Abstract
Reactive carbonyl compounds (RCCs) are ubiquitous in the environment and are generated endogenously as a result of various physiological and pathological processes. These compounds can react with biological molecules inducing deleterious processes believed to be at the basis of their toxic effects. Several of these compounds are considered to be implicated in neurotoxic processes, aging disorders, and cancer. Therefore, a method characterizing exposures to these chemicals will provide insights into how they may influence overall health and contribute to disease pathogenesis. Here, we have developed a high resolution accurate mass (HRAM) screening strategy allowing simultaneous identification and relative quantitation of DNPH-derivatized carbonyls in human biological fluids. The screening strategy involves the diagnostic neutral loss of hydroxyl radical triggering MS3 fragmentation, which is only observed in positive ionization mode of DNPH-derivatized carbonyls. Unique fragmentation pathways were used to develop a classification scheme for characterizing known and unanticipated/unknown carbonyl compounds present in saliva. Furthermore, a relative quantitation strategy was implemented to assess variations in the levels of carbonyl compounds before and after exposure using deuterated d3-DNPH. This relative quantitation method was tested on human samples before and after exposure to specific amounts of alcohol. The nano-electrospray ionization (nano-ESI) in positive mode afforded excellent sensitivity with detection limits on-column in the high-attomole levels. To the best of our knowledge, this is the first report of a method using HRAM neutral loss screening of carbonyl compounds. In addition, the method allows simultaneous characterization and relative quantitation of DNPH-derivatized compounds using nano-ESI in positive mode.
Keywords: high resolution accurate mass data-dependent MS3 neutral loss screening, exposome, DNPH-derivatization, aldehydes/carbonyl compounds, relative quantitation
Introduction
Carbonyl compounds are a major component of the exposome, a term that was introduced to describe the totality of chemical exposures in an individual’s life-time [1]. These compounds are ubiquitous in the environment and are generated endogenously in the body through a variety of metabolic and physiological processes. Exposure to carbonyl compounds can result from numerous sources: air pollution, fossil fuel combustion, industrial processes as well as foods and beverages, in which they are used as flavorings [2]. Endogenously, carbonyl compounds can originate from the body’s metabolism, from processes such as oxidative stress and lipid peroxidation and from the interaction of the body with the microbiome [3]. These electrophilic compounds are reactive and can covalently bind to the nucleophilic sites of peptides, proteins, lipids, DNA, and RNA, inducing modifications, which are at the base of their mechanism of toxicity. Exposure to RCCs is associated with a wide variety of aging- and oxidative stress-related diseases. Among these compounds formaldehyde and acetaldehyde have been classified as carcinogenic to humans by IARC [4, 5]. Exposure to these compounds results from a combination of exogenous sources and endogenous processes, and little is known about the overall identity and abundance of RCCs in relevant biological fluids and consequently the levels of overall human exposure. A method to screen and characterize RCCs in biological samples accounting for both endogenous and exogenous sources is important to gain insights into how specific exposures influence overall health and disease etiology.
Characterization and quantitation of RCCs are typically accomplished by derivatization with 2,4-dinitrophenylhydrazine (DNPH) to form their corresponding hydrazones. The carbonyl-hydrazones are then analyzed using HPLC-UV and LC/MS [6–8]. Derivatization is performed to increase the molecular weight of small and polar aldehydes, thereby increasing their retention and ionization during LC-UV and LC/MS analyses, respectively [9]. DNPH derivatization of RCCs has been widely used in environmental monitoring of air and water quality [9–14], in the food industry [15–17], and in biomedical research [18–20]. Although HPLC-UV is routinely used to quantify these carbonyls, the identity of unanticipated and unknown compounds cannot be determined using this method without authentic internal standards. In addition, co-eluting peaks during LC separation can further confound the identification and quantitation of carbonyl compounds via UV.
Mass spectrometry-based approaches are used extensively to measure DNPH-derivatized carbonyl compounds because of the molecular weight and structural information obtained during analysis. Different ionization techniques have been employed including atmospheric pressure chemical ionization (APCI) [12, 14, 21–23], atmospheric pressure photoionization (APPI) [13], and electrospray ionization (ESI) [10, 11, 18, 19, 24]. The MS/MS and MSn spectra have been used to identify unique fragment ions for structural elucidation [12, 25, 26]. These studies described the MS and MS/MS behavior of DNPH-derivatized carbonyls in negative ionization mode, whereas the behavior of these compounds in positive mode is poorly understood. Recent studies reported characterization of DNPH-derivatized malondialdehyde using ESI in positive mode [27, 28]. In addition, ESI (+) has been used to characterize carbonyl compounds derivatized with various labeling agents including 4-(2-(trimethylammonio)ethoxy)benzenaminium halide (4-APC) [29] and 2,4-bis(diethylamino)-6-hydrazino-1,3,5-triazine [30]. The relatively high-flow rates used in these studies, however, limit the sensitivity for detection and quantitation of trace level analytes.
Here, we have developed a high resolution accurate mass MS3 neutral loss screening strategy for all DNPH-derivatized carbonyls in biological fluids, allowing the simultaneous detection and quantitation of suspected and unknown/unanticipated carbonyl compounds. The characteristic neutral loss of the hydroxyl radical, •OH from DNPH-derivatized carbonyl compounds in positive ionization mode, allows unambiguous identification of RCCs. The neutral loss of •OH is not observed in negative mode ESI of the compounds investigated in this study. Furthermore, a relative quantitation strategy using isotope labeling with deuterated d3-DNPH was implemented to determine the relative levels of carbonyl exposures. Limits of detection (LOD) in the high-attomole range on-column were obtained. The MS-based workflow presented here provides an accurate, rapid, and robust method to identify and quantify toxic and carcinogenic carbonyls in various biological matrices. This is in contrast to previous work, which used relatively high flow rates (0.2 mL min−1 - 1.5 mL min−1) and low resolution MS analysis, limiting their sensitivity and confident identification for trace analyte levels.
Experimental
Materials
2,4-Dinitrophenylhydrazine (DNPH) was purchased from Tokyo Chemical Industry (TCI, Portland, OR); d3-labeled DNPH (>98% isotopic purity) and malondialdehyde (MDA) was obtained from Santa Cruz Biotechnology (Dallas, TX); Ketones Mix (Propanone, 2-Butanone, 4-Methyl-2-Pentanone, 2-Hexanone, Cyclohexanone) and TO11/IP-6A Aldehyde/Ketone-DNPH Mix (Acetaldehyde-DNPH, Propanone-DNPH, Propanal-DNPH, Acrolein-DNPH, Benzaldehyde-DNPH, Butanal-DNPH, Crotonaldehyde-DNPH, 2,5-Dimethylbenzaldehyde-DNPH, Formaldehyde-DNPH, Hexanal-DNPH, Pentanal-DNPH, Isopentanal-DNPH, o-Tolualdehyde-DNPH, m-Tolualdehyde-DNPH, p-Tolualdehyde-DNPH), 4-hydroxy-3-methoxybenzaldehyde (Vanillin), Indole-3-carboxaldehyde, Hexanophenone, Heptanophenone, 2,3-Butanedione, 2,3-Pentanedione, trans-2-methyl-2-butenal, 3-methyl-2-butenal (methylcrotonaldehyde), sodium acetate, and concentrated HCl were obtained from Sigma Aldrich (Milwaukee, WI); 13C2-Acetaldehyde was obtained from Cambridge Isotope Laboratories (Tewksbury, MA); Methylglyoxal (MG) was obtained from Thermo Fisher (Morris Plains, NJ); Strata-X polymeric reverse-phase cartridge was purchased from Phenomenex (Torrance, CA). All acids and organic solvents were HPLC grade or better.
