Abstract
Amyloid precursor protein (APP) has been reported to be highly expressed in acute myeloid leukemia (AML)1-eight-twenty one (ETO)-positive AML. In the present study, the clinical and prognostic significance of APP expression was assessed in 65 patients with AML1-ETO-positive AML using reverse transcription-quantitative polymerase chain reaction. The patients were divided into an APP-high expression (APP-H) group (n=32) and an APP-low expression (APP-L) group (n=33) according to the cut-off value of APP relative expression, which was calculated by receiver operating characteristic curve analysis. It was observed that C-KIT mutations (14/32 vs. 3/33, P=0.009), white blood cell count (median, 23.2×109 vs. 12.4×109 cells/l; P=0.011) and bone marrow cellularity (median, 91.0 vs. 84.0%; P=0.039) and incidence of extramedullary leukemia (11/32 vs. 3/33, P=0.013) were all significantly increased in the APP-H group compared with the APP-L group. Furthermore, significantly lower rate of cumulative two-cycle complete remission (83.9 vs. 100%, P=0.016), major molecular remission following two courses of consolidation (34.5 vs. 71.4%, P=0.005), and poorer relapse-free survival (RFS) (33.5±5.2% vs. 76.3±6.9%, P<0.001) and overall survival (OS) (44.5±7.0% vs. 81.9±5.8%, P=0.002) were associated with APP overexpression. Multivariate analysis revealed that APP overexpression was a significant adverse factor affecting both RFS and OS. Taken together, these data suggest that APP may be correlated with C-KIT mutations and involved in leukemia cell proliferation, and its overexpression has an adverse effect on the prognosis in AML1-ETO-positive AML.
Keywords: amyloid precursor protein, AML1-ETO, acute myeloid leukemia, clinical significance, prognosis
Introduction
The chromosomal translocation t(8;21)(q22;q22), which generates the acute myeloid leukemia (AML)1-eight-twenty-one (ETO) fusion gene, represents one of the most common cytogenetic abnormalities in AML (1). It is usually associated with a relatively low risk of relapse and long survival (2,3). However, AML1-ETO-positive AML is a highly heterogeneous disease that harbors a high rate of C-KIT mutations (6–31%) (4,5), which have a negative effect on the relapse rate (≤70%) and survival of AML patients (6,7), and lead to high incidence of extramedullary leukemia (EML) (15.0–26.7%), which also adversely affects prognosis (8). Amyloid precursor protein (APP), a type I integral membrane protein that is implicated in synapse formation and plasticity, and whose cleavage is directly associated with the pathogenesis of neurodegenerative disorders such as Alzheimers disease (9), has been observed to be highly expressed in AML1-ETO-positive leukemia but not in other subtypes of leukemia (10,11). It has been reported that APP promotes cancer cell proliferation and metastasis, and has an adverse effect on prognosis in oral squamous cell carcinoma, pancreatic cancer and colorectal cancer (12–15). In our previous study, APP was observed to be positively correlated with EML, and its overexpression enhanced the migration of Kasumi-1 cells via matrix metalloproteinase (MMP)-2 (16). However, APP does not have an obvious clinical and prognostic significance in other subtypes of leukemia (11). Therefore, APP is important in AML1-ETO-positive AML. With the exception of the correlation between APP and leukemia cell migration, whether there is another important role of APP in AML1-ETO-positive AML remains to be investigated. The present study describes the clinical and prognostic significance of APP in 65 patients with AML1-ETO-positive AML.
Materials and methods
Patients
From February 2006 to June 2013, a total of 65 patients from Nanfang Hospital (Guangzhou, China), who were diagnosed according to the World Health Organization 2008 classification (17) as AML1-ETO-positive AML, were enrolled in the present study. The median observation time was 24 months (range, 3–96 months). Of all patients, 58 were AML subtype M2, 5 were AML subtype M4 and 2 were AML subtype M5, according to the French-American-British classification (18,19). The study included 13 pediatric cases (≤18 years of age; median, 13 years) and 52 adult cases (>18 years of age; median, 36.5 years). The main characteristics of the patients are listed in Table I. From the 65 patients, 64 completed 1–2 cycles of induction chemotherapy followed by ‘3+7’ regimens consisting of daunorubicin at a daily dose of 40–60 mg/m2, idarubicin at a daily dose of 8–10 mg/m2 or other anthracyclines for 3 days, and arabinosylcytosine (Ara-C) at 100–150 mg/m2/day for 7 days. A total of 62 cases who were followed up subsequent to complete remission (CR) were divided into two groups according to the consolidation protocols: i) Standard-dose Ara-C (SDAC)-based regimen group, in which the patients were treated by the aforementioned conventional induction regimens with or without 1–2 cycles of Ara-C (1–2 g/m2 every 12 h 6–8 times); and ii) median-dose Ara-C (MDAC)-based regimen group, in which the patients were treated with ≥3 cycles of Ara-C (1–2 g/m2 every 12 h 6–8 times) with or without autologous hematopoietic stem cell transplantation.
Table I.
