Abstract
Carbapenemase-producing ‘super bacteria’, particularly NDM-1 and its variants, have become a major public health concern worldwide. The present study aimed to explore the molecular mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Seventy-eight non-repeated Enterobacteriaceae strains resistant to any carbapenem were screened at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) between December 2011 and December 2015. Outer membrane porin (OMP) proteins were detected using SDS-PAGE. Carbapenemases, extended-spectrum β-lactamases (ESBLs) and plasmid AmpC enzyme genes were detected using polymerase chain reaction (PCR). PCR and SDS-PAGE demonstrated that 60.3% (47/78) of the strains produced carbapenemases, of which 33.3% (26/78) produced KPC-2 carbapenemase, suggesting that the strains resisted carbapenems primarily through carbapenemases. SDS-PAGE showed that the OMP proteins in the majority of carbapenem-resistant Enterobacteriaceae (CRE) strains were deleted or decreased compared with those in the sensitive strains. Of the 44 Klebsiella strains, 59.1% (26/44) did not express or expressed less OmpK35 or OmpK36. Among the 34 strains of other enterobacteria, 97.1% (33/34) did not express or expressed less OmpC or OmpF. Of all CRE strains, 35.9% (28/78) lost at least one OMP protein, indicating that the strains resisted carbapenems also by producing ESBLs and/or plasmid AmpC enzyme, as well as by losing OMP proteins. The resistance of clinically isolated CRE strains may primarily be attributed to the production of carbapenemases, and may also involve the deletion of OMP proteins or mutation of OMP genes.
Keywords: carbapenem-resistant enterobacteriaceae, polymerase chain reaction, SDS-PAGE
Introduction
Enterobacteriaceae, such as Escherichia coli, Klebsiella pneumoniae, Citrobacter freundii and Enterobacter cloacae, are the most common pathogenic bacteria for nosocomial infection (1). According to the results of national bacterial resistance monitoring over the years, Enterobacteriaceae accounted for 60–70% of all Gram-negative bacteria. Carbapenems, such as imipenem, meropenem and ertapenem, are the most effective antibiotics for the clinical treatment of infection caused by Enterobacteriaceae, especially multi-drug-resistant strains producing extended-spectrum β-lactamases (ESBLs) and AmpC enzyme (2). However, carbapenem-resistant Enterobacteriaceae (CRE) are emerging as carbapenems have been widely used in clinical practice. For example, the resistance rates of E. coli and K. pneumoniae have risen from 0 to 1%, and those of some other members of the Enterobacteriaceae family have already reached 7.0%. Some non-fermenters also have the resistance rates of 20–30% (3). Since most carbapenem-resistant bacteria also resist many commonly used antibiotics, they have become pan-resistant strains, posing a great threat to the life of patients and challenging the clinical application (4). Therefore, it is imperative to clarify the resistance mechanism of CRE strains and to effectively control the resulting nosocomial infection.
Production of carbapenemases is one of the main mechanisms for the resistance of CRE strains. Besides, these strains can also produce ESBLs and/or AmpC enzyme as well as lose outer membrane porin (OMP) proteins (5). Carbapenemases are β-lactamases using carbapenems as hydrolysis substrates, including Ambler classes A, B and C enzymes. For Enterobacteriaceae, KPC, as a class A carbapenemase, has been highlighted. KPC enzymes can hydrolyze almost all β-lactams including penicillins, cephalosporins and carbapenems, but can be easily inhibited by clavulanic acid and tazobactam. Meanwhile, they are resistant to other types of antibiotics (6). From 2006 to 2016, KPC-2-producing K. pneumoniae, C. freundii and Serratia marcescens were isolated from the Zhejiang province of China, indicating that KPC enzymes had been spread to bacteria of Enterobacteriaceae other than K. pneumoniae (7).
The permeability of outer membrane is vital to drug entrance into Gram-negative bacteria. OMP proteins can mainly be classified into OmpC with high molecular weights and OmpF with low molecular weights. Either deletion or decrease of OMP proteins leads to bacterial drug resistance. As to Enterobacteriaceae, the resistance to carbapenems may be ascribed to OMP protein deletion or reduction in the presence/absence of production of β-lactamases (e.g., AmpC enzymes and ESBLs). The OMP proteins of K. pneumoniae are mainly OmpA and OmpK. Carbapenemase-producing ‘super bacteria’, especially NDM-1 and its variants, has become a major public health concern worldwide. NDM-1 can hydrolyze a wide range of β-lactam antibiotics including carbapenems, as the last resort of antibiotics for treating infections induced by resistant bacteria (8).
