown triggering mechanisms.6
Although serous macular detachments form rapidly and last for 3 to 6 months in most cases,7 visual acuity is usually preserved, suggesting a good preservation of photoreceptors function and structure.8 This differs strikingly with macula-off rhegmatogenous retinal detachment (RRD), in which detachments lasting more than 3 to 5 days lead to irreversible visual impairment.9 Mechanisms of photoreceptor cell death after RRD are multiple, triggered by oxidative and metabolic stress, complement alternative pathway activation, immune response, and inflammation.10,11 In RRD, SRF originates mainly from liquefied vitreous, which diffuses through a retinal tear under the neuroretina. In response to neuroretinal detachment, RPE cells proliferate and migrate into the subretinal space,12 which may contribute to SRF accumulation. Whether SRF composition influences the differential photoreceptor survival and visual prognosis in different types of neuroretinal detachment is not known.
The aim of this study was to investigate the molecular composition of subretinal fluid in CSCR and RRD using proteomics and metabolomics. We compared the SRF profile from one case with severe CSCR and two cases with chronic RRD. Due to the rarity of SRF samples from CSCR, we opted for an untargeted proteomic approach, with independent cross-assessment by metabolomics analysis.
Methods
Study Subjects
This study involving human subjects adhered to the tenets of the Declaration of Helsinki, and was approved by the local Ethics Committee of the Swiss Department of Health on research involving human subjects (CER-VD N°340/15 and CER-VD N°19/15). Patients signed an informed consent. A 48-year-old male patient with persistent bullous CSCR underwent vitrectomy for subretinal fibrinous clot removal. Two patients underwent vitrectomy for long-standing macula-off RRD repair: an 82-year-old Caucasian female (Patient RRD-1), and a 58-year-old African male (Patient RRD-2). In all cases, 23-G trocars were inserted at the pars plana and SRF was collected using a back flush cannula connected to a syringe, through retinal tears in RRD, and through a retinotomy in the CSCR case, as described in the Results section. Samples were directly frozen and stored at −80°C in our institutional biobank, a platform similar to the biorepository described by Skeie at al.13 Coded samples and their associated clinical data were then sent to the laboratories in charge of the proteomics and metabolomics analyses. The CSCR patient underwent multimodal retinal imaging at regular intervals, consisting in spectral-domain optical coherence tomography (SD-OCT), fundus autofluorescence, fluorescein, and indocyanine green (ICG) angiography on Spectralis (Heidelberg Engineering, Heidelberg, Germany).
Proteomic Analysis
The proteome of the SRF was compared between samples from the CSCR patient and the two patients with chronic RRD. The total protein concentration in each sample was determined using a Bradford protein assay (Protein assay Dye reagent concentrate, Bio-Rad, Hercules, CA) according to the manufacturer's instructions.
Sample Preparation
The two control RRD samples were pooled to homogenize experimental conditions and filter small interindividual differences, while increasing the concentration of proteins or metabolites related to the pathogenic process, as previously described for the omics analysis of ocular fluids.14 In order to reduce the impact of potential blood contamination during sample collection, 100 μg of SRF from the CSCR patient and 100 μg of SRF pooled from the two RRD patients (a 1:1 vol/vol ratio) were filtered using a commercial resin according to the manufacturer's recommendations (Proteome Purify 12 Human Serum Protein Immunodepletion Resin; R&D Systems, Abingdon Science Park, UK) to deplete the classical top-12 most abundant serum proteins (alpha-1-acid glycoprotein, alpha-1-antitrypsin, alpha-2-macroglobulin, albumin, apolipoprotein A-I, apolipoprotein A-II, fibrinogen, haptoglobin, immunoglobulin (Ig)A, IgG, IgM, transferrin). Using the Bradford method, the total protein concentration after depletion was estimated at 9.5 μg/mL in the CSCR sample and 9.6 μg/mL in the pooled RRD samples. Subsequently, 5 μg of samples (CSCR and RRD) were reduced using 33 uL of 6-M urea (Merk, BoisÎle-de-France, France) and 2 uL of 50-mM tris-(2-carboxyethyl)phosphine (TCEP; Sigma-Aldrich, Lesquin, France). After incubation at 37°C during 1 hour, 1 μL of 400-mM iodoacetamid (Sigma-Aldrich) was added and samples were incubated for 30 minutes and 67 μL of 0.1-M triethylammonium bicarbonate buffer (TEAB; Sigma-Aldrich) were added before trypsin (1 μg/50 μg of proteins, porcine origin; Promega Corporation, Charbonnières-les-Bains, France) digestion overnight at 37°C. Samples were labeled with one of the 6 Tandem Mass Tag reagents (TMT; isobaric label reagent sets; ThermoFisher Scientific, Steinhausen, Switzerland) applying the Simultaneous Marker discovery And verification for the Rapid Translation of Exogeneous Reference material (SMARTER)-based approach,15 according to manufacturer's instructions. Briefly, tag 128 was attributed to pooled SRFs (used as clinical controls) from RRD patients and tag 130 to the case study CSCR. Tags 126 and 127 were used to label vitreous pool of RRD patients and commercial tears (HMTEARS; Seralab, West Sussex, UK) both used as biological controls. Finally, the 2 remaining tags (129 and 131) were used as internal technical controls. The total quantity of each labeled sample (30 μg) was pooled and dried in a speed vacuum.
Off-Gel Electrophoresis (OGE)
Previously dried samples were resuspended in 5% CAN and 0.1%FA and purified under Macrospin columns (Harvard Apparatus, Holliston, MA). A 3100 OFFGEL Fractionator (Agilent Technologies, Les Ulis, France) was then used to separate peptides according to their pI, as reported previously16 with a 13 cm IPG strip (immobiline Dry strip pH 3–10, 13 cm; GE Healthcare, Little Chalfont, UK) and 12 OGE wells. The focusing parameters were 20 Kvh, 800v, 50 uA, 200 mW, and 100 s. The hold parameters were 500 V, 20 uA, and 50 mW. After overnight fractionation, microspin columns (Harvard Apparatus) were performed according to manufacturer's recommendations and the 12 fractions were dried under speed vacuum.
Mass Spectrometry Analysis
For each fraction, 0.5 μg of sample was injected four times (final amount injected: 2 μg) and then analyzed in gas-phase fractionation (GPF4) mode17 by tandem mass spectrometry (MS) (liquid chromatography (LC)-MS/MS) using a LTQ Orbitrap Velos Pro (Thermo Fisher Instrument, Steinhaussen, Switzerland) coupled to a nanoflow high pressure liquid chromatography (HPLC, nanoaquity system; Waters, Milford, MA). LC-MS/MS analyses were performed as described elsewhere.18
Data Analyses
Peak lists and resulting files were searched against the UniProt-Swiss-Prot database (2014_10 version Homo sapiens taxonomy) using Phenyx 2.6 software (Gene Bio, Geneva, Switzerland). Variable amino acid modifications were oxidized methionine, glutamine, and asparagine deamidation, and TMT-labeled peptides amino terminus (+229.1629 Da if required). TMT-labeled lysine (if concerned) and carbamidomethylation of cysteines were set as fixed modification.18 Trypsin was selected as the enzyme, with one potential missed cleavage. Only proteins matching two different and unique peptide sequences were selected for identification and quantification. A false discovery rate of 1% was selected. Protein quantification was obtained from Mascat method in Easyprot (version 2.3) as statistical tools in order to calculate the proteins ratios between the CSCR SRF (tag 130) and the RRD SRFs (tag 128).19 Briefly, to obtain the ratio of a protein, Mascat computes the geometric mean of all peptide ratios linked to this protein. Proteins were considered as significantly different if the 130/128 ratio were above 1.50 or below 0.67, with a P value < 0.05.20 For more details about quantification and statistical criteria, please refer to the previously reported methods.19,20 The 12 proteins that were initially resin depleted were removed from the final lists, if identified.
