Abstract
Epidemiological and mechanistic evidence on the causative role of human papillomaviruses (HPV) in esophageal squamous cell carcinoma (ESCC) is unclear. We retrieved alcohol- and formalin-fixed paraffin-embedded ESCC tissues from 133 patients seropositive for antibodies against HPV early proteins, from high-incidence ESCC regions; South Africa, China, and Iran. With rigorous care to prevent nucleic acid contamination, we analyzed these tissues for the presence of 51 mucosotropic human alpha-papillomaviruses by two sensitive, broad-spectrum genotyping methods, and for the markers of HPV-transformed phenotype: (i) HPV16/18 viral loads by quantitative real-time PCR, (ii) type-specific viral mRNA by E6*I/E6 full-length RT-PCR assays and (iii) expression of cellular protein p16INK4a. Of 118 analyzable ESCC tissues, 10 (8%) were positive for DNA of HPV types: 16 (four tumors); 33, 35, 45 (one tumor each); 11 (two tumors); and 16, 70 double infection (one tumor). Inconsistent HPV DNA+ findings by two genotyping methods and negativity in qPCR indicated very low viral loads. A single HPV16 DNA+ tumor additionally harbored HPV16 E6*I mRNA but was p16INK4a negative (HPV16 E1 seropositive patient). Another HPV16 DNA+ tumor from an HPV16 E6 seropositive patient showed p16INK4a up-regulation but no HPV16 mRNA. In the tumor tissues of these serologically preselected ESCC patients, we did not find consistent presence of HPV DNA, HPV mRNA or p16INK4a up-regulation. These results were supported by a meta-analysis of 14 other similar studies regarding HPV-transformation of ESCC. Our study does not support the etiological role of the 51 analyzed mucosotropic HPV types in the ESCC carcinogenesis.
INTRODUCTION
Esophageal cancer is the eighth most common malignancy worldwide and the sixth most common cause of death from cancer1. Of various histological subtypes esophageal squamous cell carcinoma (ESCC) is the dominant histological subtype worldwide2.
ESCC incidence varies over 100-fold between low-incidence regions such as western Africa and eastern China, and high-incidence regions such as southern Africa, northern China, and the Caspian littoral of Iran1. While tobacco smoking and alcohol consumption are the main risk factors for ESCC in many countries3–5, additional factors play a role such as consumption of hot tea in South America and Iran, diet deficiencies in parts of South Africa and China, and the consumption of opiates in Iran (6 and references therein).
A potential association of mucosotropic human papillomavirus (HPV) types of the alpha-papillomavirus genus with ESCC, was first proposed in 1982 based on the histological identification of condylomatous lesions in 38% of ESCC7, 8. Since then, more than 100 epidemiological and laboratory studies have reported HPV DNA presence in ESCC, with positivity varying from 0 – 100%9, 10, even among studies conducted within the same geographical region11–13. Some of the highest HPV DNA prevalences have been continuously reported from the high-incidence regions in South Africa (39%)9, 10, 14, China (44%)10, and Iran (37%)10. However, in the three most recent studies that documented significant efforts to prevent potential nucleic acid cross-contamination during esophageal tissue sectioning and HPV DNA analysis, these high prevalence values for South Africa, China and Iran could not be reproduced (prevalence of HPV DNA positivity was <1%)15–17.
While epidemiological data are an important starting point in ascertaining causation, mechanistic data are considered essential to establish a biological causal link, especially for infection-cancer associations. Cervical squamous cell carcinoma (CSCC) is the best-understood model for HPV-transformation by one of the 12 high-risk (HR-)HPV types that are considered carcinogenic to humans by the International Agency for Research on Cancer (IARC)3, 18–22. In addition to HPV DNA presence, CSCC is characterized by: (i) at least 1 viral genome copy present in each tumor cell20, 23, (ii) expression of viral oncogenes E6 and E719, 24, and (iii) alteration of steady state levels of cellular proteins, most consistently up-regulation of p16INK4a 24, 25. Such biological evidence has been provided for the 12 HR-HPV types, but not for the low risk (LR-)HPV types in CSCC3. Studies on head-and-neck cancers have provided further evidence that the same criteria are valid for a subset of malignant head-and-neck lesions26–29. Most importantly, these studies demonstrated that the presence of HPV DNA alone in invasive tumor tissues is insufficient proof of viral causality and could result in misclassification of malignant lesions. For HPV DNA-positive ESCC, very limited direct evidence for viral transforming activity exists: just one study demonstrated in vitro transformation of esophageal epithelium by HPV1830, while two other reports that analyzed the presence of HPV genomes in esophageal cell lines had contradictory outcomes31, 32. Scarce evidence has been reported on HPV viral load33–36 or expression of viral transcripts37. And while 14 studies have reported on expression of p16INK4a protein in ESCC tissues they used varying cut-offs to define p16INK4a positivity and reported contradicting results15, 34, 38–49. As a result of inconsistent HPV DNA data and very limited direct functional evidence provided in the last 30 years of research, the worldwide expert panel of the 100th IARC Monograph concluded that the evidence for a causal association between HPV and ESCC remains inconclusive3.
In addition to HPV functional markers that can be assessed in tumor tissues, antibodies to HPV early proteins, especially E6 and E7, have been demonstrated to be markers for HPV-driven SCC of the cervix50, 51, penis52, and oropharynx51, 53. Recently InterSCOPE, the largest sero-epidemiological study on HPV in ESCC, compared (in a blinded fashion) 1,561 ESCC case, with 2,502 control subjects from six geographical areas, for antibodies against sixteen early proteins of the eight HR-HPV types most prevalent in HPV-driven cancers, and LR-HPV types 6 and 11 which are also known to infect the upper aerodigestive tract. HPV early protein antibodies were rare; the highest prevalence in cases was 2.6% for HPV6 E6. Only two significant but weak ESCC associations were found for antibodies to E6 proteins; these were for HPV16 (OR=1.89, 95%CI=1.09–3.29, p=.023) and HPV6 (OR=2.53, 95%CI=1.51–4.25, p<.001)6.
In order to identify ESCC cases which were most likely to be HPV-driven, we selected patients who were seropositive for HPV early proteins in the InterSCOPE study and analyzed their tumor tissues for the presence of HPV DNA and a combination of HPV functional markers (viral load, HPV mRNA, p16INK4a up-regulation) known to characterize HPV-driven cervical and oropharyngeal cancers, and used state-of-the-art methods, and stringent conditions, to prevent nucleic acid cross-contamination.
MATERIALS AND METHODS
Ethical clearance
All analyses were approved by the appropriate national or institutional ethics committees or review boards. Written or witnessed oral informed consent was obtained from study participants.
Study population
Sera from 1,811 ESCC patients were analyzed in the sero-epidemiological InterSCOPE study, of which 1,561 had sufficient covariate data to be included in the case-control analysis6. Of these 1,561 ESCC patients, 357 were seropositive to at least one of the sixteen HPV early proteins from the eight most prevalent HR-HPV types: HPV16 (E1, E2, E6, E7); HPV18 (E6, E7); HPV31, 33, 35, 45, 52, and 58 (E6); or LR-HPV types 6 and 11 (E6, E7). These 357 patients originated from six world regions with widely varied ESCC incidence: South Africa, Northern China, Brazil, Central and Eastern Europe, Australia, and Iran6. Alcohol-fixed or formalin-fixed, paraffin-embedded (AFPE/FFPE) tumor tissue blocks could be retrieved for HPV molecular analysis from 133 patients originating from three regions with high ESCC incidence (South Africa: 58 FFPE, Northern China: 35 AFPE, and Iran: 40 FFPE).
Preparation of tissue sections and nucleic acid extraction
Tissue blocks (40 FFPE from high-incidence ESCC region in Iran and 58 FFPE from South Africa) or sections (corresponding to 35 AFPE from Shanxi Province in Northern China) were sent to Heidelberg, Germany for molecular analysis. Sections were cut for HPV DNA and HPV RNA extractions (5 µm each), and p16INK4a immunohistochemical staining (IHC) (4 µm), following established protocols24. Rigorous care was applied to control for and prevent potential nucleic acid cross-contamination during: (i) tissue sectioning, (ii) DNA and RNA extractions, (iii) PCR/Reverse-Transcription (RT)PCR/quantitative (q)PCR analysis, and (iv) hybridization of PCR products. For each patient’s specimen, a new cutting blade was used and the sectioning area was cleaned with acetone and 70% alcohol. A sectioning control (HPV DNA-free FFPE mouse liver) was cut after each 10th patient specimen and included in DNA/RNA extractions. DNA/RNA extraction controls (one lysis buffer included after each 11 patient samples), (RT-)PCR controls (one water aliquot and one PCR master-mix aliquot after each 14 patient samples on the 96 well-plate), and hybridization controls (two hybridization buffer samples per 96 well-plate) were included. All controls yielded HPV DNA−/RNA− results. For each 11 ESCC cases, tissue sections of pretested HPV16 DNA+/RNA+ CSCC were included in DNA/RNA extraction, (RT-)PCR runs and on hybridization plate as a positive control and to control for data reproducibility. These controls showed reproducible HPV16 DNA+ and RNA+ results in all assay runs. Detailed data of individual molecular assays per patient sample, in combination with HPV serology data from patient blood, are summarized in Supplementary Table S1.
Additional sections above and below the DNA/RNA/IHC sections were examined and verified by a pathologist (CF) for the presence of: (i) squamous cell carcinoma, (ii) ≥25% non-necrotic tumor cells (23/118 ESCC tissues had 25 – 50% of tumor cells in the biopsies, and 95/118 had >50% tumor cells), (iii) muscular wall (in surgical specimens only) to confirm esophageal origin, and (iv) for the degree of keratinization. DNA was extracted from sections by overnight Proteinase K digestion at 56oC as described24. RNA was extracted from sections using the PureLink FFPE Total RNA Isolation Kit (Invitrogen) with overnight incubation at 56oC and QIAGEN DNase digestion as described24.
HPV genotyping, viral load and mRNA analysis
For genotyping, 5 µl of DNA extract was used from each of the 133 specimens. Two sensitive genotyping assays targeting HPV L1 and E7 gene sequences were applied. The broad spectrum BSGP5+6+-PCR/Multiplex Papillomavirus Genotyping (BSGP5+6+-PCR/MPG) assay homogenously amplifies a ~150 bp fragment from the L1 region of 51 defined mucosotropic human alpha-papillomavirus types including all HPV types classified by IARC/WHO as carcinogenic, probably carcinogenic, or possibly carcinogenic3. These 51 HPV types hereafter are called mucosal HPV types. The assay further amplifies a 208 bp cellular β-globin sequence. The detection limits per reaction are between 10 to 1,000 copies for the viral genomes and 300 copies for β-globin23.
