Abstract
A significant number of proteins in all living species contains amino acid repeats (AARs) of various lengths and compositions, many of which play important roles in protein structure and function. Here, I have surveyed select homopolymeric single [(A)n] and double [(AB)n] AARs in the human proteome. A close examination of their codon pattern and analysis of RNA structure propensity led to the following set of empirical rules: (1) One class of amino acid repeats (Class I) uses a mixture of synonymous codons, some of which approximate the codon bias ratio in the overall human proteome; (2) The second class (Class II) disregards the codon bias ratio, and appears to have originated by simple repetition of the same codon (or just a few codons); and finally, (3) In all AARs (including Class I, Class II, and the in-betweens), the codons are chosen in a manner that precludes the formation of RNA secondary structure. It appears that the AAR genes have evolved by orchestrating a balance between codon usage and mRNA secondary structure. The insights gained here should provide a better understanding of AAR evolution and may assist in designing synthetic genes.
Keywords: Genetics, Computational biology, Structural biology, Bioinformatics
1. Introduction
Amino acid repeats (AARs) of various degrees of complexity are found in essentially all organisms [1, 2]. Much attention has been paid to relatively complex and degenerate repeats such as WD, HEAT, and tetratricopeptide repeat (TPR), most of which are involved in protein-protein interaction [3, 4, 5, 6, 7]. In contrast, simple AARs, such as single or double amino acids repeats (SAAR and DAAR, respectively), have remained intriguing in terms of both protein structure and genetic organization [8, 9]. Specifically, these repeats take the form (A)n for SAAR and (AB)n for DAAR, where A and B are nonidentical amino acids, and ‘n’ is the number of times they are repeated. While the complex repeats form conserved secondary structures, such as the β-strand arrangements of the 40-amino acid WD40 repeats and the tandem α-helical repeats of the 34-residue TPR domains [4, 5, 6, 7], the single and double AARs clearly constitute a single patch of a continuous and repetitive physical property that does not generally coincide with a defined secondary structural element. For example, a run of a nonpolar aliphatic amino acid in SAAR, such as (Ala)n, would form a highly hydrophobic patch that needs to be internalized in the protein structure, whereas a poly-Glu repeat will form a strongly acidic region, to be neutralized by a counterion at physiological pH. At the DNA level, SAARs pose the risk of expansion due to further repeats of the same codon. Although the molecular mechanism of such expansions are not clear, these changes underlie several genetic diseases of the “trinucleotide repeat disorder” family, such as the polyQ (poly-Gln) diseases, caused by expansion of the CAG (one of two Gln codons) repeats above a normal threshold number. Common examples include Huntington Disease (HD), Spinocerebellar ataxia-8, X-linked spinal tubular muscular atrophy (SBMA, or Kennedy disease), and certain forms of breast and prostate cancers, caused by polyQ-tract expansion in the AIB1 (“amplified in breast cancer gene 1”) and AR (androgen receptor) genes, respectively [10, 11, 12, 13]. In the recent past, one AAR study investigated the physical properties and intracellular localization of 30-mer repeat polypeptides, recombinantly expressed in primate cells [14]. Another study focused on the length and location of repeats in the newly mined proteome of Mycobacterium tuberculosis [15]. Nevertheless, the overall codon and amino acid patterns of AARs in general and their potential ramification on mRNA structure have remained unexplored.
In this bioinformatic study, I have interrogated the processes that govern the evolution and operation of the common AARs, and specifically, asked the following questions: (a) Do the repeats follow a specific codon or nucleotide pattern? (b) Is there a preference among the synonymous codons used in the repeats? (c) Does the choice of codons in amino acid repeats affect mRNA folding, which may affect translation? As elaborated later, these queries not only apply to SAARs but also to DAARs, and hence both were investigated. Unexpectedly, the search for answers to these questions led to a set of empirical and interesting rules of codon choice in the AARs.
2. Results
2.1. Single amino acid repeats (SAARs)
In recent searches, including our own (unpublished), of protein databases of various species, the SAARs have shown bias for certain amino acids [14, 16]. Overall, homopolymeric repeats of 11–20 residues in length commonly consisted of (in no particular order) Ala (A), Arg (R), Gln (Q), Glu (E), Gly (G), His (H) and Pro (P), whereas the underrepresented amino acids included Ile (I), Met (M), Val (V), Trp (W) and Tyr (Y). My search was, therefore, focused on repeats of four amino acids, namely A, Q, P and R, since the primary goal of the study was to identify any pattern as proof-of-concept rather than perform a comprehensive analysis of all repeats in the human proteome, which is estimated to be a few thousands, the exact number depending on how the repeat is defined in terms of length and allowance of interruption [16, 17]. Regardless, when the SAAR search of one amino acid retrieved a protein that contained the SAAR of another amino acid elsewhere in the polypeptide, both stretches were considered.