Thereby motivated, we aimed to study the incidence of carbapenemase-producing strains in clinical isolates of Enterobacteriaceae and the mechanism by which CRE strains resisted carbapenems. Carbapenemases, ESBLs and AmpC enzyme genes in 78 clinically isolated strains were tested by β-lactamase detection and DNA sequencing. The types and genotypes of these enzymes were confirmed. OMP proteins in CRE strains were detected to clarify the relationship between carbapenem resistance and deletion of these proteins.
Materials and methods
Experimental and quality control strains
A total of 6,584 Enterobacteriaceae strains clinically isolated from December 2011 to December 2015 in our hospital were selected, from which 78 strains that resisted any carbapenem were screened. All strains were identified by API or ATB (BioMérieux, Craponne, France). E. coli ATCC 25922 was used as the quality control strain for the drug susceptibility test, and clinically isolated C. freundii known to produce IMP-8 metalloenzyme was utilized as the positive quality control strain in modified Hodge test.
E. coli producing TEM and SHV β-lactamases and E. coli producing CTX-M and TOHO ESBLs were used as positive control strains for ESBLs detection by polymerase chain reaction (PCR). Clinically isolated strains were used as the sensitive controls for SDS-PAGE of OMP proteins, and Lambda Marker was employed as the standard for bacterial typing using pulsed-field gel electrophoresis. The bacteria were all preserved by our hospital.
Screening of CRE strains by the KB test for drug susceptibility
Bacterial suspension at the concentration of 1.5×108/ml was uniformly coated onto the entire surface of Mueller-Hinton agar culture medium and dried at room temperature for several min. Then antibiotics-loaded filter paper discs were uniformly stuck onto the agar surface by a sterile tweezer and cultured in a 35°C incubator for 18–24 h. Afterwards, the inhibitory circle diameter (mm) including the disc diameter was measured by a vernier caliper. The experiment was performed and the drug susceptibility was determined according to the standards (2010 edition) of the Clinical Laboratory Standards Institute (CLSI). The susceptibilities to imipenem, meropenem and ertapenem were detected by the disc diffusion method (Table I).
Table I.
Standards for determining drug susceptibility using the Kirby-Bauer method (mm).
| Drug | Drug content of disc (µg) | Susceptible (S) | Intermediate (I) | Resistant (R) |
|---|---|---|---|---|
| Imipenem | 10 | ≥16 | 14–15 | ≤13 |
| Meropenem | 10 | ≥16 | 14–15 | ≤13 |
| Ertapenem | 10 | ≥19 | 16–18 | ≤15 |
Detection of carbapenemases by modified Hodge test
The experiment was conducted according to the method recommended by CLSI. E. coli ATCC 25,922 with a 0.5 McFarland turbidity standard was diluted and spread on an MH plate with a paper strip of ertapenem (10 µg) stuck in the middle. Bacteria were inoculated by an inoculating loop through streaking from the outer edge of the paper strip to the edge of the plate, during which particular attention should be paid not to scratch the plate surface. The bacteria were then cultured overnight at 35°C. On the next day, the bacteria exhibiting sagittal growth in the inhibition zone of ertapenem were identified to produce carbapenemases.
Detection of OMP proteins in 78 CRE strains by SDS-PAGE
The strains stored at −80°C were thawed at room temperature, inoculated on MH agar plates, and incubated at 37°C for 18–20 h. A single colony was picked up, inoculated in LB medium, cultured while shaking for 20 h, and centrifuged at 10,000 rpm and 4°C for 10 min to obtain the bacterial bodies that were then washed twice with PBS, resuspended, ultrasonically fragmented, and centrifuged at 20,000 rpm and 4°C for 10 min. After the supernatant was discarded, the precipitate was suspended in PBS containing 2% sodium dodecylamine and centrifuged at 16,000 rpm and room temperature for 30 min, and OMP proteins were resuspended in 100 µl of PBS solution. The gel was fixed in an electrophoresis device and added Tris-glycine electrophoresis buffer. The samples were added in a predetermined order using a Hamilton microinjector, 15 µl for each sample. Attention should be paid to positive and negative electrodes when the electrophoresis device was connected with the power supply. Firstly, 50 V of voltage was applied, which was then increased to 80 V after the front edge of dye entered the separating gel. The electrophoresis was continued until the indicating line of bromophenol blue reached the bottom of the separating gel (about 2–3 h). The gel was thereafter removed, soaked in a 5-fold volume of Coomassie brilliant blue staining solution, and stained by gentle shaking on a horizontal shaker for over 4 h at room temperature. Afterwards, the gel was taken out, soaked in a destaining solution, and destained by gentle shaking for 4–8 h, during which the destaining solution was refreshed 3–4 times. After destaining, the gel was photographed by Bio-Rad GelDocXR gel imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and the relative expressions of OMP protein bands in sensitive and resistant strains were measured by Quantity One image analysis software (Bio-Rad Laboratories, Inc.).