Sequence of the polymerase chain reaction primers for each gene.
| Gene | Primers | Sequences |
|---|---|---|
| FLT3-ITD | Forward | 5′-GCAATTTAGGTATGAAAGCCAGC-3 |
| Reverse | 5′-CTTTCAGCATTTTGACGGCAACC-3 | |
| FLT3-TKD | Forward | 5′-CCAGGAACGTGCTTGTC-3 |
| Reverse | 5′-TCAAAAATGCACCACAGTGAG-3 | |
| NPM1 | Forward | 5′-TTAACTCTCTGGTGGTAGAATGAA-3 |
| Reverse | 5′-CAAGACTATTTGCCATTCCTAAC-3 | |
| C-KIT | ||
| Exon 8 | Forward | 5′-CTCCCTGAAAGCAGAAAC-3 |
| Reverse | 5′-CAGAAAGATAACACCAAAATAG-3 | |
| Exon 17 | Forward | 5′-GCAAAGGCATATTAGGAACTC-3 |
| Reverse | 5′-GTTGTAGTAATGTTCAGCATACC-3 | |
| NRAS | ||
| Exon 2 | Forward | 5′-AGAACCAAATGGAAGGTCACA-3 |
| Reverse | 5′-TGGGTAAAGATGATCCGACA-3 | |
| Exon 3 | Forward | 5′-GCAATTTGAGGGACAAACCA-3 |
| Reverse | 5′-CCCTAGATTCTCAATGTCAAACAA-3 | |
| KRAS | ||
| Exon 2 | Forward | 5′-CTTAAGCGTCGATGGAGGAG-3 |
| Reverse | 5′-AGAATGGTCCTGCACCAGTAA-3 | |
| Exon 3 | Forward | 5′-TTTTTGAAGTAAAAGGTGCACTG-3 |
| Reverse | 5′-TGCATGGCATTAGCAAAGAC-3 | |
| JAK2 (V617F) | Forward | 5′-ATCTATAGTCATGCTGAAAGTAGGAGAAAG-3 |
| Reverse | 5′-CTGAATAGTCCTACAGTGTTTTCAGTTTCA-3 |
FLT3, Fms related tyrosine kinase 3; NPM1, nucleophosmin; JAK2, Janus kinase 2.
Informed consent was obtained from all patients in accordance to the regulations of the Institutional Review Board of Nanfang Hospital, Southern Medical College of Medicine (Guangzhou, China) and in agreement with the Declaration of Helsinki.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
For each patient, a bone marrow sample was collected at diagnosis, and mononuclear cells were enriched by density gradient centrifugation (447 × g for 20 min at room temperature) with Ficoll solution (Fresenius Kabi Norge As, Oslo, Norway). Total RNA extraction, complementary DNA synthesis, design of PCR primer sequences for APP and β-actin, and APP expression assessment by RT-qPCR were performed as described earlier (16).
The levels of AML1-ETO messenger RNA (mRNA) transcripts expressed in bone marrow, which served as the index of minimal residual disease (MRD) monitoring at the time of diagnosis, following induction therapy, subsequent to every consolidation chemotherapy, at 3-month intervals for the first 2 years of follow-up and at relapse, were quantified using RT-qPCR as previously described (20). Early treatment response was assessed as the rate of major molecular remission (MMR) following two courses of consolidation [which was defined as a >3-log reduction in AML1-ETO mRNA transcript levels compared with those during pre-treatment (20)] and the MRD negative rate during the period of complete continuous remission (CCR) [which was defined as PCR negativity according to the Europe Against Cancer criteria (21)].
Morphology, immunophenotyping and cytogenetics
Bone marrow cell morphology was studied using Wright-Giemsa staining, and cytochemical, peroxidase-based, specific and non-specific esterase tests were performed for each patient at diagnosis.
Flow cytometric analysis of samples obtained at diagnosis was performed using antibodies (BD Biosciences, Franklin Lakes, NJ, USA) against the following: Human leukocyte antigen-antigen D-related (anti-HLA-DR APC-Cy™7; #335814), cluster of differentiation (CD)13 (PE mouse anti-human CD13; #347837), CD33 (APC mouse anti-human CD33; #340474), CD34 (FITC mouse anti-human CD34; #348053), CD19 (PerCP mouse anti-human CD19; #347544), CD56 (FITC mouse anti-human CD19; #340410) and CD117 (PE-Cy™7 mouse anti-human CD117; #339195). All antibodies were diluted according to the manufacturer's instructions and incubated with patient samples for 15–20 min at room temperature prior to examination. Expression rates of ≥20% were referred to as positive for all membrane antigens, with the exception of CD117, which was considered to exhibit a positive expression when the expression rate was ≥10% (22).
Chromosome banding analysis combined with fluorescence in situ hybridization was performed in all patients following standard methods (23). Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (24).
Molecular analysis
DNA from mononuclear cells, which were isolated from bone marrow samples obtained at diagnosis, was extracted using a DNA extraction kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturers protocol. The screening of Fms related tyrosine kinase 3 (FLT3)-ITD, FLT3-TKD, C-KIT (exons 8 and 17), nucleophosmin (NPM1), KRAS (exons 2 and 3), NRAS (exons 2 and 3) and Janus kinase 2 (JAK2) (V617F) mutations was performed by PCR and direct Sanger sequencing, according to previously described protocols (25). PCR primers are shown in Table I. The total reaction volume of 25 µl contained 2 µl DNA (100 ng), 1 µl of each primer (50 pmol), 12.5 µl PCR mix (Takara Biotechnology Co., Ltd., Dalian, China) and 8.5 µl double-distilled H2O. Purified PCR products were sequenced by Sanger sequencing. The results were analyzed with Chromas software version 2.3 (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Statistical analysis
SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA) was used for data analysis. To assess the prognostic effect of APP in AML1-ETO-positive AML, receiver operating characteristic (ROC) curve analysis was used to calculate the cut-off value of APP relative expression levels, which served to divide the patients into an APP-high expression (APP-H) group (with levels of APP ≥ cut-off value) and an APP-low expression (APP-L) group (with levels of APP < cut-off value). Not normally distributed data were described by the median. A comparison of distribution between two groups of data was performed by a Mann-Whitney nonparametric U test, and correlations between two variables were performed by Spearman rank correlation analysis. A nonparametric test was performed using the χ2 test, and survival analyses were calculated by Kaplan-Meier survival curves and the log-rank test. Relapse-free survival (RFS) was measured from the CR date to relapse or last follow-up. Overall survival (OS) was calculated from the diagnosis to the last observation or mortality. Univariate analysis with the log-rank test and multivariate analysis on categorized data were performed using Cox regression. P<0.05 was considered to indicate a statistically significant difference.
