Abstract
Natural compounds derived from plants have been an important source of numerous clinically useful anticancer agents. Nevertheless, limited studies indicate that xanthohumol (XN), a major prenylated flavonoid in hop plants (Humulus lupulus), may possess anticarcinogenic properties. The purpose of the present study was to clarify the antitumorigenic effects and the underlying mechanism of XN on breast cancer in vivo and in vitro. A 4T1 breast tumor mouse model was used in the present study to investigate XN suppression of tumor growth as detected by tumorigenicity assays in vivo. In addition, in vitro studies revealed that XN significantly decreased cell viability, induced G0/G1 cell cycle arrest and apoptosis in MCF-7 and MDA-MB-231 cells, as confirmed by an MTT assay, flow cytometry and western blot analysis, indicating anticarcinogenic activity of XN against breast cancer. Furthermore, immunohistochemistry was performed to confirm the inactivation of the Notch signaling pathway, Notch 1 and Ki-67, in vivo; consistently, XN caused decreased activation of the Notch signaling pathway and apoptotic regulators B-cell lymphoma-2 (Bcl-2), Bcl-extra large and caspase 3, as determined by western blot analysis in vitro. This study suggests that XN may potentially be useful as a chemopreventive agent during breast hyperplasia and carcinogenesis, acting via the regulation of Notch associated apoptotic regulators in vivo and in vitro.
Keywords: xanthohumol, Notch signaling pathway, apoptosis, cell cycle, migration, breast cancer
Introduction
Breast cancer is the most common type of cancer in females and one of the most devastating cancers worldwide (1,2). Although the underlying mechanism that causes breast cancer remains uncertain, substantial evidence suggests that the Notch signaling pathway may serve an important role in breast cancer pathogenesis and, thus, may be a novel therapeutic target (3,4).
The Notch pathway serves an important role in normal breast cell development, cell fate determination and stem cell self-renewal (3). It is also implicated in breast cancer development and progression as the aberrant activation of this pathway is associated with breast cancer (3–5). Inhibition of Notch signaling by gamma secretase inhibitors, anti-Notch1 or anti-delta-like 4 (DLL4) monoclonal antibodies have been revealed to result in antitumor activity in a variety of tumors, including T-cell acute lymphoblastic leukemia (T-ALL) and solid tumors, through multiple mechanisms inclusive of the induction of cell cycle arrest or apoptosis, and also the disruption of angiogenesis (4–10). Notch signaling serves important roles in various physiological and pathological processes including cell differentiation, proliferation, invasion, angiogenesis, tumor metastasis and apoptosis, which contribute to the development of various types of human cancer (5,11). Furthermore, an activated Notch signaling pathway and its target genes are commonly observed in breast cancer, and the upregulation of Notch1 expression has been revealed to protect breast cancer cells from apoptosis (12). Currently known gene targets for Notch signaling include the hairy enhancer of split (Hes) genes, p21, cyclinD1, c-Myc, nuclear factor κB, B-cell lymphoma-2 (Bcl-2) and Bcl-extra large (Bcl-xl) (13–18).
Previous studies suggest that xanthohumol (XN), a prenylated chalcone derived from hops (Humulus lupulus), may inhibit cell growth and induce apoptosis in numerous types of human cancer, including breast, prostate, leukemia and colon cancer cells (19–23); however, the underlying mechanism of this inhibition remains unknown. In the present study, the therapeutic potential of XN in breast cancer cell lines was investigated, focusing on its ability to inhibit breast cancer cell proliferation, cell cycle arrest, apoptosis induction in vitro and slowing tumor growth in vivo. Additionally, possible XN-mediated inhibition in human breast cancer growth via the regulation of the Notch1 signaling pathway was investigated. To the best of our knowledge, the present study was the first to observe that XN suppressed breast carcinoma growth by inhibiting the Notch signaling pathway in vitro and in vivo. Therefore, blocking of the Notch signaling pathway may be a novel therapeutic approach for the treatment of breast cancer, by inhibiting tumor development, progression and metastasis.
Materials and methods
Cell lines and cell culture
Human MCF-7, MDA-MB-231 and HEK-293T cells, h-TERT-BJ, MCF-10A and murine 4T1 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). They were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and maintained at 37°C in a humidified atmosphere of 5% CO2 in air.
Animals
All studies evaluating in vivo toxicity and therapeutic effectiveness were performed using 26 female BALB/c mice (17–18 g; 8 weeks of age) and obtained from Lanzhou Veterinary Research Institute, Chinese Academic of Agricultural Sciences (Lanzhou, China). The Institutional Animal Care and Use Committee of Lanzhou University approved use of BALB/c mice for the present study, and procedures involving animals and their care complied with the Guide for the Care and Use of Laboratory Animals. Mice were maintained in a temperature-controlled (23±4°C) environment with a strict 12 h light/dark cycle. Food was purchased from Wanqianjiaxing Biotech Corp (Wuhan, China) and water was autoclaved. Food and water were freely available to the mice.
Reagents and antibodies
XN (purity 98.6%) was provided by Yumen Tuopu Science Development and Technology Co., Ltd. (Yumen, China). Duration of Dual Antiplatelet Therapy (DAPT) was purchased from Sigma-Aldrich (Merck Millipore, Darmstadt, Germany). Primary antibodies against Notch 1 (120 kDa; cat. no., 3268; dilution, 1:1,000), p21 (cat. no., 2947; dilution, 1:1,000), cyclin-dependent kinase 4 (CDK4) (cat. no., 12790; dilution, 1:1,000), c-Myc (cat. no., 5605; dilution, 1:1,000), survivin (cat. no., 2808; dilution, 1:1,000), Bcl-2 (cat. no., 15071; dilution, 1:500), Bcl-xL (cat. no., 2764; dilution, 1:1,000), cyclin D1 (cat. no., 2978; dilution, 1:1,000), caspase-3 (cat. no., 9665; dilution, 1:1,000) and poly (ADP-ribose) polymerase (PARP; cat. no., 9532; dilution, 1:1,000) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Hes1 (cat. no., ABIN2779597; dilution, 1:1,000) was acquired from Abnova Biotechnology (Taipei, Taiwan ROC). GAPDH (cat. no., sc-47724; dilution, 1:5,000) was supplied by Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). The secondary antibodies, peroxidase-conjugated AffiniPure goat anti-rabbit (cat. no., ZB-2301; dilution, 1:500) and anti-mouse (cat. no., ZB-2305; dilution, 1:500) IgG (H+L), were purchased from ZSGB-Bio (Beijing, China).
