Abstract
Purpose
Pediatric sarcomas provide a unique diagnostic challenge. There is considerable morphologic overlap between entities, increasing the importance of molecular studies in the diagnosis, treatment, and identification of therapeutic targets. We developed and validated a genome-wide DNA methylation–based classifier to differentiate between osteosarcoma, Ewing sarcoma, and synovial sarcoma.
Methods
DNA methylation status of 482,421 CpG sites in 10 Ewing sarcoma, 11 synovial sarcoma, and 15 osteosarcoma samples were determined using the Illumina Infinium HumanMethylation450 array. We developed a random forest classifier trained from the 400 most differentially methylated CpG sites within the training set of 36 sarcoma samples. This classifier was validated on data drawn from The Cancer Genome Atlas synovial sarcoma, TARGET-Osteosarcoma, and a recently published series of Ewing sarcoma.
Results
Methylation profiling revealed three distinct patterns, each enriched with a single sarcoma subtype. Within the validation cohorts, all samples from The Cancer Genome Atlas were accurately classified as synovial sarcoma (10 of 10; sensitivity and specificity, 100%), all but one sample from TARGET-Osteosarcoma were classified as osteosarcoma (85 of 86; sensitivity, 98%; specificity, 100%), and 14 of 15 Ewing sarcoma samples were classified correctly (sensitivity, 93%; specificity, 100%). The single misclassified osteosarcoma sample demonstrated high EWSR1 and ETV1 expression on RNA sequencing, although no fusion was found on manual curation of the transcript sequence. Two additional clinical samples that were difficult to classify by morphology and molecular methods were classified as osteosarcoma; one had been suspected of being a synovial sarcoma and the other of being Ewing sarcoma on initial diagnosis.
Conclusion
Osteosarcoma, synovial sarcoma, and Ewing sarcoma have distinct epigenetic profiles. Our validated methylation-based classifier can be used to provide diagnostic assistance when histologic and standard techniques are inconclusive.
INTRODUCTION
Pediatric sarcomas constitute a rare and diverse group of mesenchymal malignancies of soft tissue and bone. Ewing sarcoma (EWS), synovial sarcoma (SS), and osteosarcoma (OS) are among the most common malignant solid tumors in children.1 Although these tumors can occur in similar anatomic locations, optimal management and treatment strategies differ substantially depending on the tumor type.2,3 Accurate diagnosis is thus paramount for clinical management, but can be challenging.
Histologically, EWS is mainly composed of small round blue cells4 but occasionally consists of larger, more pleomorphic cells, making the diagnosis of EWS based solely on histopathological analysis unreliable.5 The discovery of the EWSR1-FLI1 fusion detected by fluorescent in situ hybridization (FISH) has significantly improved diagnostic accuracy, but the fusion is only present in approximately 85% of samples that are histologically consistent with EWS.6 In the remainder of cases, most tumors harbor a fusion of the EWSR1 gene with a different member of the E-26 transformation specific family of transcription factors.7,8,9,10,11
SS typically has a biphasic appearance consisting of epithelioid and fibroblast-like spindle cell components; however, a monophasic spindle cell variant is also commonly seen. Furthermore, a poorly differentiated (ie, round cell) variant exists that is histologically indistinct from other poorly differentiated tumors, which complicates diagnosis.12 Analogous to EWS, SS is characterized by a pathognomonic t(X;18)(p11.2;q11.2) translocation that can be detected by cytogenetics and may aid making the diagnosis.13 Nonetheless, the clinical behavior of SS is varied, indicating biologic heterogeneity.14
Similar to SS and EWS, OS also has a variety of histologic appearances, with subtypes including fibroblastic, osteoblastic, chondroblastic, giant cell, telangiectatic, and small cell. Morphologic variants include spindle-cell OS (resembling fibrosarcoma or monophasic synovial sarcoma), high-grade pleomorphic OS (resembling undifferentiated pleomorphic sarcoma), or small round blue cell OS (resembling classic EWS).12 The presence of osteoid deposition is helpful in the histologic diagnosis, but this deposition may not be present in very poorly differentiated specimens or small biopsy specimens. In contrast to the characteristic gene fusions found in EWS and SS, there is no pathognomonic molecular aberration that has been recognized in OS. A wide range of copy number changes, most frequently including chromosomes 6p, 8q, 13q, and 17p, have been observed.15-18 Additionally, the presence of the EWSR1-FLI1 translocation in a rare subset of small-cell OS further complicates diagnostics.19-22
Previous work has demonstrated the feasibility of classifying small round cell tumors using machine-learning techniques and artificial neural networks trained on gene expression data,23 or support vector machine–based classifiers to distinguish between sarcoma subtypes with variable sensitivity and specificity (range, 50% to 100%).24 A new approach to aid the diagnosis of solid tumors is based on the molecular signatures of genome-wide DNA methylation profiles. This technique has been pioneered as a powerful diagnostic tool in pediatric brain tumors and has been shown to be superior for risk stratification compared with standard histopathology.25-27 For the purposes of classification alone, the advantage of methylation is resolution. Older studies relied on complementary DNA microarrays with far fewer probes compared with methylation arrays exceeding 480,000 probes. There is also an advantage of decreased noise, because methylation is more invariant to formalin fixation, time to fixation, cold ischemia time, temperature out of the body, immune status of the host, and several other factors that affect gene expression. Integrative DNA-methylation analysis has been examined as a tool to classify high-grade soft-tissue sarcoma, including SS, but not OS or EWS.28 The aim of the current study was to determine if methylation profiling can be used to accurately distinguish among SS, OS, and EWS.