Metabolomics Analysis
Similarly, independent untargeted metabolomics was performed to compare SRF metabolome in the CSCR patient with the two RRD patients. Metabolites were extracted from 50 μL of SRF samples using cold methanol/ethanol (1:1, vol/vol) in a 1:3 ratio.21
Untargeted Ultra-High Performance Liquid Chromatography (UHPLC) – High-Resolution Mass Spectrometry (HRMS) Analysis
For comparison of CSCR and RRD, two independent analyses were performed on two different days. These analyses were performed on UHPLC (UltiMate 3000 RSLCnano System; Thermo Scientific) hyphenated with HRMS (Q Exactive Plus MS; Thermo Scientific). The metabolites were separated by reversed phase chromatography on a Kinetex C18 (2.6 μm, 50 mm × 2.1 mm internal diameter [ID]) column (Phenomenex, Los Angeles, CA) using MeOH:H2O 0.1% formic acid solvent in a gradient elution mode with fixed flow rate at 0.3 mL/min. Quality controls (i.e., representative pool of samples) and internal standards were used to assess the over-batch repeatability. Data acquisition was performed in full scan mode in both negative and positive polarities considering suitable tuning methods.21 Subsequently, MS/MS spectra were acquired in a data-dependent acquisition mode. Resolutions were fixed at 70,000 and 17,000 for full scan and MS/MS acquisitions, respectively.
Chemometric Analysis
Raw UHPLC-HRMS data were converted to appropriate format to be processed by XCMS online software (The Scripps Research Institute, San Diego, CA) for peak detection, chromatogram alignment, and isotope annotation.22 Preprocessed data were normalized by using sample-wise mean normalization as well as feature-wise Pareto scaling. The assessment of the metabolic patterns was driven by the use of unsupervised and supervised learning approaches, including prediction analysis for microarrays data mining (PAM) into the R software (Version 3.3.0, R Foundation for Statistical Computing, R Core Team, 2016, Vienna, Austria. http://www.R-project.org) and the ‘pamr' package, as well as the online Metaboanalyst tool (http://www.metaboanalyst.ca/). PAM classifier gives the opportunity to keep only the subset of features that maximized the model performance in predicting class membership.23
Metabolite Identification
Differential metabolites were confirmed based on their retention time and MS2 fragmentation pattern21 using open-access libraries, including the HMDB database (www.hmdb.ca), lipidmaps (www.lipidmaps.org), metlin (http://metlin.scripps.edu/index.php), and mzcloud (www.mzcloud.org).24
Biological Process and Pathway Analysis
The Panther software (Protein Analysis Through Evolutionary Relationships, version 10.0; www.pantherdb.org) was used to analyze the protein lists in order to generate the gene ontology categories (protein class).25 The overrepresentation of the pathways in which the differential proteins of the CSCR SRF compared with the RRD SRF are involved was performed with QIAGEN's Ingenuity Pathway Analysis (IPA; QIAGEN Redwood City, CA).
Overrepresentation analysis on the metabolomics data was performed by Consensuspathdb (http://consensuspathdb.org) to obtain accurate insight into the underlying biology of differentially expressed metabolites exploiting the KEGG database (www.genome.jp/kegg/pathway.html). Enrichment set analysis was appreciated by 4150 identifications for human species included in the KEGG library. Cytoscape software (Version 3.2.1, http://cytoscape.org) and the MetScape plugin (Version 3.1.2; metscape.ncibi.org/) were used to build the compound-gene metabolic network.26 From the set of genes obtained in the compound-gene network, gene ontology terms were integrated and functionally organized in a gene ontology network function using the ClueGO plugin (Version 2.2.5; apps.cytoscape.org/apps/cluego).27
Results
Clinical History of the CSCR Patient
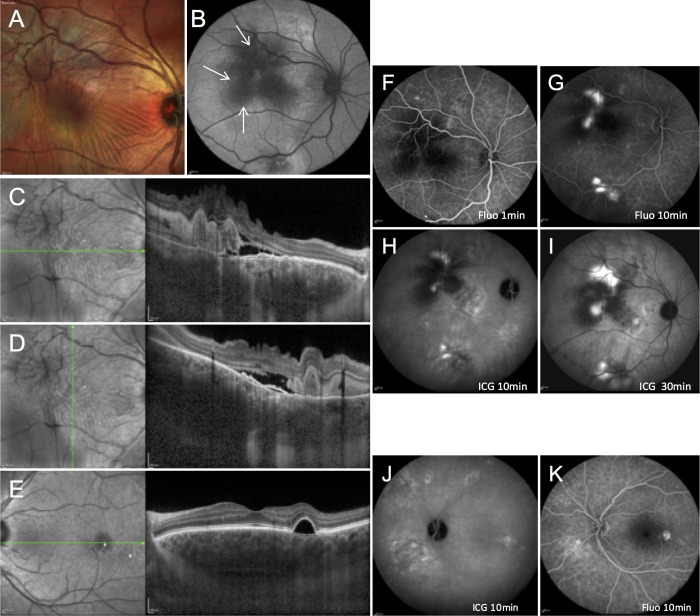
A 48-year-old male was referred with right eye vision loss for 3 months. He reported steroid nasal spray use for allergic rhinitis, and no remarkable medical history. Best-corrected visual acuity of his right eye was decreased to 20/200. No ocular inflammation was observed, and intraocular pressure was normal. Fundus examination showed a posterior serous retinal detachment with multiple retinal folds and presence of focal subretinal yellowish material (Fig. 1A). On fundus autofluorescence, extensive hyperautofluorescent gravitational tracks contrasted with central hypoautofluorescence due to subretinal material masking effect (Fig. 1B, arrows). SD-OCT showed a serous retinal detachment with retinal folds, hyperreflective material in the subretinal space (Figs. 1C, 1D) and flat irregular pigment epithelial detachments (Figs. 1C, 1D). Subfoveal choroidal thickness, using enhanced-depth imaging SD-OCT was 548 μm in the right eye and 661 μm in the left eye, superior to choroidal thickness reported in healthy subjects.8 In the left eye a regular pigment epithelial detachment temporal to the fovea was observed on SD-OCT (Fig. 1E). On fluorescein angiography, multiple leakage sites were observed in the right eye, temporal to the fovea, and along the superior and inferior temporal vessels (Figs. 1F, 1G), and pigment epithelial alterations, suggestive of previous CSCR episodes, were visible in the left eye (Fig. 1K). On ICG angiography, choroidal hyperpermeability was clearly observed in both eyes (Figs. 1H, 1J).