Results
APP expression in AML1/ETO-positive AML patients
Previous studies reported that the APP gene is overexpressed in AML1-ETO positive AML patients (9,10). In the present study, the relative APP levels assessed by RT-qPCR exhibited a median value of 0.12422 (range, 0.00041–3.18640), and a mean ± standard deviation of 0.23131±0.05359.
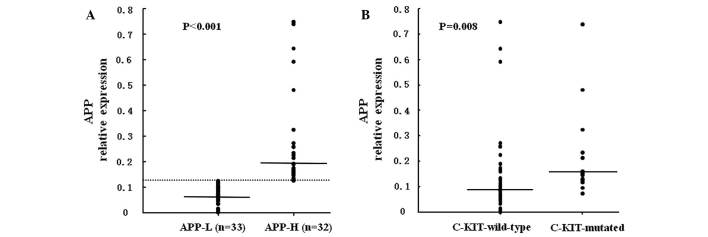
ROC curve analysis revealed that APP had a negative effect on RFS [area under the ROC curve (AZ)=0.772, P<0.001] or OS (AZ=0.673, P=0.029). According to the coordinates of the ROC curve, a value of 0.12424, which had optimal sensitivity and specificity in RFS prediction, was considered as the cut-off value, while 0.12455 was selected for OS prediction. In order to balance the deviation, 0.12440, which is the mean value of 0.12424 and 0.12455, was considered as the cut-off value (Fig. 1). Among the 65 patients, 32 cases, whose APP expression levels were ≤0.12440, were included in the APP-H group, and the other 33 cases, whose APP levels were <0.12440, were included in the APP-L group. The median APP levels in the APP-H group were significantly higher than those in the APP-L group [0.19022 (range, 0.12484–3.18640) vs. 0.06890 (range, 0.00041–0.12422), respectively; AZ=−6.928, P<0.001] (Fig. 2A).
Figure 1.
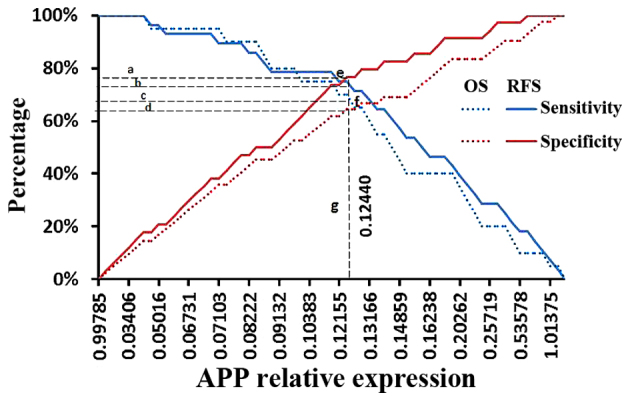
Cut-off value of high and low APP relative expression levels. The sensitivity and specificity of different APP expression levels in RFS and OS prediction were evaluated by receiver operating characteristic curve analysis. According to the coordinates of the curve, the cut-off value of APP expression levels was calculated for both RFS (point e, where the APP level was 0.12424 and the values of sensitivity and specificity were both 75.6%] and OS prediction (point f, where the APP level was 0.12455 and the sensitivity and specificity values were both 66.0%). The value 0.12440 (line g), which is the mean value of 0.12424 and 0.12455, was considered the cut-off value, with sensitivity and specificity values of 76.5% (line a) and 73.2% (line b), respectively, in RFS prediction, and of 67.5% (line c) and 64.3% (line d), respectively, in OS prediction. APP, amyloid precursor protein; OS, overall survival; RFS, relapse-free survival.
Figure 2.
The incidence of C-KIT mutations is positively correlated with APP expression. (A) APP relative expression level was evaluated by reverse transcription-quantitative polymerase chain reaction. The dashed line represents the cut-off value (0.12440) of high and low APP expression level. APP levels were significantly higher in the APP-H group (with relative levels of APP ≥0.12440) than in the APP-L group (with relative levels of APP <0.12440). (B) Significantly increased APP expression levels were observed in C-KIT-mutated patients. APP-H, APP-high expression; APP-L, APP-low expression; APP, amyloid precursor protein.
Distinct clinical features according to APP expression levels in patients with AML1-ETO-positive AML
With respect to gene mutations in AML1-ETO-positive AML patients, C-KIT mutations were the most common ones. Of the 65 investigated patients, 17 displayed C-KIT mutations, while 7 NRAS, 1 FLT3-ITD, 1 JAK2 (V617F) and 3 NPM1 mutations were detected (Table II). Furthermore, no gene mutations, with the exception of C-KIT, were observed to be correlated with APP expression levels. In total, 13 out of the 17 cases with C-KIT mutations belonged to the APP-H group (13/32), whereas only 4 mutations were observed in the APP-L group (4/33, P=0.009) (Table II). Additionally, APP was highly expressed in the C-KIT-mutated patients with a median of 0.15427 (range, 0.07432–3.18640) as compared with the C-KIT wild-type cases (median, 0.09132; range, 0.00041–1.19080; Az=−2.672, P=0.008) (Fig. 2B).
Table II.