The detailed data from searches for homopolymers of Ala, Gln, Pro, and Arg, which also contained data of other amino acid repeats found in the same proteins, are presented in Supplementary Material 1. For size constraint and easy visualization, the major results are also pictorially summarized in a color-coded fashion (Fig. 1), whereby each codon of a given amino acid is assigned a different color as shown (Fig. 1F). Although I have presented a small cohort of data here, they are illustrative of essentially all major types of pattern observed in a larger number of proteins and SAARs that are not shown to save space.
Fig. 1.
Synonymous codon patterns of selected single amino acid repeats (SAARs). The repeats were retrieved from the human proteome as described in Materials and Methods. In each example, the protein name (e.g. Fibrosin-1) or its HGNC (HUGO Gene Nomenclature Committee)-approved symbol (e.g. SKIDA1) is followed by the residue and its number of repeats (e.g. Ala19). Panels A, B, C, D, E show, respectively, representative repeats of Ala, Gln, Pro, miscellaneous amino acids, and Arg; panel F shows the color codes used for the repeat codons and percentage of each synonymous codon use for a given amino acid in the human proteome, acquired from the Codon Usage Database, http://www.kazusa.or.jp/codon/[27]. Note that in panel F the percentage values for the six-codon amino acids, Ser and Arg, will not add up to 100, as two codons of each have been omitted to conserve space. The vast majority of examples are uninterrupted SAARs; in a few cases, the interrupting amino acids/codons are written overhead (when space permitted) and indicated by white color. Two relatively complex repeat runs, SPT20HL1 (panel A) and RUNX2 (panel B) are marked with apparent microrepeat units and expansions. In each panel, the repeats are listed from the most Class I type (i.e. diverse, multi-colored codons) to the most Class II type (i.e. identical, single-color codons) from top to bottom, by qualitative visual inspection.
Even a cursory look at the schematic (Fig. 1) indicates that there are two fundamental extremes of repeats, which I have arbitrarily designated Class I and Class II. The Class I repeats are multicolored, because they are composed of multiple synonymous codons. The Class II repeats, in sharp contrast, use a single color, because they repeat a single codon, ignoring the other synonymous codons. However, most repeats fall somewhere in between, as one can easily see from the various mosaic patterns of multiple colors (Fig. 1), and therefore, I have used a grayscale to indicate a gradual transition from Class I to Class II for each SAAR panel (from top to bottom). Thus, the designations of I and II primarily serve as conceptual benchmarks. In what follows, I summarize the multiple patterns found in these homopolymer tracts, including generalizations and exceptions (Fig. 1 and Supplementary Material 1).
2.2. Compliance and defiance of codon bias: Class I versus Class II
First of all, it should be realized that the codon usage ratio in a SAAR of a single protein cannot be expected to exactly match the ratio averaged from the whole human proteome (Fig. 1F), as the latter is a much larger sample size. Thus, many repeats with multiple synonymous codon usage were designated close to Class I, even when the usage ratio deviated from the proteomewide codon bias for that amino acid (see comments in Supplementary Material 1). For example, Ala has four synonymous codons (GCN), but in the Fibrosin-1 Ala19 repeat, only three were used. The GCU and GCA codons were used in the ratio of 3:1, whereas in the whole proteome they are found in nearly equal number (1.04:1). The GCC and GCU codons, in contrast, are closer to their proteomewide ratio. Overall, since three different codons of Ala were used in this repeat, it was considered closer to the Class I end of the scale (Fig. 1A). An overall pattern, noticed in Ala repeats, is that the GCG codon is often favored, although GCG is the least used Ala codon in the proteome.