Figure 1.
Retinal imaging findings in a 48-year-old patient with severe fibrinous central serous chorioretinopathy. Right eye: (A) multicolor fundus imaging showing a posterior serous retinal detachment with multiple folds and focal subretinal protein deposits. (B) Fundus autofluorescence showing a large hyperautofluorescent area covering the posterior pole and extending toward the superior and inferior temporal vascular arcades, due to recent exudative detachment, with central autofluorescence masking due to protein deposits (arrows). (C, D) SD-OCT showing a serous retinal detachment with retinal folds, and hyperreflective material partially occupying the subretinal space. (F, G) Fluorescein angiography showing leakage sites, temporal to the fovea and along the superior and inferior temporal vessels. (H, I) Mid- and late-phase ICG angiography showing choroidal vascular hyperpermeability. Left eye: (E) SD-OCT showing a serous pigment epithelial detachment temporal to the fovea. (J) Midphase ICG angiography demonstrating choroidal hyperpermeability. (K) Fluorescein angiography showing multifocal retinal pigment epithelium alterations and progressive dye filling of the pigment epithelial detachment.
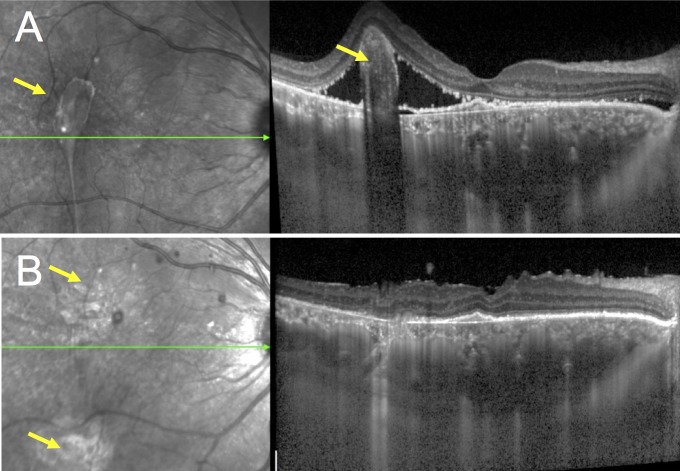
After a 2-month observation period without any SRF resolution, this refractory CSCR patient with signs of chronic epitheliopathy was treated with the oral mineralocorticoid-receptor antagonist spironolactone (25 mg/day for a week and then with 50 mg/day) as previously reported.28–30 After 3 months, visual acuity had improved to 20/50. SRF and subretinal protein exudates decreased dramatically (Figs. 2A, 2B), but a globular fibrous clot persisted just temporal to the fovea, preventing complete macular reattachment (Fig. 2A, arrow). Pars plana vitrectomy with retinotomy and subretinal clot removal was performed, and SRF was collected prior to clot removal. Visual acuity had improved to 20/25 1 month later, and the macular detachment had resolved without remnants of subretinal material (Figs. 2C, 2D). Visual acuity remained unchanged without CSCR recurrence over an additional 2-year follow-up.
Figure 2.
Resolution of residual retinal detachment after surgical subretinal fibrin removeal in a 48-year-old patient with severe fibrinous central serous chorioretinopathy. (A) SD-OCT showed persistence of subretinal material preventing complete retinal reattachment 3months after initiation of oral spironolactone, which prompted to perform vitrectomy with retinotomy and internal removal of a subretinal fibrin clot. (B) SD-OCT showed disappearance of the subretinal deposit and complete macular reattachment 1 month after the procedure (arrow).
Clinical History of the RRD Patients
Patient RRD-1 had no remarkable medical or ocular history, except for mild myopia. She presented with subtotal macula-off RRD 16 days since central vision loss, and underwent pars plana vitrectomy, laser photocoagulation, and gas tamponade. Final visual acuity was 20/50. Patient RRD-2 had no remarkable medical history. His mother had a history of RRD. He presented 20 days after central vision loss with total macula-off RRD and stage B proliferative vitreoretinopathy, and underwent pars plana vitrectomy, laser photocoagulation, and silicone tamponade. After silicone removal 3 months later, final visual acuity was 20/100.
Subretinal Fluid Proteome
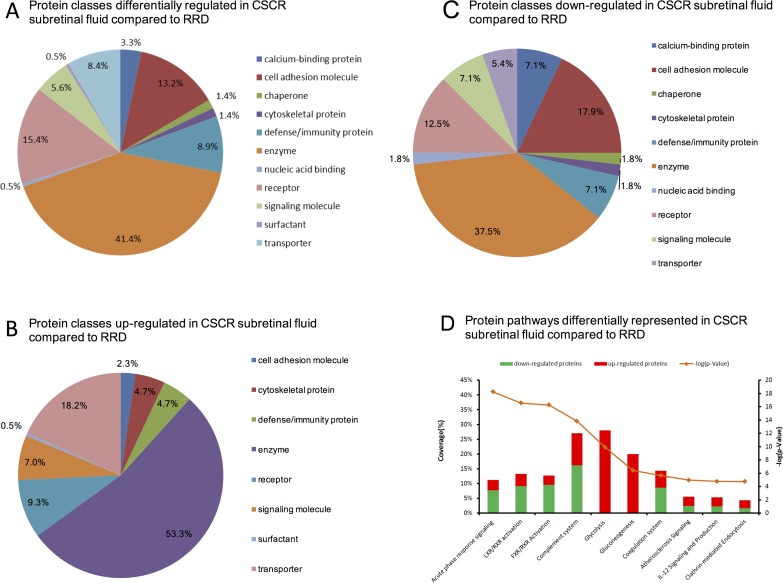
The quantitative proteomics analysis identified 291 proteins in all SRF samples, among which 128 were differentially regulated between the CSCR and RRD samples (see Supplementary Table S1 for the complete list of identified proteins). Of proteins, 77 were downregulated (<0.67) and 51 were upregulated (>1.5) in CSCR compared with RRD (see Supplementary Table S2 for the list of up- and downregulated proteins). Using gene ontology, these differentially regulated proteins were categorized as enzymes (41.5%, 55 proteins including complement factors, peptidases, and protease inhibitors), receptors (15.4%, 32 proteins), cell adhesion molecules (15%, 29 proteins), defense/immunity proteins (8.9%, 19 proteins), and transporters (8.4%, 17 proteins) (Fig. 3A). Protein classes of the upregulated and of the downregulated proteins in CSCR SRF as compared with RRD are, respectively, represented in Figures 3B and 3C. These figures show that among upregulated proteins the most represented classes are enzymes (53.3%) and transporters (18.2%), and that among downregulated proteins the most represented classes are enzymes (37.5%), cell adhesion molecules (17.9%), and receptors (12.5%).