Clinical characteristics of 65 patients with AML1-eight-twenty-one-positive AML.
| Characteristics | Total | APP-H | APP-L | P-value |
|---|---|---|---|---|
| Patients, n | 65 | 32 | 33 | |
| Median age (range), years | 30 (4–69) | 33 (5–69) | 27 (4–58) | 0.435 |
| Adult/pediatric patients, n (ratio) | 52/13 (4.0) | 26/6 (4.3) | 26/7 (3.7) | 0.804 |
| Male/female patients, n (ratio) | 40/25 (1.6) | 20/12 (1.7) | 20/13 (1.5) | 0.875 |
| Blood counts | ||||
| Median WBC count (range), ×109 cells/l | 18.6 (1.7–97.6) | 23.2 (3.1–97.6) | 12.4 (1.7–70.3) | 0.011 |
| Median hemoglobin level (range), g/dl | 73.0 (39.0–154.0) | 73.0 (39.0–154.0) | 7.0 (41.0–123.0) | 0.984 |
| Median PLT count (range), ×109 cells/l | 23.0 (5.0–137.0) | 22.5 (5.0–93.0) | 24.0 (7.0–137.0) | 0.728 |
| EML, n/total (%) | 14/65 (21.5) | 11/32 (34.4) | 3/33 (9.1) | 0.013 |
| Median marrow blasts (range), % | 35.0 (12.0–94.0) | 38.0 (17.0–94.0) | 34.0 (12.0–93.0) | 0.446 |
| Median bone marrow cellularity (range), % | 88.0 (47.0–99.0) | 91.0 (47.0–99.0) | 84.0 (52.0–98.0) | 0.039 |
| Immunophenotype, n/total (%) | ||||
| HLA-DR-positive | 55/64 (85.9) | 28/31 (90.3) | 27/33 (12.8) | 0.328 |
| CD13-positive | 55/64 (85.9) | 25/31 (80.4) | 30/33 (90.9) | 0.238 |
| CD33-positive | 51/64 (79.7) | 26/31 (83.9) | 25/33 (75.8) | 0.420 |
| CD34-positive | 60/64 (93.8) | 30/31 (96.8) | 30/33 (90.9) | 0.333 |
| CD19-positive | 26/64 (40.6) | 14/31 (45.2) | 12/33 (36.4) | 0.474 |
| CD56-positive | 41/64 (64.1) | 19/31 (61.3) | 22/33 (66.7) | 0.654 |
| CD117-positive | 60/64 (93.8) | 30/31 (96.8) | 30/33 (90.9) | 0.333 |
| Cytogenetic aberrations in addition to t(8;21)(q22;q22) (n=52) | ||||
| Solely t(8;21)/ACA, n | 26/26 | 14/13 | 12/13 | 0.781 |
| Loss of sex chromosomes, n/total (%) | 19/52 (36.5) | 9/27 (33.3) | 10/25 (40.0) | 0.618 |
| Molecular mutations in addition to RUNX1-RUNX1T1, n/total (%) | ||||
| C-KITa | 17/65 (26.2) | 13/32 (40.6) | 4/33 (12.1) | 0.009 |
| JAK2 (V617F) | 1/65 (1.5) | 1/32 (3.1) | 0/33 (0.0) | 0.306 |
| NRAS | 7/65 (10.8) | 4/32 (12.5) | 3/33 (9.1) | 0.658 |
| KRAS | 0/65 (0.0) | 0/32 (0.0) | 0/33 (0.0) | – |
| FLT3-ITD | 1/65 (1.5) | 1/32 (3.1) | 0/33 (0.0) | 0.306 |
| FLT3-TKD | 0/65 (0.0) | 0/32 (0.0) | 0/33 (0.0) | – |
| NPM1 | 3/65 (4.6) | 1/32 (3.1) | 2/33 (6.1) | 0.573 |
| Induction therapy and regimens, n | ||||
| DA | 22 | 14 | 8 | 0.153 |
| IA | 34 | 16 | 18 | 0.153 |
| Other | 8 | 2 | 6 | 0.153 |
| Consolidation therapy and regimens, n | ||||
| SDAC-based/MDAC-based | 31/31 | 18/12 | 13/19 | 0.127 |
| Allo-HSCT/no allo-HSCT | 12/50 | 5/25 | 7/25 | 0.604 |
| Outcome, n/total (%) | ||||
| Cumulative two-cycle CR | 59/64 (92.2) | 26/31 (83.9) | 33/33 (100.0) | 0.016 |
| MMR following two courses of consolidation | 30/57 (52.6) | 10/29 (34.5) | 20/28 (71.4) | 0.005 |
| MRD negative | 36/57 (63.2) | 13/29 (44.8) | 23/28 (82.1) | 0.004 |
| Relapse | 28/62 (45.2) | 22/30 (73.3) | 6/32 (18.8) | <0.001 |
| Mortality subsequent to CR | 20/62 (32.3) | 15/30 (50.0) | 5/32 (15.6) | 0.004 |
D816 (n=10), N822 (n=6), c.1253delACG (exon 8; n=1). AML, acute myeloid leukemia; WBC, white blood cell; PLT, platelet; EML, extramedullary leukemia; HLA-DR, human leukocyte antigen-antigen D related; CD, cluster of differentiation; ACA, additional chromosomal abnormality; RUNX1, Runt-related transcription factor 1; RUNX1T1, RUNX1 translocation partner 1; JAK2, Janus kinase 2; FLT3, Fms related tyrosine kinase 3; NPM1, nucleophosmin; DA, daunorubicin + cytarabine; IA, idarubicin + cytarabine; SDAC, standard-dose Ara-C; MDAC, median-dose Ara-C; Ara-C, arabinosylcytosine; HSCT, hematopoietic stem cell transplantation; CR, complete remission; MMR, major molecular remission; MRD, minimal residual disease; APP-H, APP-high expression; APP-L, APP-low expression; APP, amyloid precursor protein.