The Class II repeats, i.e. the pure, single-codon repeats, by definition exhibit total disregard for codon diversity, oblivious of the other synonymous codons of that amino acid. Again, such pure Class II repeats are relatively rare, and are exemplified by the Ala17 repeat in RPL4 (Fig. 1A), His8 in POU3F3 (Fig. 1D), Arg20 in FMR1 (Fig. 1E), and Gln repeats in ATXN8 (Fig. 1B). The ATXN8 protein, an 80-residue long polypeptide, is in fact all Gln with just an initiator Met, i.e., has the sequence Met-(Gln)79, where all Gln codons are CAG. Clearly, the ATXN8 polyQ repeat is committed to repeating the CAG triplet, regardless of codon usage bias. Of note, the polyQ repeats, in general, tend to be Class II type, with preference for the CAG codon of Gln (blue colors in Fig. 1B). Those using both the synonymous codons of Gln (CAG, CAA) are less common (mixture of blue and red in Fig. 1B); some of them are close to the proteome ratio of ∼2.7CAG:1CAA (e.g. both repeats of TRAP230, i.e. Gln26, Gln33; Fig. 1B), but even in them, the CAG repeat tends to occur in longer stretches. In other words, whether in Class I or Class II, the Gln SAARs prefer the codon with a higher bias (CAG). Due to the scattered nature of the CAG-CAA distribution in the Class I Gln repeats (Fig. 1B), no consistent pattern could be drawn. However, the Gln23 repeat of RUNX2 is notable since it appears to be an expansion of the di-codon CAA(CAG)n unit with ‘n’ increasing stepwise from 3 to 6 (see Microrepeats later). Also to mention, a common pattern in polyQ SAARs is a single penultimate CAA codon in a run of CAG codons (e.g. in TBP, EP400, and the two Huntington repeats) (Fig. 1B).
While most amino acids have four codons, three amino acids (Arg, Leu, Ser) have six codons each. Ser, for example, is coded for by four UCN codons as well as two AGN codons, viz. AGU and AGC. However, in two patches of Ser repeats in Fibrosin-1 (Ser6, Supplementary Material 1; and Ser15, Fig. 1D), only the UCN codons were used, not AGU or AGC. The complete absence of AGC/U codons in a total of 21 Ser codons is significant, especially when AGC/U codons constitute a large percentage of usage in the general proteome. AGC, for example, constitutes 24% of all six Ser codons in the proteome, which is actually larger than the UCC usage of 22%; regardless, the 21 Ser repeats in Fibrosin-1 did not use a single AGC codon while using 10 UCC codons. Even UCG, the least used Ser codon in the human proteome (only 5% of all Ser codons; Fig. 1F) was used 6 times (Supplementary Material 1). Clearly, although I have designated Fibrosin-1 Ser15 as close to Class I due to its use of multiple Ser codons of the UCN family, the overall usage ratio of synonymous Ser codons in Fibrosin-1 runs afoul of the codon usage bias.
Another form of codon bias violation in SAARs is what can be called ‘overcompliance’ of the usage ratio. In such cases, the codon with higher proteomic bias is used more frequently, but exceeding the bias. An example is the TBP Gln38 repeat (Fig. 1B), in which the CAG:CAA ratio is 12:1, whereas the proteomewide ratio is only 3:1. Similarly, the Androgen Receptor (AR) Gln23 repeat consists of 22CAG codons in a row, followed by a single CAA (Supplementary Material 1), decidedly violating the 3:1 ratio. In the same protein (AR), the Gly23 SAAR (Supplementary Material 1) contains 17 GGC codons, all in an uninterrupted repeat run, but only one GGG codon and no GGA codon. Although GGC indeed has the highest codon bias of 34% in the Gly codon family, GGG and GGA are close second, both at 25%. Clearly, the codon usage in this SAAR is a significant departure from codon bias.
2.3. Microrepeats
In several mixed-codon SAARs (i.e. the Class I type), a small set of codons appears to be repeated, and hence, I have designated them as microrepeats. For example, in the Glu16 repeat in SKIDA1, GAA1-GAG4 may be considered a microrepeat (Fig. 1D). Another interesting example is the large and complex Ala55 repeat of SPT20HL1 (Fig. 1A), which is essentially Class II, because it repeats a single Ala codon, GCU, while the GCA, GCC, GCG codons are each used only once. However, the Ala runs are frequently interrupted by single Pro and Leu codons, CCU and CUA, respectively, and as a result, portions of it appear to be a repeat of CCU-GCU-CUA-(GCU)4 (i.e. peptide PALAAAA). Some variations of this microrepeat, with Val instead of Leu, are also found downstream (not shown). RUNX2 (Fig. 1B) can also be viewed as composed of four microrepeats of CAA-(CAG)n.