Detection of carbapenemases, ESBLs and plasmid AmpC enzyme genes in 78 CRE strains by PCR
Extraction of bacteria by boiling: A colony of bacteria purely cultured overnight was suspended in 500 µl of TE buffer, heated at 100°C for 13 min and centrifuged at 10,000 rpm for 3 min. Then 2 µl of the supernatant was used as the template for PCR reaction. The reaction system was 50 µl, including 5 µl of 10X buffer, 4 µl of dNTP, 1 µl of each primer, 0.25 µl of Tag enzyme, 37 µl of ddH2O and 2 µl of template. Reaction conditions: Pre-denaturation at 94°C for 5 min, 30 cycles of reaction at 55°C for 45 sec and at 72°C for 1 min, and finally extension at 72°C for 10 min. The primer sequences for PCR are listed in Table II. The electrophoresis of PCR products was conducted using 1.5% agarose gel at 130 V for 35 min. Subsequently, the gel was stained with ethidium bromide for 20 min and ultimately photographed by the gel imaging system.
Table II.
Primer sequences for PCR.
| Primer | Sequence (5′-3′) |
|---|---|
| KPC-R | TCGCTAAACTCGAACAGG |
| KPC-F | TTACTGCCCGTTGACGCCCAATCC |
| IMP-R | CTACCGCAGCAGAGTCTTTG |
| IMP-F | AACCAGTTTTGCCTTACCAT |
| IMI-R | ATAGCCATCTTGTTTAGCTC |
| IMI-F | TCTGCGATTACTTTATCCTC |
| NMC-R | TGCAGCTTAATTATTTTCAGATTAG |
| NMC-F | ATTTTTTTCATGATGAAGTTAAGCC |
| GIM-F | AGAACCTTGACCGAACGCAG |
| GIM-R | ACTCATGACTCCTCACGAGG |
| VIM-F | TCTACATGACCGCGTCTGTC |
| VIM-R | TGTGCTTTGACAACGTTCGC |
| SEM-IRS1 | AACGGCTTCATTTTTGTTTAG |
| SEM-IRS2 | GCTTCCGCAATAGTTTTATCA |
| SEM-IRS5 | AGATAGTAAATTTTATAG |
| SEM-IRS6 | CTCTAACGCTAATAG |
| IND-R | GCCCAGGTTAAAGATTTTGTAAT |
| IND-F | CATGGCCACCGCCTTTCCATTC |
| IND-R1 | GGTTTGCATATCTATCTGCC |
| IND-F1 | ATCCAAAGAGAGGCTGGAGT |
| OXA-69-F | CTAATAATTGATCTACTCAAG |
| OXA-69-R | CCAGTGGATGGATGGATAGATTATC |
| OXA-55-F | CATCTACCTTTAAAATTCCC |
| OXA-55-R | AGCTGTTCCTGCTTGAGCAC |
| OXA-48-F | TTGGTGGCATCGATTATCGG |
| OXA-48-R | GAGCACTTCTTTTGTGATGGC |
| OXA-50-F | AATCCGGCGCTCATCCATC |
| OXA-50-R | GGTCGGCGACTGAGGGGG |
| OXA-60-F | AAAGGAGTTGTCTCATGCTGTCTCG |
| OXA-60-R | AACCTACAGGCGCGCGTCTCACGGTG |
PCR, polymerase chain reaction; F, forward; R, reverse.
Statistical analysis
All data were analyzed by GraphPad (GraphPad Software, Inc., La Jolla, CA, USA). All experiments were performed in triplicate, and the results were expressed as number and percentage.
Results
Distribution of 78 CRE strains
The 78 CRE strains were isolated from urine, respiratory tract, blood, cerebrospinal fluid and wound samples, with the isolation rates of 55.1 (43/78), 41.0 (32/78), 1.3 (12/78), 1.3 (12/78) and 1.3% (12/78) respectively.
Besides, 71.8% (56/78) of the CRE strains were isolated from wards in neurosurgery department, and the remaining was from ICU [10.3% (8/78)], internal medicine department [6.4% (5/78)], VIP wards [5.1% (4/78)], outpatient department [2.6% (2/78)], foreigner department [2.6% (2/78)] and surgery department [1.3% (1/78)].
CRE strains could be isolated from all patients hospitalized in the same bed at different times from 2011 to 2015. For example, three C. freundii strains and one K. pneumoniae strain resisting carbapenems were isolated from four patients hospitalized in January 2012, May and September 2013 and February 2015. For the same patient, CRE strains with different species could also be isolated. In the beginning, K. pneumoniae strains were susceptible to carbapenems, but became resistant after treatment for a period of time.