MTT assay
The h-TERT-BJ, MCF-10A, MCF-7 and MDA-MB-231 cells (5×103) were seeded in a 96-well culture plate; following a 24-h incubation the cells were treated with 5, 10, 15 and 25 µmol/l XN for 24 and 48 h in incubator. Control cells were treated with 0.1% dimethyl sulfoxide (DMSO) in culture medium. Following treatment, the cells were incubated with MTT reagent (0.5 mg/ml) at 37°C for 4 h. The resulting formazan crystals were solubilized by the addition of 200 µl DMSO to each well. The absorbance was read at 490 nm in a Vector3 Multilevel Plate Counter (PerkinElmer, Inc., Waltham, MA, USA) and all MTT experiments were performed in triplicate and repeated ≥3 times.
Transient transfection and luciferase reporter assay
Transient transfections were performed with Lipofectamine® (Invitrogen; Thermo Fisher, Inc.), according to the manufacturer's protocol. Briefly, HEK-293T cells were plated in 24-well plates at a density of 1×105 cells/well. Following a 24-h incubation at 37°C, cells were transfected with plasmids of 0.8 µg promoter-linked luciferase vector (23A, 4xCBF1 binding element plasmid) and 0.2 µg pGL4.20 vector for 4 h in DMEM (Sigma-Aldrich; Merck Millipore). Following DLL-4 (100 ng/ml) stimulation, cells were treated with XN for 12 h at 37°C. The cell lysates were evaluated in a luciferase assay using a dual luciferase reporter assay kit (E1910; Promega, Madison, WI, USA), and the emitted light was determined with a luminometer (Wallac 1420 VICTOR, Inc., Waltham, MA, USA) as previously described (24). Luciferase activity was normalized to β-galactosidase and plotted as relative light units.
Flow cytometry analysis of cell cycle
The cell cycle was analyzed by flow cytometry. The MCF-7 and MDA-MB-231 cells (1×106) were collected and washed in PBS, prior to being fixed in 75% alcohol at 20°C overnight. Following washing in cold PBS three times, cells were resuspended in 1 ml PBS solution with 50 µg propidium iodide (PI; Sigma-Aldrich; Merck Millipore) and 100 µg RNase A (Sigma-Aldrich; Merck Millipore), for 30 min at 37°C. Samples were then analyzed for their DNA content by fluorescence-activated cell sorting (FACS; BD Biosciences, San Jose, CA, USA). Each experiment was repeated ≥3 times.
Flow cytometric analysis of apoptosis
Cell apoptosis was assessed by flow cytometry using an Annexin-V-fluorescein isothiocyanate/PI apoptosis detection kit (BD Biosciences, San Jose, CA, USA), according to the manufacturer's instructions. The pretreated MCF-7 and MDA-MB-231 cells were harvested and washed twice with PBS (4°C). Following this, they were resuspended in 1X binding buffer at a concentration of 1×106 cells/ml, stained with Annexin V/PI and kept on ice for 30 min in the dark. The cells were then analyzed on a FACSCalibur™ flow cytometer (BD Biosciences). Assays were performed three times in triplicate.
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from cell lines using an RNeasy kit as described by the manufacturer (Qiagen Inc., Valencia, CA, USA). The reverse transcription reaction was performed using the SuperScript® First-Strand Synthesis System (Invitrogen; Thermo Fisher Scientific, Inc.) in a final volume of 20 µl, containing 5 µg total RNA, 200 ng random hexamers, 1X reverse transcription buffer, 2.5 mM MgCl2, 1 mM deoxynucleotide triphosphate mixture, 10 mM DTT, RNaseOUT recombinant ribonuclease inhibitor (Invitrogen; Thermo Fisher Scientific, Inc.), 50 U superscript reverse transcriptase and diethylpyrocarbonate-treated water. Following incubation at 42°C for 50 min, the reverse transcription reaction was terminated by heating to 85°C for 5 min. The newly synthesized cDNA was amplified by PCR. The reaction mixture contained 2 µl cDNA template, 1.5 mM MgCl2, 2.5 U Taq polymerase and 0.5 µM primers (primers are presented in Table I). The reactions were performed under the following conditions: 5 min at 94°C, 30 sec at 94°C, 30 sec at 55°C, 30 sec at 72°C, 25 cycles of 30 sec at 94°C, 5 min at 72°C and 30 min at 4°C. The mRNA levels of Hes1 and Hes related family BHLH transcription factor with YRPW motif 1 (Hey1) were normalized to GAPDH.
Table I.
Primer sequences for reverse transcription-polymerase chain reaction.
| Gene | Upstream primer | Downstream primer |
|---|---|---|
| Hes1 | 5′-TTGGAGGCTTCCAGGTGGTA-3′ | 5′-GGCCCCGTTGGGAATG-3′ |
| Hey1 | 5′-CGAGGTGGAGAAGGAGAGTG-3′ | 5′-CTGGGTACCAGCCTTCTCAG-3′ |
| GAPDH | 5′-TCTCATCACCATCTTCCA-3′ | 5′-CATCACGCCACAGTTTCC-3′ |
Hes1, hairy enhancer of split 1; Hey1, Hes related family BHLH transcription factor with YRPW motif 1.