2.4. Double amino acid repeats (DAARs) avoid mRNA secondary structures
Multiple studies have analyzed AARs for their effect on translation rate and protein folding. However, strong secondary structure of the mRNA can also impede translation and even promote termination [18, 19]. A major goal of this study was to examine the AARs for propensity to form mRNA secondary structure. Basically, the plan was to test if one part of the repeat region can base pair with another part by RNA folding (i.e. antiparallel), and thus forming the stem of a hairpin of sufficient thermodynamic stability. To this end, it was realized that mRNA sequences in SAARs, such as the ones presented above, would not fold into a structure because neither identical nor synonymous codons are self-complementary. For example, GCN codons (coding for Ala), regardless of the identity of N, will not pair with another GCN codon. In a specific example 5′-GCA-3′ will not pair with itself or with 5′-GCC-3′, 5′-GCG-3′ or 5′-GCU-3′, even when G:U base pair (in addition to canonical Watson-Crick A:U and G:C) is allowed. Attention was, therefore, paid to the next higher level of repeats, i.e. double amino acid repeats or DAARs.
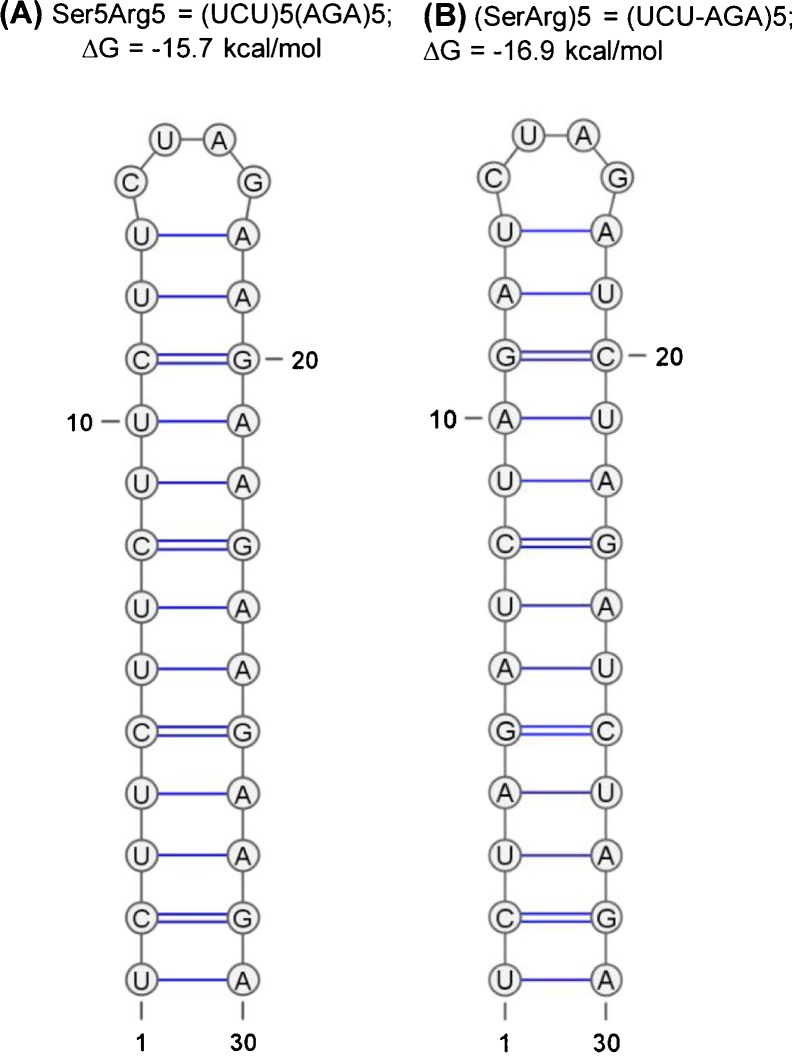
To predict if any DAAR is capable of base-pairing at the mRNA level, all codons corresponding to all 20 amino acids were manually examined for their theoretical complementarity with other codons. The results, showing all possible hairpin compatibility of codon pairs, were tabulated (Supplementary Material 2). The GenBank protein database was then queried for either (aa1)n(aa2)n or (aa1aa2)n. The optimal value of ‘n’ was considered to be 5 (i.e., pentamer of either arrangement), based on the prospect of finding mRNAs that can fold into hairpins. A thermodynamically viable hairpin needs ∼6 base-pairs of stem plus ∼4–6 nt of loop, totaling to ∼18 nt; however, extra nucleotides (total of 30 nt) were queried to make allowance for larger loops and one or two insertions or bulges in the stem region for imperfect repeats. Such imperfections would affect hairpin stability, but this may be compensated by longer stems. In other words, the goal here was to not eliminate the slightly imperfect matches in the amino acid BLAST, in case such imperfect amino acid repeats would still generate a common RNA fold. For illustration purposes, two hypothetical hairpins of perfect complementarity are shown for the two types of Ser-Arg DAARs (Fig. 2), which have essentially the same ΔG because the H-bonds are identical, resulting from base-pairing between UCU and AGA triplets, with 4 nt allotted for the loop. Lastly, the sliding nature of the BLAST alignment ensured that the search for pentameric DAARs would also include all longer DAARs (n > 5). In addition, when n = 5 search failed to find any DAAR, I searched for several double amino acid strings of shorter length, such as n = 3. In summary, the various iterations of the search covered essentially all DAARs except the very short, insignificant ones.