Susceptibility of CRE strains to antibacterial agents
Drug susceptibility test (Fig. 1 and Table III) showed that 83.3% (63/78) of the CRE strains were resistant to imipenem, meropenem and ertapenem simultaneously.
Figure 1.
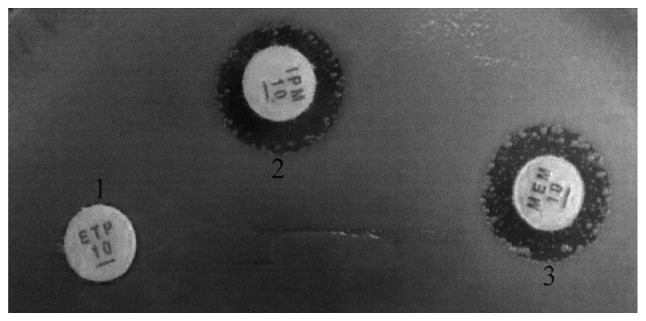
Drug susceptibility test results using the Kirby-Bauer method.
Table III.
Drug susceptibility test results using the Kirby-Bauer method (n%).
| Drug | Number of strains | % |
|---|---|---|
| Imipenem | 71 | 91.0 |
| Meropenem | 70 | 89.7 |
| Ertapenem | 72 | 92.3 |
| Imipenem + meropenem | 66 | 84.6 |
| Imipenem + ertapenem | 65 | 83.3 |
| Meropenem + ertapenem | 67 | 85.9 |
| Imipenem + meropenem + ertapenem | 63 | 83.0 |
Detection results of carbapenemase genes
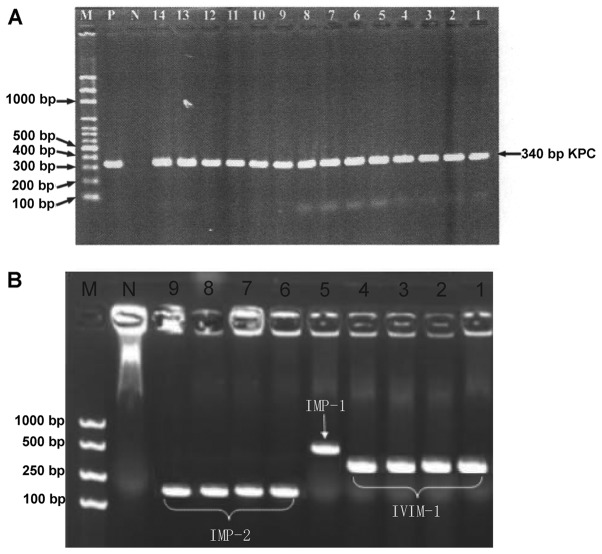
PCR showed that the 78 CRE strains had negative detection results of VIM-2, SPM, NMC, IMI, IND, OXA-48, OXA-50, OXA-55, OXA-60 and OXA-24 like carbapenemase genes. As shown in Fig. 2A, 33.3% (26/78) of the strains produce KPC-2 carbapenemase, of which 7.7 (2/26), 7.7 (2/26) and 3.8% (1/26) simultaneously produce VIM-1, IMP-2 and IMP-1 carbapenemases respectively. In addition, 6.4 (5/78), 5.1 (4/78), 3.8 (3/78), 6.4 (5/78) and 12.8% (10/78) of the strains produced VIM-1, IMP-1, IMP-2, GIM and OXA-69 carbapenemases respectively (Fig. 2B). As to OXA carbapenemases, 20.5% (16/78) of the strains produced OXA-23 like, OXA-51 like or OXA-58 like carbapenemases.
Figure 2.
(A) PCR results of CRE strains producing KPC carbapenemases. 1–6, K. pneumoniae; 7–14, C. freundii; N, negative control E. coli ATCC 25922; P, positive control KPN2528; M, molecular weight marker. (B) PCR results of CRE strains producing metallocarbapenemases. 1–4, K. pneumoniae (VIM-1); 5, 1551K. pneumoniae (IMP-1); 6–9, C. freundii (IMP-2); N, negative control E. coli ATCC 25922; M, molecular weight marker. PCR, polymerase chain reaction; CRE, carbapenem-resistant Enterobacteriaceae; K. pneumoniae, Klebsiella pneumoniae; C. freundii, Citrobacter freundii; E. coli, Escherichia coli.
PCR results of OMP genes
PCR revealed that the detection rates of OmpK35, OmpK36 and Ompk37 in the 78 strains were 37.2 (29/78), 6.4 (5/78) and 3.8% (3/78) respectively, but 61.5% of the strains had negative detection results for these three genes (Fig. 3 and Table IV).