The type-specific E7-PCR/MPG assay (TS-E7-PCR/MPG) utilizes HPV type-specific primer pairs targeting the E7 region of 21 genital HPV types plus primers for the amplification of a β-globin sequence54, 55. The cycling conditions and the sequences of the primers have been previously described56. Here, a modified protocol for the amplification of shorter (~100 bp) fragments for ten HPV types: HPV16, 18, 31, 33, 35, 52, 56, 66; 6 and 11, and 117 bp for β-globin, was applied. Modified, shorter primer sequences are listed in Supplementary Table S2.
To measure viral load, a multiplex HPV16/18 quantitative real-time PCR (qPCR) with very short amplicons was developed to best suit the analysis of DNA extracted from fixed tissues (Schmitt et al., in preparation). The multiplex HPV16/18 qPCR amplifies 104 bp of HPV16 E6, 110 bp of HPV18 E7, and 110 bp of β-globin sequence with a detection limit of 10 HPV plasmid and 10 β-globin copies per reaction. One µl of DNA extract was used for viral load measurements. The primer and probes sequences are listed in Supplementary Table S3.
The HPV type-specific E6*I mRNA assays developed for 20 HR/pHR-HPV types24 and the E6 full length (fl) mRNA assay developed for HPV11, were applied for detection of viral transcripts. These assays amplify 65 – 75 bp HPV and 81 bp ubiquitin C (ubC) cDNA and were extensively validated on cervical and head-and-neck SCC FFPE samples, deep fresh frozen specimens (DFT) and exfoliated cells24, 29. Analytical sensitivity of each assay is 10 to 100 copies per reaction for 19 HPV types and for ubC, 1,000 copies for HPV67 and 10,000 copies for HPV7024. All 133 ESCC patients’ tissues were analyzed for the presence of: (i) HPV16 E6*I mRNA, (ii) ubC mRNA as a cellular mRNA positive control, and (iii) mRNA of the non-HPV16 types determined by genotyping and/or serological assays. We did not test for mRNA of LR-HPV6 since the mRNA assay for HPV6 could not be thoroughly validated on HPV6 DNA+ tissues.
Each ESCC specimen that had ≥25% non-necrotic tumor cells prior and after sectioning for HPV DNA, RNA and IHC, and yielded HPV DNA and/or β-globin DNA-positive (DNA+) signal in at least one of the three methods used in the DNA analysis, was considered DNA valid. Specimens that were HPV and/or ubC mRNA-positive (RNA+) in RNA analysis were considered RNA valid.
P16INK4a immunohistochemistry
Expression of p16INK4a was assessed in all 133 samples using the primary p16INK4a antibody (clone No. E6H4, Roche MTM Laboratories, Heidelberg, Germany) in a manual IHC procedure as previously described24, 29, 57. Each staining batch included tissue sections for HPV16 DNA+/RNA+ cervical cancer, and HPV DNA-negative (HPV DNA−) normal oral epithelium, which served to control for intra- and inter-day staining reproducibility and protocol performance. Expression of p16INK4a was evaluated separately by well-defined criteria for protein down- and up-regulation by two experienced investigators blinded of HPV results (GH and DH). Evaluation involved semi-quantitative scoring of the staining intensity (0=no expression, 1 or 2=low intensity, and 3=high intensity), estimation of the percentage of stained tumor cells (<10%, 11–25%, 26–50%, 51–75% and >75%) and of the staining pattern (focal or diffuse). This thorough evaluation criterion for p16INK4a up-regulation in relation to HPV was defined on 321 HPV DNA+/RNA+ cervical cancer specimens in collaboration with two experienced pathologists as described22. For final protein expression only two categories were applied: (i) up-regulation (diffuse p16INK4a expression in >25% of tumor cells with intensity 3+), or (ii) down-regulation (focal or patchy p16INK4a expression in ≤25% of tumor cells with intensity ≤3+)29. Evaluation was discordant in 1 case for which a new tissue section was cut and stained for p16INK4a, re-evaluated by both investigators (GH and DH) and consensus was reached.
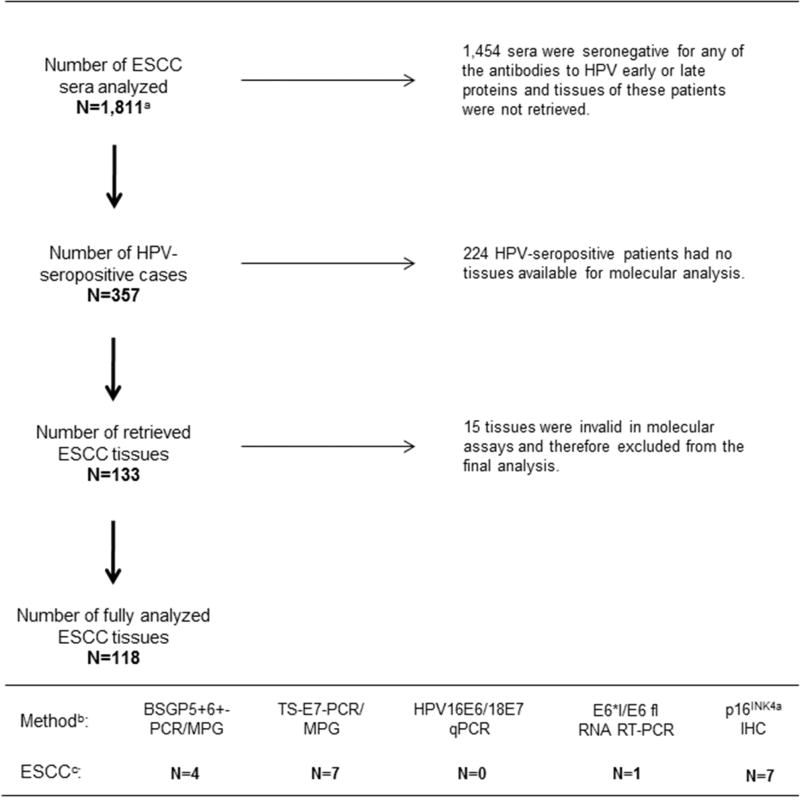
Overview of total number of ESCC sera and ESCC tissues analyzed in the InterSCOPE study, and overview of the molecular methods applied for tissue analysis, is depicted in Figure 1.
Figure 1. Overview of serological and tissue analyses in InterSCOPE study.
a 1,565 of the 1,811 sera had sufficient covariate data that were included in a previous case-control analysis (Sitas et al., 2012). b 118 ESCC tissues were analyzed using five molecular methods to assess: (i) presence of HPV DNA (BSGP5+6+-PCR/MPG and TS-E7-PCR/MPG), (ii) viral load (HPV16/18 E6 qRT-PCR), (iii) HPV mRNA (E6*I/E6 fl RNA RT-PCR), and (iv) expression of p16INK4a protein as a surrogate marker of HPV-transformed phenotype (p16INK4a IHC). c Of total 118 ESCC tissues, 10 (8%) were HPV DNA+ when results of both genotyping methods were combined, none of the HPV16 DNA+ tissues showed high viral load, and only a single HPV16 DNA+ case expressed viral mRNA (HPV16 DNA+/mRNA+/p16INK4a−). Seven ESCC tissues showed up-regulation of p16INK4a and only one tissue with p16INK4a up-regulation was HPV16 DNA+ but showed no positivity for HPV16 mRNA (HPV16 DNA+/mRNA−/p16INK4a +).
Meta-analysis of studies with HPV DNA and p16INK4a data
A literature search of the PUBMED and MEDLINE databases was performed to identify studies in English language published in peer-reviewed journals by December 2014, which addressed association of HPV and esophageal cancer by including both HPV DNA analyses and analyses of p16INK4a expression in ESCC tissues. We did not include studies that addressed presence of HPV DNA and expression of p16INK4a in Barrett’s esophagus, esophageal adenocarcinoma or esophageal papilloma.
Fourteen previous studies which reported data on both HPV DNA testing and expression of p16INK4a as an HPV functional marker were identified for possible inclusion in the meta-analysis15, 34, 38–49. Of these fourteen studies, five were excluded for the following reasons: Antonsson and colleagues38, and Koshiol and colleagues15, assessed p16INK4a in HPV DNA+ tumors only; Malik and colleagues47 tested only p16INK4a positive (p16INK4a +) tumors for HPV DNA expression; Bellizzi and colleagues identified no HPV DNA+ cases in their ESCC series and hence the study provides no information about relative probabilities of HPV DNA+ and p16INK4a+39 ; and Vaiphei and colleagues49 did not report specific frequencies of p16INK4a + tumors. The remaining nine studies, and our own findings (i.e. 10 studies in total), were included in the meta-analysis. Study-specific relative risks (RR) were calculated as the proportion of HPV DNA+ cases that were p16INK4a + relative to the proportion of HPV DNA− cases that were p16INK4a +. The pooled RR was estimated using the DerSimonian and Laird random effects model58 and heterogeneity was assessed using the residual heterogeneity statistics from the inverse-variance fixed-effect model59. A continuity correction of 0.2 was added to the single zero cell in Teng et al., 201448. A funnel plot and Egger’s test60 were used to assess the likelihood of publication bias. In additional analysis, for the 8 out of 10 studies that provided a p16INK4a + cut-off, a meta-regression was performed with study-specific (log) RR regressed against p16INK4a + cut-off values.
RESULTS
Patient samples
Of the 133 ESCC tissues analyzed, 118 contained sufficient non-necrotic tumor cells (at least 25%) in sections above and below the DNA/RNA/IHC sections. Validity of DNA in DNA extracts varied between PCR methods and increased with shorter β-globin DNA amplicon size (Table 1). BSGP5+6+-PCR/MPG yielded 74/118 (63%), TS-E7-PCR/MPG 115/118 (97%), and HPV16/18 qRT-PCR 106/118 (90%) DNA valid samples (Table 1, Supplementary Table S1). All 118 tissues were positive for ubiquitin C (100%) and thus valid in RNA analysis. Two DNA extracts were invalid in all three HPV DNA assays (HPV DNA− and β-globin DNA−) but valid in RNA analysis and therefore not excluded.
Table 1.