Fig. 2.
Hypothetical perfect RNA hairpins formed in DAARs. Nucleotide (mRNA) sequences consisting of complementary codons (UCU, AGA) for the two types of DAARs are illustrated by Ser-Arg repeats, (S)5(R)5 and (SR)5. RNA structure prediction was conducted by the MFE method as described in Materials and Methods. In this display, the A:U base pair bonding is indicated by a single line, and G:C pair, by a double line. Note the very similar thermodynamic stability (ΔG) of the two structures, primarily because of identical base-pairs and lengths.
As summarized (Supplementary Material 2), multiple searches of various DAARs of type (aa1)n(aa2)n did not find any repeat of respectable size (n > 3). The search was then conducted for the (aa1aa2)n type DAAR and again no such DAAR could be found (Supplementary Material 2). Although these are negative results, it appears that in the evolution of a DAAR, a large number of amino acid pairs, whose codons can base pair to form secondary structures in mRNA, have been selected against.
2.5. Strategies of avoidance of RNA structure in DAAR regions
I then paid a closer look at the few (aa1aa2)n-type DAARs that were actually found, imperfect as they were, in order to understand how they may have evaded mRNA structures. Specifically, for each such amino acid repeat, I retrieved the nucleotide sequence of the mRNA of that region, and subjected it to RNA folding algorithms. I also tabulated all codons for their base pairing complementarity with one another, which served as a guideline for finding the AARs with the potential to form RNA folds (Supplementary Material 3). These investigations revealed three kinds of strategies that averted base-paring, which are described below.
-
(i)
Shorter repeats: While searching for Trp-containing DAARs, such as WP, WS, WL, I found several very short and spaced WX (X is any amino acid) repeats in relatively unusual proteins, such as an alternate prion, keratin 9, and unnamed proteins. The WX lengths were too short to form a structure. This pattern was observed in a few other protein dimers such as PR (Pro-Arg). This is in fact the reason I queried the original search with pentamers.
-
(ii)
Replacement by a conservative amino acid: A much more common scenario was where an amino acid in the repeat was substituted with a conservative replacement, thereby providing a non-pairing codon. For example, a search for (CI)5 could not find this repeat, but instead found (CV)5 in an ankyrin repeat LEM domain protein, such that the hydrophobic Ile is replaced with similarly hydrophobic Val or Leu (CVCVCVCVCVCVCLCVCVCV), no codon of which could pair with the Cys codons. As a result, RNA folding was precluded while the amino acid repeat likely retained its structure and function in the protein. In another example, search for (HV)5 led to (HI)5 instead, whereby Ile replaced Val, both residues being hydrophobic. The replacement averted base pairing between complementary CAU (His) and GUG (Val) codons, and none of the three Ile codons (AUU, AUC, AUA) was complementary to CAU (His) due to the mismatch between the first base C of CUA with any of the three third bases of Ile (U, C, A). Although many amino acids have four codons that differ only in the third (wobble) base, Ile has only these three, as the possible fourth codon AUG codes for Met, which could pair with CAU. In other words, a His-Met repeat mRNA, consisting of CUA-AUG repeat could form an RNA structure, and therefore, it is probably not a coincidence that His-Met repeats do not exist (Supplementary Material 2). Many GX repeats (X being any amino acid) avoided the Gly-Ile pair with codons GGU and AUC, which are complementary, and instead replaced Ile with a variety of other hydrophobic amino acids, such as in GMGIGVGTGVDAGMGIGVGTG in XP_016886049.1, producing an RNA sequence that would not fold. Similarly, the search for IY repeat that could pair AUA (Ile) with UAU (Tyr) led instead to short repeats of IX (Ile-any amino acid) with diverse X, such as the sequence IYIYIHTYIHICIYIYMYFYIYVY in XP_016864375.1. In fact, in cases of extreme diversity, this type of DAAR may not even qualify as a repeat.