Figure 3.
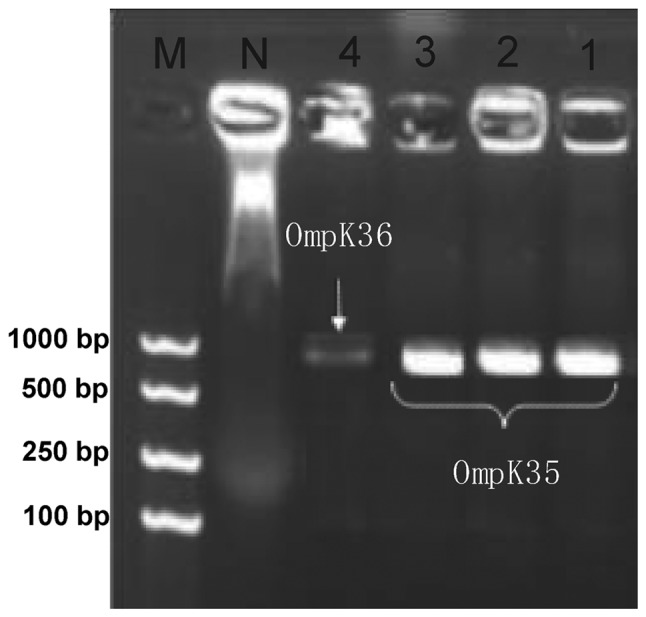
PCR results of OMP genes. 1–3, K. pneumoniae (OmpK35); 4, K. pneumoniae (OmpK36); N, negative control; M, molecular weight marker.
Table IV.
Polymerase chain reaction results of OMP genes.
| Type | Number of strains | % |
|---|---|---|
| All negative | 48 | 61.5 |
| OmpK35-positive | 23 | 29.5 |
| OmpK37-positive | 1 | 1.3 |
| OmpK35- and OmpK36-positive | 4 | 5.1 |
| OmpK35- and OmpK37-positive | 1 | 1.3 |
| OmpK35-, OmpK36- and OmpK37-positive | 1 | 1.3 |
| Total | 78 | 100.0 |
OMP, outer membrane porin.
SDS-PAGE results of OMP proteins
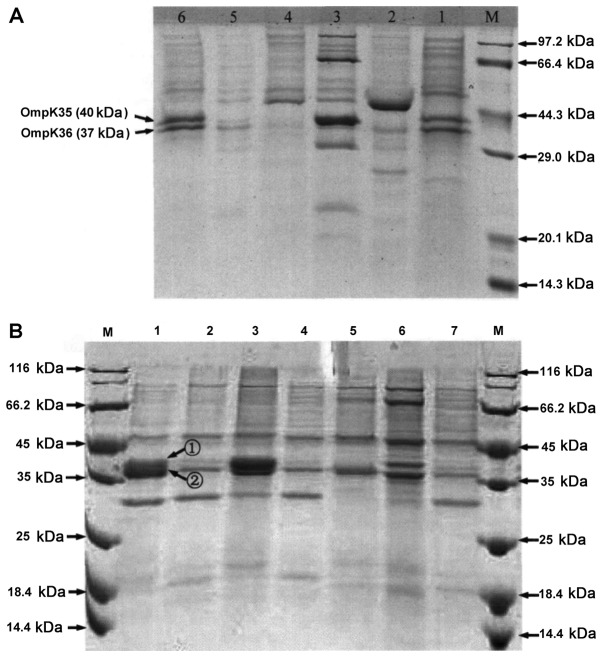
SDS-PAGE showed that 40.9% of 44 K. pneumoniae strains had normal OMP protein expressions, and the remaining 59.1% had downregulated or deleted expression of OmpK35 or OmpK36 (Fig. 4 and Table V). Of 34 C. freundii strains, only 2.9% had normal OMP protein expression, and the remaining 97.1% had downregulated or deleted expression of OmpC or OmpF. The OMP proteins in most CRE strains were deleted or decreased compared with those in the sensitive ones, indicating that they played crucial roles in the resistant of CRE strains to carbapenems.
Figure 4.
(A) SDS-PAGE results of OMP proteins in K. pneumoniae. 1 and 6, carbapenem-sensitive strains; 2–5, carbapenem-resistant strains; M, molecular weight marker. (B) SDS-PAGE results of OMP proteins in C. freundii. 1, carbapenem-sensitive strain; 2–7, carbapenem-resistant strains; M, molecular weight marker. (1) OmpC protein (42 kDa); (2) OmpF protein (39 kDa). OMP, outer membrane porin; K. pneumoniae, Klebsiella pneumoniae; C. freundii, Citrobacter freundii.
Table V.