Characteristics of ESCC patients and tissues stratified by HPV status
| Parameter | All
analyzed (N=133)a |
Valid (N=118)b |
HPV
DNA− (N=108)c |
HPV DNA+ (N=10)d |
p-valuee | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | (%) | N | (%) | N | (%) | N | (%) | |||
| Gender | ||||||||||
| Male | 82 | (65) | 71 | (64) | 68 | (67) | 3 | (30) | 0.034 | |
| Female | 44 | (35) | 40 | (36) | 33 | (33) | 7 | (70) | ||
| No data | 7 | 7 | 7 | 0 | ||||||
| Age (years) | ||||||||||
| Median | 62 | 61 | 61 | 61 | ||||||
| Alcohol use | ||||||||||
| Ever drinker | 48 | (38) | 42 | (38) | 37 | (37) | 5 | (50) | 0.502 | |
| Never | 77 | (62) | 68 | (62) | 63 | (63) | 5 | (50) | ||
| No data | 8 | 8 | 8 | 0 | ||||||
| Tobacco use | ||||||||||
| Currentf | 58 | (46) | 53 | (48) | 48 | (48) | 5 | (50) | 1.000 | |
| Former g | 19 | (15) | 15 | (14) | 14 | (14) | 1 | (10) | ||
| Never | 48 | (39) | 42 | (38) | 38 | (38) | 4 | (40) | ||
| No data | 8 | 8 | 8 | 0 | ||||||
| Other: hot teag | ||||||||||
| Warm or lukewarm | 19 | (48) | 17 | (49) | 16 | (48) | 1 | (50) | 1.000 | |
| Hot | 15 | (38) | 12 | (34) | 11 | (33) | 1 | (50) | ||
| Very hot | 6 | (15) | 6 | (17) | 6 | (18) | 0 | (0) | ||
| No data | 93 | 83 | 75 | 8 | ||||||
| Histology | ||||||||||
| Keratinizing | 121 | (97) | 114 | (97) | 104 | (96) | 10 | (100) | 1.000 | |
| Basaloid | 3 | (2) | 3 | (3) | 3 | (3) | 0 | (0) | ||
| Mixed form | 1 | (1) | 1 | (1) | 1 | (1) | 0 | (0) | ||
| NA | 8 | 0 | 0 | 0 | ||||||
| p16INK4a status (IHC) | ||||||||||
| Negative | 111 | (94) | 111 | (94) | 102 | (94) | 9 | (90) | 0.471 | |
| Positive | 7 | (6) | 7 | (6) | 6 | (6) | 1 | (10) | ||
| NA | 15 | 0 | 0 | 0 | ||||||
All tumor tissues analyzed. Tumor tissues that are:
valid in molecular/immunohistochemical analyses,
HPV DNA−/RNA−,
HPV DNA+ only (N=9) or DNA+/RNA+ (N=1).
Fisher’s exact test for differences in proportions of HPV DNA− and HPV DNA+ cases.
Smoking within 5 years of the interview.
Quit smoking prior to 5 years before the interview.
Information about hot tea consummation was available for ESCC patients from Iran only.
NA - not analyzable
HPV DNA, viral load and mRNA expression in ESCC tissues
Of the 118 tissues, 10 (8%) were HPV type DNA+ but always by a single HPV DNA assay; four (3%) by BSGP5+6+-PCR/MPG, seven (6%) by TS-E7-PCR/MPG, and none by HPV16/18 qPCR, indicating low viral loads (Table 2). HPV types identified were: HPV16 (four tumors); HPV33, 35, 45 (one tumor each), HPV11 (two tumors); and HPV16, 70 double infection (one tumor) (Table 2).
Table 2.
Molecular characteristics of HPV DNA+ ESCC tissues
| Patient Number |
Patient Seropositivity |
HPV type DNA | HPV type RNAd |
p16INK4a IHC |
||
|---|---|---|---|---|---|---|
|
| ||||||
| BSGP5+6+- PCR/MPGa |
TS-E7- PCR/MPGb |
HPV16/18 qPCRc | ||||
| 1 | 16 E6 | − | 16 | − | − | + |
| 2 | 6 E6 | − | 16 | − | 16 | − |
| 3 | 45 E6 | − | 16 | − | − | − |
| 4 | 16 E2 | − | 16 | − | − | − |
| 5 | 18 E6 | 33 | − | invalid | − | − |
| 6 | 16 E6 | − | 35 | − | − | − |
| 7 | 16 E1 | 45 | − | − | − | − |
| 8 | 18 E6 | − | 11 | − | − | − |
| 9 | 16 E1 | 11 | − | − | − | − |
| 10 | 16 E1 | 70 | 16 | invalid | − | − |
“−“, negative finding; “+“, positive finding; invalid, negative for HPV and β-globin DNA.
Available for all currently defined 51 mucosal HPV types (12 HR-, 8 pHR- and 31 LR-HPV); amplicon size: 150 bp HPV, 208 bp β-globin (total 51×118 = 6,018 reactions).
Available for 21 mucosal HPV types (12 HR-, 7 pHR- and 2 LR-HPV); amplicon size: ~100 bp HPV (for HPV6, 11, 16, 18, 31, 33, 35, 52, 56 and 66), 117 bp β-globin (total 21×118 = 2,478 reactions). For other HPV types the amplicon size was as previously published by Gheit and colleagues.
Available for HPV16 and 18; amplicon size: 104 bp HPV16, 110 bp HPV18, 110 bp β-globin (total 2×118 = 236 reactions).
Available for 21 mucosotropic HPV types (12 HR-, 8 pHR- and LR-HPV11); amplicon size: 65–75 bp HPV, 81 bp ubiquitin C. All samples were tested for the presence of HPV16 E6*I mRNA and for the presence of E6*I mRNA of the HPV type defined by genotyping assays and/or serology (total 170 reactions). E.g. for Patient Number 5, the RNA sample was analyzed for the presence of HPV16, HPV18 and HPV33 E6*I mRNA, respectively. No mRNA assay was available for LR-HPV type 6.
Of the 118 tissues analyzed in a total of 170 RNA reactions (118 reactions for HPV16 and ubC and 52 additional reactions performed for a non-HPV16 type determined by genotyping or serological assays), a single tissue positive for HPV16 DNA exclusively by TS-E7-PCR/MPG also expressed HPV16 E6*I mRNA (Table 2). None of the other HPV DNA+ or HPV DNA− tissue samples contained viral mRNA.
Expression of p16INK4a in ESCC tissues
All 118 tissue samples were valid in immunohistochemical analysis. Up-regulation of p16INK4a was found in 7/118 (6%) tissues (Supplementary Table S1). Only one of the 7 tissues with up-regulated p16INK4a was HPV16 DNA+ by TS-E7-PCR/MPG only, but it had a low viral load and lacked HPV16 transcripts (Table 2).
Meta-analysis of studies with HPV DNA and p16INK4a data
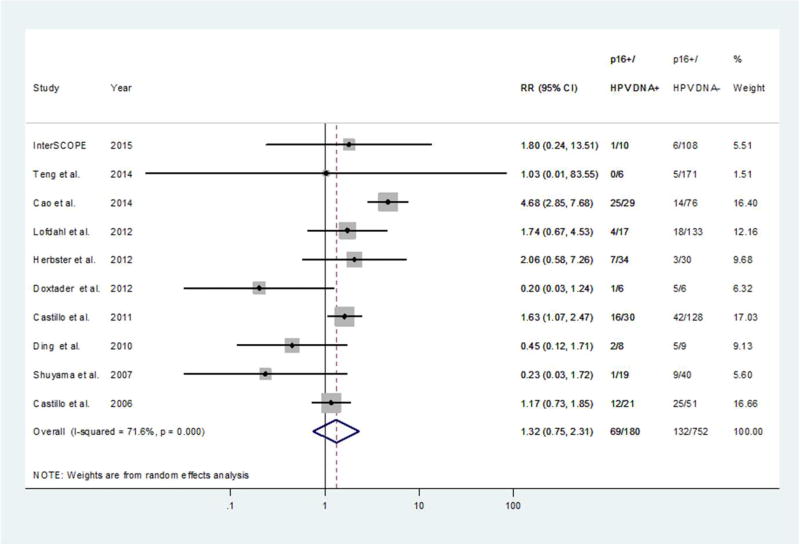
We identified 14 studies in the current literature that evaluated both tumor tissue HPV DNA positivity and expression of p16INK4a as a functional marker in their ESCC patient series (Table 3, Figure 2), and performed a meta-analysis including our own findings. Ten of the total 15 studies were valid for the meta-analysis (see Material and Methods for details on study inclusion). Significant heterogeneity was observed among the 10 study-specific relative risks (RR) (p for heterogeneity<0.001) (Table 3, Figure 2), but removal of the outlier RR estimate from Cao and colleagues40 left a considerably more homogenous set of estimates (p for heterogeneity=0.17). In two of the 10 studies40, 43, HPV DNA+ cases were significantly more likely to be p16INK4a + than HPV DNA− cases. No significant associations between HPV DNA and p16INK4a were found in any of the remaining eight studies or overall (pooled RR=1.32, 95%CI 0.75 to 2.31). Neither the funnel plot nor Egger’s test suggested evidence of publication bias (p=0.19). Meta-regression suggested no evidence of a relationship between study-specific (log) RR and p16INK4a + cut-off values (p=0.65).
Table 3.