Boyden chamber assay
For Boyden chamber assays, 1×105 MDA-MB-231 cells were seeded per well in a Boyden chamber with a pore size of 8 µM (Corning Incorporated, Corning, NY, USA), without fetal bovine serum. For the control and treated samples (following XN and DAPT treatment), the lower compartment was filled with medium containing 10% FBS. Following 6 or 12 h, cells were fixed with 4% paraformaldehyde solution and stained with crystal violet (Beyotime Institute of Biotechnology, Haimen, China; C0121). Cells on top of the filter were removed with a cotton bud and the bottom sides were photographed using a bright field light microscope (4× objective). For quantification, the membranes sectioned, and 0.1 ml 33% acetic acid elution was used to dissolve the cells, then 40 µl was added into 96 plates and the values were determined at 570 nm.
Western blot analysis
Western blotting was performed as described in a previous study (24). Briefly, total proteins from the two human breast cancer cell lines were lysed in lysis buffer (cat. no., P00138; Beyotime Institute of Biotechnology) and incubated for 15 min at 4°C. The concentrations of total proteins were determined using a bicinchoninic acid assay protein assay kit (Thermo Fisher Scientific, Inc.). Total proteins were fractionated using 10% SDS-PAGE and the gels were then transferred onto a nitrocellulose membrane. The membranes were blocked with 5% non-fat milk in Tris-buffered saline containing 0.1% Tween-20 prior to incubation with the appropriate primary antibodies overnight at 4°C. Horseradish peroxidase-conjugated anti-goat IgG was used as the secondary antibody, and the protein bands were detected using the electrochemiluminescence (ECL) method (cat. no., RPN2134; Western Blotting Detection Reagent, GE Healthcare Life Sciences, Chalfont, UK). Western blot analysis was quantified by laser densitometry, and the results are presented as the mean of three independent experiments with error bars representing the standard deviation.
In vivo tumorigenicity assays and immunohistochemistry
Firstly, 2×105 4T1 tumor cells from cultures, suspended in 0.2 ml DMEM without FBS, were injected into the right flank of 8-week-old BALB/c mice. Tumor-bearing mice were randomly assigned in three groups [n=10 control mice; n=16 XN mice (8 received 100 mg/kg XN and 8 received 200 mg/kg XN)]. Following 24-h, the mice were gavaged with 200 µl vehicle or 200 µl XN at the appropriate concentrations (200 and 100 mg/kg). Intragastric administration was performed once a day for two weeks and the mice were weighed daily. Following two weeks, all of the mice were sacrificed by breaking of the neck and the stripped subcutaneous sarcorna tumors were subsequently weighed to evaluated the antitumor effect. Tumors treated with 200 mg/kg XN were stored in formaldehyde for immunohistochemistry as described in a previous study (25).
Statistical analysis
The statistical analysis was performed using SPSS, Inc., version 12.0 software (Chicago, IL, USA). All data were expressed as the mean ± SD. Student's t-test was performed for comparisons between the two treatment groups. The three treatment groups were compared by one-way analysis of variance, followed by the significant difference method for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference and P<0.01 was considered to indicate a highly statistically significant difference.
Results
XN regulates Notch1 signal pathway activity and downstream targets
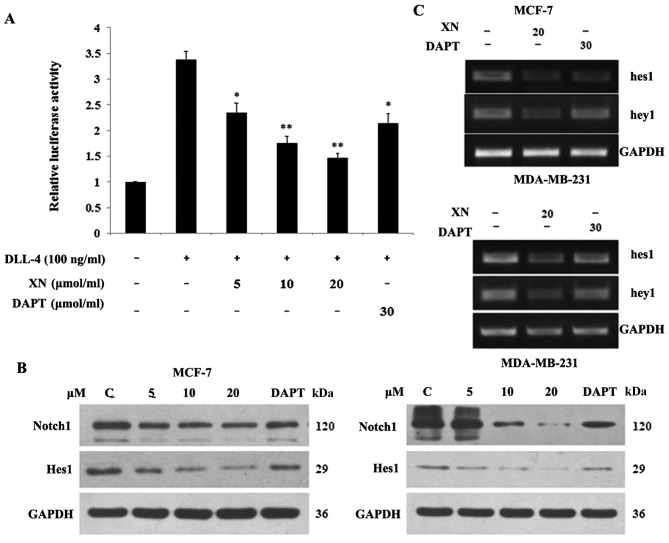
In order to determine if the Notch pathway was targeted by XN, a Notch1 functional assay and DAPT (gamma secretase inhibitor) was used as a positive control. Upon binding of Notch1 to the CBF1 transgene, luciferase activity may be determined in the documented reporter assay. The aim of the present study was to determine if XN reduced the binding activity of Notch1 to CBF1. The cells were stimulated with/without DLL-4 (100 ng/ml). The stimulated groups were treated with XN (5, 10, 20 µM) and DAPT (30 µM). Relative to the control, a 62% and a 34% decrease in the luciferase signal were identified following treatment with XN (20 µM) and DAPT (30 µM), respectively, for 24 h in 293T cells (Fig. 1A). Subsequently, the level of XN, which was associated with a reduction in Notch 1 protein expression, was investigated. Cells were harvested following 24 h of treatment and analyzed by western blotting with antibodies specific for Notch 1 and Hes1 (Fig. 1B). Notch 1 and Hes1 expression levels decreased in a dose-dependent manner in both cell lines. This reduction in Notch 1 activity led to an evaluation into whether XN was functioning similar to a γ-secretase inhibitor, or if it is operating at the transcriptional level. RNA was isolated following one day of XN or DAPT treatment, and RT-PCR was performed. Lastly, the downstream target genes of Notch were investigated. It was revealed that treatment with XN resulted in decreased Hes1 and Hey1 transcription in MCF-7 and MDA-MB-231 cells. Concomitantly, a decrease in Hes1 and Hey1 transcription in MCF-7 and MDA-MB-231 cells was observed following treatment with DAPT (Fig. 1C).