Sample collection and DNPH derivatization
Human saliva samples were collected from the ongoing research study “Acetaldehyde-derived DNA adducts for the investigation of alcohol hangover” at the University of Minnesota, approved by the Institutional Review Board (IRB). In this study, healthy non-smoking volunteers were exposed to a specific amount of alcohol. Pre-exposure saliva samples were collected before alcohol consumption. Subjects were then administered a specific amount of vodka to reach a BAC of 0.11%. After 1 h, post-exposure saliva samples were collected. The samples were frozen and stored at −20°C until further analysis. Frozen samples were thawed, centrifuged to remove debris, and derivatized with DNPH to form their corresponding hydrazones. The reaction was performed and optimized as previously described with minor modifications [31]. Briefly, 100 μL of saliva was derivatized with excess amounts of DNPH (400 μL of 1 mg/mL in 6N HCl) for 1 h with shaking at room temperature. After which, the reaction was quenched by adding 1.2 mL of 3 M sodium acetate. The resulting hydrazones were partially purified by Strata-X reverse-phase solid phase extraction (SPE). The cartridge was conditioned with 1 mL methanol then equilibrated with 1 mL water. The sample was then loaded into the cartridge and allowed to flow through by gravity. The cartridge was washed with 1 mL water and 4 mL 60% methanol. The hydrazone products were finally eluted using 4 mL 100% acetonitrile. One hundred microliter (100 μL) aliquot was dried and resuspended in 100 μL 20% methanol. One microliter (1 μL) of the sample was used for LC/MS analysis.
LC/MS Parameters
A data-dependent neutral loss acquisition method was used to screen for all DNPH-derivatized carbonyl compounds. The analysis was performed on a Thermo LTQ Orbitrap Velos (Thermo Scientific, Waltham, MA) mass spectrometer coupled to a Dionex RSLC HPLC (Thermo Scientific, Sunnyvale, CA). Reverse phase chromatography on a home-packed Acclaim C8 column (5 μm, 120 Å, 100 mm × 75 μm, Thermo Scientific, Waltham, MA) was performed at room temperature and flow rate of 0.3 μL min−1 using 0.1% formic acid as mobile phase A and methanol as mobile phase B. One microliter (1 μL) of the sample was injected on-column. Gradient elution was carried out starting with 5% B for the first 5 min with a flow rate of 1.0 μL min−1. The flow rate was reduced to 0.3 μL min−1 over 2 minutes, with switching of the injection valve position at 6 min into the run to take the 5 μL injection loop out of the flow path. A linear gradient from 5% to 98% B over 23 min was employed followed by a constant 98% B for 10 min. Finally, a linear gradient from 95% to 5% B over 3 min was carried out and the flow rate was increased to 1.0 μL min−1 within 2 min. The column was re-equilibrated at 5% B for another 5 min. The total run time was 50 min. The nano-electrospray voltage was set to 2.2 kV with a source temperature of 350 °C. An automated instrument tuning feature while monitoring the background ion signal m/z 371.10 (decamethylcyclopentasiloxane) was used to generate the tune file for all LC/MS analysis. The ion focusing and transfer elements were optimized for maximum signal intensity, which resulted in an S-Lens RF setting of 60%. The LTQ Orbitrap Velos mass spectrometer was routinely calibrated using the Pierce LTQ ESI Positive Ion Calibration Solution (Thermo Scientific, Rockford, IL). In addition, the mass accuracy (< 5 ppm) of the signal of an abundant background ion, decamethylcyclopentasiloxane (m/z 371.10124), was evaluated before analysis. Instrument sensitivity and mass accuracy was checked using a mixture of synthetic DNPH-derivatized carbonyls (TO11/IP-6A Aldehyde/Ketone-DNPH Mix, Sigma-Aldrich) before all LC/MS analysis.
Data-Dependent MS3 Neutral Loss Screening
Data-dependent MS3 neutral loss screening was performed in positive ionization mode with full scan detection (m/z 170 - m/z 500), followed by data-dependent CID fragmentation of the top 5 most intense ions, and MS3 CID fragmentation triggered upon observation of the neutral loss of the •OH. For the full scan event, the effective resolution of the Orbitrap mass analyzer was 43,000 (at m/z 240), an automatic gain control (AGC) setting of 1 × 106, and a maximum injection time of 100 ms. MS2 CID fragmentation was performed in the ion trap of the top 5 most intense ions in the full scan. The fragment ions were then detected in the Orbitrap at a resolution of 7500, AGC of 2 × 105 with full scan injection waveforms enabled. MS2 parameters include isolation width of 2.0 m/z units, normalized collision energy of 35%, activation Q of 0.25, and activation time of 10 ms. The data-dependent acquisition parameters include: triggering threshold of 500 counts, repeat count of 1, dynamic exclusion for 30 s, and exclusion mass width of ± 5 ppm. Fragment ions in the MS2 spectra that have a characteristic loss of 17.0027 ± 0.0001 Da (± 5 ppm) undergo MS3 fragmentation in the ion trap and detection in the Orbitrap at a resolution of 7500. MS3 scan event parameters include isolation width of 3 m/z units, normalized collision energy of 35%, activation Q of 0.25, and activation time of 10 ms.
Relative Quantitation Strategy
Pre-exposure samples (before drinking) were derivatized as described above with unlabeled DNPH (d0-DNPH) while post-exposure samples (after drinking) were derivatized with d3-labeled DNPH (Supporting Figure 1). A 1:1 ratio of d0-DNPH (derivatized pre-exposure saliva, 100 μL) and d3-DNPH (derivatized post-exposure saliva, 100 μL) were combined, dried, and resuspended in 100 μL 20% methanol before LC/MS analysis. One microliter (1 μL) of the sample was injected on column. Mass spectral data were analyzed using Xcalibur 3.0.63 (Thermo Scientific, San Jose, CA), with manual evaluation of the spectra for structural characterization of the DNPH-derivatized carbonyls. For quantitative analysis, extracted ion current (EIC) chromatograms (5 ppm mass tolerance) of d0-DNPH-labeled and d3-DNPH-labeled hydrazone products were generated and peak areas integrated and expressed as fold change (d3-DNPH /d0-DNPH). For the LOD determination, we spiked the matrix (pooled saliva samples, n=4) with synthetic d0-DNPH-derivatized RCC standards at different levels and analyzed by LC-MS. Because endogenous RCCs are present in saliva, samples were derivatized with d3-DNPH prior to spiking so that the endogenous RCCs would have different masses (3 Da) than the spiked synthetic d0-DNPH-derivatized RCCs and therefore would not contribute to their extracted ion signals.