Figure 3.
Differential subretinal fluid proteome in central serous chorioretinopathy compared with rhegmatogenous retinal detachment. (A) Circle chart of differentially regulated protein classes obtained with the Panther software. Some classes contain several subclasses (cell adhesion and extracellular matrix protein: cell adhesion molecule, cytoskeletal protein, extracellular matrix protein, structural protein, surfactant; enzyme: enzyme modulator, hydrolase, isomerase, kinase, lyase, oxidoreductase, phosphatase, protease, transferase; transporter: membrane traffic protein, transfer/carrier protein, transporter). From the list, Panther did not take 29 proteins into account. (B) Protein pathways differentially represented using overrepresentation pathway analysis in CSCR subretinal fluid compared with RRD. The 10 most significant pathways provided by the Ingenuity Pathway Analysis software were selected based on right-tailed Fisher's exact test significance levels. Red and green bars represent up- and downregulated proteins, respectively. The orange line represents the P value corresponding to each pathway.
Among the 15 most upregulated proteins in the CSCR sample (ratio >10), 10 were involved in glycolysis and gluconeogenesis: fructose-bisphosphate aldolase A, beta-enolase, glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic L-lactate dehydrogenase A chain, phosphoglucomutase-1, alpha-enolase, pyruvate kinase, phosphoglycerate kinase-1, triosephosphate isomerase, and glyceraldehyde-3-phosphate dehydrogenase. The most downregulated proteins were mainly involved in cell adhesion or extracellular matrix interaction, including opticin, myocilin, calsyntenin-1, versican, metalloproteinase-2, osteopontin, neurexin-3, cell-adhesion molecule 1 and 2, and insulin-like growth factor-binding protein 7 (see Supplementary Table S2 for P values). Among proteins involved in cell adhesion, galectin-3 binding protein was the only upregulated molecule.
The overrepresentation analysis of differentially regulated pathways is displayed in Figure 3B, and the corresponding up- and downregulated proteins within overrepresented pathways are displayed in the Table. The following pathways were overrepresented in CSCR, as compared with RRD: (1) the immune and inflammatory response (acute phase response signaling, P = 5.85E−19; complement system, P = 1.49E−14), with a majority of downregulated proteins in CSCR (68% and 60% respectively); (2) lipid transport and macrophage activation (liver X receptor/retinoid X receptor pathway, liver X receptor/retinoid X receptor [LXR/RXR], with 69% of downregulated proteins in CSCR, P = 2.86E−18; (3) metabolism of biliary acids (farnesoid X receptor/retinoid X receptor pathway [FXR/RXR], with 75% of downregulated proteins in CSCR, P = 5.57E−17); (4) the glycolysis (P = 1.16E−10) and gluconeogenesis (P = 3.74E−7) pathways, in which all proteins were upregulated in CSCR compared with RRD (aldolase, fructose-bisphosphate A, enolase 1, 3, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase 1, pyruvate kinase, muscle, triosephosphate isomerase 1); (5) coagulation system (P = 2.18E−6); (6) atherosclerosis signaling (P = 1.1E−5; upregulation of apolipoprotein B, C-III, lysozyme, and paraoxonase 1, and downregulation of apolipoprotein A-IV, retinol binding protein 3, and serpin family A member 1); (7) interleukin (IL)-12 signaling (P = 1.74E−5); and (8) clathrin-mediated endocytosis (P = 1.81E−5).
Table.
Proteins Identified in the Subretinal Fluid and Differentially Expressed in Central Serous Chorioretinopathy Compared With Rhegmatogenous Retinal Detachment

Subretinal Fluid Metabolome
After matching against known metabolites in the HMDB database using the mass to charge ratio (m/z), the putative metabolite list was reduced to 651 single molecules. The supervised learning approach was successful in reconstructing sample classes, based on the differentially expressed metabolites between CSCR and RRD. In the SRF sample from the CSCR patient, 76 metabolites were differentially regulated as compared with RRD of which 43 were downregulated and 33 were upregulated (see Supplementary Table S3 for a list of differentially regulated metabolites).
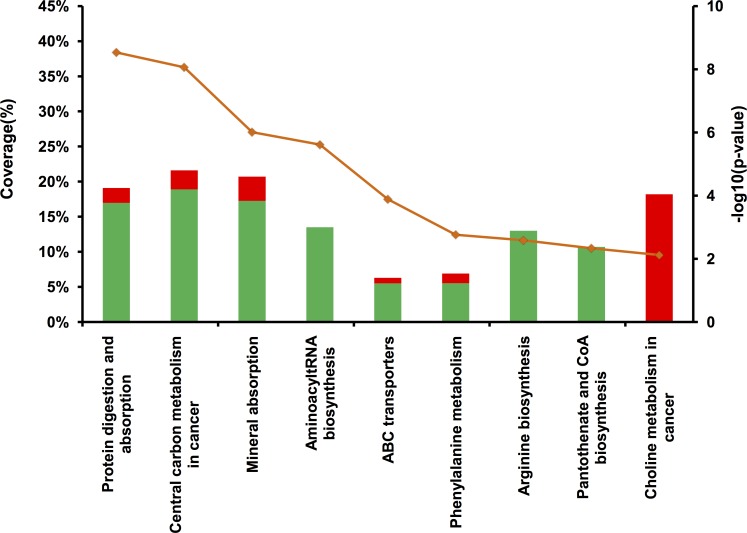
Overrepresentation pathway analysis was performed on the deregulated metabolites (Fig. 4). It confirmed the proteomics results with the protein digestion and absorption pathway (P = 3E−09) defined by approximately 20% coverage of inputted metabolites (specifically methionine, phenylalanine, valine, tyrosine, leucine, glutamine, arginine, isovalerylcarnitine, and tyramine-o-sulfate), which is in accordance with the peptidase and protease inhibitor activity differentially regulated within the enzyme in the proteomics analysis.
Figure 4.
Overrepresentation pathway analysis of differentially represented metabolites in the central serous chorioretinopathy subretinal fluid compared with rhegmatogenous retinal detachment. A minimum of two metabolites per pathway and a significant association cut-off (P < 0.01 and q value <0.05) were used. Red and green bars represent metabolites up- and downregulated, respectively. The orange line represents the associated P value corresponding to each pathway.
Interestingly, additional pathways were also highlighted in CSCR compared with RRD samples, including: (1) the choline pathway, containing exclusively overregulated metabolites, such as phosphatidylcholine (16:0/18:4) (fold change [FC] = 5.8, P = 4E−5) and phosphatidylglycerol (18:0/18:1) (FC = 1.58, P = 6E−3), with 18% metabolite coverage; (2) the aminoacyl tRNA biosynthesis, arginine biosynthesis and pantothenate/CoA biosynthesis pathways, determined exclusively by downregulated metabolites; (3) the mineral absorption and ABC transporters pathways were mostly downregulated, which is consistent with the reduced expression of transporter proteins in the CSCR SRF; and (4) phosphoric acid, also involved in ATP production, was the only upregulated metabolite in the mineral absorption pathway (FC = 4.22).