-
(iii)
Use of alternative synonymous codons: By far the most intriguing strategy to avoid RNA structure through codon choice was found in multiple examples in which a pairable codon was substituted by a non-pairable codon of the same amino acid (i.e. a synonymous codon). For example, the purest RA (Arg-Ala) repeat found was RARARARARATRARRAVQKRA in NP_057691.1 (armadillo repeat X-linked protein), but the RA repeat length was not long enough for a hairpin. More interestingly, this repeat avoided the two codons that could base-pair, namely CGU of Arg and GCG for Ala, and used several other, synonymous codons for both amino acids (e.g. AGN for Arg), which could not base-pair (Supplementary Material 3). In another example, a number of Gly and Val codons are complementary with each other, such as (written in the order Gly-Val) GGC-GUU, GGU-GUU, GGC-GUC, and GGU-GUC. However, the few Gly-Val repeats, which were in fact found (such as in XM_017030560.1, coding for a fibroin heavy chain-like protein, rich in various repeats), used GGU for Gly and GUG for Val, which are noncomplementary. The GGU codon was chosen over GGC, even though in the overall human proteome, GGU is the rarest codon for Gly, and GGC is the most frequent (GGU:GGC = 0.16:0.34) (Supplementary Material 2).
Polymeric runs of GU present a unique situation, an example of which was encountered in a DAAR search. The mRNA of NP_001265373.1 (human ankyrin repeat and LEM domain containing 1, ANKLE1) contained a repeat of GUG-UGU, corresponding to a heptamer stretch of the Val-Cys dipeptide, located at the very C-terminus of the polypeptide. However, in a long GU/UG run, most G:U hydrogen bonds do not actually form for thermodynamic reasons; accordingly, the calculated ΔG of this hypothetical hairpin was only – 6 kcal by Mfold, and attempts to fold this sequence in RNAstructure returned the message “This structure contains no pairs”. In this particular example, even if the hairpin were to form in vivo, any ribosomal slowdown promoted by it would actually help the translation termination process at the natural stop codon that follows the hairpin. In general, DAARs with purely GU-containing codons are unlikely to impede mRNA translation through RNA structural folds.
3. Discussion
In this communication, I have surveyed the various design formats of amino acid repeats (AARs) and their mRNA sequences in terms of codon usage. Based on the representative results, I have presented a schematic model for the evolution of the AARs (Fig. 3), whereby two major forces shape the design of a repeat: codon bias and RNA fold. While adherence to codon bias is considered favorable for optimal speed of translation and folding of the nascent polypeptide [19][reviewed in 19], formation of a RNA hairpin is generally detrimental to translation [18]. I have presented examples of amino acid repeats that apparently comply with codon bias (majority being in Class I SAARs) and those that are robustly noncompliant (all Class II type SAARs). In contrast, and regardless of the nature of the repeat or its codon usage, there was not a single instance of any AAR mRNA that could form RNA secondary structure. In SAARs, this is because of the way codons have been created, such that all synonymous codons are noncomplimentary, which also holds true for amino acids with six synonymous codons (Arg, Leu, Ser). The best-known examples are the so-called trinucleotide repeat diseases, such as the polyQ expansion diseases, where the coding sequence consists of mainly CAG repeats. Since RNA structure is not a concern in SAARs, the functional abnormality of these sequences may be determined primarily at the protein level, such as protein conformation and solubility. In fact, SAAR of hydrophobic residues, such as polyQ and polyA are widely known to oligomerize and aggregate [14, 20].
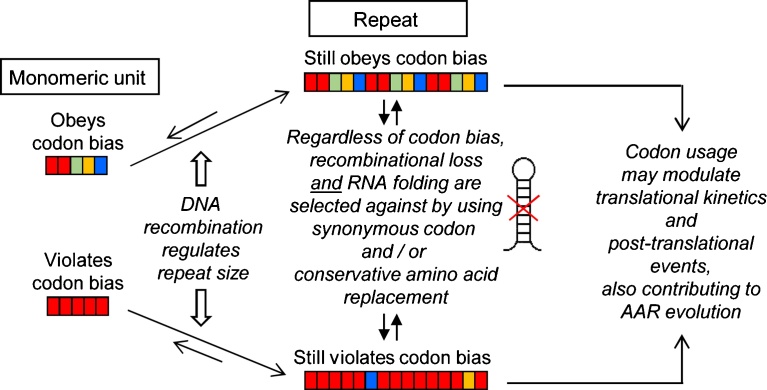
Fig. 3.