METHODS
Tissue Collection and DNA Extraction
Eighty tissue samples from patients newly diagnosed with OS, EWS, and SS were obtained from the archives of the Departments of Pathology at the New York University Langone Medical Center (NYULMC), Memorial Sloan Kettering Cancer Center, and Montefiore Medical Center. The study was conducted at NYULMC and approved by the institutional review board in accordance with all local and federal regulations. EWSs were screened by histology and the diagnosis confirmed by the presence of an EWSR1 rearrangement by FISH. SSs were diagnosed on the basis of histologic features and, when available, by the presence of t(X;18) on FISH. OSs were diagnosed on the basis of histologic features and, where available, absence of fusions characteristic of EWS and SS.
To develop the classifier, we selected histopathologically classic (OS, SS, EWS) and molecularly confirmed (SS, EWS) reference samples. The final cohort of samples included 36 total samples, including one secondary malignancy (OS). DNA was extracted from nondecalcified formalin-fixed paraffin-embedded (FFPE) specimens with the Maxwell 16 FFPE Plus LEV DNA Purification Kit (NYULMC and Montefiore samples; Promega, Madison, WI), or from tumor lysate (Memorial Sloan Kettering Cancer Center samples) using a Promega Maxwell 16 instrument, following manufacturer's instructions. The DNA was bisulfite converted using the EZ-96 DNA Methylation Kit from Zymo Research (Irvine, CA). DNA from FFPE samples subsequently underwent restoration using the Infinium HD FFPE DNA Restore Kit (Illumina, San Diego, CA).
Methylation Profiling, Preprocessing of Methylation Data, and Unsupervised Hierarchical Clustering
The HumanMethylation450 array (Illumina) was used to determine the DNA methylation status of 482,421 CpG sites, following manufacturer’s instructions. as previously reported.29 Standard β-mixture quantile normalization, background correction, quality control, and rule-based filtering of samples of probes were implemented using the RnBeads (http://rnbeads.mpi-inf.mpg.de/) pipeline to calculate final β values.30 β value is defined as the ratio of fluorescence intensity of the methylated probe over the overall intensity and was used in all visualization. Unsupervised hierarchical clustering was done with Euclidean measure for distance matrix and complete agglomeration method for clustering.
Statistical Analysis
All statistical tests and modeling were completed in the open-source software R (https://www.r-project.org/). M values were calculated from β values and used for statistical tests. For the ith probe, the M value was calculated as a log2 transform of a ratio of the β value.
A one-way analysis of variance via empirical Bayes method (R package limma) was conducted to select for the most differentially methylated probes among the different sarcoma subtypes. An α cutoff of 0.01 after Bonferroni correction was applied to the F test P values to correct for multiple testing of 482,421 CpG sites. More than 8,556 probes were found to be significantly differentially methylated; however, many were very highly correlated (Data Supplement); thus, we selected the top 400 probes with the greatest median absolute deviation in β values (Data Supplement).
Random Forest Classifier
The random forest algorithm was used for classification. We used the randomForest package in R for the implementation of the classifier (version 4.6-7).31 Classifier results were relatively insensitive to parameter choice as long as the number of trees was sufficiently large (> 200). The number of trees was set at 400 with a number of variables tried at each node of 305 (tuned to reduce out-of-bag error) and minimum node size of 1.
Validation Samples
We validated our classifier using The Cancer Genome Atlas (TCGA), TARGET-Osteosarcoma (TARGET-OS; from Children's Oncology Group and The Hospital for Sick Children, Toronto, Canada), and EWS samples from a recently published series. Raw signal intensity data (IDAT) files from TCGA SSs (n = 10) were downloaded from the legacy archives of the Genomic Data Commons.32 Similarly, IDAT files and normalized gene quantification from mRNA sequencing belonging to the discovery cohort of TARGET-OS (n = 86) were accessed through the TARGET data matrix (https://ocg.cancer.gov/programs/target/data-matrix). IDAT files from Huertas-Martinez et al (Institut d'Investigacio Biomedica de Bellvitge [IDIBELL]) were obtained from the corresponding author.33
Genomic Pathway Analysis
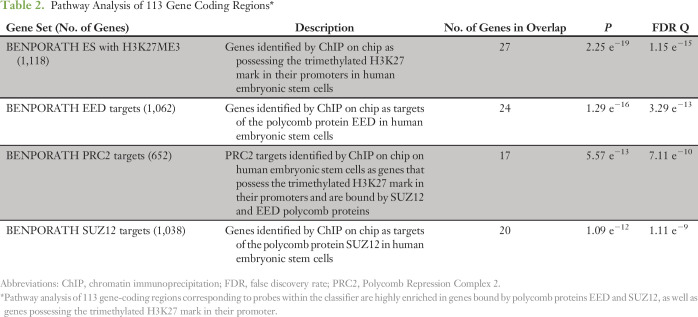
Probes in the classifier were annotated using the HumanMethylation450 manifest (version 1.2; Illumina). Genomic information including DNA sequence and coordinates of gene coding regions were obtained from the University of California Santa Cruz Genome Browser database.34 We accessed 113 probes corresponding to enhancers and gene coding regions, using the Molecular Signatures Database, a gene-set based pathway analysis. In total, we interrogated the overlap of classifier genes with 4,729 curated gene sets, which include known chemical and genetic perturbations (n = 3,500), gene sets derived from the KEGG (n = 186), BIOCARTA (n = 217), and REACTOME (n = 674) pathways, and canonical pathways curated by domain experts (n = 1,329).35
Data Access
IDAT files of the training set have been deposited in Gene Expression Omnibus, accession number GSE97529. R-script for classifier is available on Github (https://github.com/spw08536/Methylation).