The chemical subclassification of metabolites deregulated in CSCR, as compared with RRD, was obtained from the HMDB library. The largest subgroup (30%) belonged to the amino acid/peptide subclass, confirming the overrepresentation pathway analysis (see Supplementary Fig. S1 for a chart of deregulated metabolites). Using this classification system, the next highly susceptible metabolite classes were phosphosphingolipids (12%), fatty acids/conjugates (7%), and fatty acid esters (6%), which demonstrated alterations in lipid metabolism, in agreement with the proteomic results showing deregulation of the LXR/RXR pathway.
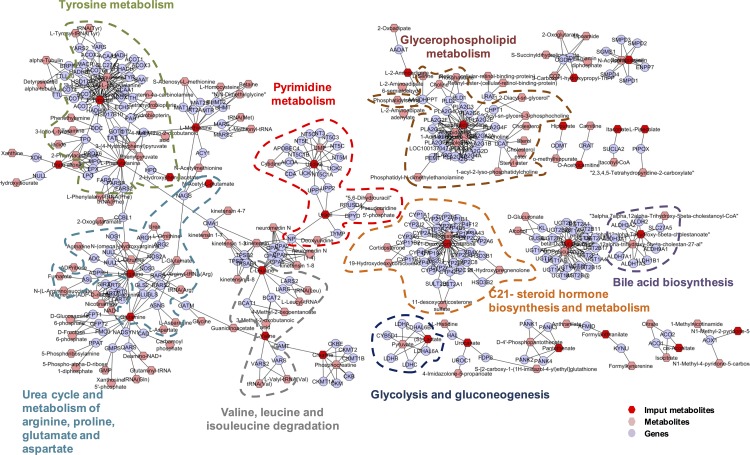
Finally, the pathway-based network was explored to create a compound-gene network on the MetScape software (Fig. 5), providing a comprehensive overview of the specific SRF metabolic signature in the CSCR sample. Among subnetworks, branch-chain amino acid degradation pathway, glycolysis and gluconeogenesis, steroid hormone biosynthesis and metabolism, and bile acid biosynthesis were remarkably deregulated, showing a high consistency with the proteomics results.
Figure 5.
Pathway-based network of differentially expressed metabolites in the subretinal fluid between CSCR and RRD. In the compound-gene networks, detected metabolites are shown in red. Blue nodes correspond to the genes identified by enrichment analysis performed using the differentially detected metabolites.
Discussion
The present pilot study reports the first proteomic and metabolomic investigation of SRF, to the best of our knowledge. Applied comparatively with CSCR and RRD, this approach provided unique molecular information, given the rarity of bullous variants of CSCR requiring subretinal surgery,1,2 the difficulty of SRF collection, and the limited volume of SRF available. Although proteomics results were based on a single CSCR case, they were supported by an independent and orthogonal metabolomics analysis, and by the biological consistence with current understanding of CSCR pathophysiology.
Our results are based on the assumption that the molecular composition of the subretinal material that we collected was related to the disease process. Indeed, because there is no fluid between the retina and the RPE in normal healthy conditions, the presence of SRF itself constitutes a pathological process. Other factors than CSCR or RRD may influence the proteome/metabolome of SRF described here, but the existence of these samples is the sole consequence of the causing disease, CSCR or RRD, which should be therefore considered as the main factor influencing their characteristics.
In addition to demonstrating the feasibility of multiomics exploration of SRF, this study suggests that inflammatory and immune responses in the subretinal space are differentially regulated in CSCR and RRD. Regarding the complement pathway, the upregulation of complement factor H (CFH) confirms its involvement in CSCR pathogenesis. This finding is consistent with previous reports that genetic variants of CFH are associated to a higher risk of developing CSCR,31–33 and that other variants could protect from the disease.33 The upregulation of CFH and CFH-related protein-1, and the decrease in complement factors B and 4B in the SRF of CSCR may indicate a lower activation of the alternative complement pathway, subsequent reduction of membrane attack complex and reduced photoreceptor cell death.34,35 Noticeably, the C4B genomic copy number is related to the risk of developing chronic CSCR, with absent C4B conferring a higher risk and presence of three copies decreasing this risk.36 On the other hand, the alternative complement pathway contributes to physiologic transport of ions and macromolecules through pores formation, such as C5b-8 and C5b-9.35 This mechanism involves CD59 protein, also found in our study as downregulated. Impaired activation of this mechanism could result in fluid and macromolecule accumulation in the subretinal space of patients with CSCR.
Among proteins differentially regulated in the acute phase response-signaling pathway, Serum amyloid P component (SAP or pentraxin-2) is of specific interest. SAP interacts with complement factors37 and inhibits the recruitment of profibrotic macrophages.38 In addition, it protects from cell damage induced by histone H3,39 a potential mechanism contributing to photoreceptor cell death.40
Regarding proteins related to gluco- and mineralocorticoid hormonal systems, the CSCR sample showed a moderate elevation of galectin-3, a biomarker linked to mineralocorticoid activation and vascular fibrosis,41,42 and a reduction in cortisol binding globulin and angiotensin (Supplementary Table S2). In addition, the identification of the C21 steroid hormone biosynthesis metabolic subnetwork (Fig. 4), which includes critical gluco- and mineralocorticoid molecules, such as cortisol and aldosterone (both having 21 carbons), reflects the deregulation of steroid metabolism, as previously identified in CSCR patients.43,44 This increase in C21 steroids and the reduced cortisol binding globulin in the SRF of CSCR could also modulate photoreceptor cell death and survival.45
Clathrin-mediated endocytosis, one of the differentially regulated pathways between CSCR and RRD, could be an additional mechanism for controlling C5b-8 or C5b-9 levels at the RPE cell membrane, and regulating their lytic threshold.34 Interestingly, five of seven proteins of the clathrin-mediated endocytosis pathway have functions in lipid transport and are involved in vascular diseases (apolipoprotein A-IV, retinol binding protein 3, serpin family A member 1, apolipoprotein B, C-III, and paraoxonase 1). The LXR/RXR pathway, involved in lipid transport, also regulates macrophage-mediated inflammatory response in atherosclerosis,46 and induces microglial activation in the subretinal space.47 Taken together, these results indicate that microglia/macrophage and lipid metabolism are differentially regulated during rhegmatogenous or serous retinal detachments, and are potentially involved in CSCR pathophysiology.