Results and Discussion
Characterization of DNPH-derivatized carbonyls using nano-ESI mass spectrometry
Previous studies on LC/MS analysis of DNPH-derivatized carbonyls were carried out using APCI, APPI, and ESI in negative ionization mode at relatively high-flow rates (0.2 mL min−1 – 1.5 mL min−1). To date, no literature has described the behavior of these compounds in nano-electrospray and positive ionization mode. To increase the sensitivity of detecting and quantifying trace level analytes, we have investigated nano-LC separation (300 nL min−1) of DNPH-derivatized carbonyls and studied their behavior using nano-ESI in both positive and negative ionization modes.
The nano-ESI spectra of DNPH-derivatized carbonyls in positive mode yielded higher ion abundances of the protonated precursor ions [M + H]+ compared to the deprotonated precursor ions [M - H]− in negative mode (Figure 1a). Except for formaldehyde-DNPH, all of the aldehydes tested exhibited signal intensities 2–16 times higher in positive mode than in negative mode. For instance, the protonated precursor ion of acetaldehyde-DNPH (m/z 225.0618) is 8 times more abundant than its deprotonated precursor ion (m/z 223.0472). While nitro aromatic compounds are best ionized by ESI or APCI in negative mode, amino-containing nitro aromatic compounds, such as DNPH, ionize well using electrospray in positive mode [32]. In fact, we observed higher ion abundances for most, if not all DNPH-derivatized carbonyls investigated in this study. Moreover, the CID or HCD fragmentation of the protonated precursor ion of DNPH-derivatized carbonyls yielded a base peak corresponding to the loss of a common functional group (Figure 1b). The CID or HCD fragmentation of the deprotonated precursor ions [M - H]−, on the other hand, gave extensive fragmentation, which is consistent with the findings of Kolliker and co-workers [25, 26](Figure 1c). Since our focus was on developing a screening strategy, the neutral loss observed in the MS2 spectra was used to trigger MS3 for all derivatized carbonyls. High resolution accurate mass analysis confirmed that this neutral loss is due to the loss of the hydroxyl radical (•OH, 17.0027 Da). Without accurate mass spectral data, this neutral loss could also be attributed to the loss of NH3 (17.0266 Da).
Figure 1.
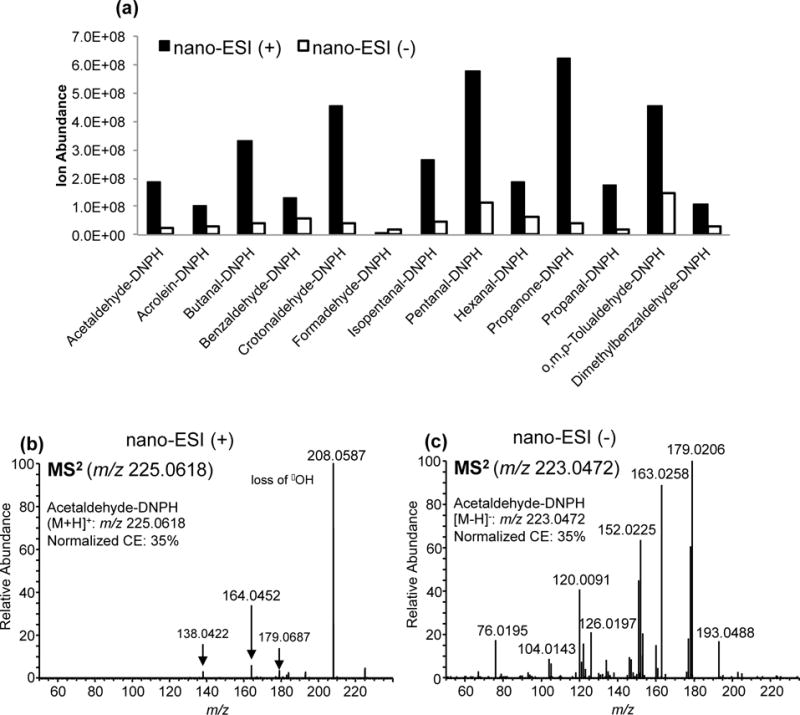
(a) Relative abundances of carbonyl-DNPH standards analyzed using nano-ESI in positive and negative modes. (b) MS2 spectrum of acetaldehyde-DNPH in positive mode and (c) MS2 spectrum of acetaldehyde-DNPH in negative mode.
To confirm that the loss of the hydroxyl radical is from the DNPH portion of DNPH-derivatized carbonyls, we performed CID fragmentation of the protonated precursor ion [M + H]+ (m/z 199.0461) of DNPH. The MS2 spectrum of DNPH yielded a base peak at m/z 182.0434, which is consistent to the loss of •OH observed in acetaldehyde-DNPH (Figure 2a). Several less intense ions were also observed in the MS2 of DNPH. MS3 fragmentation of [DNPH + H - OH]+ yielded fragment ions m/z 78.0334 and m/z 164.0327 (Figure 2b). The structures and elemental compositions based on high resolution accurate mass analysis of fragment ions corresponding to the loss of •OH in the MS2 and the fragment ions in the MS3 are shown in Figure 2a,b. The analogous negative ions, m/z 76 and m/z 163, respectively, were previously observed in MS2 of DNPH using negative ion APCI [25].
Figure 2.
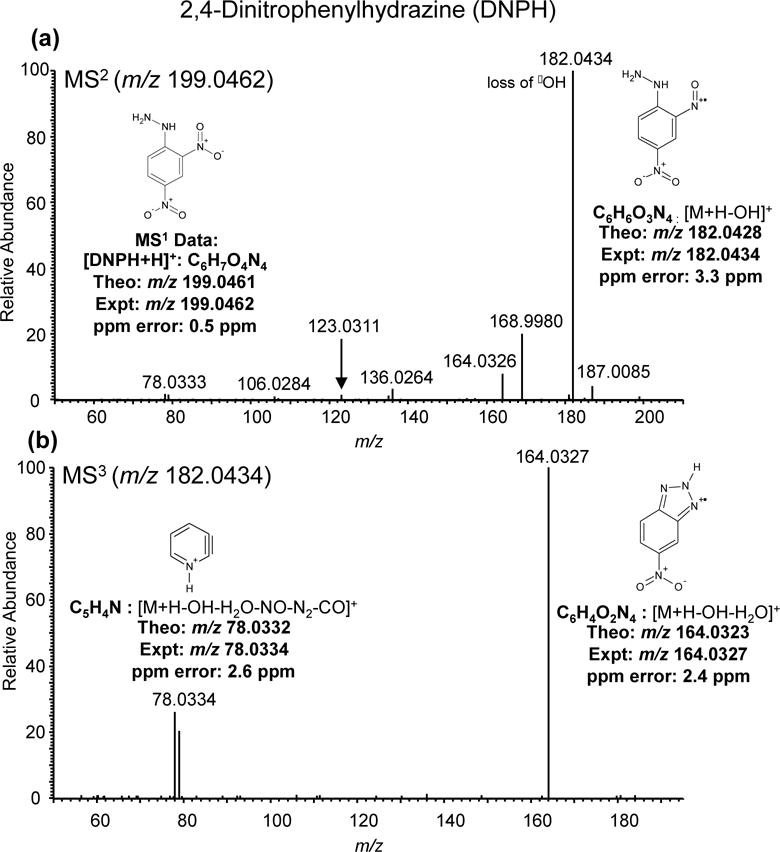
MS2 spectrum of the derivatizing agent, 2,4-Dinitophenylhydrazine (DNPH) in positive ionization mode and proposed structures of fragment ions. (a) The dominant base peak ion in the MS2 is due to the loss of •OH. (b) While specific fragment ions m/z 78.0334 and m/z 164.0327 were dominant in the MS3 spectrum.