According to other clinical parameters, a significantly higher white blood cell (WBC) count (median, 23.2×109 vs. 12.4×109 cells/l; P=0.011) and bone marrow cellularity (median, 91.0 vs. 84.0%; P=0.039) were detected in APP-H patients compared with APP-L patients. Furthermore, EML was correlated with APP expression levels, since 11/32 patients exhibited EML in the APP-H group, while only 3/33 cases exhibited EML in the APP-L group (P=0.013). With respect to age, sex, peripheral hemoglobin levels, platelet counts, bone marrow blasts, immunophenotype and karyotype, no significant differences were observed between the two groups (Table II).
Patient outcomes. The median follow-up time was 24.0 months (range, 3.0–96.0 months) among the surviving patients. Of the 64 patients analyzed, 59 received CR following 1–2 cycles of induction chemotherapy, and 62 patients achieved CR following induction. In total, 28 patients experienced a relapse, 20 succumbed to disease (15 due to relapse and 5 due to treatment-associated mortality) and 44 survived during the follow-up time. For the study population that could be analyzed, the cumulative two-cycle CR rate was 92.2%, the cumulative incidence of relapse was 45.2%, the RFS rate was 54.8% and the OS rate was 67.7%.
APP overexpression indicates poor disease prognosis in AML1-ETO-positive AML patients
Poor early response was correlated with APP overexpression. By subgroup analysis, significantly lower cumulative two-cycle CR rate was identified in the APP-H group compared with that in the APP-L group (83.9 vs. 100.0%, P=0.016). APP overexpression also had an adverse effect on early molecular response, since the cumulative MMR rate following two courses of consolidation [which is a significant predictor of relapse (20,26)] and the MRD negative rate during the CCR period in the APP-H group were statistically lower than those in the APP-L group (34.5 vs. 71.4%, P=0.005 and 44.8 vs. 82.1%, P=0.004, respectively) (Table II).
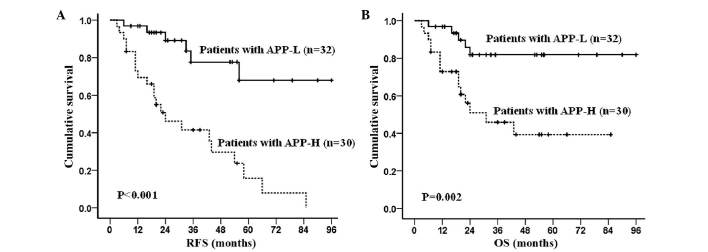
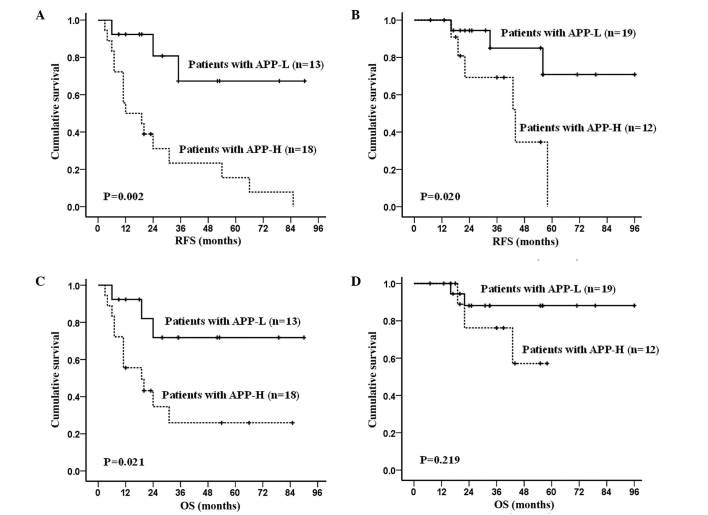
APP overexpression had a significantly negative effect on RFS and OS. By subgroup analysis, in the APP-H group, the RFS rate was 33.5±5.2% and the OS rate was 44.5±7.0%, with medians of 10.0 months (range, 2.0–64.0 months) and 20.0 months (range, 3.0–85.0 months), respectively. The 3-year RFS and OS in the APP-H group were 31.9±5.5% and 39.4±7.1%, respectively. In the APP-L group, the RFS rate was 76.3±6.9% (P<0.001) and the OS rate was 81.9±5.8% (P=0.002), with medians of 24.5 months (range, 3.0–94.0 months) and 31.0 months (range, 6.0–96.0 months), respectively. In the APP-L group, the 3-year RFS and OS were 73.5±7.6% (P<0.001) and 77.3±7.3% (P=0.005), respectively (Fig. 3). To avoid the influence of therapeutic regimens, the data in the patients receiving either SDAC-based or MDAC-based regimens were also analyzed: RFS was significantly increased (P=0.002 and P=0.020, respectively; Fig. 4A and B) in the APP-L group compared with the APP-H group; whereas OS was significantly increased in patients receiving SDAC-based therapy (P=0.021), but was not significantly different (P=0.219) in patients receiving MDAC-based therapy in the APP-L group vs. the APP-H group (Fig. 4C and D).
Figure 3.
APP as an indicator of disease outcome in AML1-eight-twenty one-positive AML patients. (A) RFS and (B) OS according to APP expression levels. APP, amyloid precursor protein; APP-H, APP-high expression; APP-L, APP-low expression; OS, overall survival; RFS, relapse-free survival; AML, acute myeloid leukemia.
Figure 4.
Survival analysis of AML1-eight-twenty one-positive AML patients according to different chemotherapy regimens and APP expression. Significant differences in RFS between the APP-L and APP-H groups were observed in patients receiving (A) standard-dose Ara-C-based regimen or (B) median-dose Ara-C-based regimen. (C) A significant difference in OS, according to high or low APP expression, was observed in patients receiving standard-dose Ara-C-based regimen. (D) No statistically significant difference in OS between the APP-L and APP-H groups was observed in those receiving median-dose Ara-C-based regimen. APP, amyloid precursor protein; APP-H, APP-high expression; APP-L, APP-low expression; Ara-C, arabinosylcytosine; OS, overall survival; RFS, relapse-free survival; AML, acute myeloid leukemia.