RESULTS
Patient Characteristics
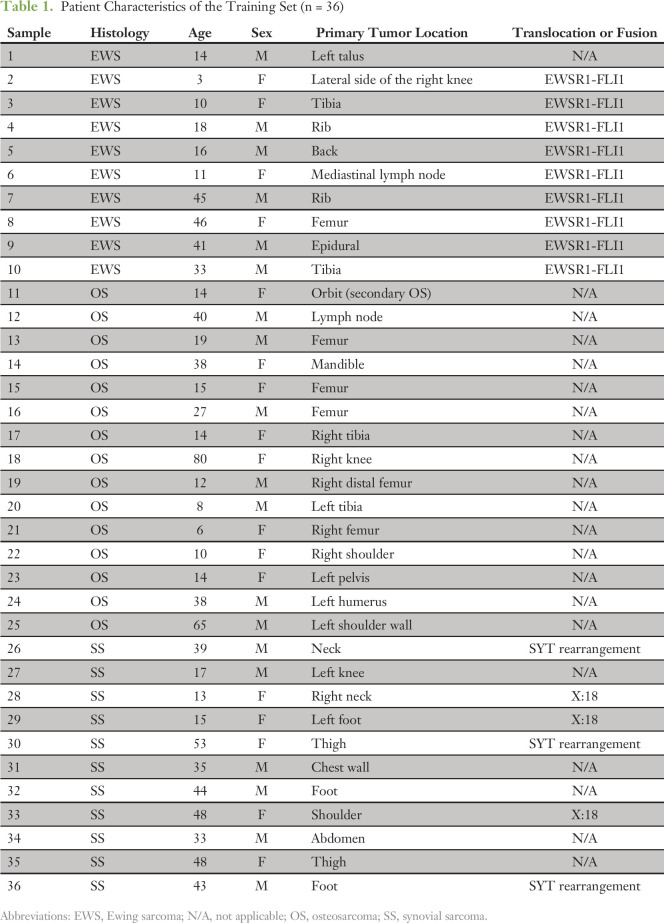
From the 80 initial patients, 36 representative samples were chosen to construct the training set: EWS (n = 10), SS (n = 11), and OS (n = 15). These are detailed in Table 1. The median patient age in the training cohort was 23 years (range, 3 to 80 years); 53% of the patients were male. Most of the samples (78%) were obtained from the primary tumor site. Sample location included the appendicular (11 of 36; 31%) and axial skeleton (23 of 36; 64%); biopsy sites of two samples (5%) were unknown.
Table 1.
Patient Characteristics of the Training Set (n = 36)
Sarcomas Showed Distinct Patterns of Methylation
Unsupervised hierarchical clustering using the 400 most differentially methylated probes among the training set demonstrated three distinct molecular phenotypes corresponding to the pathologic diagnoses (Fig 1A). We observed that probes that were methylated in OS were generally unmethylated in SS. The methylation signature of EWS tumors showed a third, distinct pattern of methylation. None of the 400 probes are colocalized with known genetic probes of interest. For EWS, two probes (cg21242508 and cg06516502) are located on chromosome 22q, approximately 1 Mb away from EWSR1 within a tetratricopeptide repeat domain, TTC28. The closest probe to FLI1 is 5 Mb away (cg13153466), corresponding to the promoter region of ASAM, a Coxsackie- and adenovirus-receptor–like membrane protein. No probes lie near the cytogenetic band 18q11.2 belonging to the SYT translocation partner of synovial sarcoma; the closest four probes in chromosome 18 lie in a 0.1-Mb strip at 18q23, notably hypomethylated in SS. Pathway analysis identified targets of polycomb group proteins SUZ12 and EED, possessing the H3K27 trimethylated mark, as highly enriched within classifier genes (Table 2).
Fig 1.
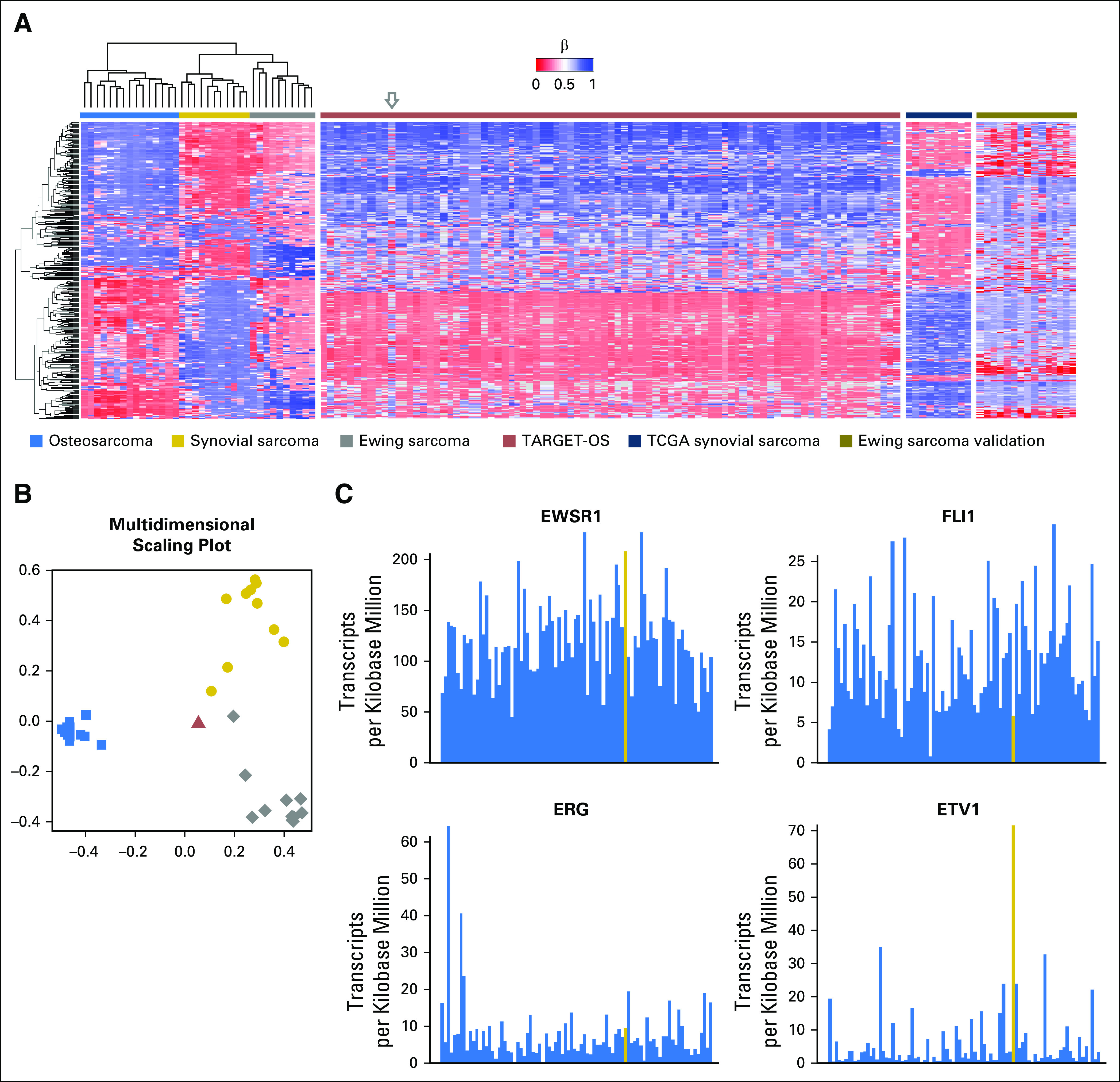
(A) Unsupervised hierarchical clustering of validation and training sets shows three distinct clusters for osteosarcoma, synovial sarcoma, and Ewing sarcoma. The gray arrow indicates the sample from the TARGET-Osteosarcoma (TARGET-OS) dataset (TARGET-40-PASEFS) that demonstrated hypermethylation in several CpG islands uncharacteristic of other osteosarcomas. (B) Multidimensional scaling plot of random forest classifier samples demonstrates this TARGET-OS sample (red triangle) was classified as Ewing sarcoma (EWS). (C) RNA sequence analysis of the sample TARGET-OS sample identified as EWS by methylation classifier. The TARGET-40-PASEFS sample shows overexpression of EWSR1 and ETV1. Other common fusion partners of EWSR1, including FLI1 and ERG, do not show increased transcription. Transcripts of interest are highlighted in gold. RNA expression data are consistent with the methylation sarcoma classifier diagnosis of EWS.