The difference in glycolysis and gluconeogenesis pathways between CSCR and RRD is also emphasized by both proteomics and metabolomics approaches. Compared with RRD, all proteins involved in glycolysis and gluconeogenesis pathways were upregulated in the SRF from CSCR, which could point to differential neuronal survival mechanisms. Indeed, gluconeogenesis products contribute to protecting the retina against oxidative and nitrosative stress,48,49 and increased glucose levels may promote cone photoreceptor survival by stimulation of aerobic glycolysis.50
The FXR pathway identified by proteomics analysis is directly linked to the bile acid pathway identified by metabolomics. Bile acids, increased in the SRF of CSCR as compared with RRD are potent antiapoptotic components for photoreceptors in various models.51 For example, tauroursodeoxycholic acid (TUDCA) prevents photoreceptors cell death after experimental retinal detachment.52 Similarly, the pentothenate and CoA pathways have a protective effect against retinal degeneration, and were also activated in CSCR, pointing out another potential neuroprotective mechanism.53
Proteins involved in cell migration and adhesion, essential to cellular movement, accounted for 15% of the differentially regulated proteins in the SRF of CSCR, compared with RRD. Consistently, most proteins usually found in the vitreous54,55 were less abundant in the SRF from CSCR, which is not expected to be contaminated by vitreous fluid, such as opticin,56 osteopontin,57 and versican.58 Proteins involved in retinal structure were also reduced in the SRF of CSCR as compared with RRD (such as contactin 1,59 SYNCAM, and lumican60), which may be related to the better architecture preservation of the detached retina in CSCR. A clear reduction in the choline metabolic pathway, which intervenes in the photoreceptor outer segment/RPE microvilli interaction,61 may also be related to the better preservation of photoreceptor outer segments in CSCR as compared with RRD.62
Very few studies have analyzed the proteome of SRF in patients with RRD. Shitama et al.63 have performed the proteomic analysis of 10 SRF samples from RRD, compared with 13 control vitreous samples. These results should be compared with caution to our findings, because the authors used a different strategy, by not removing abundant plasma proteins from ocular samples. The most increased proteins in RRD SRF identified by Shitama et al.63 were albumin, transferrin, transthyretin, and cathepsin D. Other proteins, identified as potential markers of RRD were α-1 antitrypsin, clusterin (also known as apolipopretein J), pigment epithelium-derived factor (PEDF), and apolipoprotein A-IV (APOA4). In contrast, our proteome analysis of SRF in RRD could not identify albumin, transferrin or α-1 antitrypsin that had been depleted among the 12 major plasma proteins removed at the initial step of our protocol. Yet, we identified transthyretin, clusterin, cathepsin D, PEDF, and APOA4 in the SRF from patients with RRD (See Supplementary Table S1). Moreover, as proteomic methods differ between studies,54,55,64 direct comparison of the SRF proteomes to published datasets is not possible. However, in one of our preliminary experiments, the vitreous and SRF from three patients presenting with RRD were analyzed using the same proteomics strategy (unpublished data). We identified 70% of proteins that were common to the SRF and vitreous from these patients (257 of 367 proteins). Among these proteins, several known to be enriched in the vitreous such as opticin, ostoepontin, versican, retinoschisin, transthyretin, or hyaluronan-binding protein 2 (related to hyaluronic acid), or collagen alpha-2 were found. These data are consistent with the notion that RRD involves the creation of a retinal tear, leading to passage of liquefied vitreous humor through the tear into the subretinal space. Unfortunately, the proteomic strategy used in the present study does not allow us to separate the raw protein lists specific to CSCR or RRD SRF. Therefore, we cannot compare the composition of the CSCR SRF with the vitreal proteome. Moreover, we can assume that, because no retinal tear occurs in CSCR, it is unlikely that vitreal proteins with high molecular weight could migrate through the retina.
Finally, a recent proteomics analysis of intraretinal schisis fluid from two subjects with X-linked retinoschisis highlighted canonical pathways also identified in the present study, such as LXR/RXR activation, complement system, and acute phase response signaling.65 Because chronic CSCR may also manifest with retinoschisis-like cavities,66 similar biochemical changes may occur in both diseases. These convergent results support the involvement of the identified pathways in a range of retinal disorders, and show the emerging potential of ocular proteomics.
To summarize, this work demonstrated the feasibility and power of proteomics, combined to metabolomics, for the analysis of SRF. This approach opens perspectives for the comprehensive analysis of ocular biomarkers in retinal diseases, which is a current trend aiming at establishing early diagnosis and personalized therapies.67,68 This study demonstrates in particular the interest of the omics approach for the screening of neuroprotective therapeutic targets in retinal detachment. Its main weakness was the limited number of SRF samples analyzed. Although results from proteomics and metabolomics were largely consistent with each other, and highlighted key molecular players in CSCR pathophysiology previously reported in the literature, they must be interpreted with caution. Results also relied on the consistency of the control group, for which two cases with different clinical characteristics were pooled in order to highlight their common RRD-related molecular characteristics, and filter individual variability while preserving sample representativeness.
Although the indication of subretinal surgical procedures in CSCR is exceptional, the larger-scale feasibility and repeatability of this multiomics approach, and the translational results obtained will require confirmation with additional samples. This first comparative molecular analysis of SRF in CSCR and RRD improved the understanding of both disorders, and identified pathways involved in the better photoreceptor preservation in CSCR.
Supplementary Material
Acknowledgments
Authors thank Cindy Salvisberg for technical assistance.
Supported by grants from the Swiss National Science Foundation (NT: #PMPDP3_158370; AT: #310030-156771; and FBC: #320030_156401), from the Agence Nationale de la Recherche (FBC: NR-15-CE18-0032 “ROCK SUR MER”), and from the Faculty of Biology and Medicine Research Commission Fund, University of Lausanne (AM).