Overview of HPV DNA and p16INK4a studies in ESCC tissues
| Nr studies |
Author | Country | Years of diagnosis |
Biopsy type |
Genotyping method |
N (ESCC patients enrolled) |
N (ESCC valid DNA)a |
N (HPV DNA+ ESCC)(%) |
HPV types identified |
VL [viral copies per cell] |
N (HPV RNA+ among HPV DNA+) |
N (p16INK4a+ among HPV DNA+) |
N (p16INK4a+ among HPV DNA−) |
Cut off
for p16INK4a positivity |
Cross- contamination controls |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| InterSCOPE study | 133 | 118 | 10 (8%) | 16, 33, 35, 45, 70; 11 | 0.00 | 1 (10%) | 1 (10%) | 6 (6%) | |||||||
| South Africa | 1995–2000 | AFPE | BSGP5+6+ PCR & TS-E7-PCR | 58 | 48 | 7 (6%) | 16, 35, 45, 70; 11 | 0.00 | 1 (10%)b | 1 (10%)c | 2 (2%) | ≥25% | yes (A, B, C)d | ||
| North China (Shanxi) | 1997–2005 | FFPE | 35 | 35 | 1 (1%) | 16 | 0.00 | 0 (0%) | 0 (0%) | 3 (3%) | |||||
| Iran | 2005–2007 | FFPE | 40 | 35 | 2 (2%) | 16, 33 | 0.00 | 0 (0%) | 0 (0%) | 1 (1%) | |||||
| 1 | *Antonsson et al., 2011 | Australia | 2002–2005 | FFPE | GP5+6+ PCR + seq. | 222 | 222 | 8 (4%) | 16, 35 | nt | nt | 4 (50%)e | nte | ns | yes (A, B, C) |
| 2 | *Bellizzi et al., 2009 | USA | 1994–2007 | FFPE | ISH | 31 | 29 | 8 (4%) | none | nt | nt | 0 (0%) | 7 (24%)f | ≥25% | yes (C) |
| 3 | Cao et al., 2014 | North China (Shandong) | 2006–2008 | FFPE | ISH | 105 | 105 | 29 (28%) | 16 | nt | nt | 25 (86%) | 14 (18%) | ≥50% | nsg |
| 4 | Castillo et al., 2011 | 166 | 166 | 31 (19%) | 16, 18, 35, 45, 51, 68; 6 | 0.121 (0.05 – 0.27)h | 16 (53%)i | 42 (33%)i | |||||||
| Pakistan | 1996–2002 | FFPE | SPF1/2 InnoLiPA | 42 | 42 | 11 (26%) | 16, 18; 6 | 0.124h | nt | ns | ns | ≥10% | nsj | ||
| Columbia | 1996–2001 | 49 | 49 | 9 (18%) | 16, 18, 35, 45; 6 | 0.251h | ns | ns | |||||||
| Japan | 1987–2005 | 75 | 75 | 11 (15%) | 16, 18, 51, 68 | 0.072h | ns | ns | |||||||
| 5 | Castillo et al., 2006 | South America | 73 | 73 | 21 (29%) | 16, 18 | 12 (57%) | 25 (49%)k | |||||||
| Columbia | 1996–2001 | FFPE | GP5+6+ PCR & seq. & Southern blot | 47 | 47 | 16 (34%) | 16, 18 | nt | nt | ns | ns | ≥10% | ns | ||
| Chile | 1996–2000 | 26 | 26 | 5 (19%) | 18 | ns | ns | ||||||||
| 6 | Ding et al., 2010 | North China (Henan) | 2005–2008 | FFPE | HPV16 E6 PCR | 17 | 17 | 8 (47%) | 16 | nt | nt | 2 (25%) | 5 (56%) | ≥10% | ns |
| 7 | Doxatader et al., 2011 | USA | ns | FFPE | ISH | 20 | 20 | 1 (17%)l | ns | nt | nt | 1 (17%)l | 5 (83%)l | ≥50% | ns |
| 8 | Herbster et al., 2012 | Brazil | 2000–2009 | FFPE & DFT | GP5+6+ PCR & ISH & seq. | 264 | 264 | 34 (13%) | 16, 18, 66 | nt | nt | 7 (21%) | 3 (10%) | ns | yes (A, B, C) |
| 9 | *Koshiol et al., 2009 | North China (Linxian) | 2006–2007 | FFPE & DFT | PCR (PGMY) & SPF10 LiPA | 272 | 267 | 3 (1%) | 16, 31; 89 | nt | nt | 0 (0%) | ntm | ns | yes (A, B, C) |
| 10 | Lofdahl et al., 2012 | Sweden | 1999–2006 | FFPE | GP5+6+ PCR & Luminex | 204 | 204 | 20 (10%) | 16, 33, 45, 51, 52, 73, 82, 66; 42 | nt | nt | 4 (24%)n | 18 (16%)o | nsp | yes (C) |
| 11 | *Malik et al., 2011 | USA | 2001–2009 | FFPE | ISH | 25 | 25 | 0 (0%)q | none | nt | nt | 0 (0%)q | ntq | nsr | yes (C) |
| 12 | Shuyama et al., 2007 | China | 59 | 59 | 19 (32%) | 16, 18, 51; 6 | <0.00001 – 1.7 | 1 (5%) | 10 (25%) | ||||||
| Gansus | 1994–2005 | FFPE | SPF10 InnoLiPA & GP5+6+ PCR & Southern blot | 26 | 26 | 17 (65%) | 16 | <0.00001 | nt | nsu | 3 (12%) | ≥80% | ns | ||
| Shandogs | 33 | 33 | 2 (6%) | 16, 18 | <0.0001 – 1.7 | nsu | 7 (21%) | ||||||||
| 13 | Teng et al., 2014 | East China (Shanghai) | 1999–2011 | FFPE | PCR + RDB | 177 | 177 | 6 (3%) | 16, 35; 11 | nt | nt | 0 (0%) | 5 (3%) | ≥80% | yes (B, C) |
| 14 | *Vaiphei et al., 2013 | India | ns | DFT | PCR | 23 | 23 | 20 (87%) | 16, 18, 39, 52, 59; 6, 11 + 11 more | nt | nt | nsv | nsv | ≥40% | yes (Bw, C) |
Studies that could not be included in meta-analyses for p16INK4a due to limited tissue analyses (see specific footnotes).
Number of valid ESCC biopsy equals number of patients enrolled in each of the studies.
the single HPV16 mRNA+ case was HPV16 DNA+ by TS-E7-PCR only, did not show upregulation of p16INK4a (<10% p16INK4a+ tumor cells), and was not seropositive for HPV16 antibodies.
A single HPV DNA+/p16INK4a+ case (>75% p16INK4a+ cells) was HPV16 mRNA-. HPV DNA was identified by TS-E7-PCR only; patient was seropositive for HPV16 E6 antibodies.
Use of a new blade for sectioning of each tissue specimen (A), different laboratories for DNA extraction, PCR set up and PCR products analyses (B), use of (pretested) HPV DNA+ and HPV DNA− tissues as a control (C).
Only HPV DNA+ tumors (N=8) were tested for p16INK4a expression by IHC.
6/7 tumors were scored as 25–50%, and 1/7 tumors was scored as 50–75% p16INK4a+ tumor cells.
CSCC was used as a positive control during ISH.
Geometric mean of HPV genome copies per cell across HPV16 DNA+ tumors analyzed.
158 of 166 samples had enough material for p16INK4a analysis (30/31 HPV DNA+ and 128/135 HPV DNA− cases).
For 11 ESCC cases from Japan authors additionally analyzed tissue biopsies of tumor adjacent epithelia of which none showed HPV DNA+.
72/73 samples had enough material for p16INK4a analysis (21/21 HPV DNA+ and 51/52 HPV DNA− cases).
only p16INK4a+ cases (N=6), and the equal number of p16INK4a− cases, were tested for HPV by ISH. Only 1/6 p16INK4a+ cases was HPV DNA+.
Only HPV DNA+ ESCC were tested for p16INK4a.
17/20 HPV DNA+ ESCC had enough material for p16INK4a evaluation.
p16INK4a was evaluated in 113/184 HPV DNA− cases (6.5 HPV DNA− controls per HPV DNA+ case).
Samples were considered positive if there was homogeneous p16INK4a staining of a clear majority of tumor cells.
Only p16INK4a+ tumors (N=11) were tested for HPV DNA by ISH.
Percentage of p16INK4a+ cells was not specified. Authors stated that only biopsies with nuclear or nuclear and cytoplasmic, diffuse and strong staining were considered p16INK4a+.
Gansu is a high-incidence ESCC region in NW China and Shandong is a low-incidence ESCC region in E China.
HPV51 and HPV6 were present only as a co-infection with HPV16 in two respective cases for which region of origin was not specified.
Authors did not specify if the one p16INK4a+ case was from Gansu or from Shandong.
Number of p16INK4a+ tumors was not specified (12 tumors were positive for p16 and p63). Authors concluded that p16INK4a+ and p63 positivity did not correlate with HPV status.
Authors report using sterile forceps and blades but do not specify changing them for sectioning of each tissue sample.
Abbreviations: AFPE - alcohol-fixed paraffin-embedded, FFPE - formalin-fixed paraffin-embedded, DFT - deep frozen tissue, ISH - in situ hybridization, RDB - reverse dot blot, CSCC – cervical squamous cell carcinoma, seq. – sequencing, nt – not tested, ns - not specified
Figure 2. Study-specific and pooled relative risks (RR) corresponding to the proportion of HPV DNA+ cases that were p16INK4a + relative to the proportion of HPV DNA− cases that were p16INK4a +.
Clinical characteristics of ESCC patients and histopathological tumor patterns according to the HPV status
ESCC patients with HPV DNA− tumors (N=108) did not differ statistically from the patients with HPV DNA+ tumors (N=10) with respect to age, alcohol consumption, smoking habits, or degree of tumor keratinization (Table 1). Interestingly, only 33% of patients with HPV DNA− tumors were female, in contrast to the 70% females among patients with HPV DNA+ tumors (p=0.019).
Pathology review of the 118 tumors revealed 114 (97%) keratinizing, 3 (3%) basaloid, and one (1%) tumor of mixed histology (keratinizing/basaloid). All HPV DNA+ (N=9) and HPV DNA+/RNA+ (N=1) tumors were keratinizing.
DISCUSSION
In order to comprehensively address the question of causal HPV involvement in the pathogenesis of esophageal squamous cell carcinoma, we used advanced molecular methods on ESCC tissue sections and searched for key molecular markers indicative of HPV-association (HPV DNA with viral load, HPV RNA and a surrogate cellular protein p16INK4a) in a large series of 118 ESCC tissues. These analyzed 118 tumor tissues were enriched for potentially HPV-driven cancers by pre-selection among 1,561 ESCC patients who were positive for HPV early protein antibodies in their blood. Using stringent precautions to prevent nucleic acid contamination, we found HPV DNA positivity in 10 (8%) of these cases, using a combination of two highly sensitive HPV genotyping assays. One of these sensitive assays targeted the L1 region of all defined 51 mucosal HPV types, while the other evaluated the E7 region of 21 mucosal HPV types (12 HR-, 7 possibly (p)HR- and 2 LR-HPV types). All HPV DNA+ findings were inconsistent between the two genotyping assays and all samples were negative by HPV16/18 qPCR, suggesting that viral loads in HPV DNA+ ESCC tissues were extremely low and/or close to the lower limits of detection. All ESCC tissues were also analyzed for HPV16 E6*I mRNA but only one (an HPV16 DNA+ tumor) showed this marker. None of the tumors that were positive for other HPV DNA types (HPV33, 35, 45, 70, or 11) showed type-concordant E6*I or E6 fl mRNA. Of seven tumors with p16INK4a up-regulation, only one contained HPV DNA (type 16), and all seven were negative for HPV16 mRNA. Thus, among the 118 ESCC tissues analyzed for three HPV functional markers in addition to HPV DNA (i.e. (i) viral load 1 copy per cell, (ii) presence of HPV type-concordant E6*I mRNA, and (iii) up-regulation of the cellular surrogate marker p16INK4a), none was positive for all three or even two functional markers. Of the two HPV16 DNA+ tissues that displayed at least one functional marker, one harbored HPV16 mRNA but without p16INK4a up-regulation and the other showed up-regulated p16INK4a in the absence of HPV16 transcription. Based on the absence of HPV functional markers in our evaluation here, we conclude that ESCC does not follow the HPV-transformation pathogenetic model of cervical cancer, and that there is no evidence for a role of mucosal alpha-papillomavirus types investigated here, in the etiology of ESCC in the geographic high-incidence regions studied.