The loss of the hydroxyl radical is diagnostic of nitro aromatic compounds after CID of the [M + H]+ ion [32]. We did not observe this neutral loss in negative mode. Moreover, the amino group in DNPH makes it ideal for protonation and thus, easily detectable during MS analysis in ESI positive ionization mode, consistent with our observations and those of Levsen et al. (2007) [32]. Therefore, analysis of the DNPH-derivatized carbonyls in positive mode provides additional specificity for DNPH-derivatized carbonyl compound identification. The high mass accuracy acquisition of our method, typically less than 5 ppm mass error, allows unambiguous identification of elemental compositions of all precursor ions as well as fragment ions (restricting elements to C, N, O, H) (Figure 2a,b).
Different collision energies (CE) were then investigated using both HCD and CID to determine the optimal CE to obtain maximum base peak due to the loss of •OH. The optimal normalized collision energies obtained were 40% and 35% for HCD and CID, respectively. To further investigate whether the loss of •OH is common to all DNPH-derivatized carbonyls, the MS2 and MS3 fragmentation behavior of mixture of standards containing saturated aldehydes, unsaturated aldehydes, aromatic aldehydes, ketones, and dicarbonyls were investigated. The same loss of •OH was observed for saturated aldehydes (Formaldehyde, Acetaldehyde, Propanal, Butanal, Isopentanal, Pentanal, Hexanal), unsaturated aldehydes (Acrolein and Crotonaldehyde), aromatic aldehydes (Benzaldehyde, o,m,p-Tolualdehyde, Dimethylbenzaldehyde, 4-hydroxy-3-methoxybenzaldehyde/Vanillin, Indole-3-carboxaldehyde), and ketones (Propanone, 2-Butanone, 4-Methyl-2-Pentanone, 2-Hexanone, Cyclohexanone, Hexanophenone, Heptanophenone) (Supporting Figures 2–6). The unique and characteristic structural feature of nitro aromatic compounds such as DNPH (loss of hydroxyl radical) was used to develop the neutral loss screening strategy for the analysis of DNPH-derivatized carbonyls. The observation of specific fragment ions in the MS3 due to the derivatizing agent (m/z 78.0332 and m/z 164.0323) was used to confirm the detection of DNPH-derivatized carbonyls.
Principles of the neutral loss screening strategy
The high resolution accurate mass (HRAM) data-dependent MS3 neutral loss screening strategy involves selection of the top 5 protonated precursor ions [M + H]+ in the full scan (m/z 170 – m/z 500) for fragmentation. The fragment ions are then interrogated for the loss of the high resolution accurate mass (17.0027 Da) due to the loss of •OH in the MS2. Observation of ions with characteristic neutral loss of •OH triggers another fragmentation event (MS3). Specific fragment ions (m/z 78.0332 and m/z 164.0323) in the MS3 are used to confirm the identity of DNPH-derivatized carbonyls (Figure 3a).
Figure 3.
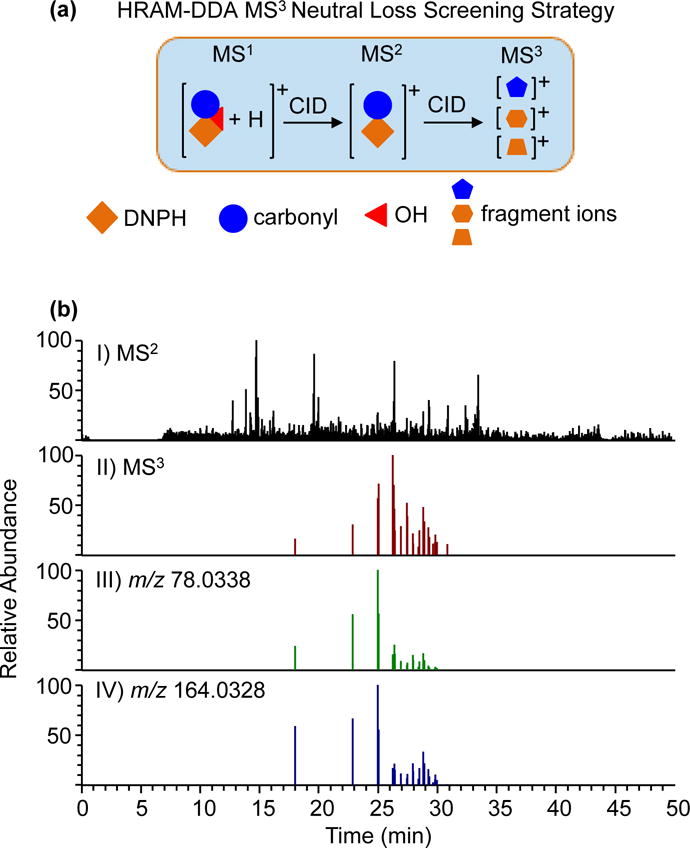
Development of a high resolution accurate mass data-dependent MS3 neutral loss screening strategy. The high resolution accurate mass of •OH (17.0027 Da) was used to screen for all DNPH-derivatized aldehydes. In addition, monitoring specific fragment ions (m/z 78.0332 and m/z 164.0323) minimizes possible false positive identification.
The neutral loss screening strategy simplifies the data analysis and structural characterization of DNPH-derivatized carbonyls. For instance, LC/MS analysis of mixture of standards in water yielded ~3,600 MS2 spectra while the additional MS3 triggering upon loss of •OH yielded 28 MS3 spectra, all corresponding to standards spiked in the water (Figure 3b, Panel I and Panel II). Importantly, the appearance of the fragment ions (m/z 78.0332 and m/z 164.0323) in the MS3 spectrum, except for unsaturated carbonyls, minimizes false positive identification (Figure 3b, Panel III and Panel IV).