Disclosure: L. Kowalczuk, None; A. Matet, None; M. Dor, None; N. Bararpour, None; A. Daruich, None; A. Dirani, None; F. Behar-Cohen, None; A. Thomas, None; N. Turck, None
References
- 1. Balaratnasingam C, Freund KB, Tan AM,et al. Bullous variant of central serous chorioretinopathy: expansion of phenotypic features using multimethod imaging. Ophthalmology. 2016; 123: 1541– 1552. [DOI] [PubMed] [Google Scholar]
- 2. Bondalapati S, Pathengay A, Chhablani J. . External drainage for exudative retinal detachment secondary to central serous chorioretinopathy. Eye Sci. 2015; 30: 204– 208. [PubMed] [Google Scholar]
- 3. Prunte C, Flammer J. . Choroidal capillary and venous congestion in central serous chorioretinopathy. Am J Ophthalmol. 1996; 121: 26– 34. [DOI] [PubMed] [Google Scholar]
- 4. Ferrara D, Mohler KJ, Waheed N,et al. En face enhanced-depth swept-source optical coherence tomography features of chronic central serous chorioretinopathy. Ophthalmology. 2014; 121: 719– 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spitznas M. . Pathogenesis of central serous retinopathy: a new working hypothesis. Graefes Arch Clin Exp Ophthalmol. 1986; 224: 321– 324. [DOI] [PubMed] [Google Scholar]
- 6. Marmor MF. . New hypotheses on the pathogenesis and treatment of serous retinal detachment. Graefes Arch Clin Exp Ophthalmol. 1988; 226: 548– 552. [DOI] [PubMed] [Google Scholar]
- 7. Daruich A, Matet A, Marchionno L,et al. Acute central serous chorioretinopathy: factors influencing episode duration. Retina. 2017; 37: 1905– 1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daruich A, Matet A, Dirani A,et al. Central serous chorioretinopathy: recent findings and new physiopathology hypothesis. Prog Retin Eye Res. 2015; 48: 82– 118. [DOI] [PubMed] [Google Scholar]
- 9. van Bussel EM, van der Valk R, Bijlsma WR, La Heij EC. . Impact of duration of macula-off retinal detachment on visual outcome: a systematic review and meta-analysis of literature. Retina. 2014; 34: 1917– 1925. [DOI] [PubMed] [Google Scholar]
- 10. Murakami Y, Notomi S, Hisatomi T,et al. Photoreceptor cell death and rescue in retinal detachment and degenerations. Prog Retin Eye Res. 2013; 37: 114– 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sweigard JH, Matsumoto H, Smith KE,et al. Inhibition of the alternative complement pathway preserves photoreceptors after retinal injury. Sci Transl Med. 2015; 7: 297ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson DH, Stern WH, Fisher SK, Erickson PA, Borgula GA. . The onset of pigment epithelial proliferation after retinal detachment. Invest Ophthalmol Vis Sci. 1981; 21: 10– 16. [PubMed] [Google Scholar]
- 13. Skeie JM, Tsang SH, Zande RV,et al. A biorepository for ophthalmic surgical specimens. Proteomics Clin Appl. 2014; 8: 209– 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Escoffier P, Paris L, Bodaghi B, Danis M, Mazier D, Marinach-Patrice C. . Pooling aqueous humor samples: bias in 2D-LC-MS/MS strategy? J Proteome Res. 2010; 9: 789– 797. [DOI] [PubMed] [Google Scholar]
- 15. Azurmendi L, Degos V, Tiberti N,et al. Measuring serum amyloid A for infection prediction in aneurysmal subarachnoid hemorrhage. J Proteome Res. 2015; 14: 3948– 3956. [DOI] [PubMed] [Google Scholar]
- 16. Dayon L, Turck N, Kienle S,et al. Isobaric tagging-based selection and quantitation of cerebrospinal fluid tryptic peptides with reporter calibration curves. Anal Chem. 2010; 82: 848– 858. [DOI] [PubMed] [Google Scholar]
- 17. Scherl A, Shaffer SA, Taylor GK, Kulasekara HD, Miller SI, Goodlett DR. . Genome-specific gas-phase fractionation strategy for improved shotgun proteomic profiling of proteotypic peptides. Anal Chem. 2008; 80: 1182– 1191. [DOI] [PubMed] [Google Scholar]
- 18. Dayon L, Pasquarello C, Hoogland C, Sanchez JC, Scherl A. . Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J Proteomics. 2010; 73: 769– 777. [DOI] [PubMed] [Google Scholar]
- 19. Gluck F, Hoogland C, Antinori P,et al. EasyProt–an easy-to-use graphical platform for proteomics data analysis. J Proteomics. 2013; 79: 146– 160. [DOI] [PubMed] [Google Scholar]
- 20. Salvisberg C, Tajouri N, Hainard A, Burkhard PR, Lalive PH, Turck N. . Exploring the human tear fluid: discovery of new biomarkers in multiple sclerosis. Proteomics Clin Appl. 2014; 8: 185– 194. [DOI] [PubMed] [Google Scholar]
- 21. Dunn WB, Broadhurst D, Begley P,et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011; 6: 1060v1083. [DOI] [PubMed] [Google Scholar]
- 22. Wolfender J-L, Marti G, Thomas A, Bertrand S. . Current approaches and challenges for the metabolite profiling of complex natural extracts. J Chromatogr A 2015; 1382: 136– 164. [DOI] [PubMed] [Google Scholar]
- 23. Guo Y, Graber A, McBurney RN, Balasubramanian R. . Sample size and statistical power considerations in high-dimensionality data settings: a comparative study of classification algorithms. BMC Bioinformatics. 2010; 11: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas A, Deglon J, Lenglet S,et al. High-throughput phospholipidic fingerprinting by online desorption of dried spots and quadrupole-linear ion trap mass spectrometry: evaluation of atherosclerosis biomarkers in mouse plasma. Anal Chem. 2010; 82: 6687– 6694. [DOI] [PubMed] [Google Scholar]
- 25. Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. . PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016; 44: D336– D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karnovsky A, Weymouth T, Hull T,et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012; 28: 373– 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bindea G, Mlecnik B, Hackl H,et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25: 1091– 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bousquet E, Beydoun T, Zhao M, Hassan L, Offret O, Behar-Cohen F. . Mineralocorticoid receptor antagonism in the treatment of chronic central serous chorioretinopathy: a pilot study. Retina. 2013; 33: 2096– 2102. [DOI] [PubMed] [Google Scholar]
- 29. Singh RP, Sears JE, Bedi R, Schachat AP, Ehlers JP, Kaiser PK. . Oral eplerenone for the management of chronic central serous chorioretinopathy. Int J Ophthalmol. 2015; 8: 310– 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Daruich A, Matet A, Dirani A,et al. Oral mineralocorticoid-receptor antagonists: real-life experience in clinical subtypes of nonresolving central serous chorioretinopathy with chronic epitheliopathy. Transl Vis Sci Technol. 2016; 5 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miki A, Kondo N, Yanagisawa S, Bessho H, Honda S, Negi A. . Common variants in the complement factor H gene confer genetic susceptibility to central serous chorioretinopathy. Ophthalmology. 2014; 121: 1067– 1072. [DOI] [PubMed] [Google Scholar]
- 32. Moschos MM, Gazouli M, Gatzioufas Z,et al. Prevalence of the complement factor H and Gstm1 genes polymorphisms in patients with central serous chorioretinopathy. Retina. 2016; 36: 402– 407. [DOI] [PubMed] [Google Scholar]
- 33. de Jong EK, Breukink MB, Schellevis RL,et al. Chronic central serous chorioretinopathy is associated with genetic variants implicated in age-related macular degeneration. Ophthalmology. 2015; 122: 562– 570. [DOI] [PubMed] [Google Scholar]