SDS-PAGE results of OMP proteins.
| A, Klebsiella (44 strains) | ||
|---|---|---|
| Protein change | Number of strains | % |
| Normal | 18 | 40.9 |
| Downregulated OmpK35 + deleted OmpK36 | 5 | 11.4 |
| Downregulated OmpK35 + OmpK36 | 3 | 6.8 |
| Normal OmpK35 + downregulated OmpK36 | 3 | 6.8 |
| Deleted OmpK35 + normal OmpK36 | 1 | 2.3 |
| Normal OmpK35 + deleted OmpK36 | 4 | 9.1 |
| Deleted OmpK35 + OmpK36 | 4 | 9.1 |
| Downregulated OmpK35 + normal OmpK36 | 3 | 6.8 |
| Deleted OmpK35 + downregulated OmpK36 | 3 | 6.8 |
| B, Other Enterobacteriaceae (34 strains) | ||
| Protein change | Number of strains | % |
| Downregulated OmpC + OmpF | 21 | 61.8 |
| Normal OmpC + OmpF | 1 | 2.9 |
| Deleted OmpC + downregulated OmpF | 5 | 14.7 |
| Normal OmpC + downregulated OmpF | 1 | 2.9 |
| Deleted OmpC + OmpF | 3 | 8.8 |
| Downregulated OmpC + deleted OmpF | 3 | 8.8 |
OMP, outer membrane porin.
PCR results of ESBLs and plasmid AmpC enzyme genes
PCR revealed that 83.3% (65/78) of the strains produced CTX-M ESBLs (Fig. 5 and Table VI), and 1.3 (1/78), 70.5 (55/78) and 25.6% (20/78) produced CTX-M-2, CTX-M-14 and CTX-M-15 ESBLs respectively. Moreover, 39.7% (31/78) of the strains produced DHA-1 or CMY-2 plasmid AmpC enzyme (Fig. 6). Although carbapenemase genes were not detected in 39.7% (31/78) of the strains, 87.9% (29/33) produced CTX-M ESBLs (CTX-M-15, 48.3%; CTX-M-14, 72.4%; CTX-M-14L and CTX-M-15 simultaneously, 20.7%). Of all CRE strains, 35.9% (28/78) at least lose one band of OMP protein.
Figure 5.
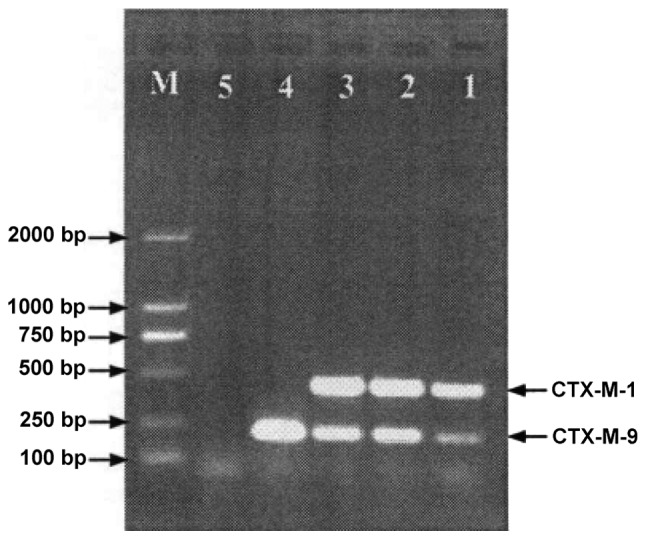
PCR results of CTX-M ESBL. 1–3, K. pneumoniae; 4, positive control; 5, negative control; M, molecular weight marker. PCR, polymerase chain reaction; ESBL, extended-spectrum β-lactamase; K. pneumoniae, Klebsiella pneumoniae.
Table VI.
ESBL genes in 78 CRE strains.
| KPN (43 strains) | CFR (25 strains) | Others (10 strains) | All strains | |||||
|---|---|---|---|---|---|---|---|---|
| Resistant genes | Number of strains | % | Number of strains | % | Number of strains | % | Number of strains | % |
| Negative | 0 | 0.0 | 0 | 0.0 | 3 | 30.0 | 3 | 3.8 |
| ESBLs | 16 | 37.2 | 2 | 8.0 | 2 | 20.0 | 20 | 25.6 |
| ESBLs + AmpC | 4 | 9.3 | 4 | 16.0 | 8 | 10.3 | ||
| Carbapenemase | 7 | 16.3 | 1 | 4.0 | 4 | 40.0 | 12 | 15.4 |
| Carbapenemase + ESBLs | 10 | 23.3 | 3 | 12.0 | 13 | 16.7 | ||
| Carbapenemase + ESBLs + AmpC | 6 | 14.0 | 15 | 60.0 | 1 | 10.0 | 22 | 28.2 |
| Total | 43 | 100.0 | 25 | 100.0 | 10 | 100.0 | 78 | 100.0 |
ESBLs, extended-spectrum β-lactamases; CRE, carbapenem-resistant Enterobacteriaceae.