Figure 1.
XN inhibited Notch1 activity. (A) XN inhibited Notch1-dependent luciferase activity. Luciferase data were normalized to β-galactosidase and are presented as the means ± SD. *P<0.05, **P<0.01 vs. control. (B) Notch 1 and Hes1 protein expression levels were detected by western blotting. (C) XN inhibited Notch1-specific downstream gene Hes1 and Hey1 mRNA expression levels. DLL-4; anti-delta-like 4; DAPT, Duration of Dual Antiplatelet Therapy; Ctrl, control; XN, xanthohumol; Hes1, hairy enhancer of split 1; hey1, Hes related family BHLH transcription factor with YRPW motif 1.
XN inhibits cell growth and migration in breast cancer cell lines
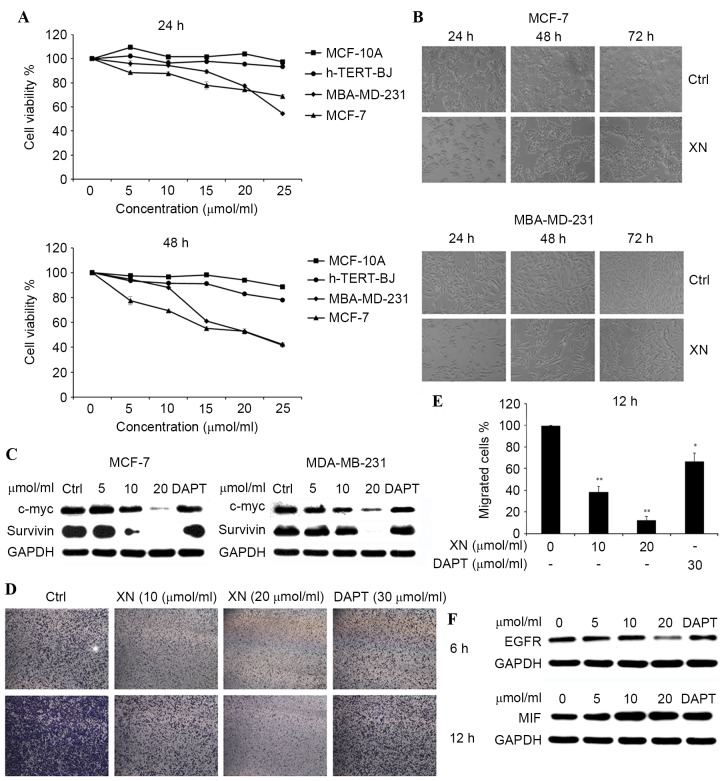
The treatment of MCF-7 and MDA-MB-231 breast cancer cells for 24–48 h with 5, 10, 15, 20 and 25 µM XN resulted in cell growth inhibition in a dose- and time-dependent manner, and the inhibition was revealed to be more pronounced with 25 µM XN treatment by an MTT assay (Fig. 2A, left). Comparison of the effects on cell viability among MCF-7, MDA-MB-231, hTERT-BJ and MCF-10A cells following 24 and 48 h drug treatments (Fig. 2A, right), demonstrated that XN induced a higher level of apoptosis for DU145 and MDA-MB-231 cells compared with hTERT-BJ and MCF-10A cells, subsequent to treatment (Fig. 2A). The inhibition of cell growth was also identified by viewing with an inverted microscope (×10 magnification; Fig. 2B).
Figure 2.
XN inhibited breast cancer cell proliferation in a dose- and time-dependent manner. (A) h-TERT-BJ, MCF-10A, MCF-7 and MDA-MB-231 were plated equally (six replicates) and treated with XN. MTT assays were performed daily for two days. DMSO was a vehicle control. (B) MDA-MB-231 and MCF-7 cells were treated with 20 µM XN for 24–72 h and visualized by light microscopy. (C) The c-Myc and survivin protein expression levels were detected by western blotting. (D) The Boyden chamber transwell assay demonstrated MDA-MB-231 cell migration. MCF-7 and MDA-MB-231 cells were treated with XN (data are presented as the mean ± SEM of three independent experiments). (E) Quantification of migrated MDA-MB-231 cells. Data are presented as the mean ± SEM of three independent experiments. *P<0.05, **P<0.01 vs. control. (F) EGFR and MIF expression levels were determined by western blot analysis. SEM; standard error of the mean; DMSO, dimethyl sulfoxide; EGFR, epidermal growth factor receptor; MIF, tumor metastasis-associated protein; CDK4, cyclin-dependent kinase 4; DAPT, Duration of Dual Antiplatelet Therapy; XN, xanthohumol; Ctrl, control.
Western blot analysis of c-Myc and survivin was also performed. It has been observed that expression of c-Myc and survivin were gradually decreasing compared with the control in both cell lines, following treatment with XN for 24 h (Fig. 2C). These results indicated that XN may inhibit breast cancer cell growth and migration by blocking the Notch 1 signaling pathway.
The effect of XN on the transmigration of MDA-MB-231 cells was determined with a Boyden chamber assay. Cell migration was inhibited significantly by XN treatment at 10 or 20 µM, down to 40 or 30%, respectively. Similar effects (83%) were observed with DAPT treatment, which was used as positive control (Fig. 2D and E). The effect of XN on epidermal growth factor receptor (EGFR) and tumor metastasis-associated protein (MIF) was determined by using a western blot assay. There was a notable decrease in EGFR expression levels and an increase in MIF expression levels, as the concentration of XN increased in the control and treatment groups in MDA-MB-231 cells (Fig. 2F).