Classification scheme as a guide for the structural analysis of known and unknown DNPH-derivatized carbonyls
A classification scheme for the structural analysis of known and unknown DNPH-derivatized carbonyls was developed using the high resolution accurate masses of the MS2 and MS3 mass spectral data of all DNPH-carbonyl standards used for our method development (Figure 4). The scheme and the differences in fragmentation pathways are used to differentiate between saturated aldehydes, unsaturated aldehydes, aromatic aldehydes, and ketones. A similar scheme was developed using the MS2 and MSn fragmentation data obtained using APCI in negative ionization mode [25]. In our current approach, however, the high resolution accurate mass data (typically < 5 ppm mass error) were used to unambiguously assign the molecular formula of both precursor ions and fragment ions. For instance, high molecular weight (HMW) carbonyls differ from low molecular weight ones (MW > 260 Da) by the loss of 183.0274 Da (C6H5O4N3) due to the loss of DNPH resulting from the cleavage of the N-N bond of the hydrazine portion during MS2 fragmentation (Supporting Figure 8a,b). We observed that as the MW of the carbonyl compound increases, the intensity of the signal due to the loss of 183 Da increases (data not shown). In addition, we observed the fragmentation behavior/pattern is somewhat different with HMW carbonyl compounds, in particular, fragment ions coming from the carbonyl portion are apparent in the MS3 spectra of these compounds (Supporting Figure 3). The loss of 163.0250 Da (C6H3N + N2 + NO2) and 29.9980 Da (NO) from [M + H - OH]+ in the MS3 spectra further supports the presence of aromatic carbonyls. The proposed fragmentation pathways, structures of fragment ions, and neutral losses of aromatic carbonyls are shown in Supporting Figure 8a,b. Although the MS1 ion signal determines the molecular weight of the carbonyl, the loss of 183 Da in the classification scheme allows easier evaluation of MSn data for structural elucidation.
Figure 4.
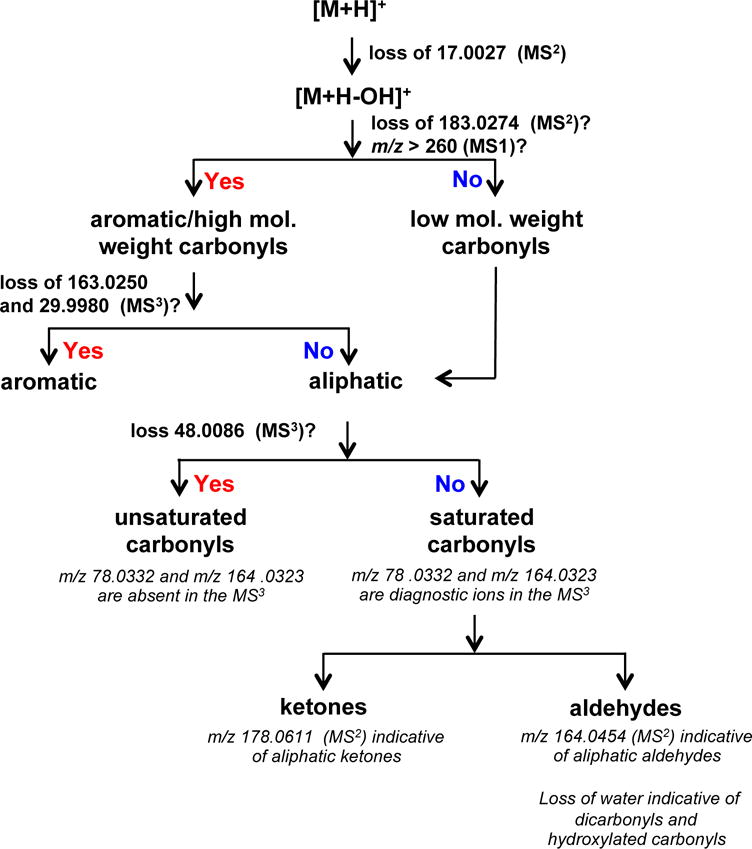
Classification scheme for characterizing known and unknown DNPH-derivatized carbonyls. Proposed fragmentation pathways and structures of neutral losses and characteristic ions used for structural elucidation are shown in Supporting Figure 8 and Supporting Figure 9.
Aliphatic aldehydes are differentiated from unsaturated carbonyls by unique fragment ions in the MS3. The MS3 spectra of aldehydes and ketones after the loss of •OH showed characteristic and intense fragment ions coming from the derivatizing agent (m/z 78.0332, C5H4N and m/z 164.0323, C6H4O2N4) (Supporting Figure 2). Unsaturated aldehydes such as acrolein-DNPH and crotonaldehyde-DNPH, on the other hand, showed a loss of 48.0086 Da in the MS3, which is due to the neutral loss of the molecular formula corresponding to H2NO2 (Supporting Figure 4). The APCI (−) spectra of unsaturated aldehydes showed similar fragment ion due to the loss of 47 (HNO2) [25]. Interestingly, the appearance of specific ions, m/z 78.0332 and m/z 164.0323 in the MS3 spectra of saturated aldehydes is prominent, while these ions are absent or less intense in the MS3 spectra of unsaturated aldehydes. Thus, the presence and intensity of diagnostic fragment ions (m/z 78.0332 and m/z 164.0323) could be used to differentiate saturated/aliphatic carbonyls from unsaturated/aliphatic carbonyls.
Aliphatic aldehydes can be differentiated from aliphatic ketones by the appearance of characteristic ions in the MS2 spectra. Aliphatic aldehydes showed a characteristic ion, m/z 164.0454, while aliphatic ketones showed a characteristic ion at m/z 178.0611 (Supporting Figure 2, Supporting Figure 5). The general scheme for differentiating aliphatic aldehydes from aliphatic ketones and the proposed fragmentation pathways leading to the ions m/z 164.0454 and m/z 178.0611 are shown in Supporting Figure 9a. For instance, the isobaric carbonyls, hexanal-DNPH, 2-hexanone-DNPH, and 4-methyl-2-pentanone-DNPH can be differentiated using the appearance of the ions, m/z 164.0454 and m/z 178.0611 in the MS2 spectra (Supporting Figure 9b). The m/z 164.0454 is only present in hexanal-DNPH, while m/z 178.0611 is only present in 2-hexanone and 4-methyl-2-pentanone (Supporting Figure 2 and Supporting Figure 5).
The m/z 190.0483 in an MS3 spectra can be used to further differentiate hexanal from its isobaric compounds, 2-hexanone and 4-methyl-2-pentanone. The m/z 190.0483, which corresponds to the molecular formula C8H6O2N4, results due to the loss of the alkyl and hydroxyl groups as shown in Supporting Figure 9c. This ion is also formed in butanal, pentanal, and isopentanal (Supporting Figure 2d–g), however, it is not observed in the MS3 spectra of aliphatic ketones, 2-butanone, 2-hexanone, and 4-methyl-2-pentanone. Thus, the presence of m/z 190.0483 in an MS3 spectrum can be used to classify an isobaric aliphatic aldehyde from its aliphatic ketone counterpart. However, at this point, we are not able to differentiate the isomeric ketones, 2-hexanone from 4-methyl-2-pentanone based on the MS2 and MS3 fragmentation data for these compounds.