Figure 6.
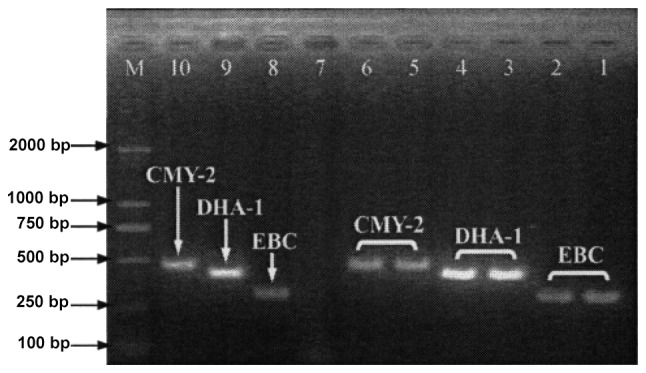
PCR results of plasmid AmpC enzyme in E. coli and K. pneumoniae. 1 and 2, EBC positive control strains; 3 and 4, DHA-1 gene-positive; 5 and 6, CMY-2 gene-positive; 7, negative control; 8–10, EBC, DHA-1 gene and CMY-2 gene positive controls; M, molecular weight marker. PCR, polymerase chain reaction; E. coli, Escherichia coli; K. pneumoniae, Klebsiella pneumoniae.
Based on the modified Hodge test, 93.6% (73/78) of the strains produced carbapenemases, of which 35.6% (26/73) did not contain carbapenemase genes.
Discussion
Enterobacteriaceae are Gram-negative, facultative anaerobic bacilli or coccobacilli widely distributed in the environment, mainly including Escherichia, Klebsiella, Enterobacter, Citrobacter, Serratia and Salmonella (9,10). As important pathogenic bacteria for community-acquired infections and nosocomial infections, Enterobacteriaceae can infect the respiratory tract and urethra. Of all clinically isolated Gram-negative bacilli, Enterobacteriaceae account for 80%, and they also account for 50% of the bacteria detected in clinical laboratories. Additionally, approximately 50% of septicemia and over 70% of urinary infections are caused by Enterobacteriaceae (11–13). Carbapenems (imipenem, meropenem and ertapenem), which have broad antibacterial spectra as well as high antibacterial activities and stability to β-lactamases, are potent antibiotics for treating infections induced by Gram-negative bacteria (14). However, Enterobacteriaceae have become increasingly resistant to carbapenems owing to extensive and irrational use in clinical practice. As a result, the treatment outcomes are affected, increasing the economic burden of patients. CRE directly endanger human life and health by elevating the mortality rate. Recently, humans have been threatened by KPC enzyme-producing Gram-negative bacilli (4). Steinmann et al reported that from July 2010 to January 2011, four patients died of KPC-2-producing K. pneumoniae in a German hospital (15). Hu et al detected KPC carbapenemases in 77 Enterobacteriaceae strains from Huashan Hospital, Fudan University (16). In March 2011, NDM-1-producing E. coli was isolated in Hong Kong, China (17). Of 46 Enterobacteriaceae strains with decreased susceptibility to carbapenems collected from 2004 to 2008 (18), 8 produced IMP-4 and 2 produced IMP-8. Antibiotics enter bacterial cells via OMPs as porous channels. When these proteins are deleted or reduced, the outer membrane permeability is decreased, leading to drug resistance by hindering their entrance. Enterobacteriaceae (e.g., E. cloacae and Enterobacter aerogenes) not only have OmpK35 and OmpK36 analogous to OmpF and OmpC, but also express OmpD (19). One of the mechanisms for bacterial drug resistance is that antibacterial agents fail to permeate into cells. For Gram-negative bacteria, the permeability of OMPs is key to the entrance of exit of antibiotics. OMPs related to the drug resistance of Enterobacteriaceae mainly function through synthesis reduction or deletion, mutation, blockage of channels and replacement by other proteins. In this study, PCR and SDS-PAGE showed that 60.3% (47/78) of the 78 strains produced carbapenemases, and 33.3% (26/78) produced KPC-2 carbapenemase. Accordingly, carbapenemases may be responsible for the resistance of CRE strains to carbapenems. Compared with sensitive strains, OMPs of most CRE strains were deleted or decreased. Of the 44 Klebsiella strains, 59.1% (26/44) did not express or expressed less OmpK35 or OmpK36. Among the 34 strains of other enterobacteria, 97.1% (33/34) did not express or expressed less OmpC or OmpF. Of all CRE strains, 35.9% (28/78) lost at least one OMP protein, suggesting that the strains resisted carbapenems also by producing ESBLs and/or plasmid AmpC enzyme as well as by losing OMP proteins.