In line with other studies15, 38, 44, 48 we identified HPV DNA of mucosal alpha-papillomaviruses in a few ESCC tissues, albeit at very low copy numbers and almost always with none or only a single marker of HPV-transformed phenotype. Our findings suggest four possible sources for HPV DNA positivity in ESCC: (i) HPV DNA in the oral cavity, which could pass into the esophagus after swallowing HPV-containing saliva (it has been shown that oral rinses can contain HPV DNA)61, (ii) non-transforming HPV infection in neighboring normal esophageal tissue or even parts of the tumor (where HPV acts as a passenger and not as a transforming agent)37 ; (iii) cross-contamination from other HPV-infected tissues in routine pathology tissue processing62 ; or (iv) cross-contamination from HPV PCR products or HPV plasmids in the laboratory analysis63. Furthermore, Schaffer and colleagues have demonstrated increased uptake of HPV virions under certain conditions: HPV18 pseudovirion uptake by esophageal cells can be increased in vitro when the cells are treated with benzo-α-pyrene, an abundant carcinogen in tobacco smoke64. In line with these functional data, epidemiological studies showed that tobacco users have higher prevalence of HPV16 DNA in their oral cavity65, and clinical studies demonstrated that OPSCC patients who smoke have worse survival irrespective of HPV tumor status66–68. Also, several studies suggested that there is a high risk of HPV vaginal contamination during use of routine endocavity vaginal ultrasound probes, an underestimated route of nosocomial infection69–71. Similarly, contamination with DNA of HPV types from oral cavity could occur during esophagoscopy or esophageal tissue sampling.
In support of above described data, Gillison and colleagues demonstrated that the prevalence of oral infection with mucosal HPV types from the five HR-species among healthy men and women aged 14 to 69 years was 6.9% in the United States61. Similar data were shown in Iran where 6.1% of 114 healthy individuals age 16 – 66 years, had oral HPV DNA72. Bottalico and colleagues showed that 37% of 117 immunocompetent men, harbored HPV DNA in oral washes of which 28% were alpha-, 64% were beta- and 8% were gamma-papillomavirus infections73. The authors have concluded that the oral cavity contains a wide spectrum of HPV types predominantly from the beta- and gamma-papillomavirus genera, which were previously considered to be cutaneous types. Similar results were found by a recent study for alpha- (72%), beta- (27%), and gamma-papillomaviruses (27%) in the anal canal74. The presence of beta- and gamma-papillomaviruses at mucosal sites leaves an open question of their tissue tropism and their biological role e.g. biological role of beta- and gamma-papillomaviruses in ESCC, which was not examined here. However, none of these questions can be answered based on HPV DNA data only. Beta- and gamma-papillomaviruses are frequently found both in skin lesions and in healthy skin3. Further, the functional data on transforming activity for beta-papillomaviruses are scarce and limited to HPV5 and HPV3875–80. Therefore, to substantiate a biological contribution of HPV types (alpha-, beta-, or gamma-) to the transformed phenotype of tumor cells, viral RNA or protein, representing viral activity, and expression levels of cellular proteins altered by viral oncoproteins, need to be demonstrated in addition to viral DNA.
It should be noted that in contrast to high prevalences for mucosal alpha-papillomaviruses ranging from 37% to 44% previously reported for Africa, China, and Iran10, the most recent studies that also documented significant efforts to prevent potential nucleic acid cross-contamination during tissue sectioning and HPV DNA analysis found less than one percent of HPV DNA+ ESCC cases15–17. In agreement with these latest studies, our combination of two highly sensitive HPV genotyping methods, capable of identifying the broadest spectrum of mucosal HPV types analyzed so far, detected traces of HPV DNA in 3% to 15% of ESCC from the same geographic regions.
Our observation of low viral load in the few ESCC cases that were HPV DNA+ is in agreement with Guo and colleagues81 who in 29 Chinese ESCC tissues found a median of 0.04 HPV16 copies per cell, and only a single case with close to one copy per cell. In contrast, four other studies identified cases with HPV loads >1 genome copy per cell33–36 with only one study34 analyzing additional HPV functional markers and demonstrating p16INK4a up-regulation (≥80% tumor cells p16INK4a positive) in one of 19 HPV16 DNA+ cases.
The type-specific HPV RNA assays applied here have been extensively validated on FFPE tissues and shown to reliably and sensitively detect HPV E6*I mRNA of 20 mucosal alpha-papillomaviruses in cervical cancer tissues22, 24. We found a single HPV16 DNA+ ESCC tumor that expressed HPV16 mRNA, however, without p16INK4a up-regulation. HPV16 E6*I transcripts can be abundantly expressed in the cervix in the absence of malignant transformation, as well as in some non-HPV-driven oropharyngeal and laryngeal cancers27, 29. The E6*I transcripts are, therefore, markers of active viral infection but are not transformation-specific; thus we did not consider this single low viral load HPV DNA+/RNA+ ESCC an HPV-driven tumor. Our RNA data disagree with one HPV16 transcription study which used in situ hybridization to identify HPV16 RNA in 19 of 31 Chinese ESCC and six of 23 normal adjacent mucosa biopsies37.
Though we tested for RNA of all HPV types identified by two HPV DNA assays and also, all HPV types for which HPV antibodies were detected in patients’ sera (see Supplementary Table S1), we did not test for RNA of LR-HPV6. The HPV6 RNA assay could not be validated on HPV6 DNA+ tumor tissues in contrast to the HPV11 RNA assay29, 82. Among the 118 patients with the analyzable ESCC tissues, 20 HPV6, and 127 non-HPV6 type-specific antibody responses were detected (see Supplementary Table S1). In only 3/127 (2%) of these HPV type-specific antibody responses (in three separate cases) could we also detect HPV DNA in the corresponding ESCC tissue (three of the five HPV16 DNA+ cases), and in none of them could we detect both HPV DNA and RNA, or HPV RNA only. Therefore, it is unlikely that testing for HPV6 RNA would yield crucial functional data in any of these 20 HPV6 DNA−/p16INK4a - ESCC tissues of patients seropositive for HPV6.
In a setting of HPV-infection, p16INK4a up-regulation is considered the best biomarker to define tumors with HPV-transformed phenotype both in research and in clinical studies27, 57, 83–85. P16INK4a up-regulation in HPV-infected cells is a result of a cellular defense mechanism referred to as oncogene-induced senescence induced by expression of the viral E7 oncoprotein86, 87. Lack of CDKN2a mutations in combination with p16INK4a up-regulation was demonstrated for HPV RNA+ cervical cancers and cervical cancer cell lines88–91, as well as for the HPV RNA+ tumors of the head-and-neck including oral, oropharyngeal and laryngeal tumors92. Furthermore, immunohistochemical studies demonstrated that p16INK4a expression gradually increases from 5% in normal cervical epithelium, to 10% in low grade squamous intraepithelial lesions (LSIL), 25 – 80% in high (H)SIL and 90 – 100% in cervical cancers93–95. Together, these studies have established p16INK4a up-regulation as an excellent biomarker to define HR-HPV-associated lesions and cancers in addition to HPV DNA and/or HPV RNA positivity.
With a single tumor among 10 HPV DNA+ ESCC, and six tumors among 108 HPV DNA− ESCC, that showed p16INK4a up-regulation, we found no significant association between p16INK4a up-regulation and HPV DNA positivity (p=0.57). For the one case with HPV16 DNA+ ESCC with p16INK4a up-regulation in our study, the low viral load and absence of HPV16 E6*I mRNA do not support classification of this case as HPV16-driven. Nine other studies have also addressed question of p16INK4a expression in HPV DNA+ and HPV DNA− tumors of which seven found no significant associations34, 41, 42, 44–46, 48. Two studies found p16INK4a positivity statistically significantly associated with HPV DNA+ ESCC40, 43. We do not find an explanation for the high detection of p16INK4a up-regulation in 86% of HPV DNA+ and 18% of HPV DNA− tissues by Cao and colleagues that applied a cut-off as high as 50% to define p16INK4a positivity40. Castillo and colleagues used 10% as a cut-off to define p16INK4a positivity and reported high p16INK4a positivity in both HPV DNA+ (56%) and HPV DNA− tissues (33%)43. In summary, seven of nine studies specified cut-offs for definition of p16INK4a positivity/up-regulation, however this cut-off varied from ≥10% (three studies), ≥50% (two studies) to ≥80% (two studies) indicating that evaluation of p16INK4a in tumor tissues in relation to HPV is still a non-standardized method. Our statement is supported by, and in agreement with, a recent study that reviewed p16INK4a and HPV DNA studies in ESCC96. The use of p16INK4a expression as an indicator of virally induced deregulation of the cell cycle97 in HPV DNA+(/RNA+) tissues, requires standardization of methods and cut-off values in different anatomical sites. Only then can it be reliably and consistently used to define HPV-driven cancers.
Our study is limited by the relatively small number of tumors analyzed from each of the three high-incidence regions. Other limitations include the absence of full functional data on all samples tested, and the fact that our samples all came from high-incidence ESCC regions and may not be representative of other geographic regions. Also, if HPV was supposed to contribute to the cancer development by a never fully convincingly demonstrated “hit and run mechanism”98, we were not able to detect this ill-defined possibility by our approach here. Further, we did not test in the tissues for DNA of human beta- and gamma-papillomavirus types whose DNA occasionally has been isolated from mucosal sites.
The major strengths of our study include the application of multiple sensitive and established technologies for the analysis of HPV DNA, the use of an array of functional markers for active HPV infection, and the use of stringent precautions to prevent and monitor laboratory contaminations. A further strength, counteracting the overall small number of tumors analyzed from each of the three individual high-incidence regions, is the unique preselection of potentially HPV-associated tumors by HPV serology. Our study also demonstrates how detailed knowledge about HPV-transformation in cervical cancer can be used to clarify the potential transforming role of HPV in other tumor sites in which HPV DNA is found.
Recently, HPV serology has been shown to be highly specific for the definition of potentially HPV-associated oropharyngeal squamous cell carcinomas (OPSCC) in which HPV16 E6 antibodies can be detected as early as 13 years prior to cancer diagnosis53. More precisely, HPV16 E6 seropositivity was present in prediagnostic samples of 35% of OPSCC patients and 0.6% controls (OR, 274; 95% CI 110 to 681) but was not associated with cancer at other sites including esophagus53. Here we confirm that positive HPV serology does not define an HPV-driven ESCC, and we base this conclusion not only on serology but also on a broad set of tissue biomarkers, the combination of which have been analyzed for the first time in ESCC. The only previous study that has provided data on such a broad biomarker panel was a recent study by Anantharaman and colleagues who similarly demonstrated no association of HPV with cancer of the lung despite the occurrence of HPV seropositivity in some of the lung cancer patients82.