XN treatment is associated with cell cycle arrest and apoptosis
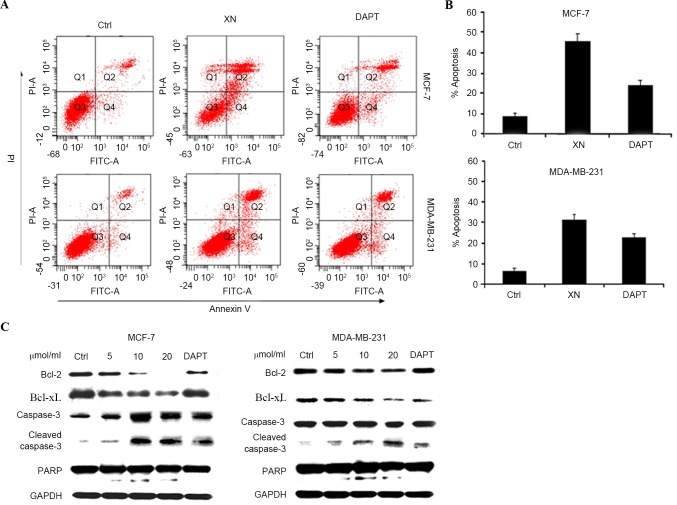
To determine whether cell cycle arrest was an underlying mechanism by which XN may be effecting reduced cell growth in breast cancer, the present study performed western blot analysis of certain cell cycle associated proteins. There was a notable increase in p21WAF1/CIP1 expression levels in MCF-7 cells as the concentration of XN treatment increased, in addition to an increase in p21WAF1/CIP1 expression levels between the control and treatment groups in MDA-MB-231 cells (Fig. 3A). As anticipated, from an increase in p21WAF1/CIP1 expression levels, flow cytometry experiments revealed that cells responded in time-dependent and dose-dependent manners to XN, through arresting cells in the G0/G1 phase of the cell cycle (Fig. 3B). Cells were treated with XN for 24 or 48 h, and the percentage of control cells in G0/G1 phase was 51.1 and 10%; however, following treatment with 20 µM XN, 70.6 and 24.8% of the cells were in the G0/G1 phase. Cell cycle arrest was confirmed by a decrease in CDK4 and cyclin D1 expression levels in the MCF-7 and MDA-MB-231 cells (Fig. 3A). As a consequence of G0/G1 phase cell cycle arrest, the present study investigated whether XN inhibits breast cancer cells by inducing apoptosis; this was determined by an Annexin V/PI assay and cleavage of PARP-1, caspase-3 and Bcl-xL and Bcl-2 expression levels. As presented in Fig. 4 (a representative experiment), following exposure to various media (DMEM, XN or DAPT for 48 h, the percentage of apoptotic cells in the XN treated groups increased significantly in comparison with the control groups. DAPT was also revealed to induce the apoptosis of breast cancer cells; however, the XN treatment groups contained a higher number of apoptotic cells compared with the DAPT treated groups. Apoptosis assessed by western blot assay demonstrated the same trends as the Annexin-V/PI assay (Fig. 4A and B). An increase in cleaved caspase-3 and cleaved PARP was revealed with increasing doses of XN and DAPT, suggesting that the cells went through the caspase cascade-mediated apoptotic pathway (Fig. 4C). These results indicated that XN exerts an inhibitory effect on cell proliferation and also promotes cell apoptosis.
Figure 3.
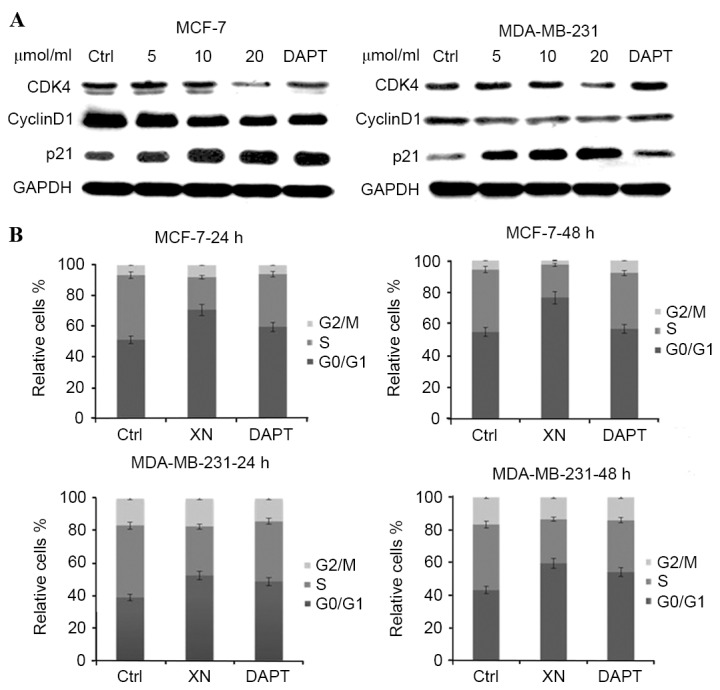
XN arrests the cell cycle in the G0/G1 phase. (A) XN regulated cell cycle protein expression levels were detected by western blotting. (B) MCF-7 and MDA-MB-231 cells were harvested for cell cycle analysis using PI staining. PI, propidium iodide; CDK4, cyclin-dependent kinase 4; DAPT, Duration of Dual Antiplatelet Therapy; Ctrl, control; XN, xanthohumol.
Figure 4.
XN promote apoptosis of breast cells. (A) MCF-7 and MDA-MB-231 cells were harvested for apoptotic analysis using Annexin V-FITC staining. (B) Quantification of apoptosis. Data are presented as the mean ± SD error of the mean of three independent experiments. (C) Apoptosis associated proteins Bcl-2, Bcl-xL and caspase-3 and cleaved PARP were analyzed by western blotting. GAPDH was used as the control. FITC, fluorescein isothiocyanate; FITC-A, FITC-annexin; Ctrl, control; XN, xanthohumol; DAPT, Duration of Dual Antiplatelet Therapy; PI-A, propidium iodide annexin; Bcl-2, B cell lymphoma-2; Bcl-xL, B cell lymphoma extra 1; PARP, poly (ADP-ribose) polymerase.