Schematic model of AAR formation. In this scheme (as in Fig. 1), the colored boxes indicate different but synonymous codons for the same amino acid of an AAR. All AARs may start with a short sequence of a few amino acids, operationally defined as a “monomeric unit”. The starting unit as well as the final repeat, produced by imperfect replication, may have various degrees of agreement or disagreement with the organismic codon bias. DNA repeats can promote excision of units (indicated by the shorter arrow at the recombination step), but this is minimized by the imperfection of the repeats, produced by conservative amino acid replacement and substitution with synonymous codons, which at the same time prevents formation of secondary structural folds in the mRNA. The resultant codon distribution may regulate translational kinetics and folding of the AAR protein, also contributing to the overall evolution of the AAR sequence.
It can be envisaged that all repeats began life as a smaller unit (Fig. 3), perhaps as a primal motif, which gradually increased in length through DNA replication and/or recombination, with some form of “proof-reading” at every step to fine tune RNA folding and codon usage. Although the forward evolution takes precedence in creating a repeat, dynamic homologous recombination may occur at each step, causing partial loss of the repeats, depending on the degree of homology and the evolutionary fitness of the intermediate repeat lengths. Indeed, continuing deletion and expansion by internal DNA recombination is a general concern in homopolymeric genome sequences, as in SAARs. This has been considered the reason behind the instability of polyQ sequences in neurodegenerative diseases, such as Huntington disease and Spinocerebellar ataxia-8 (Fig. 1B). The CAG repeats in the AIB1 polyQ gene, sequenced in a large collection of breast cancer cell lines, were also highly polymorphic in length, but unlike those in the neurodegenerative diseases, were clonally stable in length [12]. Interestingly, the major difference between the two kinds of polyQ genes is that the CAG repeats in AIB1 are frequently interrupted by CAA, the other Gln (Q) codon, whereas those in neurodegenerative diseases, such the huntingtin and ataxin-8 (ATXN8), contain pure CAG repeats (Fig. 1B). Since RNA folding is not an issue in SAARs, interruption by synonymous codons, especially in the Class II SAARs (many examples in Fig. 1), most likely serves to evade homologous DNA recombination, as previously hypothesized [12]. Nevertheless, the significance of Class I (interrupted) and Class II (uninterrupted) types of SAARs remains an open question; stated in terms of polyQ repeats, it is unclear why the polyQ sequences in the two disease types are designed differently and whether recombinational plasticity in the uninterrupted CAG repeats may actually have an evolutionary advantage in the neurodegenerative polyQ genes.
The highly variable and polymorphic sequences in various AARs suggest that such sequences are dynamic in nature and in a constant evolutionary flux, offering a spectrum of functionality between health and disease. Nonetheless, each sequence variant in a family of AAR, such as the polyQ, must have attained a metastable evolutionary equilibrium, maintaining its sequence long enough to exert a biological consequence [12]. Due to the complexity of this dynamism, the molecular checkpoints of the evolution of an AAR remains undefined. Clearly, how the needs of compliance with codon bias and protein folding co-evolved with the apparent avoidance of RNA secondary structure in an AAR sequence would constitute an interesting study of the future.
4. Materials and methods
4.1. Sequence retrieval
First of all, to avoid variability between species and to obtain results that may have future clinical benefits, I focused exclusively on human repeats. All repeats were retrieved from GenBank by BLASTP search of the Homo sapiens protein database. For greater authenticity, only confirmed proteins were included in the study, and ‘hypothetical’, ‘unnamed’ and ‘probable’ proteins were excluded. For the same reason, only those proteins, for which validated RefSeq mRNA sequences were available, were acquired. The default parameters of BLASTP were used with full stringency with the goal to obtain perfect or at least nearly perfect repeats, since the insertion of other amino acids would interrupt and hence disqualify a repeat.
For the SAAR search, a 12-mer single amino acid repeat sequence, such as (Ala)12 (queried as AAAAAAAAAAAA) was used; for DAARs, 5-mer repeat of the dipeptide was used, such as (Phe-Lys)5, queried as FKFKFKFKFK. The rationale behind the repeat length is described in appropriate places under Results. The protein sequences were screened to retain only the best matches (high similarity scores with little or no gaps). The corresponding nucleotide sequences were retrieved and the codons of the repeat region were identified, cataloged, and analyzed.
4.2. RNA structure prediction
In predicting the RNA structural folds, well-established structure-modeling approaches were employed, which computed structure either on the basis of Minimum Free Energy (MFE) [21] or Maximum Expected Accuracy (MEA) [22]. In general, the RNAstructure suite, version 5.8.1 (University of Rochester), was used first [23]. For MFE, the maximum % energy difference was 10, and window size 0. The MEA model was built on RNA single strand partition function, also computed at the RNAstructure suite, leading to CT files that were used to draw the secondary structures, if found, using the Java applet VARNA (Visualization Applet for RNA) [24]. Additionally, independent confirmation was obtained by using the RNA Mfold web server [25], although all RNA folding suites currently share the modified Turner parameters or some variations thereof [21, 26]. In all cases, the different folding programs predicted the same structure.