Table 2.
Pathway Analysis of 113 Gene Coding Regions*
Accurate Methylation-Based Classification of Bone Sarcomas
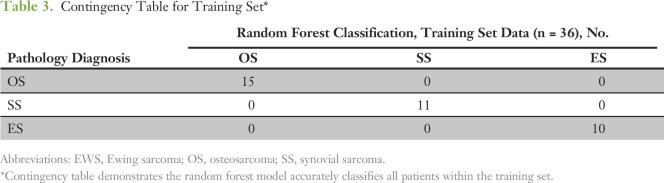
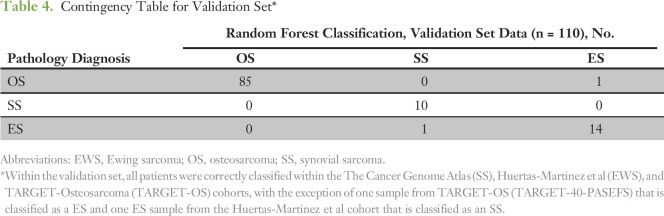
When the classifier was applied to the training cohort, all samples were accurately classified with a minimum margin score of 0.2, thus confirming internal validity (Table 3). Margin is defined as the proportion of votes given to the correct class minus maximum proportion of votes for the other classes. Thus, positive margin means correct classification. Within the validation cohorts, all samples (10 of 10) from TCGA were accurately classified as SS, 14 of 15 EWS samples were accurately classified as EWS, and all tumors from TARGET-OS (85 of 86) were classified as OS except one sample, which was classified as EWS (Table 4). A single case from the EWS validation set that classified as a SS did not have RNA data available for confirmatory studies.
Table 3.
Contingency Table for Training Set*
Table 4.
Contingency Table for Validation Set*
The discrepant OS sample, TARGET-40-PASEFS (highlighted by the arrow in Fig 1A and displayed in Fig 1B), was submitted from an 18-year-old white man who was enrolled and treated in Children’s Oncology Group clinical trial AOST0331. The tumor was located in the proximal tibia, a common location for both EWS and OS. To determine whether the tumor might have been misclassified as EWS at the time of original diagnosis, we investigated EWSR1, FLI1, ERG, and ETV1 mRNA transcript counts, available in the TARGET-OS dataset, to examine the common gene products of oncogenic EWS fusions. Although the expression of FLI1 was similar to other samples in the cohort, we noted a striking overexpression of ETV1 (n = 70 transcripts per kilobase million) and EWSR1 (n = 200 transcripts per kilobase million) compared with the other OS samples in the dataset (Fig 1C). Among all the TARGET-OS samples, this sample had the highest expression of ETV1 and third highest expression of EWSR1. We hypothesized that the tumor harbored a EWSR1/ETV1 rearrangement consistent with EWS; however, after manually curating the transcript sequence reads, there was no evidence of an ETV1 fusion, suggesting an alternative mechanism leading to ETV1 upregulation.