Univariate analysis revealed that ≥90% bone marrow cellularity (P=0.023), high APP expression (P<0.001) and C-KIT mutations (P=0.001) were unfavorable predictors for RFS, while EML (P=0.047), high APP expression (P=0.005) and C-KIT mutations (P=0.007) were the adverse factors for OS. MDAC as a consolidation regimen and MMR following two courses of consolidation represented protective factors for both RFS (P=0.011 and P<0.001, respectively) and OS (P=0.007 and P<0.001, respectively). Multivariate analysis indicated that high APP expression was a significant adverse prognostic factor for both RFS and OS in the present series, while acquiring MMR following two courses of consolidation was beneficial for both RFS and OS, and MDAC as consolidation regimen benefited OS (Table III).
Table III.
Risk factors for RFS and OS in patients with AML1-eight-twenty-one-positive AML.
| RFS | OS | |||||
|---|---|---|---|---|---|---|
| Risk factors | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Univariate analysisa | ||||||
| Sex (male vs. female) | 0.76 | 0.35–1.67 | 0.497 | 0.82 | 0.33–2.05 | 0.655 |
| Age (adult vs. pediatric) | 0.64 | 0.28–1.46 | 0.288 | 0.87 | 0.31–2.39 | 0.781 |
| WBC (≥20×109 vs. <20×109 cells/l) | 1.21 | 0.56–2.62 | 0.622 | 1.29 | 0.51–3.24 | 0.591 |
| EML (with vs. without) | 2.08 | 0.91–4.76 | 0.083 | 2.54 | 1.01–6.38 | 0.047 |
| Bone marrow cellularity (≥90 vs. <90%) | 2.46 | 1.13–5.33 | 0.023 | 2.38 | 0.97–5.86 | 0.059 |
| CD19 expression (positive vs. negative) | 0.69 | 0.31–1.52 | 0.350 | 0.49 | 0.18–1.36 | 0.173 |
| CD56 expression (positive vs. negative) | 1.09 | 0.49–2.42 | 0.836 | 1.33 | 0.51–3.47 | 0.558 |
| Additional chromosomal abnormality (present vs. absent) (n=52)b | 0.82 | 0.36–1.85 | 0.633 | 0.75 | 0.29–1.95 | 0.55 |
| Loss of sex chromosomes (present vs. absent) (n=52)b | 1.13 | 0.49–2.57 | 0.777 | 1.11 | 0.42–2.95 | 0.834 |
| APP expression (low vs. high) | 5.73 | 2.31–14.19 | <0.001 | 4.23 | 1.53–11.70 | 0.005 |
| C-KIT (mutation vs. wild-type) | 3.58 | 1.66–7.70 | 0.001 | 3.33 | 1.38–8.01 | 0.007 |
| Chemotherapy (SDAC- vs. MDAC-based regimens) | 0.36 | 0.16–0.79 | 0.011 | 0.25 | 0.09–0.69 | 0.007 |
| MMR following two courses of consolidation (present vs. absent) (n=56)b | 0.04 | 0.01–0.16 | <0.001 | 0.06 | 0.01–0.24 | <0.001 |
| Multivariate analysisc | ||||||
| APP expression (low vs. high) | 3.55 | 1.22–10.32 | 0.020 | 4.95 | 1.42–17.29 | 0.012 |
| MMR following two courses of consolidation (present vs. absent) (n=56)b | 0.04 | 0.01–0.22 | <0.001 | 0.06 | 0.01–0.31 | 0.001 |
| Chemotherapy (SDAC- vs. MDAC-based regimens) | 0.45 | 0.17–1.21 | 0.113 | 0.25 | 0.07–0.83 | 0.024 |
Factors with P<0.10 in the univariate analysis were next subjected to multivariate analysis.
Patients with an unknown variable were included in the analysis using a dummy variable indicating missing data.
Backward stepwise Cox proportional hazard model was used in multivariate analysis of risk factors. RFS, relapse-free survival; OS, overall survival; AML, acute myeloid leukemia; WBC, white blood cell; EML, extramedullary leukemia; CD, cluster of differentiation; APP, amyloid precursor protein; MMR, major molecular remission; SDAC, standard-dose arabinosylcytosine; MDAC, median-dose arabinosylcytosine; HR, hazard ratio; CI, confidence interval.
Discussion
It has been reported that APP promotes cancer cell proliferation and metastasis in solid cancer and has an adverse effect on the prognosis of patients (12–15). The present study demonstrates that APP has a clinical significance and an adverse effect on disease outcome in AML1-ETO-positive AML, suggesting that APP also is important in certain subtypes of leukemia.
APP has been reported to be positively correlated with leukemia cell metastasis, and its overexpression enhanced the migration of Kasumi-1 cells by MMP-2 in our previous study (16). In the current study, significantly higher incidence of EML was identified in APP-H patients compared with that in APP-L patients, which further confirmed the close association of APP with leukemia cell metastasis, and could explain the high incidence of EML in AML1-ETO-positive AML.
Peripheral WBC count and bone marrow cellularity were correlated with APP expression in the present study, since elevated peripheral WBC count and higher bone marrow cellularity were observed in APP-H patients as compared with APP-L cases, indicating that APP may promote leukemia cell proliferation. These findings are in accordance with the notion that the APP gene may be involved in hematopoiesis (27) and the fact that APP promotes cancer cell proliferation in solid cancer (15).