The resistance of clinically isolated CRE strains can primarily be ascribed to production of carbapenemases, also involving deletion of OMP proteins or mutation of OMP genes. In clinical practice, it is of great significance to monitor and to control carbapenemase-producing CRE strains by PCR. In addition, the emergence of resistant bacterial strains is directly associated with irrational use of antibiotics, so it is crucial to timely find them by predicting the probability of drug resistance and performing drug susceptibility test when carbapenems are used. In summary, severe infections can be prevented by rationally using antibiotics and avoiding long-term contact between antibiotics and bacteria.
References
- 1.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbier F, Pommier C, Garrouste-Orgeas M, Schwebel C, Ruckly S, Dumenil AS, Lemiale V, Mourvillier B, Clec'h C, Darmon M, et al. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in critically iii patients: Impact of carriage and infection on carbapenem consumption, duration of Icu stay, and mortality. Intensive Care Med Exp. 2015;3(Suppl 1):A1. doi: 10.1186/2197-425X-3-S1-A1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mushtaq S, Woodford N, Hope R, Adkin R, Livermore DM. Activity of BAL30072 alone or combined with β-lactamase inhibitors or with meropenem against carbapenem-resistant Enterobacteriaceae and non-fermenters. J Antimicrob Chemother. 2013;68:1601–1608. doi: 10.1093/jac/dkt050. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol Med. 2012;18:1–272. doi: 10.2119/molmed.2012.00299. [DOI] [PubMed] [Google Scholar]
- 5.Satlin MJ, Chen L, Patel G, Gomez-Simmonds A, Weston G, Kim AC, Seo SK, Rosenthal ME, Sperber SJ, Jenkins SG, et al. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother. 2017;61:pii: e02349–16. doi: 10.1128/AAC.02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 7.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017;17:101. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2014;20:831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 10.Vasoo S, Barreto JN, Tosh PK. Emerging issues in gram-negative bacterial resistance: An update for the practicing clinician; Mayo Clin Proc; 2015; pp. 395–403. [DOI] [PubMed] [Google Scholar]
- 11.Brust K, Evans A, Plemmons R. Favourable outcome in the treatment of carbapenem-resistant Enterobacteriaceae urinary tract infection with high-dose tigecycline. J Antimicrob Chemother. 2014;69:2875–2876. doi: 10.1093/jac/dku185. [DOI] [PubMed] [Google Scholar]
- 12.Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 13.Surgers L, Boyd A, Boelle PY, Lalande V, Jolivot PA, Girard PM, Arlet G, Cambier C, Homor A, Decre D, Meynard JL. Clinical and microbiological determinants of severe and fatal outcomes in patients infected with Enterobacteriaceae producing extended-spectrum β-lactamase. Eur J Clin Microbiol Infect Dis. 2017;36:1261–1268. doi: 10.1007/s10096-017-2932-9. [DOI] [PubMed] [Google Scholar]
- 14.Paul M, Carmeli Y, Durante-Mangoni E, Mouton JW, Tacconelli E, Theuretzbacher U, Mussini C, Leibovici L. Combination therapy for carbapenem-resistant Gram-negative bacteria. J Antimicrob Chemother. 2014;69:2305–2309. doi: 10.1093/jac/dku168. [DOI] [PubMed] [Google Scholar]
- 15.Steinmann J, Kaase M, Gatermann S, Popp W, Steinmann E, Damman M, Paul A, Saner F, Buer J, Rath P. Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German university hospital, July 2010 to January 2011. Euro Surveill. 2011;16:pii: 19944. [PubMed] [Google Scholar]
- 16.Hu F, Chen S, Xu X, Guo Y, Liu Y, Zhu D, Zhang Y. Emergence of carbapenem-resistant clinical Enterobacteriaceae isolates from a teaching hospital in Shanghai, China. J Med Microbiol. 2012;61:132–136. doi: 10.1099/jmm.0.036483-0. [DOI] [PubMed] [Google Scholar]
- 17.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One. 2011;6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: Results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54:573–577. doi: 10.1128/AAC.01099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JY, Hong YK, Lee H, Ko KS. High prevalence of non-clonal imipenem-nonsusceptible Enterobacter spp. isolates in Korea and their association with porin down-regulation. Diagn Microbiol Infect Dis. 2017;87:53–59. doi: 10.1016/j.diagmicrobio.2016.10.004. [DOI] [PubMed] [Google Scholar]