Clinical Application
To illustrate the clinical utility of our classifier, we present two challenging clinical cases. The first is that of a 16-year-old girl with a past medical history of rhabdomyosarcoma of the right orbit initially diagnosed in 2007 and treated with chemotherapy and radiation therapy. She had recurrence of the tumor in 2009 and was treated with orbital exenteration and maxillectomy, chemotherapy, and re-irradiation. In mid-2016, recurrence was noted in the diploic cavity (Fig 2A). A biopsy specimen demonstrated a predominantly spindled neoplasm with fascicular growth, necrosis, frequent mitotic activity, and nuclear pleomorphism (Fig 2B). Immunohistochemical stains were positive for vimentin, CD56, and desmin, but tumor cells were negative for myogenin, Myo-D1, S-100, CAM 5.2, AE 1/3, BCL-2, CD99, CD34, and EMA. INI-1/BAF47 expression was preserved. FISH testing using a dual-color break-apart probe showed a rearrangement involving SS18 (SYT; 88.0% of cells; data not shown), raising the concern that the tumor may represent SS. However, SatB2 was positive by immunohistochemistry, suggesting osteoblastic differentiation.36 SSX1 and SSX2, the usual fusion partners for SYT in synovial sarcoma, had no abnormalities.
Fig 2.
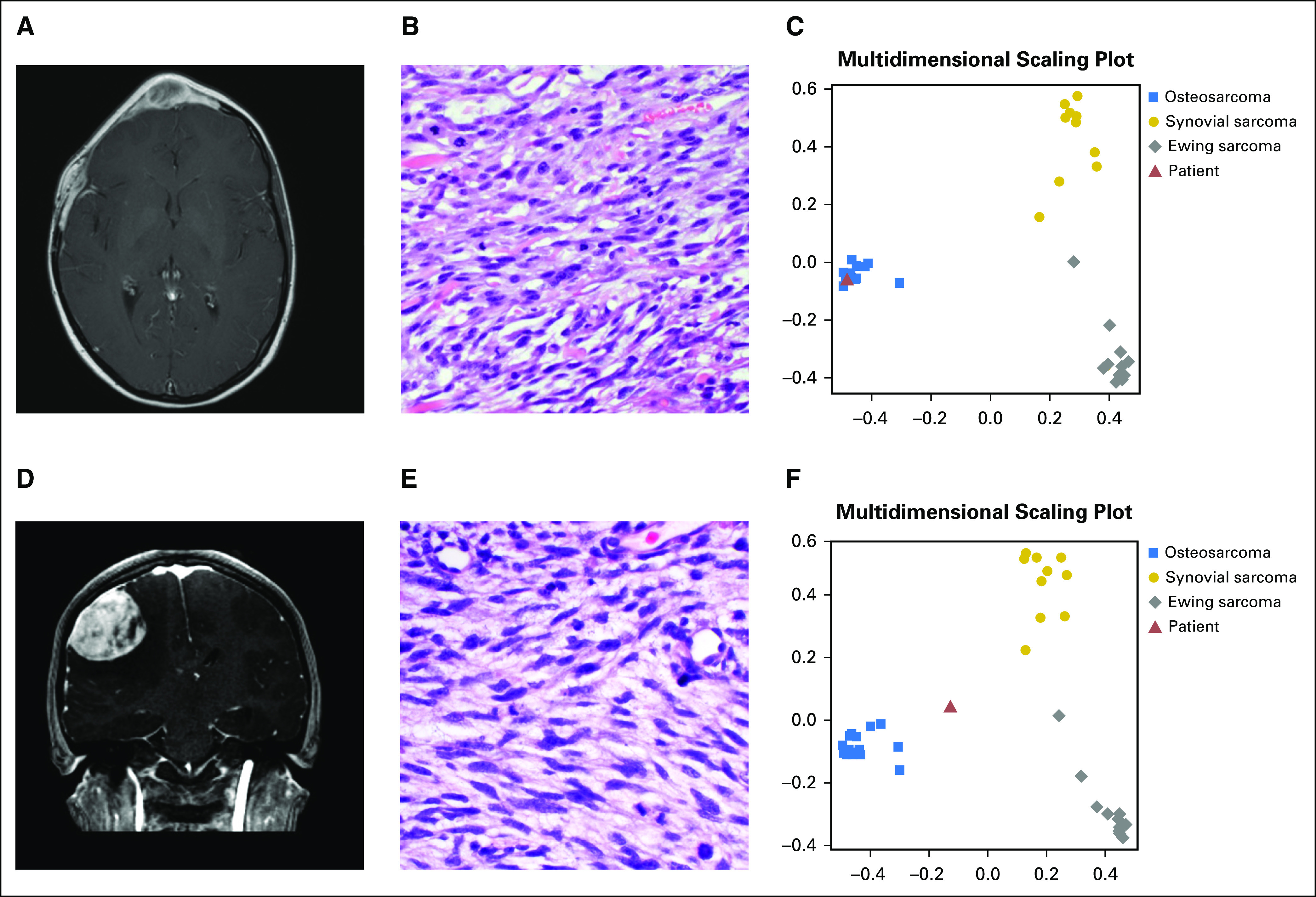
Case examples. (A) T1-weighted postcontrast magnetic resonance image demonstrating a sharply circumscribed, homogeneously enhancing soft-tissue mass. (B) The tumor consists of alternating epithelioid and plump spindled cells with interspersed coarse collagen fibers. On the basis of positive fluorescent in situ hybridization analysis, the tumor was initially diagnosed as a synovial sarcoma; however, multidimensional scaling plot of the classifier including the patient's tumor (red triangle) (C) shows that the tumor classifies with osteosarcoma. (D) T1-weighted postcontrast magnetic resonance image of the second clinical case, demonstrating a sharply circumscribed, homogeneously enhancing soft-tissue mass. (E) Histology shows sheets of poorly differentiated spindled tumor cells notably lacking distinctive morphologic features of synovial sarcoma, Ewing sarcoma, or osteosarcoma. (F) The primary tumor was originally diagnosed as Ewing sarcoma; however, EWSR1 rearrangement was never identified, and the multidimensional scaling plot of the metastasis shows the patient sample (red triangle) grouping most closely with osteosarcoma.