The present study also identified that patients with APP-H had higher incidence of C-KIT mutations than APP-L patients (40.6 vs. 12.1% in the APP-H and APP-L groups, respectively; P=0.009). APP was highly expressed in C-KIT-mutated cases, suggesting that C-KIT mutations are correlated with APP expression. This could explain the high incidence of C-KIT mutations in AML1-ETO-positive AML. It is known that C-KIT mutation is one of the most important events subsequent to presentation of the RUNX1-RUNXT1 fusion gene in AML1-ETO cells (4–7,28). Mutated C-KIT increases leukemia cell proliferation and cooperates with AML1-ETO to induce AML (8,29). Thus, it can be inferred from the clinical point of view that there is a cooperation between the APP gene and C-KIT mutations in the regulation of AML1-ETO cell proliferation.
Importantly, prognostic analysis demonstrated that APP is an independent indicator of poor disease outcome; thus, it may be considered an adverse prognostic biomarker. Poor early response, including hematologic and molecular response, was correlated with APP overexpression, since significantly lower cumulative two-cycle CR rate, MMR rate following two courses of consolidation and MRD negative rate during the period of CCR were observed in the APP-H group compared with the APP-L group. Furthermore, patients with APP-H had worse RFS and OS than APP-L patients, since the 3-year RFS and OS were much lower in patients with APP-H (31.9±5.5% and 39.4±7.1%, respectively) than in patients with APP-L (73.5±7.6%, P<0.001 and 77.3±7.3%, P=0.005, respectively). This was also observed in the patients receiving MDAC-based regimen, which has been reported to improve the prognosis of this leukemia subtype (30,31). Multivariate analysis further demonstrated that APP overexpression is an independent prognostic factor for RFS and OS.
In summary, our results suggested that APP may be correlated with C-KIT mutations and involved in leukemia cell proliferation. In addition, the present data demonstrated that APP overexpression has an adverse effect on prognosis, indicating that APP could be a novel biomarker for targeting therapy in AML1-ETO-positive AML.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China (grant no. 81500138), Natural Science Foundation of Guangdong Province, China (grant no. 2014A030313270), Medical Research Foundation of Guangdong Province, China (grant no. B2014250), Science and Technology Program of Guangzhou, China (grant no. 2013J4100109), National High Technology Research and Development Program of China (863) (grant no. 2012AA02A505), the Clinical Research Initiative of Southern Medical University (grant no. LC2016YM006), and the President Funding of Nanfang Hospital (grant no. 2015C025).
Glossary
Abbreviations
- ACA
additional chromosomal abnormality
- AML
acute myeloid leukemia
- APP
amyloid precursor protein
- CCR
complete continuous remission
- CI
confidence interval
- CR
complete remission
- EML
extramedullary leukemia
- MDAC
median-dose Ara-C
- MMR
major molecular remission
- MRD
minimal residual disease
- OS
overall survival
- RFS
relapse-free survival
- HR
hazard ratio
- ROC
receiver operating characteristic
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- SDAC
standard-dose Ara-C
- WBC
white blood cell
References
- 1.Rowley JD. Identification of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973;16:109–112. [PubMed] [Google Scholar]
- 2.Marcucci G, Mrózek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, Mayer RJ, Pettenati MJ, Powell BL, Edwards CG, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): A Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK, National Cancer Research Institute Adult Leukaemia Working Group Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5,876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 4.Shimada A, Taki T, Tabuchi K, Tawa A, Horibe K, Tsuchida M, Hanada R, Tsukimoto I, Hayashi Y. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): A study of the Japanese Childhood AML Cooperative Study Group. Blood. 2006;107:1806–1809. doi: 10.1182/blood-2005-08-3408. [DOI] [PubMed] [Google Scholar]
- 5.Krauth MT, Eder C, Alpermann T, Bacher U, Nadarajah N, Kern W, Haferlach C, Haferlach T, Schnittger S. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: Frequency and impact on clinical outcome. Leukemia. 2014;28:1449–1458. doi: 10.1038/leu.2014.4. [DOI] [PubMed] [Google Scholar]
- 6.Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW, Carroll AJ, et al. Cancer and Leukemia Group B Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 7.Schnittger S, Kohl TM, Haferlach T, Kern W, Hiddemann W, Spiekermann K, Schoch C. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107:1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 8.Byrd JC, Weiss RB, Arthur DC, Lawrence D, Baer MR, Davey F, Trikha ES, Carroll AJ, Tantravahi R, Qumsiyeh M, et al. Extramedullary leukemia adversely affects hematologic complete remission rate and overall survival in patients with t(8;2 1)(q22;q22): Results from Cancer and Leukemia Group B 8461. J Clin Oncol. 1997;l5:1–475. doi: 10.1200/JCO.1997.15.2.466. [DOI] [PubMed] [Google Scholar]