- 34. Georgiannakis A, Burgoyne T, Lueck K, Futter C, Greenwood J, Moss SE. . Retinal pigment epithelial cells mitigate the effects of complement attack by endocytosis of C5b-9. J Immunol. 2015; 195: 3382– 3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Farkas I, Baranyi L, Ishikawa Y,et al. CD59 blocks not only the insertion of C9 into MAC but inhibits ion channel formation by homologous C5b-8 as well as C5b-9. J Physiol. 2002; 539: 537– 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Breukink MB, Schellevis RL, Boon CJ,et al. Genomic copy number variations of the complement component C4B gene are associated with chronic central serous chorioretinopathy. Invest Ophthalmol Vis Sci. 2015; 56: 5608– 5613. [DOI] [PubMed] [Google Scholar]
- 37. Bottazzi B, Inforzato A, Messa M,et al. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J Hepatol. 2016; 64: 1416– 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horgan SJ, Watson CJ, Glezeva N,et al. Serum amyloid P-component prevents cardiac remodeling in hypertensive heart disease. J Cardiovasc Transl Res. 2015; 8: 554– 566. [DOI] [PubMed] [Google Scholar]
- 39. Iba T, Hamakubo T, Nagaoka I, Sato K, Thachil J. . Physiological levels of pentraxin 3 and albumin attenuate vascular endothelial cell damage induced by histone H3 in vitro. Microcirculation. 2016; 23: 240– 247. [DOI] [PubMed] [Google Scholar]
- 40. Kawano H, Ito T, Yamada S,et al. Toxic effects of extracellular histones and their neutralization by vitreous in retinal detachment. Lab Invest. 2014; 94: 569– 585. [DOI] [PubMed] [Google Scholar]
- 41. Calvier L, Miana M, Reboul P,et al. Galectin-3 mediates aldosterone-induced vascular fibrosis. Arterioscler Thromb Vasc Biol. 2013; 33: 67– 75. [DOI] [PubMed] [Google Scholar]
- 42. Martinez-Martinez E, Calvier L, Fernandez-Celis A,et al. Galectin-3 blockade inhibits cardiac inflammation and fibrosis in experimental hyperaldosteronism and hypertension. Hypertension. 2015; 66: 767– 775. [DOI] [PubMed] [Google Scholar]
- 43. Garg SP, Dada T, Talwar D, Biswas NR. . Endogenous cortisol profile in patients with central serous chorioretinopathy. Br J Ophthalmol. 1997; 81: 962– 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haimovici R, Rumelt S, Melby J. . Endocrine abnormalities in patients with central serous chorioretinopathy. Ophthalmology. 2003; 110: 698– 703. [DOI] [PubMed] [Google Scholar]
- 45. Cubilla MA, Castaneda MM, Bachor TP, Suburo AM. . Glucocorticoid-dependent mechanisms in photoreceptor survival. Adv Exp Med Biol. 2012; 723: 101– 106. [DOI] [PubMed] [Google Scholar]
- 46. Lee SD, Tontonoz P. . Liver X receptors at the intersection of lipid metabolism and atherogenesis. Atherosclerosis. 2015; 242: 29– 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choudhary M, Malek G. . A brief discussion on lipid activated nuclear receptors and their potential role in regulating microglia in age-related macular degeneration (AMD). Adv Exp Med Biol. 2016; 854: 45– 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mamczur P, Mazurek J, Rakus D. . Ubiquitous presence of gluconeogenic regulatory enzyme, fructose-1,6-bisphosphatase, within layers of rat retina. Cell Tissue Res. 2010; 341: 213– 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Frenzel J, Richter J, Eschrich K. . Pyruvate protects glucose-deprived Muller cells from nitric oxide-induced oxidative stress by radical scavenging. Glia. 2005; 52: 276– 288. [DOI] [PubMed] [Google Scholar]
- 50. Ait-Ali N, Fridlich R, Millet-Puel G,et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 2015; 161: 817– 832. [DOI] [PubMed] [Google Scholar]
- 51. Lawson EC, Bhatia SK, Han MK,et al. Tauroursodeoxycholic acid protects retinal function and structure in rd1 mice. Adv Exp Med Biol. 2016; 854: 431– 436. [DOI] [PubMed] [Google Scholar]
- 52. Mantopoulos D, Murakami Y, Comander J,et al. Tauroursodeoxycholic acid (TUDCA) protects photoreceptors from cell death after experimental retinal detachment. PLoS One. 2011; 6: e24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hayflick SJ. . Defective pantothenate metabolism and neurodegeneration. Biochem Soc Trans. 2014; 42: 1063– 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aretz S, Krohne TU, Kammerer K,et al. In-depth mass spectrometric mapping of the human vitreous proteome. Proteome Sci. 2013; 11: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skeie JM, Roybal CN, Mahajan VB. . Proteomic insight into the molecular function of the vitreous. PLoS One. 2015; 10: e0127567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reardon AJ, Le Goff M, Briggs MD,et al. Identification in vitreous and molecular cloning of opticin, a novel member of the family of leucine-rich repeat proteins of the extracellular matrix. J Biol Chem. 2000; 275: 2123– 2129. [DOI] [PubMed] [Google Scholar]
- 57. Deeg CA, Eberhardt C, Hofmaier F, Amann B, Hauck SM. . Osteopontin and fibronectin levels are decreased in vitreous of autoimmune uveitis and retinal expression of both proteins indicates ECM re-modeling. PLoS One. 2011; 6: e27674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keenan TD, Clark SJ, Unwin RD, Ridge LA, Day AJ, Bishop PN. . Mapping the differential distribution of proteoglycan core proteins in the adult human retina, choroid, and sclera. Invest Ophthalmol Vis Sci. 2012; 53: 7528– 7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. D'Alessandri L, Ranscht B, Winterhalter KH, Vaughan L. . Contactin/F11 and tenascin-C co-expression in the chick retina correlates with formation of the synaptic plexiform layers. Curr Eye Res. 1995; 14: 911– 926. [DOI] [PubMed] [Google Scholar]
- 60. Ribic A, Liu X, Crair MC, Biederer T. . Structural organization and function of mouse photoreceptor ribbon synapses involve the immunoglobulin protein synaptic cell adhesion molecule 1. J Comp Neurol. 2014; 522: 900– 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Matsumoto H, Shibasaki K, Uchigashima M,et al. Localization of acetylcholine-related molecules in the retina: implication of the communication from photoreceptor to retinal pigment epithelium. PLoS One. 2012; 7: e42841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fisher SK, Lewis GP, Linberg KA, Verardo MR. . Cellular remodeling in mammalian retina: results from studies of experimental retinal detachment. Prog Retin Eye Res. 2005; 24: 395– 431. [DOI] [PubMed] [Google Scholar]
- 63. Shitama T, Hayashi H, Noge S,et al. Proteome profiling of vitreoretinal diseases by cluster analysis. Proteomics Clin Appl. 2008; 2: 1265– 1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Murthy KR, Goel R, Subbannayya Y,et al. Proteomic analysis of human vitreous humor. Clin Proteomics. 2014; 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sudha D, Kohansal-Nodehi M, Kovuri P,et al. Proteomic profiling of human intraschisis cavity fluid. Clin Proteomics. 2017; 14: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee JH, Park HY, Baek J, Lee WK. . Alterations of the lamina cribrosa are associated with peripapillary retinoschisis in glaucoma and pachychoroid spectrum disease. Ophthalmology. 2016; 123: 2066– 2076. [DOI] [PubMed] [Google Scholar]
- 67. Velez G, Roybal CN, Colgan D, Tsang SH, Bassuk AG, Mahajan VB. . Precision medicine: personalized proteomics for the diagnosis and treatment of idiopathic inflammatory disease. JAMA Ophthalmol. 2016; 134: 444– 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Villani E, Vujosevic S. . Foreword: biomarkers and surrogate endpoints in ophthalmic clinical research. Invest Ophthalmol Vis Sci. 2017; 58: BIOi– BIOii. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.