We performed methylation profiling on the 2016 recurrence sample. We analyzed the tumor using our sarcoma classifier, revealing a match with OS (Fig 2C). This case highlights the diagnostic value of our sarcoma classifier in samples that are difficult to diagnose using standard-of-care molecular methods. Moreover, it indicates robustness of a methylation-based sarcoma classifier not only in de novo but also in radiation-induced OS.
The second difficult case was a 2-year-old girl who had a mass involving T7-9 of the thoracic spine with radiographic evidence of pulmonary metastases. Given the histologic appearance and diffuse CD99 membranous staining, a diagnosis of EWS was made, although molecular confirmation of an EWSR1 rearrangement was never established. She received intensive multiagent chemotherapy and involved field radiation therapy to the spine.
Three years later at the age of 5, she presented with recurrent disease, including a left-sided pleural mass and hilar adenopathy. She received salvage chemotherapy including irinotecan and temozolomide, followed by a left thoracotomy and radical wedge resection. Pathology revealed viable disease with similar morphologic findings as her initial disease.
At the age of 15, the patient presented with headache, pupillary asymmetry, and slurred speech. A brain magnetic resonance image revealed a frontoparietal mass, which was subsequently resected (Fig 2D). Pathology revealed a high-grade pleomorphic and spindle-cell sarcoma (Fig 2E). CD99 staining was focally membranous. Targeted DNA (MSK-IMPACT37) and RNA (Archer FusionPlex; ArcherDX, Boulder, CO) were nondiagnostic but revealed a genomically unstable tumor with numerous mutations, which would be unusual for translocation-driven sarcomas such as EWS. Quantitative predictive probabilities derived from the random forest model were used to classify this sample, with a predicted probability of 48% OS, 32% SS, and 20% EWS. In the scenario in which OS and EWS compose the differential diagnosis, OS was the most likely and EWS the least likely grouping (Fig 2F).
DISCUSSION
In sarcomas lacking pathognomonic gene fusions, diagnostic differentiation can be extremely challenging, even with the help of modern diagnostic tools including immunohistochemistry and cytogenetics.38-41 Additionally, tumor heterogeneity and sampling errors can significantly confound the diagnosis. Because therapeutic decisions depend on sarcoma subtype, novel methods are needed to improve diagnostic accuracy.
The least invasive histologic sampling method is fine-needle aspiration biopsy42; more invasive techniques to obtain larger amounts of tumor tissue include open or core biopsy.43,44
However, even with an adequate specimen, a diagnosis based solely on histopathology and detailed clinical information is often inconclusive. Ancillary cytogenetic studies such as FISH can assist in differentiating between morphologically similar tumor specimens45; however, these tests frequently fail to identify a pathognomonic genetic abnormality and can occasionally produce false-positive results.
The HumanMethylation450 array is a rapid and cost-effective method for genome-wide quantitative profiling of the methylation of CpG loci.29 To explore the utility of DNA-based methylation profiling in patients with sarcoma, we used a similar approach that has been shown to be highly accurate and reproducible in subclassifying other histologically similar tumors (ie pediatric brain tumors).46 Using random forest modeling, we developed a classifier that we successfully validated using SS and OS samples from two publicly available datasets and an EWS validation set obtained from a recently published series.33
In our training set, our classifier correctly identified all 36 samples as the tumor type determined by a combination of clinical, histologic, and molecular genetic factors. When applied to our validation cohorts (TCGA and TARGET-OS), all 10 SS samples and 85 of 86 of the OS samples (99%) were correctly classified. Of particular interest is the sample that was classified incorrectly from the TARGET OS cohort as EWS. Our analysis shows that the case TARGET-40-PASEFS may represent a tumor that is biologically related to EWS,47 as indicated by overexpression of EWSR1 and ETV1, illustrating the power and usefulness of methylation-based profiling for discovery.
Our study highlights the epigenetic heterogeneity present in EWS despite a single recurrent oncogenic fusion driver. Some samples within the EWS cohort seem to be truly misclassified, and, qualitatively, the methylation pattern of the training and validation sets of EWS seem to be the most heterogeneous, even after hierarchical clustering. As demonstrated recently, a larger number of EWS samples is needed to fully characterize the epigenetic heterogeneity.48
A key advantage of methylation-based analysis is the interrogation of multiple diagnoses using a single test, thereby expediting the diagnosis. Methylation also provides insights into sarcoma biology; for example, OS seems to be characterized by a consistently hypomethylated strip along chromosome 1q43, among other features. Furthermore, with development and integration of additional sarcoma classifiers, such as rhabdomyosarcoma,49 molecular diagnostics of all major sarcoma subgroups could be cost-efficiently performed using a single, methylation-based platform. A prospective study comparing the performance and cost-effectiveness of our methylation classifier and traditional immunohistochemical and FISH testing could represent a fruitful avenue of research.
In summary, we developed and validated a DNA methylation–based classifier that accurately differentiated three of the most common subtypes of bone sarcomas. Given their clinically and histologically overlapping features and markedly different clinical management, this methylation-based classifier may provide a useful tool in the differential diagnosis of bone sarcomas.
ACKNOWLEDGMENT
We thank Dr. Oscar M. Tirado for providing Ewing sarcoma data.