- 9.Zhang MY, Zheng CY, Zou MM, Zhu JW, Zhang Y, Wang J, Liu CF, Li QF, Xiao ZC, Li S, et al. Lamotrigine attenuates deficits in synaptic plasticity and accumulation of amyloid plaques in APP/PS1 transgenic mice. Neurobiol Aging. 2014;35:2713–2725. doi: 10.1016/j.neurobiolaging.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Baldus CD, Liyanarachchi S, Mrózek K, Auer H, Tanner SM, Guimond M, Ruppert AS, Mohamed N, Davuluri RV, Caligiuri MA, et al. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: Amplification discloses overexpression of APP ETS2, and ERG genes; Proc Natl Acad Sci USA; 2004; pp. 3915–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Meng FY, Huang ZF, Huang M, Liu LX. Expression and role of amyloid precursor protein gene in acute myeloid leukemia. Zhonghua Xue Ye Xue Za Zhi. 2010;31:309–314. (In Chinese) [PubMed] [Google Scholar]
- 12.Hansel DE, Rahman A, Wehner S, Herzog V, Yeo CJ, Maitra A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63:7032–7037. [PubMed] [Google Scholar]
- 13.Ko SY, Lin SC, Chang KW, Wong YK, Liu CJ, Chi CW, Liu TY. Increased expression of amyloid precursor protein in oral squamous cell carcinoma. Int J Cancer. 2004;111:727–732. doi: 10.1002/ijc.20328. [DOI] [PubMed] [Google Scholar]
- 14.Krause K, Karger S, Sheu SY, Aigner T, Kursawe R, Gimm O, Schmid KW, Dralle H, Fuhrer D. Evidence for a role of the amyloid precursor protein in thyroid carcinogenesis. J Endocrinol. 2008;198:291–299. doi: 10.1677/JOE-08-0005. [DOI] [PubMed] [Google Scholar]
- 15.Takayama K, Tsutsumi S, Suzuki T, Horie-Inoue K, Ikeda K, Kaneshiro K, Fujimura T, Kumagai J, Urano T, Sakaki Y, et al. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Res. 2009;69:137–142. doi: 10.1158/0008-5472.CAN-08-3633. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Yu G, Meng W, Wang Z, Meng F, Ma W. Overexpression of amyloid precursor protein in acute myeloid leukemia enhances extramedullary infiltration by MMP-2. Tumour Biol. 2013;34:629–636. doi: 10.1007/s13277-012-0589-7. [DOI] [PubMed] [Google Scholar]
- 17.Swerdlow SH, International Agency for Research on Cancer, and World Health Organization, corp-author, editor. WHO classification of tumours of haematopoietic and lymphoid tissues. International Agency for Research on Cancer; Lyon, France: 2008. p. 439. [Google Scholar]
- 18.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 19.Gralnick HG, Galton DAG, Catovsky D, Sultan D, Bennett JM. Classification of acute leukemia. Ann Intern Med. 1977;87:740–753. doi: 10.7326/0003-4819-87-6-740. [DOI] [PubMed] [Google Scholar]
- 20.Zhu HH, Zhang XHs, Qin YZ, Liu DH, Jiang H, Chen H, Jiang Q, Xu LP, Lu J, Han W, et al. MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: Results from the AML05 multicenter trial. Blood. 2013;121:4056–4062. doi: 10.1182/blood-2012-11-468348. [DOI] [PubMed] [Google Scholar]
- 21.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) - a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 22.Reuss-Borst MA, Bühring HJ, Schmidt H, Mülle CA. AML: Immunophenotypic heterogeneity and prognostic significance of c-kit expression. Leukemia. 1994;8:258–263. [PubMed] [Google Scholar]
- 23.Haferlach C, Rieder H, Lillington DM, Dastugue N, Hagemeijer A, Harbott J, Stilgenbauer S, Knuutila S, Johansson B, Fonatsch C. Proposals for standardized protocols for cytogenetic analyses of acute leukemias, chronic lymphocytic leukemia, chronic myeloid leukemia, chronic myeloproliferative disorders, and myelodysplastic syndromes. Genes Chromosomes Cancer. 2007;46:494–499. doi: 10.1002/gcc.20433. [DOI] [PubMed] [Google Scholar]
- 24.Shaffer LG, McGowan-Jordan J, Schmid M. ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013) S. Karger AG; Basel: 2013. [DOI] [Google Scholar]
- 25.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leroy H, de Botton S, Grardel-Duflos N, Darre S, Leleu X, Roumier C, Morschhauser F, Lai JL, Bauters F, Fenaux P, Preudhomme C. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21) Leukemia. 2005;19:367–372. doi: 10.1038/sj.leu.2403627. [DOI] [PubMed] [Google Scholar]
- 27.Steidl U, Kronenwett R, Rohr UP, Fenk R, Kliszewski S, Maercker C, Neubert P, Aivado M, Koch J, Modlich O, et al. Gene expression profiling identifies significant differences between the molecular phenotypes of bone marrow-derived and circulating human CD34+ hematopoietic stem cells. Blood. 2002;99:2037–2044. doi: 10.1182/blood.V99.6.2037. [DOI] [PubMed] [Google Scholar]
- 28.Park SH, Chi HS, Cho YU, Jang S, Park CJ. Effects of c-KIT mutations on expression of the RUNX1/RUNX1T1 fusion transcript in t(8;21)-positive acute myeloid leukemia patients. Leuk Res. 2013;37:784–789. doi: 10.1016/j.leukres.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Wichmann C, Quagliano-Lo Coco I, Yildiz Ö, Chen-Wichmann L, Weber H, Syzonenko T, Döring C, Brendel C, Ponnusamy K, Kinner A, et al. Activating c-KIT mutations confer oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors. Leukemia. 2015;29:279–289. doi: 10.1038/leu.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrd JC, Dodge RK, Carroll A, Baer MR, Edwards C, Stamberg J, Qumsiyeh M, Moore JO, Mayer RJ, Davey F, et al. Patients with t(8;21)(q22;q22) and acute myeloid leukemia have superior failure-free and overall survival when repetitive cycles of high-dose cytarabine are administered. J Clin Oncol. 1999;17:3767–375. doi: 10.1200/JCO.1999.17.12.3767. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen S, Leblanc T, Fenaux P, Witz F, Blaise D, Pigneux A, Thomas X, Rigal-Huguet F, Lioure B, Auvrignon A, et al. A while blood cell index as the main prognostic factor in t(8;21) acute myeloid leukemia(AML): A survey of l6l cases from the French AML Intergroup. Blood. 2002;99:3517–3523. doi: 10.1182/blood.V99.10.3517. [DOI] [PubMed] [Google Scholar]