The appearance of m/z 193.0356 and m/z 138.0424 in the MS2 spectra is also indicative of aliphatic aldehydes, while the presence of m/z 207.0513 is indicative of aliphatic ketones. The m/z 193.0356 and m/z 207.0513, however, are both observed in the MS2 spectra of propanal-DNPH and propanone-DNPH probably due to the similar structures of the fragment ions formed after the loss of the ethyl group in propanal-DNPH and the two methyl groups in propanone-DNPH upon fragmentation. The fragmentation pathways leading to these characteristic ions in aliphatic aldehydes and ketones are shown in Supporting Figure 9d.
Application of the NL screening strategy to human saliva samples after alcohol exposure
To test the applicability of the method for the simultaneous screening and quantitation of carbonyls in saliva, a human model system for alcohol exposure was used [3, 33]. In our study, healthy non-smoking volunteers were administered amounts of vodka to reach a BAC of ~0.11, after which, the identity and the levels of carbonyl compounds were determined. The classification scheme was then used for the structural analysis of detected unanticipated and unknown carbonyl compounds. For instance, an unanticipated compound was detected and identified in saliva. The MS2 spectrum of the compound exhibited a base peak due to the loss of •OH (Figure 5a). The second most abundant fragment ion is the loss of 183.0274 Da due to DNPH, consistent with a carbonyl compound of a high molecular weight (MW > 260 Da). The MS3 spectrum showed no loss of 163.0250 Da and 29.9980 Da suggesting an aliphatic carbonyl. The MS3 showed characteristic loss of 48.0086 Da (loss of H2NO2) suggesting an unsaturated aldehyde and a base peak at m/z 233.0659 Da corresponding to the loss of methyl group (Figure 5b). These results suggest the compound is unsaturated and methylated carbonyl. The high resolution accurate masses of the protonated precursor ions and both the MS2 and MS3 data are consistent with the identification of the isomeric compounds, 3-methyl-2-butenal (methyl crotonaldehyde) or trans-2-methyl-2-butenal. The identity of the compound was confirmed by DNPH-derivatization of synthetic standards of 3-methyl-2-butenal (methyl crotonaldehyde) and trans-2-methyl-2-butenal. The MS2 and MS3 fragmentation mass spectral data and retention time of the unanticipated compound are consistent with 3-methyl-2-butenal (methyl crotonaldehyde) (Figure 5c).
Figure 5.
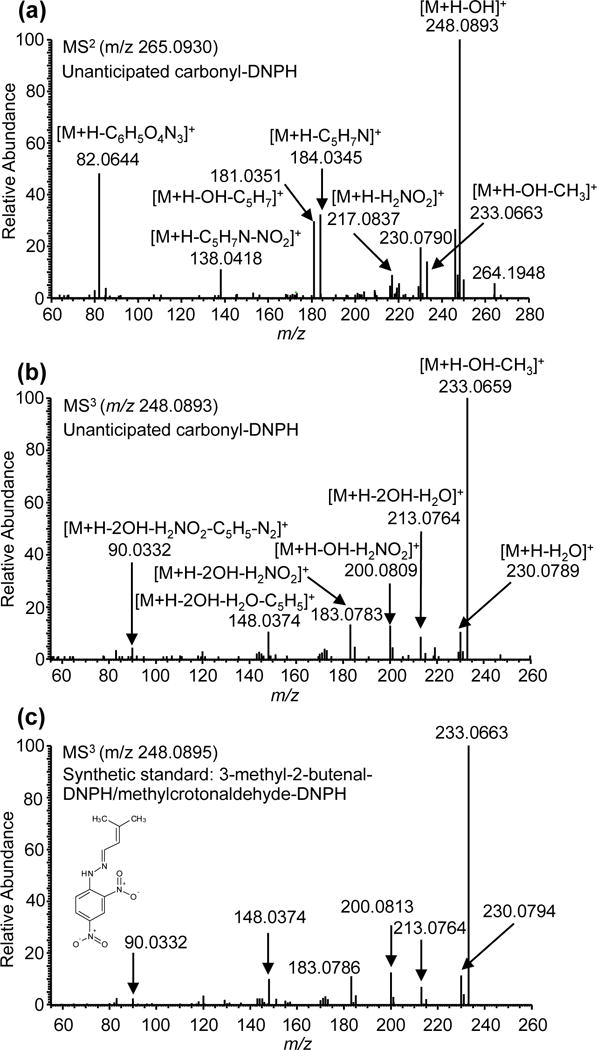
(a) MS2 and (b) MS3 spectra of an unanticipated carbonyl compound identified in saliva. (c) MS3 spectrum of DNPH-derivatized synthetic standard of 3-methyl-2-butenal. Using the classification scheme and authentic internal standards, the compound was identified as 3-methyl-2-butenal (methyl crotonaldehyde).
Next, we determined and quantified the levels of carbonyls after exposure to an alcoholic beverage (vodka). Pre-exposure (baseline) and post-exposure saliva were labeled with d0-DNPH and d3-DNPH, respectively. The relative abundances of the ions corresponding to d0-DNPH-labeled carbonyls and d3-DNPH-labeled carbonyls were determined by integrating the peak areas in the full scan EIC chromatograms and expressed as fold change (post-exposure/pre-exposure; d3-DNPH/ d0-DNPH). The EIC chromatograms of d0-DNPH-labeled acetaldehyde and d3-DNPH-labeled acetaldehyde showed co-eluting peaks at the same retention times, while the mass spectrum of d0-DNPH-labeled acetaldehyde and d3-DNPH-labeled acetaldehyde pairs showed MS signals separated by 3 Da (Figure 6a,b). Because the ionization efficiencies are similar for both d0-DNPH-labeled and d3-DNPH-labeled carbonyl compounds, accurate relative quantitation can be performed.
Figure 6.