In conclusion, the InterSCOPE study provides strong biological data against a transforming role of mucosal alpha-papillomavirus types in the pathogenesis of esophageal squamous cell carcinoma for the three high-incidence regions; China, South-Africa, and Iran. Further, our meta-analysis performed on published data on co-occurrence of HPV DNA and p16INK4a up-regulation also showed no evidence for a significant association. Together with the largely negative serological findings documented previously in the sero-epidemiological InterSCOPE study for a large population of ESCC patients from all over the world, our data provide compelling evidence that mucosal alpha-papillomaviruses have little or no role in the etiology of ESCC according to the HPV-transformation model described for cervical, other anogenital and a subset of head-and-neck cancers.
Supplementary Material
The novelty and impact of the work.
South Africa, China and Iran are countries with the highest incidences and mortalities of esophageal squamous cell carcinoma (ESCC). Epidemiological studies have suggested a role for HR-HPV types in ESCC occurring in these countries, but consistent biological evidence of viral transformation, important for vaccination program planning, is still missing. Using state-of-the-art technology, we provide comprehensive data on absence of viral biological activity for 51 mucosotropic HPV types from alpha-papillomavirus genus in ESCC from high-incidence regions.
Acknowledgments
The authors thank Mrs. Birgit Aengeneyndt for her excellent technical assistance and Dr. Stephen M. Hewitt for sharing his expertise in management of fixed-tissue biopsies. We also want to acknowledge all InterSCOPE collaborators listed below:
**InterSCOPE Collaborators
Paolo Boffetta, The Tisch Cancer Institute, Mount Sinai School of Medicine, New York, NY; International Prevention Research Institute, (IPRI) Lyon, France, Paul Brennan, Gilles Ferro International Agency for Research on Cancer, Lyon, France, David Whiteman, Adele C. Green, Nicholas K. Hayward, Population Health Department, Queensland Institute of Medical Research, Brisbane, QLD, Australia, Dianne O’Connell, Cancer Council NSW, Cancer Research Division, Sydney, NSW, Australia, David Zaridze, Institute of Carcinogenesis, Cancer Research Centre, Moscow, Russian Federation Ivana Holcatova, Institute of Hygiene and Epidemiology,1st Faculty of Medicine, Charles University in Prague, Prague, Czech Republic Dana Mates, Stephen M. Hewitt, Tissue Array Research Program, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD USA, Neonila Szeszenia-Dabrowska, Department of Epidemiology, Institute of Occupational Medicine, Lodz, Poland Vladimir Janout, Palacky University, Olomouc, Czech Republic Maria Paula Curado, International Prevention Research Institute (IPRI), Ecully, France Ana Maria Menezes, Universidade Federal de Pelotas, Pelotas, Brazil Sergio Koifman, National School of Public Health/FIOCRUZ, Rio de Janeiro, Brazil Farhad Islami, Dariush Nasrollahzadeh Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran, Nan Hu Alisa M. Goldstein Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, NCI, Bethesda, Maryland, United States of America, Ying Gao, Ti Ding, Shanxi Cancer Hospital, Taiyuan, Shanxi, PR China and Farin Kamangar School of Community Health and Policy, Morgan State University, Baltimore, MD, USA
Footnotes
Potential conflict of interest:
MP and MS have received research support through cooperation contracts of DKFZ with Roche and Qiagen in the field of development of HPV diagnostics. They are inventors on patents owned by DKFZ in the field of HPV diagnostics. Authors GH, SE, CCA, CB, SMD, CF, TG, MH, DH, RM, PRT, MT, MIU, TW and FS have no conflict.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ American Cancer S. Cancer statistics, 2004. CA: a cancer journal for clinicians. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 3.IARC. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum. A review of human carcinogens—Part B: biological agents. 2011;100B:1–475. [Google Scholar]
- 4.Freedman ND, Abnet CC, Leitzmann MF, Mouw T, Subar AF, Hollenbeck AR, Schatzkin A. A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. American journal of epidemiology. 2007;165:1424–33. doi: 10.1093/aje/kwm051. [DOI] [PubMed] [Google Scholar]
- 5.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Cogliano V Group WHOIAfRoCMW. Carcinogenicity of alcoholic beverages. The lancet oncology. 2007;8:292–3. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 6.Sitas F, Egger S, Urban MI, Taylor PR, Abnet CC, Boffetta P, O'Connell DL, Whiteman DC, Brennan P, Malekzadeh R, Pawlita M, Dawsey SM, et al. InterSCOPE study: Associations between esophageal squamous cell carcinoma and human papillomavirus serological markers. Journal of the National Cancer Institute. 2012;104:147–58. doi: 10.1093/jnci/djr499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syrjanen K, Pyrhonen S, Aukee S, Koskela E. Squamous cell papilloma of the esophagus: a tumour probably caused by human papilloma virus (HPV) Diagnostic histopathology / published in association with the Pathological Society of Great Britain and Ireland. 1982;5:291–6. [PubMed] [Google Scholar]
- 8.Syrjanen KJ. Histological changes identical to those of condylomatous lesions found in esophageal squamous cell carcinomas. Archiv fur Geschwulstforschung. 1982;52:283–92. [PubMed] [Google Scholar]
- 9.Syrjanen KJ. HPV infections and oesophageal cancer. Journal of clinical pathology. 2002;55:721–8. doi: 10.1136/jcp.55.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Li J, Diao M, Cai Z, Yang J, Zeng Y. Statistical analysis of human papillomavirus in a subset of upper aerodigestive tract tumors. Journal of medical virology. 2013;85:1775–85. doi: 10.1002/jmv.23662. [DOI] [PubMed] [Google Scholar]
- 11.Chen B, Yin H, Dhurandhar N. Detection of human papillomavirus DNA in esophageal squamous cell carcinomas by the polymerase chain reaction using general consensus primers. Human pathology. 1994;25:920–3. doi: 10.1016/0046-8177(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 12.Chang F, Syrjanen S, Shen Q, Wang L, Wang D, Syrjanen K. Human papillomavirus involvement in esophageal precancerous lesions and squamous cell carcinomas as evidenced by microscopy and different DNA techniques. Scandinavian journal of gastroenterology. 1992;27:553–63. doi: 10.3109/00365529209000119. [DOI] [PubMed] [Google Scholar]
- 13.Peixoto Guimaraes D, Hsin Lu S, Snijders P, Wilmotte R, Herrero R, Lenoir G, Montesano R, Meijer CJ, Walboomers J, Hainaut P. Absence of association between HPV DNA, TP53 codon 72 polymorphism, and risk of oesophageal cancer in a high-risk area of China. Cancer letters. 2001;162:231–5. doi: 10.1016/s0304-3835(00)00643-1. [DOI] [PubMed] [Google Scholar]
- 14.Matsha T, Donninger H, Erasmus RT, Hendricks D, Stepien A, Parker MI. Expression of p53 and its homolog, p73, in HPV DNA positive oesophageal squamous cell carcinomas. Virology. 2007;369:182–90. doi: 10.1016/j.virol.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Koshiol J, Wei WQ, Kreimer AR, Chen W, Gravitt P, Ren JS, Abnet CC, Wang JB, Kamangar F, Lin DM, von Knebel-Doeberitz M, Zhang Y, et al. No role for human papillomavirus in esophageal squamous cell carcinoma in China. International journal of cancer Journal international du cancer. 2010;127:93–100. doi: 10.1002/ijc.25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel K, Mining S, Wakhisi J, Gheit T, Tommasino M, Martel-Planche G, Hainaut P, Abedi-Ardekani B. TP53 mutations, human papilloma virus DNA and inflammation markers in esophageal squamous cell carcinoma from the Rift Valley, a high-incidence area in Kenya. BMC research notes. 2011;4:469. doi: 10.1186/1756-0500-4-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haeri H, Mardany O, Asadi-Amoli F, Shahsiah R. Human papilloma virus and esophageal squamous cell carcinoma. Acta medica Iranica. 2013;51:242–5. [PubMed] [Google Scholar]
- 18.Howley PM, Munger K, Romanczuk H, Scheffner M, Huibregtse JM. Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Princess Takamatsu symposia. 1991;22:239–48. [PubMed] [Google Scholar]
- 19.Nakagawa S, Yoshikawa H, Yasugi T, Kimura M, Kawana K, Matsumoto K, Yamada M, Onda T, Taketani Y. Ubiquitous presence of E6 and E7 transcripts in human papillomavirus-positive cervical carcinomas regardless of its type. Journal of medical virology. 2000;62:251–8. doi: 10.1002/1096-9071(200010)62:2<251::aid-jmv18>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt M, Pawlita M. The HPV transcriptome in HPV16 positive cell lines. Molecular and cellular probes. 2011;25:108–13. doi: 10.1016/j.mcp.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 21.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 22.Halec G, Alemany L, Lloveras B, Schmitt M, Alejo M, Bosch FX, Tous S, Klaustermeier JE, Guimera N, Grabe N, Lahrmann B, Gissmann L, et al. Pathogenic role of the eight probably/possibly carcinogenic HPV types 26, 53, 66, 67, 68, 70, 73 and 82 in cervical cancer. The Journal of pathology. 2014 doi: 10.1002/path.4405. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt M, Depuydt C, Benoy I, Bogers J, Antoine J, Pawlita M, Arbyn M group Vs. Viral load of high-risk human papillomaviruses as reliable clinical predictor for the presence of cervical lesions. Cancer Epidemiol Biomarkers Prev. 2013;22:406–14. doi: 10.1158/1055-9965.EPI-12-1067. [DOI] [PubMed] [Google Scholar]
- 24.Halec G, Schmitt M, Dondog B, Sharkhuu E, Wentzensen N, Gheit T, Tommasino M, Kommoss F, Bosch FX, Franceschi S, Clifford G, Gissmann L, et al. Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer. International journal of cancer Journal international du cancer. 2013;132:63–71. doi: 10.1002/ijc.27605. [DOI] [PubMed] [Google Scholar]
- 25.Sano T, Masuda N, Oyama T, Nakajima T. Overexpression of p16 and p14ARF is associated with human papillomavirus infection in cervical squamous cell carcinoma and dysplasia. Pathology international. 2002;52:375–83. doi: 10.1046/j.1440-1827.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 26.Jung AC, Briolat J, Millon R, de Reynies A, Rickman D, Thomas E, Abecassis J, Clavel C, Wasylyk B. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. International journal of cancer Journal international du cancer. 2010;126:1882–94. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- 27.Holzinger D, Schmitt M, Dyckhoff G, Benner A, Pawlita M, Bosch FX. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer research. 2012;72:4993–5003. doi: 10.1158/0008-5472.CAN-11-3934. [DOI] [PubMed] [Google Scholar]