Declarations
Author contribution statement
Sailen Barik: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Competing interest statement
The author declares no conflict of interest.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Chavali S., Chavali P.L., Chalancon G., de Groot N.S., Gemayel R., Latysheva N.S., Ing-Simmons E., Verstrepen K.J., Balaji S., Babu M.M. Constraints and consequences of the emergence of amino acid repeats in eukaryotic proteins. Nat. Struct. Mol. Biol. 2017;24:765–777. doi: 10.1038/nsmb.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mier P., Alanis-Lobato G., Andrade-Navarro M.A. Context characterization of amino acid homorepeats using evolution, position, and order. Proteins. 2017;85:709–719. doi: 10.1002/prot.25250. [DOI] [PubMed] [Google Scholar]
- 3.Andrade M.A., Perez-Iratxeta C., Ponting C.P. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura S.H., Hirano T. HEAT repeats – versatile arrays of amphiphilic helices working in crowded environments? J Cell Sci. 2016;129:3963–3970. doi: 10.1242/jcs.185710. [DOI] [PubMed] [Google Scholar]
- 5.Schüler A., Bornberg-Bauer E. Evolution of protein domain repeats in metazoa. Mol. Biol. Evol. 2016;33:3170–3182. doi: 10.1093/molbev/msw194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeytuni N., Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Luo H., Nijveen H. Understanding and identifying amino acid repeats. Brief Bioinform. 2014;15:582–591. doi: 10.1093/bib/bbt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faux N. Single amino acid and trinucleotide repeats: function and evolution. Adv. Exp. Med. Biol. 2012;769:26–40. doi: 10.1007/978-1-4614-5434-2_3. PMID:23560303. [DOI] [PubMed] [Google Scholar]
- 9.Hancock J.M., Simon M. Simple sequence repeats in proteins and their significance for network evolution. Gene. 2005;345:113–118. doi: 10.1016/j.gene.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 10.Adegbuyiro A., Sedighi F., Pilkington A.W., 4th, Groover S., Legleiter J. Proteins containing expanded polyglutamine tracts and neurodegenerative disease. Biochemistry. 2017;56:1199–1217. doi: 10.1021/acs.biochem.6b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nageshwaran S., Festenstein R. Epigenetics and triplet-repeat neurological diseases. Front. Neurol. 2015;6:262. doi: 10.3389/fneur.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai P., Wong L.J. Somatic instability of the DNA sequences encoding the polymorphic polyglutamine tract of the AIB1 gene. J Med. Genet. 2003;40:885–890. doi: 10.1136/jmg.40.12.885. PMCID:PMC1735346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orafidiya F.A., McEwan I.J. Trinucleotide repeats and protein folding and disease: the perspective from studies with the androgen receptor. Future Sci. OA. 2015;1 doi: 10.4155/fso.15.47. FSO47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oma Y., Kino Y., Sasagawa N., Ishiura S. Intracellular localization of homopolymeric amino acid-containing proteins expressed in mammalian cells. J. Biol. Chem. 2004;279:21217–21222. doi: 10.1074/jbc.M309887200. [DOI] [PubMed] [Google Scholar]
- 15.Uthayakumar M., Benazir B., Patra S., Vaishnavi M.K., Gurusaran M., Sureka K., Jeyakanthan J., Sekar K. Homopeptide repeats: implications for protein structure, function and evolution. Genom. Proteom. Bioinform. 2012;10:217–225. doi: 10.1016/j.gpb.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faux N.G., Bottomley S.P., Lesk A.M., Irving J.A., Morrison J.R., de la Banda M.G., Whisstock J.C. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mularoni L., Veitia R.A., Albà M.M. Highly constrained proteins contain an unexpectedly large number of amino acid tandem repeats. Genomics. 2007;89:316–325. doi: 10.1016/j.ygeno.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Chen C., Zhang H., Broitman S.L., Reiche M., Farrell I., Cooperman B.S., Goldman Y.E. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013;20:582–588. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 21.Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z.J., Gloor J.W., Mathews D.H. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009;15:1805–1813. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darty K., Denise A., Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. PMCID:PMC169194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seetin M.G., Mathews D.H. RNA structure prediction: an overview of methods. Methods Mol. Biol. 2012;905:99–122. doi: 10.1007/978-1-61779-949-5_8. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y., Gojobori T., Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. PMCID:PMC102460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.