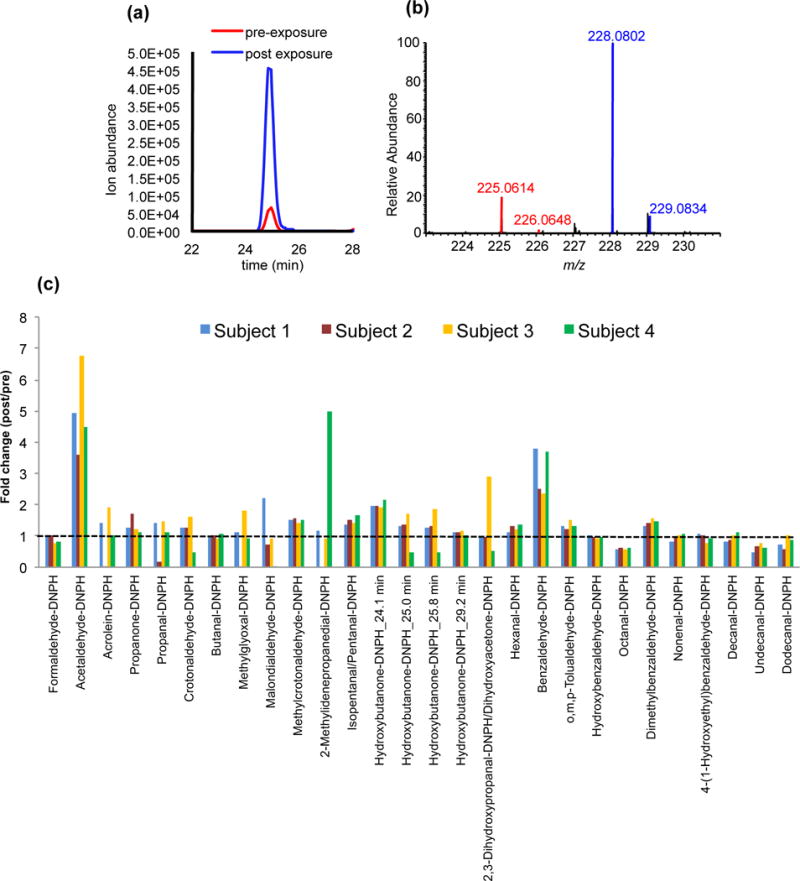
(a) Representative extracted ion current (EIC) chromatograms and (b) MS spectrum of d0-labeled acetaldehyde (pre-exposure) and d3-labeled acetaldehyde (post-exposure) before and after alcohol exposure. (c) Quantitation of DNPH-derivatized carbonyls in saliva before and after alcohol exposure.
The acetaldehyde levels in saliva samples of 4 subjects exhibited 4-7 times higher after alcohol exposure (Figure 6c), which is in agreement to previous studies [3, 33]. In fact, even a single sip of strong alcoholic beverage can instantly cause increase acetaldehyde levels in the oral cavity [3]. Benzaldehyde is also increased in the 4 subjects probably due to the flavorings present in the alcoholic beverage (vodka). Likewise, several isomers of hydroxybutanone were detected eluting at different times but the identity of each isomer cannot be deduced at this point without internal standards corresponding to the different isomers. While we expected elevated levels of acetaldehyde after alcohol exposure, most of the other carbonyls identified showed no dramatic change in the amounts (dotted line, Figure 6c), however, variability in the levels is observed among the 4 subjects due to inter individual variation (Figure 6c).
The limit of detection (LODs) of DNPH-derivatized carbonyls was determined and is defined here as the lowest concentration of the analyte where the signals corresponding to the desired analytes resulted in a clear Gaussian peak in the EIC chromatograms (5 ppm mass tolerance). The LODs on-column of most aldehydes ranged from 0.19 fmol (0.04 pg) to 3.24 fmol (0.68 pg) (Supporting Table 1 and Supporting Table 2). Although some aldehydes showed increased LODs in saliva (e.g. formadehyde-DNPH), the matrix did not adversely affect the overall LODs of most carbonyl compounds. For instance, the LOD of acetaldehyde-DNPH obtained using the current approach (LOD: 0.04 pg) is 150 times lower than that obtained by Ochs et. al (2015) (LOD : 5.97 pg) using SRM and APCI in negative ionization mode [14]. The LODs for several RCCs known to be present in biological samples were determined; however, there are potentially many other RCCs that may be present in saliva for which the LOD was not measured. There are primarily two factors determining the LOD for a given RCC using the methodology described here. First, the ion abundance in the full scan spectrum must be greater than the data dependent acquisition (DDA) threshold so that it is eligible to be selected for MS2 fragmentation. Secondly, even though the ion intensity is above the threshold, it still must be selected via DDA for MS2 fragmentation. DDA is stochastic in nature and therefore even though a given carbonyl compound ion has an intensity above the DDA threshold in the MS1 spectra, it may not be sampled for MS2 fragmentation and therefore not detected. Currently, improvements in sample clean up and the use of newer, more sensitive MS technology with faster HRAM scanning capabilities are being investigated to obtain lower LODs and improved detection of low abundant DNPH-derivatized carbonyls.
The technical and experimental reproducibility of the method was assessed using pooled saliva samples (n=4) spiked with 13C2-acetaldehyde (50 fmol on-column). The technical reproducibility was evaluated using 3 replicate preparations of the samples on the same day and analyzed using the screening method. The EICs corresponding to the 13C2-acetaldehyde-DNPH were integrated in the 3 replicate preparations and expressed as %CV. Likewise, the experimental reproducibility was evaluated using 3 different preparations of the samples on different days and analyzed using the approach. The EICs corresponding to the 13C2-acetaldehyde-DNPH were integrated in the 3 different days of sample preparation and expressed as %CV. The technical reproducibility of the quantitation was determined to be 6%, whereas the experimental reproducibility was 11%.
Although the current approach worked well with nearly all the aldehydes and ketones investigated, we found that carbonyls containing hydroxyl groups in the aliphatic chain and dicarbonyl compounds behave somewhat differently than the others. The CID MS2 fragmentation spectra of dicarbonyl-hydrazones such as 2,3-butanedione-DNPH and 2,3-pentanedione-DNPH yielded dominant m/z attributed to the loss of water (18.0106 Da) (Supporting Figure 7a,b). 2,3-butanedione with one DNPH triggered for MS3 fragmentation as the loss of •OH is still present, although the intensity is only 35% compared to the signal due to the loss of water. Similar behavior was observed for the dicarbonyls, methylglyoxal (MG) and malondialdehyde (MDA) (data not shown). The intensity of the signals due to the loss of •OH in MG and MDA is ~ 60-70% relative to the intensity of the signals due to the loss of water. In the case of 2,3-pentanedione-DNPH, the loss of •OH is no longer visible in the MS2 spectrum and therefore it is not triggered for MS3 fragmentation (Supporting Figure 7b). Further interrogation of the MS2 data found that the dominant peak for 2,3-pentanedione-DNPH due to the loss of water. In addition, hydroxylated aldehydes/carbonyls detected in saliva yielded similar MS2 spectra showing the loss of water. For dicarbonyls and hydroxylated carbonyl compounds, we are currently investigating the loss of water as an additional feature of the screening method. Because it is not as diagnostic as the loss of hydroxyl radical to screen for DNPH-derivatized compounds as it is a common feature of many molecules in biological samples, we are now investigating the fragmentation behavior of this class of compounds. Preliminary observations of analyzed samples suggest that the MS2 fragmentation of the dicarbonyls and hydroxylated carbonyl compounds results in loss of H2O and the subsequent MS3 fragmentation results in the loss of the OH, presumably from the DNPH portion of the molecule. This suggests that the neutral loss of H2O and the subsequent loss of OH could be used to trigger another fragmentation event (MS4) allowing for screening and characterization of this class of compounds. Lastly, monitoring the appearance of specific ions due to the derivatizing agent and the carbonyl portion of DNPH-derivatized carbonyls can allow confirmation and identification of dicarbonyls and hydroxylated carbonyl compounds. Additional investigations with synthetic standards of this class of compounds need to be done to confirm these observations.