XN effects mouse tumor growth and Notch 1 protein expression levels in vivo
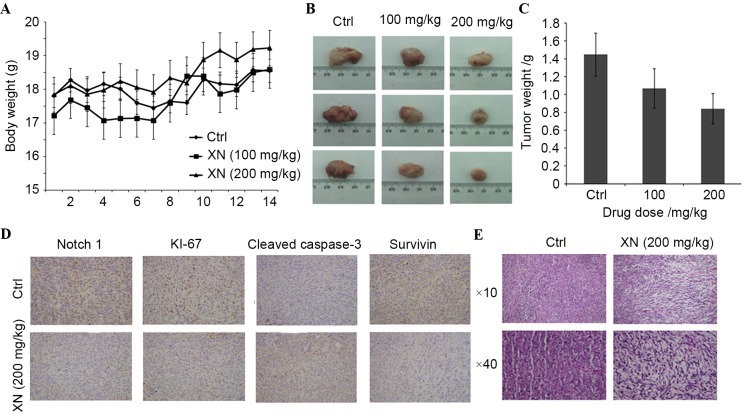
In order to evaluate whether XN downregulation affects tumor growth in vivo, BALB/c mice were used as an in vivo model of mammary carcinoma. A mouse tumor model was generated by endermic injection of 4T1 murine mammary cells. Mice treated by oral gavage with XN three times a week did not exhibit any symptoms of toxicity, and no effects were observed in the body weight profiles when XN treated mice were compared with vehicle-fed controls (data not presented). At the time of necropsy, all animals were examined for gross pathology, and no evidence of edema or abnormal organ size in target and non-target organs was observed.
There was a significant difference in mouse tumor model weight between the control and the 200 mg/kg XN-fed group (Fig. 5A). Additionally, a marked decrease in tumor size in a dose dependent manner was revealed (Fig. 5B): Tumor weight decreased 27.22 and 46.79% following various XN treatments (100 and 200 mg/kg), respectively (Fig. 5C). These results demonstrated that XN effectively inhibited breast tumor growth in vivo.
Figure 5.
XN inhibited tumor growth in vivo. (A) Total average mouse weights. 4T1 cells (1×105) were subcutaneously grafted to 8-week-old BALB/c mice. (B) Images of the tumors. (C) Tumor sizes of BALB/c mice treated with the control vehicle or XN as indicated. (D) Immunohistochemistry analysis for protein expression (×10). (E) Hematoxylin and eosin staining (×10 and ×40) revealed the tumor tissues of mice. Ctrl, control; XN, xanthohumol.
In the present study, the possibility of XN serving a role in Notch1-associated protein expression in vivo was investigated. The expression levels of Notch 1, Ki-67, survivin and caspase-3 by immunohistochemistry was determined. Fig. 5D demonstrates that XN inhibits the expression of Notch1 and Ki-67. Survivin was downregulated and cleaved caspase-3 was upregulated in XN treated tissues (Fig. 5D). Hematoxylin and eosin staining revealed that XN produced obvious cell injury in the tumor tissues of mice (Fig. 5E). These results demonstrated that XN efficiently inhibited the growth of tumors and promoted tumor cell apoptosis, which further indicates that XN has an antitumor effect.
Discussion
The Notch signaling pathway decides cell fate, an important carcinogenic factor of breast cancer (7,26). Notch1 expression levels are connected to breast cancer cell proliferation and survival (27). Farnie and Clarke (28) reported that aberrant activation of the Notch signaling pathway is an early event in breast cancer, and high expression levels of the Notch 1 intracellular domain was indicative of a reduced time to five-year post-surgical recurrence. Therefore, the Notch signaling pathway may represent a breast cancer novel therapeutic target. Notch signaling may be downregulated via inhibition of Notch ligands, inhibiting tumor cell proliferation (29). G-secretase inhibitors may be used to block Notch signaling and have previously been applied in clinical studies (30,31). However, one of the major challenges is to eliminate unwanted toxicity associated with γ-secretase inhibitors (32); thus, alternative Notch inhibitors must be identified.
XN, a product of hops, may inhibit cell growth and induce apoptosis in numerous types of human cancer, including breast cancer (19–23,33). Previous studies into the biological activity of XN have revealed their antiproliferative activity and possible antitumor activity (34). Guerreiro et al (33) suggested that XN may modulate alkaline phosphatase isoenzymes in MCF-7 breast cancer cells, and that alkaline phosphatase loss was associated with increased cell proliferation. In 2012, Cho et al (35) reported that XN induced cancer cell specific apoptosis in MCF-7 human breast cancer cells. These results raise the possibility that XN may be a potential therapeutic agent in human breast cancer. There are currently few studies that address this question directly and this appears to be an area worthy of further investigation.
Notch1 acts as an oncogene in breast cancer; therefore, inhibition of Notch1 expression may lead to inhibition of cell growth and apoptotic cell death in breast cancer cells (10,33). The present study demonstrated that proliferation inhibition and apoptosis induction by XN are specific to cancer cells with a constitutively inhibited Notch 1 signaling pathway; DAPT was used as a positive control. An MTT assay and light microscopy (Fig. 2A and B) indicated that XN inhibited cell proliferation in breast cancer cell lines. Proliferation associated proteins, including c-Myc and survivin, expression levels were determined (Fig. 2C). MD-MBA-231 cells, invasive breast cancer cells, were used to assess the ability to modulate cell migration through Boyden chamber porous membranes. As presented in Fig. 2D, cells treated with XN prevented cell migration to the lower chamber in a Boyden chamber assay, which was significantly different compared with the control. Activation of the EGFR induced signaling pathway correlated with cancer metastasis in various tumors, including breast carcinoma (36). Dai et al (37) reported that Notch and EGFR signaling pathways are positively associated with human breast cancer. The Notch signaling pathway inhibitor may also inhibit EGFR expression. Furthermore, additional factors, including MIF, which inhibits cell migration and XN, increased MIF expression.