Footnotes
Supported in part by KiDS of New York University Langone and The Friedberg Charitable Foundation and through the NIH/NCI Cancer Center Support Grant No. P30 CA008748 to Memorial Sloan-Kettering Cancer Center.
AUTHOR CONTRIBUTIONS
Conception and design: S. Peter Wu, Benjamin T. Cooper, J. Keith Killian, Twana M. Jackson, Matija Snuderl, Matthias A. Karajannis
Financial support: Matthias A. Karajannis
Administrative support: Matthias A. Karajannis
Provision of study material or patients: Shiyang Wang, Twana M. Jackson, Neerav Shukla, Richard Gorlick, Matthias A. Karajannis
Collection and assembly of data: S. Peter Wu, Benjamin T. Cooper, Fang Bu, J. Keith Killian, Jonathan Serrano, Shiyang Wang, Twana M. Jackson, Neerav Shukla, Paul A. Meyers, David J. Pisapia, Richard Gorlick, Marc Ladanyi, Kristen Thomas, Matija Snuderl, Matthias A. Karajannis
Data analysis and interpretation: S. Peter Wu, Benjamin T. Cooper, Christopher J. Bowman, Twana M. Jackson, Daniel Gorovets, Fang Bu, Paul A. Meyers, Matija Snuderl, Matthias A. Karajannis
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
S. Peter Wu
No relationship to disclose
Benjamin T. Cooper
No relationship to disclose
Fang Bu
No relationship to disclose
Christopher J. Bowman
No relationship to disclose
J. Keith Killian
No relationship to disclose
Jonathan Serrano
No relationship to disclose
Shiyang Wang
No relationship to disclose
Twana M. Jackson
No relationship to disclose
Daniel Gorovets
No relationship to disclose
Neerav Shukla
No relationship to disclose
Paul A. Meyers
Stock and Other Ownership Interests: Amgen, Bayer, E.I. Dupont, Henry Schein, Jazz Pharmaceuticals, Mednax, Novartis, Procter & Gamble, Sigma-Aldrich
Honoraria: France Foundation (I)
Consulting or Advisory Role: Boehringer Ingelheim (I)
Speakers' Bureau: France Foundation (I)
Travel, Accommodations, Expenses: Takeda, InterMune (I)
David J. Pisapia
No relationship to disclose
Richard Gorlick
Stock and Other Ownership Interests: Oncolytics
Consulting or Advisory Role: Oncolytics
Marc Ladanyi
Honoraria: Merck (I)
Consulting or Advisory Role: National Comprehensive Cancer Network/Boehringer Ingelheim Afatinib Targeted Therapy Advisory Committee, National Comprehensive Cancer Network/AstraZeneca Tagrisso Request for Proposal Advisory Committee
Research Funding: Loxo (Inst)
Kristen Thomas
No relationship to disclose
Matija Snuderl
Stock and Other Ownership Interests: Arca Biopharma, Bluebird Bio, Cellceutix, Epizyme, Geron, Isis Pharmaceuticals, Sevion Therapeutics, Acceleron Pharma, Catalyst Pharmaceutical
Travel, Accommodations, Expenses: Silicon Biosystems
Matthias A. Karajannis
No relationship to disclose
REFERENCES
- 1.Yaw KM: Pediatric bone tumors. Semin Surg Oncol 16:173-183, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Werier J, Yao X, Caudrelier JM, et al. : A systematic review of optimal treatment strategies for localized Ewing’s sarcoma of bone after neo-adjuvant chemotherapy. Surg Oncol 25:16-23, 2016 [DOI] [PubMed] [Google Scholar]
- 3.Isakoff MS, Bielack SS, Meltzer P, et al. : Osteosarcoma: Current treatment and a collaborative pathway to success. J Clin Oncol 33:3029-3035, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewing J: Diffuse endothelioma of bone. Proc NY Pathol Soc 1921;21: 17-24. [Google Scholar]
- 5.Nascimento AG, Unii KK, Pritchard DJ, et al. : A clinicopathologic study of 20 cases of large-cell (atypical) Ewing’s sarcoma of bone. Am J Surg Pathol 4:29-36, 1980 [PubMed] [Google Scholar]
- 6.Delattre O, Zucman J, Plougastel B, et al. : Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359:162-165, 1992 [DOI] [PubMed] [Google Scholar]
- 7.Sankar S, Lessnick SL: Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet 204:351-365, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorensen PH, Lessnick SL, Lopez-Terrada D, et al. : A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet 6:146-151, 1994 [DOI] [PubMed] [Google Scholar]
- 9.Jeon IS, Davis JN, Braun BS, et al. : A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 10:1229-1234, 1995 [PubMed] [Google Scholar]
- 10.Kaneko Y, Yoshida K, Handa M, et al. : Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer 15:115-121, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Peter M, Couturier J, Pacquement H, et al. : A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 14:1159-1164, 1997 [DOI] [PubMed] [Google Scholar]
- 12. Goldblum JR, Folpe AL, Weiss Sharon W: General considerations, in Goldblum JR, Folpe AL, Weiss Sharon W, eds: Enzinger and Weiss's Soft Tissue Tumors. 6th ed. Philadelphia, PA, Elsevier, 2014, p 1.