- 28.Chernock RD, Wang X, Gao G, Lewis JS, Jr, Zhang Q, Thorstad WL, El-Mofty SK. Detection and significance of human papillomavirus, CDKN2A(p16) and CDKN1A(p21) expression in squamous cell carcinoma of the larynx. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:223–31. doi: 10.1038/modpathol.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halec G, Holzinger D, Schmitt M, Flechtenmacher C, Dyckhoff G, Lloveras B, Hofler D, Bosch FX, Pawlita M. Biological evidence for a causal role of HPV16 in a small fraction of laryngeal squamous cell carcinoma. British journal of cancer. 2013;109:172–83. doi: 10.1038/bjc.2013.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Z, Cen S, Shen J, Cai W, Xu J, Teng Z, Hu Z, Zeng Y. Study of immortalization and malignant transformation of human embryonic esophageal epithelial cells induced by HPV18 E6E7. Journal of cancer research and clinical oncology. 2000;126:589–94. doi: 10.1007/PL00008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong FH, Hu CP, Chen SC, Yu YT, Chang C. Absence of genomes of DNA tumor viruses and expression of oncogenes and growth factors in two esophageal carcinoma cell lines of Chinese origin. Zhonghua Minguo wei sheng wu ji mian yi xue za zhi = Chinese journal of microbiology and immunology. 1992;25:59–68. [PubMed] [Google Scholar]
- 32.Zhang H, Jin Y, Chen X, Jin C, Law S, Tsao SW, Kwong YL. Papillomavirus type 16 E6/E7 and human telomerase reverse transcriptase in esophageal cell immortalization and early transformation. Cancer letters. 2007;245:184–94. doi: 10.1016/j.canlet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Dreilich M, Bergqvist M, Moberg M, Brattstrom D, Gustavsson I, Bergstrom S, Wanders A, Hesselius P, Wagenius G, Gyllensten U. High-risk human papilloma virus (HPV) and survival in patients with esophageal carcinoma: a pilot study. BMC cancer. 2006;6:94. doi: 10.1186/1471-2407-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuyama K, Castillo A, Aguayo F, Sun Q, Khan N, Koriyama C, Akiba S. Human papillomavirus in high- and low-risk areas of oesophageal squamous cell carcinoma in China. British journal of cancer. 2007;96:1554–9. doi: 10.1038/sj.bjc.6603765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si HX, Tsao SW, Poon CS, Wang LD, Wong YC, Cheung AL. Viral load of HPV in esophageal squamous cell carcinoma. International journal of cancer Journal international du cancer. 2003;103:496–500. doi: 10.1002/ijc.10865. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D, Zhang Q, Zhou L, Huo L, Zhang Y, Shen Z, Zhu Y. Comparison of prevalence, viral load, physical status and expression of human papillomavirus-16, -18 and, -58 in esophageal and cervical cancer: a case-control study. BMC cancer. 2010;10:650. doi: 10.1186/1471-2407-10-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou XB, Guo M, Quan LP, Zhang W, Lu ZM, Wang QH, Ke Y, Xu NZ. Detection of human papillomavirus in Chinese esophageal squamous cell carcinoma and its adjacent normal epithelium. World journal of gastroenterology : WJG. 2003;9:1170–3. doi: 10.3748/wjg.v9.i6.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonsson A, Nancarrow DJ, Brown IS, Green AC, Drew PA, Watson DI, Hayward NK, Whiteman DC, Australian Cancer S. High-risk human papillomavirus in esophageal squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:2080–7. doi: 10.1158/1055-9965.EPI-10-0033. [DOI] [PubMed] [Google Scholar]
- 39.Bellizzi AM, Woodford RL, Moskaluk CA, Jones DR, Kozower BD, Stelow EB. Basaloid squamous cell carcinoma of the esophagus: assessment for high-risk human papillomavirus and related molecular markers. The American journal of surgical pathology. 2009;33:1608–14. doi: 10.1097/PAS.0b013e3181b46fd4. [DOI] [PubMed] [Google Scholar]
- 40.Cao F, Han H, Zhang F, Wang B, Ma W, Wang Y, Sun G, Shi M, Ren Y, Cheng Y. HPV infection in esophageal squamous cell carcinoma and its relationship to the prognosis of patients in northern China. TheScientificWorldJournal. 2014;2014:804738. doi: 10.1155/2014/804738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbster S, Ferraro CT, Koff NK, Rossini A, Kruel CD, Andreollo NA, Rapozo DC, Blanco TC, Faria PA, Santos PT, Albano RM, Simao Tde A, et al. HPV infection in Brazilian patients with esophageal squamous cell carcinoma: interpopulational differences, lack of correlation with surrogate markers and clinicopathological parameters. Cancer letters. 2012;326:52–8. doi: 10.1016/j.canlet.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Castillo A, Aguayo F, Koriyama C, Torres M, Carrascal E, Corvalan A, Roblero JP, Naquira C, Palma M, Backhouse C, Argandona J, Itoh T, et al. Human papillomavirus in esophageal squamous cell carcinoma in Colombia and Chile. World J Gastroenterol. 2006;12:6188–92. doi: 10.3748/wjg.v12.i38.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castillo A, Koriyama C, Higashi M, Anwar M, Bukhari MH, Carrascal E, Mancilla L, Okumura H, Matsumoto M, Sugihara K, Natsugoe S, Eizuru Y, et al. Human papillomavirus in upper digestive tract tumors from three countries. World J Gastroenterol. 2011;17:5295–304. doi: 10.3748/wjg.v17.i48.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding GC, Ren JL, Chang FB, Li JL, Yuan L, Song X, Zhou SL, Guo T, Fan ZM, Zeng Y, Wang LD. Human papillomavirus DNA and P16(INK4A) expression in concurrent esophageal and gastric cardia cancers. World J Gastroenterol. 2010;16:5901–6. doi: 10.3748/wjg.v16.i46.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doxtader EE, Katzenstein AL. The relationship between p16 expression and high-risk human papillomavirus infection in squamous cell carcinomas from sites other than uterine cervix: a study of 137 cases. Human pathology. 2012;43:327–32. doi: 10.1016/j.humpath.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Lofdahl HE, Du J, Nasman A, Andersson E, Rubio CA, Lu Y, Ramqvist T, Dalianis T, Lagergren J, Dahlstrand H. Prevalence of human papillomavirus (HPV) in oesophageal squamous cell carcinoma in relation to anatomical site of the tumour. PloS one. 2012;7:e46538. doi: 10.1371/journal.pone.0046538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malik SM, Nevin DT, Cohen S, Hunt JL, Palazzo JP. Assessment of immunohistochemistry for p16INK4 and high-risk HPV DNA by in situ hybridization in esophageal squamous cell carcinoma. International journal of surgical pathology. 2011;19:31–4. doi: 10.1177/1066896910382005. [DOI] [PubMed] [Google Scholar]
- 48.Teng H, Li X, Liu X, Wu J, Zhang J. The absence of human papillomavirus in esophageal squamous cell carcinoma in East China. International journal of clinical and experimental pathology. 2014;7:4184–93. [PMC free article] [PubMed] [Google Scholar]
- 49.Vaiphei K, Kochhar R, Bhardawaj S, Dutta U, Singh K. High prevalence of human papillomavirus in esophageal squamous cell carcinoma: a study in paired samples. Diseases of the esophagus : official journal of the International Society for Diseases of the Esophagus / ISDE. 2013;26:282–7. doi: 10.1111/j.1442-2050.2012.01365.x. [DOI] [PubMed] [Google Scholar]
- 50.Meschede W, Zumbach K, Braspenning J, Scheffner M, Benitez-Bribiesca L, Luande J, Gissmann L, Pawlita M. Antibodies against early proteins of human papillomaviruses as diagnostic markers for invasive cervical cancer. Journal of clinical microbiology. 1998;36:475–80. doi: 10.1128/jcm.36.2.475-480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zumbach K, Hoffmann M, Kahn T, Bosch F, Gottschlich S, Gorogh T, Rudert H, Pawlita M. Antibodies against oncoproteins E6 and E7 of human papillomavirus types 16 and 18 in patients with head-and-neck squamous-cell carcinoma. International journal of cancer Journal international du cancer. 2000;85:815–8. doi: 10.1002/(sici)1097-0215(20000315)85:6<815::aid-ijc14>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 52.Heideman DA, Waterboer T, Pawlita M, Delis-van Diemen P, Nindl I, Leijte JA, Bonfrer JM, Horenblas S, Meijer CJ, Snijders PJ. Human papillomavirus-16 is the predominant type etiologically involved in penile squamous cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:4550–6. doi: 10.1200/JCO.2007.12.3182. [DOI] [PubMed] [Google Scholar]
- 53.Kreimer AR, Johansson M, Waterboer T, Kaaks R, Chang-Claude J, Drogen D, Tjonneland A, Overvad K, Quiros JR, Gonzalez CA, Sanchez MJ, Larranaga N, et al. Evaluation of human papillomavirus antibodies and risk of subsequent head and neck cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:2708–15. doi: 10.1200/JCO.2012.47.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gheit T, Landi S, Gemignani F, Snijders PJ, Vaccarella S, Franceschi S, Canzian F, Tommasino M. Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. Journal of clinical microbiology. 2006;44:2025–31. doi: 10.1128/JCM.02305-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmitt M, Dondog B, Waterboer T, Pawlita M, Tommasino M, Gheit T. Abundance of multiple high-risk human papillomavirus (HPV) infections found in cervical cells analyzed by use of an ultrasensitive HPV genotyping assay. Journal of clinical microbiology. 2010;48:143–9. doi: 10.1128/JCM.00991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gheit T, Tommasino M. Detection of high-risk mucosal human papillomavirus DNA in human specimens by a novel and sensitive multiplex PCR method combined with DNA microarray. Methods Mol Biol. 2011;665:195–212. doi: 10.1007/978-1-60761-817-1_12. [DOI] [PubMed] [Google Scholar]
- 57.Holzinger D, Flechtenmacher C, Henfling N, Kaden I, Grabe N, Lahrmann B, Schmitt M, Hess J, Pawlita M, Bosch FX. Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. International journal of cancer Journal international du cancer. 2013;133:1389–99. doi: 10.1002/ijc.28142. [DOI] [PubMed] [Google Scholar]