The results of the present study indicate that MCF-7 and MDA-MB-231 cells were inhibited in the G0/G1 phase of the cell cycle, and underwent induced apoptosis when treated with XN. Downstream target genes of the Notch signaling pathway are also involved in cell cycle arrest and apoptotic pathway (14,15,18). Cyclin D1 and CDK4 phosphorylate key cell cycle proteins, controlling the G1-S cell cycle phases; p21 is an ubiquitous inhibitor of kinase CDK, which guides cell cycle arrest (38–40). The Hes/Hey genes are Notch target genes, which are basic helix-loop-helix repressors (4). A number of additional genes, including p21 and cyclin D1, have been suggested to be direct targets of the Notch signaling pathway (13,14).
In breast cancer, the nuclear antigen Ki-67 has previously been applied widely for comparison of cell proliferation between tumor samples in immunohistochemical assessment (41,42). The immunohistochemical evaluations revealed that XN decreased Notch 1, Ki-67 and survivin expression levels, and increased caspase-3 expression. The present study demonstrated that XN may inhibit breast tumor growth and promote apoptosis (Fig. 5).
In conclusion, the present study revealed that XN possesses antiproliferative, anti-metastatic and pro-apoptotic effects in breast cancer cells. It was also revealed that XN inhibits tumor growth using an in vivo tumor growth assay. The results of the present study demonstrated that XN may be a promising chemopreventive candidate for breast cancer treatment via inhibition of the Notch signaling pathway. These results suggest that XN requires further study into its potential therapeutic role in breast cancer treatment.
Acknowledgements
The present study was supported by the Science and Technology Support Projects of Gansu Province, China (grant no. 2010GS05414). The authors would like to thank Dr Yumen Tuopu, Science Development and Technology Co., Ltd., for providing XN, and Dr. Diane Hayward (The Johns Hopkins University, Baltimore, MD, USA) for providing the 23A plasmid, which contained four copies of the CBF1-binding elements in pGL2 Luciferase Reporter Vectors.
References
- 1.Guo H, Wu F, Wang Y, Yan C, Su W. Overexpressed ubiquitin ligase Cullin7 in breast cancer promotes cell proliferation and invasion via down-regulating p53. Biochem Biophys Res Commun. 2014;450:1370–1376. doi: 10.1016/j.bbrc.2014.06.134. [DOI] [PubMed] [Google Scholar]
- 2.Keyaerts M, Xavier C, Heemskerk J, Devoogdt N, Everaert H, Ackaert C, Vanhoeij M, Duhoux FP, Gevaert T, Simon P, et al. Phase I study of 68Ga-HER2-Nanobody for PET/CT assessment of HER2-expression in breast carcinoma. J Nucl Med. 2016;57:27–33. doi: 10.2967/jnumed.115.162024. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hussaini H, Subramanyam D, Reedijk M, Sridhar SS. Notch signaling pathway as a therapeutic target in breast cancer. Mol Cancer Ther. 2011;10:9–15. doi: 10.1158/1535-7163.MCT-10-0677. [DOI] [PubMed] [Google Scholar]
- 4.Yuan X, Zhang M, Wu H, Xu H, Han N, Chu Q, Yu S, Chen Y, Wu K. Expression of Notch1 correlates with breast cancer progression and prognosis. PLoS One. 2015;10:e0131689. doi: 10.1371/journal.pone.0131689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Palmer WH, Deng WM. Ligand-independent mechanisms of Notch activity. Trends Cell Biol. 2015;25:697–707. doi: 10.1016/j.tcb.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radtke F, Raj K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor. Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 10.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao WR, Hsieh RH, Hsu KW, Wu MZ, Tseng MJ, Mai RT, Wu Lee YH, Yeh TS. The CBF1-independent Notch1 signal pathway activates human c-myc expression partially via transcription factor YY1. Carcinogenesis. 2007;28:1867–1876. doi: 10.1093/carcin/bgm092. [DOI] [PubMed] [Google Scholar]
- 12.Xia J, Li Y, Yang Q, Mei C, Chen Z, Bao B, Ahmad A, Miele L, Sarkar FH, Wang Z. Arsenic trioxide inhibits cell growth and induces apoptosis through inactivation of Notch signaling pathway in breast cancer. In J Mol Sci. 2012;13:9627–9641. doi: 10.3390/ijms13089627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling H, Jolicoeur P. Notch-1 signaling promotes the cyclinD1-dependent generation of mammary tumor-initiating cells that can revert to bi-potential progenitors from which they arise. Oncogene. 2013;32:3410–3419. doi: 10.1038/onc.2012.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomero TL, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, et al. Notch1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth; Proc Natl Acad Sci USA; 2006; pp. 18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramdass B, Maliekal TT, Lakshmi S, Rehman M, Rema P, Nair P, Mukherjee G, Reddy BK, Krishna S, Radhakrishna Pillai M. Coexpression of Notch1 and NF-kappaB signaling pathway components in human cervical cancer progression. Gynecol Onco. 2007;104:352–361. doi: 10.1016/j.ygyno.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 17.Xie M, He CS, Wei SH, Zhang L. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur J Cancer. 2013;49:3559–3572. doi: 10.1016/j.ejca.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Sionov RV, Kfir-Erenfeld S, Spokoini R, Yefenof E. A role for bcl-2 in Notch1-dependent transcription in thymic lymphoma cells. Adv Hematol. 2012;2012:435241. doi: 10.1155/2012/435241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colgate EC, Miranda CL, Stevens JF, Bray TM, Ho E. Xanthohumol, a prenylflavonoid derived from hops induces apoptosis and inhibits NF-kappaB activation in prostate epithelial cells. Cancer Lett. 2007;246:201–209. doi: 10.1016/j.canlet.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Gerhauser C, Alt A, Heiss E, Gamal-Eldeen A, Klimo K, Knauft J, Neumann I, Scherf HR, Frank N, Bartsch H, Becker H. Cancer chemopreventive activity of Xanthohumol, a natural product derived from hop. Mol Cancer Ther. 2002;1:959–969. [PubMed] [Google Scholar]