- 13.Clark J, Rocques PJ, Crew AJ, et al. : Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 7:502-508, 1994 [DOI] [PubMed] [Google Scholar]
- 14.Vlenterie M, Hillebrandt-Roeffen MH, Flucke UE, et al. : Next generation sequencing in synovial sarcoma reveals novel gene mutations. Oncotarget 6:34680-34690, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smida J, Baumhoer D, Rosemann M, et al. : Genomic alterations and allelic imbalances are strong prognostic predictors in osteosarcoma. Clin Cancer Res 16:4256-4267, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Yen CC, Chen WM, Chen TH, et al. : Identification of chromosomal aberrations associated with disease progression and a novel 3q13.31 deletion involving LSAMP gene in osteosarcoma. Int J Oncol 35:775-788, 2009 [DOI] [PubMed] [Google Scholar]
- 17.Kresse SH, Ohnstad HO, Paulsen EB, et al. : LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer 48:679-693, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Man TK, Lu XY, Jaeweon K, et al. : Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer 4:45, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dragoescu E, Jackson-Cook C, Domson G, et al. : Small cell osteosarcoma with Ewing sarcoma breakpoint region 1 gene rearrangement detected by interphase fluorescence in situ hybridization. Ann Diagn Pathol 17:377-382, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Hill DA, O’Sullivan MJ, Zhu X, et al. : Practical application of molecular genetic testing as an aid to the surgical pathologic diagnosis of sarcomas: A prospective study. Am J Surg Pathol 26:965-977, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Noguera R, Navarro S, Triche TJ: Translocation (11;22) in small cell osteosarcoma. Cancer Genet Cytogenet 45:121-124, 1990 [DOI] [PubMed] [Google Scholar]
- 22.Oshima Y, Kawaguchi S, Nagoya S, et al. : Abdominal small round cell tumor with osteoid and EWS/FLI1. Hum Pathol 35:773-775, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Khan J, Wei JS, Ringnér M, et al. : Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 7:673-679, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segal NH, Pavlidis P, Antonescu CR, et al. : Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol 163:691-700, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturm D, Orr BA, Toprak UH, et al. : New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164:1060-1072, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovestadt V, Remke M, Kool M, et al. : Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125:913-916, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pajtler KW, Witt H, Sill M, et al. : Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27:728-743, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renner M, Wolf T, Meyer H, et al. : Integrative DNA methylation and gene expression analysis in high-grade soft tissue sarcomas. Genome Biol 14:r137, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Röhrich M, Koelsche C, Schrimpf D, et al. : Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol 131:877-887, 2016 [DOI] [PubMed] [Google Scholar]
- 30. Assenov Y, Müller F, Lutsik P, et al: Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 11:1138-1140, 2014. [DOI] [PMC free article] [PubMed]
- 31.Liaw A, Wiener M: Classification and regression by randomForest. R News 2:18-22, 2002 [Google Scholar]
- 32.Weinstein JN, Collisson EA, Mills GB, et al. :: The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45:1113-1120, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huertas-Martínez J, Court F, Rello-Varona S, et al. : DNA methylation profiling identifies PTRF/Cavin-1 as a novel tumor suppressor in Ewing sarcoma when co-expressed with caveolin-1. Cancer Lett 386:196-207, 2017 [DOI] [PubMed] [Google Scholar]
- 34.Raney BJ, Dreszer TR, Barber GP, et al. : Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30:1003-1005, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, et al. : Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545-15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conner JR, Hornick JL: SATB2 is a novel marker of osteoblastic differentiation in bone and soft tissue tumours. Histopathology 63:36-49, 2013 [DOI] [PubMed] [Google Scholar]
- 37.Cheng DT, Mitchell TN, Zehir A, et al. : Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 17:251-264, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang CK, Li CW, Hsieh TJ, et al. : Characterization of bone and soft-tissue tumors with in vivo 1H MR spectroscopy: Initial results. Radiology 232:599-605, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Schulte M, Brecht-Krauss D, Heymer B, et al. : Grading of tumors and tumorlike lesions of bone: Evaluation by FDG PET. J Nucl Med 41:1695-1701, 2000 [PubMed] [Google Scholar]
- 40.Costelloe CM, Chuang HH, Chasen BA, et al. : Bone windows for distinguishing malignant from benign primary bone tumors on FDG PET/CT. J Cancer 4:524-530, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaeser B, Wiskirchen J, Wartenberg J, et al. : PET/CT-guided biopsies of metabolically active bone lesions: Applications and clinical impact. Eur J Nucl Med Mol Imaging 37:2027-2036, 2010 [DOI] [PubMed] [Google Scholar]
- 42.Kilpatrick SE, Ward WG, Chauvenet AR, et al. : The role of fine-needle aspiration biopsy in the initial diagnosis of pediatric bone and soft tissue tumors: An institutional experience. Mod Pathol 11:923-928, 1998 [PubMed] [Google Scholar]
- 43.Saghieh S, Masrouha KZ, Musallam KM, et al. : The risk of local recurrence along the core-needle biopsy tract in patients with bone sarcomas. Iowa Orthop J 30:80-83, 2010 [PMC free article] [PubMed] [Google Scholar]
- 44.Mankin HJ, Lange TA, Spanier SS: The hazards of biopsy in patients with malignant primary bone and soft-tissue tumors. J Bone Joint Surg Am 64:1121-1127, 1982 [PubMed] [Google Scholar]
- 45.Bridge RS, Rajaram V, Dehner LP, et al. : Molecular diagnosis of Ewing sarcoma/primitive neuroectodermal tumor in routinely processed tissue: A comparison of two FISH strategies and RT-PCR in malignant round cell tumors. Mod Pathol 19:1-8, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Northcott P.A., Shih DJ, Remke M, et al. : Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol 123:615-626, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith S.C., Buehler D, Choi EY, et al. : CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod Pathol 28:57-68, 2015 [DOI] [PubMed] [Google Scholar]
- 48. Sheffield NC, Pierron G, Klughammer J, et al: DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med 23:386-395, 2017. [DOI] [PMC free article] [PubMed]
- 49. Seki M, Nishimura R, Yoshida K, et al: Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 6:7557, 2015. [DOI] [PMC free article] [PubMed]