- 58.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 59.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterne JA, Egger M, Smith GD. Systematic reviews in health care: Investigating and dealing with publication and other biases in meta-analysis. Bmj. 2001;323:101–5. doi: 10.1136/bmj.323.7304.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gillison ML, Broutian T, Pickard RK, Tong ZY, Xiao W, Kahle L, Graubard BI, Chaturvedi AK. Prevalence of oral HPV infection in the United States, 2009–2010. JAMA. 2012;307:693–703. doi: 10.1001/jama.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smelov V, Eklund C, Muhr LS, Hultin E, Dillner J. 44. Detection of the human papillomavirus infection (HPV) on the internationally used environmental surfaces: ready for a 'take-off. Sexual health. 2013;10:591. [Google Scholar]
- 63.Boyd AS, Annarella M, Rapini RP, Adler-Storthz K, Duvic M. False-positive polymerase chain reaction results for human papillomavirus in lichen planus. Potential laboratory pitfalls of this procedure. J Am Acad Dermatol. 1996;35:42–6. doi: 10.1016/S0190-9622(96)90494-6. [DOI] [PubMed] [Google Scholar]
- 64.Schafer G, Kabanda S, van Rooyen B, Marusic MB, Banks L, Parker MI. The role of inflammation in HPV infection of the Oesophagus. BMC cancer. 2013;13:185. doi: 10.1186/1471-2407-13-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fakhry C, Gillison ML, D'Souza G. Tobacco use and oral HPV-16 infection. JAMA. 2014;312:1465–7. doi: 10.1001/jama.2014.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillison ML, Zhang Q, Jordan R, Xiao W, Westra WH, Trotti A, Spencer S, Harris J, Chung CH, Ang KK. Tobacco smoking and increased risk of death and progression for patients with p16-positive and p16-negative oropharyngeal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:2102–11. doi: 10.1200/JCO.2011.38.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, Wolf GT, Urba SG, Chepeha DB, Teknos TN, Eisbruch A, Tsien CI, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, smoking as indicators of response to therapy and survival in oropharyngeal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:3128–37. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoff CM, Grau C, Overgaard J. Effect of smoking on oxygen delivery and outcome in patients treated with radiotherapy for head and neck squamous cell carcinoma--a prospective study. Radiother Oncol. 2012;103:38–44. doi: 10.1016/j.radonc.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 69.Casalegno JS, Le Bail Carval K, Eibach D, Valdeyron ML, Lamblin G, Jacquemoud H, Mellier G, Lina B, Gaucherand P, Mathevet P, Mekki Y. High risk HPV contamination of endocavity vaginal ultrasound probes: an underestimated route of nosocomial infection? PloS one. 2012;7:e48137. doi: 10.1371/journal.pone.0048137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma ST, Yeung AC, Chan PK, Graham CA. Transvaginal ultrasound probe contamination by the human papillomavirus in the emergency department. Emerg Med J. 2013;30:472–5. doi: 10.1136/emermed-2012-201407. [DOI] [PubMed] [Google Scholar]
- 71.M'Zali F, Bounizra C, Leroy S, Mekki Y, Quentin-Noury C, Kann M. Persistence of microbial contamination on transvaginal ultrasound probes despite low-level disinfection procedure. PloS one. 2014;9:e93368. doi: 10.1371/journal.pone.0093368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seifi S, Asvadi Kermani I, Dolatkhah R, Asvadi Kermani A, Sakhinia E, Asgarzadeh M, Dastgiri S, Ebrahimi A, Asghari Haggi A, Nadri M, Asvadi Kermani T. Prevalence of oral human papilloma virus in healthy individuals in East azerbaijan province of iran. Iran J Public Health. 2013;42:79–85. [PMC free article] [PubMed] [Google Scholar]
- 73.Bottalico D, Chen Z, Kocjan BJ, Seme K, Poljak M, Burk RD. Characterization of human papillomavirus type 120: a novel betapapillomavirus with tropism for multiple anatomical niches. J Gen Virol. 2012;93:1774–9. doi: 10.1099/vir.0.041897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dona MG, Gheit T, Latini A, Benevolo M, Torres M, Smelov V, McKay-Chopin S, Giglio A, Cristaudo A, Zaccarelli M, Tommasino M, Giuliani M. Alpha, beta and gamma Human Papillomaviruses in the anal canal of HIV-infected and uninfected men who have sex with men. J Infect. 2015;71:74–84. doi: 10.1016/j.jinf.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Viarisio D, Mueller-Decker K, Kloz U, Aengeneyndt B, Kopp-Schneider A, Grone HJ, Gheit T, Flechtenmacher C, Gissmann L, Tommasino M. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog. 2011;7:e1002125. doi: 10.1371/journal.ppat.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viarisio D, Decker KM, Aengeneyndt B, Flechtenmacher C, Gissmann L, Tommasino M. Human papillomavirus type 38 E6 and E7 act as tumour promoters during chemically induced skin carcinogenesis. J Gen Virol. 2013;94:749–52. doi: 10.1099/vir.0.048991-0. [DOI] [PubMed] [Google Scholar]
- 77.Buitrago-Perez A, Hachimi M, Duenas M, Lloveras B, Santos A, Holguin A, Duarte B, Santiago JL, Akgul B, Rodriguez-Peralto JL, Storey A, Ribas C, et al. A humanized mouse model of HPV-associated pathology driven by E7 expression. PloS one. 2012;7:e41743. doi: 10.1371/journal.pone.0041743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saidj D, Cros MP, Hernandez-Vargas H, Guarino F, Sylla BS, Tommasino M, Accardi R. Oncoprotein E7 from beta human papillomavirus 38 induces formation of an inhibitory complex for a subset of p53-regulated promoters. J Virol. 2013;87:12139–50. doi: 10.1128/JVI.01047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wallace NA, Robinson K, Galloway DA. Beta human papillomavirus E6 expression inhibits stabilization of p53 and increases tolerance of genomic instability. J Virol. 2014;88:6112–27. doi: 10.1128/JVI.03808-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holloway A, Simmonds M, Azad A, Fox JL, Storey A. Resistance to UV-induced apoptosis by beta-HPV5 E6 involves targeting of activated BAK for proteolysis by recruitment of the HERC1 ubiquitin ligase. International journal of cancer Journal international du cancer. 2015;136:2831–43. doi: 10.1002/ijc.29350. [DOI] [PubMed] [Google Scholar]
- 81.Guo F, Liu Y, Wang X, He Z, Weiss NS, Madeleine MM, Liu F, Tian X, Song Y, Pan Y, Ning T, Yang H, et al. Human papillomavirus infection and esophageal squamous cell carcinoma: a case-control study. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:780–5. doi: 10.1158/1055-9965.EPI-11-1206. [DOI] [PubMed] [Google Scholar]
- 82.Anantharaman D, Gheit T, Waterboer T, Halec G, Carreira C, Abedi-Ardekani B, McKay-Chopin S, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Mates D, et al. No causal association identified for human papillomavirus infections in lung cancer. Cancer research. 2014;74:3525–34. doi: 10.1158/0008-5472.CAN-13-3548. [DOI] [PubMed] [Google Scholar]
- 83.Hoffmann M, Tribius S, Quabius ES, Henry H, Pfannenschmidt S, Burkhardt C, Gorogh T, Halec G, Hoffmann AS, Kahn T, Rocken C, Haag J, et al. HPV DNA, E6*I-mRNA expression and p16INK4A immunohistochemistry in head and neck cancer - how valid is p16INK4A as surrogate marker? Cancer letters. 2012;323:88–96. doi: 10.1016/j.canlet.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 84.Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1992–8. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- 85.Lassen P, Primdahl H, Johansen J, Kristensen CA, Andersen E, Andersen LJ, Evensen JF, Eriksen JG, Overgaard J, Danish H, Neck Cancer G. Impact of HPV-associated p16-expression on radiotherapy outcome in advanced oropharynx and non-oropharynx cancer. Radiother Oncol. 2014;113:310–6. doi: 10.1016/j.radonc.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 86.Munger K, Gwin TK, McLaughlin-Drubin ME. p16 in HPV-associated cancers. Oncotarget. 2013;4:1864–5. doi: 10.18632/oncotarget.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc Natl Acad Sci U S A. 2013;110:16175–80. doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.von Knebel Doeberitz M, Gissmann L. Analysis of the biological role of human papilloma virus (HPV)-encoded transcripts in cervical carcinoma cells by antisense RNA. Haematol Blood Transfus. 1987;31:377–9. doi: 10.1007/978-3-642-72624-8_80. [DOI] [PubMed] [Google Scholar]
- 89.Hirama T, Miller CW, Wilczynski SP, Koeffler HP. p16 (CDKN2/cyclin-dependent kinase-4 inhibitor/multiple tumor suppressor-1) gene is not altered in uterine cervical carcinomas or cell lines. Mod Pathol. 1996;9:26–31. [PubMed] [Google Scholar]
- 90.Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–8. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- 91.Magaldi TG, Almstead LL, Bellone S, Prevatt EG, Santin AD, DiMaio D. Primary human cervical carcinoma cells require human papillomavirus E6 and E7 expression for ongoing proliferation. Virology. 2012;422:114–24. doi: 10.1016/j.virol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yildiz IZ, Usubutun A, Firat P, Ayhan A, Kucukali T. Efficiency of immunohistochemical p16 expression and HPV typing in cervical squamous intraepithelial lesion grading and review of the p16 literature. Pathol Res Pract. 2007;203:445–9. doi: 10.1016/j.prp.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 94.Srivastava S. P16INK4A and MIB-1: an immunohistochemical expression in preneoplasia and neoplasia of the cervix. Indian J Pathol Microbiol. 2010;53:518–24. doi: 10.4103/0377-4929.68301. [DOI] [PubMed] [Google Scholar]
- 95.Cheah PL, Looi LM, Mun KS, Abdoul Rahman N, Teoh KH. Implications of continued upregulation of p16(INK4a) through the evolution from high-grade squamous intraepithelial lesion to invasive squamous carcinoma of the cervix. Malays J Pathol. 2011;33:83–7. [PubMed] [Google Scholar]
- 96.Michaelsen SH, Larsen CG, von Buchwald C. Human papillomavirus shows highly variable prevalence in esophageal squamous cell carcinoma and no significant correlation to p16INK4a overexpression: a systematic review. J Thorac Oncol. 2014;9:865–71. doi: 10.1097/JTO.0000000000000166. [DOI] [PubMed] [Google Scholar]
- 97.Carozzi F, Gillio-Tos A, Confortini M, Del Mistro A, Sani C, De Marco L, Girlando S, Rosso S, Naldoni C, Dalla Palma P, Zorzi M, Giorgi-Rossi P, et al. Risk of high-grade cervical intraepithelial neoplasia during follow-up in HPV-positive women according to baseline p16-INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. The lancet oncology. 2013;14:168–76. doi: 10.1016/S1470-2045(12)70529-6. [DOI] [PubMed] [Google Scholar]
- 98.Rautava J, Syrjanen S. Biology of human papillomavirus infections in head and neck carcinogenesis. Head Neck Pathol. 2012;6(Suppl 1):S3–15. doi: 10.1007/s12105-012-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.