- 21.Gonçalves P, Araújo JR, Pinho MJ, Martel F. In vitro studies on the inhibition of colon cancer by butyrate and polyphenolic compounds. Nutr Cancer. 2011;63:282–294. doi: 10.1080/01635581.2011.523166. [DOI] [PubMed] [Google Scholar]
- 22.Nozawa H. Xanthohumol, the chalcone from beer hops (Humulus lupulus L.), is the ligand for farnesoid X receptor and ameliorates lipid and glucose metabolism in KK-A(y) mice. Biochem Biophys Res Commun. 2005;336:754–761. doi: 10.1016/j.bbrc.2005.08.159. [DOI] [PubMed] [Google Scholar]
- 23.Pan L, Becker H, Gerhäuser C. Xanthohumol induces apoptosis in cultured 40–16 human colon cancer cells by activation of the death receptor- and mitochondrial pathway. Mol Nutr Food Res. 2005;49:837–843. doi: 10.1002/mnfr.200500065. [DOI] [PubMed] [Google Scholar]
- 24.Shi J, Chen J, Serradji N, Xu X, Zhou H, Ma Y, Sun Z, Jiang P, Du Y, Yang J, et al. PMS1077 sensitizes TNF-α induced apoptosis in human prostate cancer cells by blocking NF-κB signaling pathway. PLoS One. 2013;8:e61132. doi: 10.1371/journal.pone.0061132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Zhao L, Xu X, Liu F, Zhang W, Zhou C, Chen J, Pan Y, Du Y, Yang J, Wang Q. N6-substituted adenosine analogues, a novel class of JAK2 inhibitors, potently block STAT3 signaling in human cancer cells. Cancer Lett. 2014;354:43–57. doi: 10.1016/j.canlet.2014.07.043. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Samadi AK, Roby KF, Timmermann B, Cohen MS. Inhibition of cell growth and induction of apoptosis in ovarian carcinoma cell lines CaOV3 and SKOV3 by natural withanolide Withaferin A. Gynecol Oncol. 2012;124:606–612. doi: 10.1016/j.ygyno.2011.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pei J, Wang B. Notch-1 promotes breast cancer cells proliferation by regulating LncRNA GAS5. Int J Clin Exp Med. 2015;8:14464–14471. [PMC free article] [PubMed] [Google Scholar]
- 28.Farnie G, Clarke RB. Breast stem cells and cancer; Ernst Schering Found Symp Proc; 2006; pp. 1–153. [DOI] [PubMed]
- 29.Rasul S, Balasubramanian R, Filipović A, Slade MJ, Yagüe E, Coombes RC. Inhibition of gamma-secretase induces G2/M arrest and triggers apoptosis in breast cancer cells. Br J Cancer. 2009;100:1879–1888. doi: 10.1038/sj.bjc.6605034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih IeM, Wang TL. Notch signaling, gamma-secretase inhibitors and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 31.Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 33.Guerreiro S, Monteiro R, Martins MJ, Calhau C, Azevedo I, Soares R. Distinct modulation of alkaline phosphatase isoenzymes by 17beta-estradiol and xanthohumol in breast cancer MCF-7 cells. Clin Biochem. 2007;40:268–273. doi: 10.1016/j.clinbiochem.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Mendes V, Monteiro R, Pestana D, Teixeira D, Calhau C, Azevedo I. Xanthohumol influences preadipocyte differentiation: Implication of antiproliferative and apoptotic effects. J Agric Food Chem. 2008;56:11631–11637. doi: 10.1021/jf802233q. [DOI] [PubMed] [Google Scholar]
- 35.Cho MY, Park SY, Park S, Lee YR, Han GD, Kim JA. Geranyl derivative of phloroacetophenone induces cancer cell-specific apoptosis through Bax-mediated mitochondrial pathway in MCF-7 human breast cancer cells. Biol Pharm Bull. 2012;35:98–104. doi: 10.1248/bpb.35.98. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh CY, Tsai PC, Tseng CH, Chen YL, Chang LS, Lin SR. Inhibition of EGF/EGFR activation with naphtho[1,2-b]furan-4,5-dione blocks migration and invasion of MDA-MB-231 cells. Toxicol In Vitro. 2013;27:1–10. doi: 10.1016/j.tiv.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Dai J, Ma D, Zang S, Guo D, Qu X, Ye J, Ji C. Cross-talk between Notch and EGFR signaling in human breast cancer cells. Cancer Invest. 2009;27:533–540. doi: 10.1080/07357900802563036. [DOI] [PubMed] [Google Scholar]
- 38.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cel Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 40.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 41.Dowsett M, Nielsen TO, A'Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, et al. Assessment of Ki67 in breast cancer: Recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basu S, Combe K, Kwiatkowski F, Caldefie-Chézet F, Penault-Llorca F, Bignon YJ, Vasson MP. Cellular expression of cyclooxygenase, aromatase, adipokines, inflammation and cell proliferation markers in breast cancer specimen. PLoS One. 2015;10:e0138443. doi: 10.1371/journal.pone.0138443. [DOI] [PMC free article] [PubMed] [Google Scholar]