Conclusions
Overall, a high resolution accurate mass data dependent MS3 neutral loss screening for aldehydes/ketones in saliva was developed. The improvements provided by the new approach include: (1) the high resolution accurate mass analysis (< 5 ppm) enabling confident identification of DNPH-derivatized carbonyls; (2) NL screening (•OH) and MS3 data acquisition in positive mode allowing identification of known and structural characterization of unknown carbonyls; (3) nano-ESI affords excellent sensitivity with detection limits in the high-attomole levels; (4) a relative quantitation strategy enabling determination of the levels of compounds after exposure.
The approach developed here provides several levels of selective identification and characterization of target analytes in biological samples. First, the highly selective reaction of DNPH with carbonyl compounds such as aldehydes and ketones enables detection of derivatized carbonyl compounds. Second, the characteristic loss of hydroxyl radical used to trigger MS3 fragmentation, which is only observed in positive ionization of amino nitro aromatic compounds, allows selective detection and fragmentation of DNPH-derivatized carbonyls. Lastly, the appearance of specific fragment ions due to the derivatizing agent minimizes possible false positive identification. The approach developed here will be useful in characterizing toxic and genotoxic carbonyl compounds resulting from specific exposures and for monitoring endogenous formation of carbonyl compounds such as those derived from processes related to inflammation like lipid peroxidation and oxidative stress. To the best of our knowledge, this is the first report of the high resolution accurate mass neutral loss screening and characterization of DNPH-derivatized carbonyls using nano-ESI in positive ionization mode. Furthermore, the relative quantitation strategy using deuterated d3-DNPH provides a versatile tool for quantifying carbonyls present in a variety of biological matrices.
Supplementary Material
Acknowledgments
The authors would like to thank the volunteers of the research study “Acetaldehyde-derived DNA adducts for the investigation of alcohol hangover” at the University of Minnesota and Robert Carlson for editorial assistance. This work was supported by NIOSH-funded MCOHS ERC Pilot Research Training Program (OH008434). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official view of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention, or other associated entities. Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, University of Minnesota, funded in part by Cancer Center Support Grant CA- 077598 and S10 RR-024618 (Shared Instrumentation Grant).
References
- 1.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 2.Feron VJ, Til HP, de Vrijer F, Woutersen RA, Cassee FR, van Bladeren PJ. Aldehydes: occurrence, carcinogenic potential, mechanism of action and risk assessment. Mutat Res. 1991;259:363–385. doi: 10.1016/0165-1218(91)90128-9. [DOI] [PubMed] [Google Scholar]
- 3.Linderborg K, Salaspuro M, Vakevainen S. A single sip of a strong alcoholic beverage causes exposure to carcinogenic concentrations of acetaldehyde in the oral cavity. Food Chem Toxicol. 2011;49:2103–2106. doi: 10.1016/j.fct.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 4.Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V, et al. A review of human carcinogens–Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033–1034. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 5.IARC WorkingGroup on the Evaluation of Carcinogenic Risks to Humans. Formaldehyde, 2-butoxyethanol, and 1-tert-butoxypropan-2-ol. IARC Monogr Eval Carcinog Risks Hum. 2006;88:1–478. [PMC free article] [PubMed] [Google Scholar]
- 6.Osorio VM, Cardeal ZL. Analytical Methods to Assess Carbonyl Compounds in Foods and Beverages. J Brazil Chem Soc. 2013;24:1711–1718. [Google Scholar]
- 7.Santa T. Derivatization reagents in liquid chromatography/electrospray ionization tandem mass spectrometry. Biomed Chromatogr. 2011;25:1–10. doi: 10.1002/bmc.1548. [DOI] [PubMed] [Google Scholar]
- 8.Vogel M, Buldt A, Karst U. Hydrazine reagents as derivatizing agents in environmental analysis–a critical review. Fresenius J Anal Chem. 2000;366:781–791. doi: 10.1007/s002160051572. [DOI] [PubMed] [Google Scholar]
- 9.Richardson SD, Caughran TV, Poiger T, Guo YB, Crumley FG. Application of DNPH derivatization with LC/MS to the identification of polar carbonyl disinfection by-products in drinking water. Ozone-Sci Eng. 2000;22:653–675. [Google Scholar]
- 10.Zwiener C, Glauner T, Frimmel FH. Liquid chromatography/electrospray ionization tandem mass spectrometry and derivatization for the identification of polar carbonyl disinfection by-products. Acs Sym Ser. 2003;850:356–375. [Google Scholar]
- 11.Chi YG, Feng YL, Wen S, Lu HX, Yu ZQ, Zhang WB, et al. Determination of carbonyl compounds in the atmosphere by DNPH derivatization and LC-ESI-MS/MS detection. Talanta. 2007;72:539–545. doi: 10.1016/j.talanta.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Grosjean E, Green PG, Grosjean D. Liquid chromatography analysis of carbonyl (2,4-dinitrophenyl)hydrazones with detection by diode array ultraviolet spectroscopy and by atmospheric pressure negative chemical ionization mass spectrometry. Anal Chem. 1999;71:1851–1861. doi: 10.1021/ac981022v. [DOI] [PubMed] [Google Scholar]
- 13.van Leeuwen SM, Hendriksen L, Karst U. Determination of aldehydes and ketones using derivatization with 2,4-dinitrophenylhydrazine and liquid chromatography-atmospheric pressure photoionization-mass spectrometry. J Chromatogr A. 2004;1058:107–112. doi: 10.1016/j.chroma.2004.08.149. [DOI] [PubMed] [Google Scholar]
- 14.Ochs SDM, Fasciotti M, Netto ADP. Analysis of 31 Hydrazones of Carbonyl Compounds by RRLC-UV and RRLC-MS(/MS): A Comparison of Methods. J Spectrosc. 2015 [Google Scholar]
- 15.Wang Y, Cui P. Reactive Carbonyl Species Derived from Omega-3 and Omega-6 Fatty Acids. J Agric Food Chem. 2015;63:6293–6296. doi: 10.1021/acs.jafc.5b02376. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Li X, Shoemaker CF, Wang SC. Ultrahigh performance liquid chromatography analysis of volatile carbonyl compounds in virgin olive oils. J Agric Food Chem. 2013;61:12253–12259. doi: 10.1021/jf404368m. [DOI] [PubMed] [Google Scholar]
- 17.Faizan M, Esatbeyoglu T, Bayram B, Rimbach G. A fast and validated method for the determination of malondialdehyde in fish liver using high-performance liquid chromatography with a photodiode array detector. J Food Sci. 2014;79:C484–488. doi: 10.1111/1750-3841.12412. [DOI] [PubMed] [Google Scholar]
- 18.Banos CE, Silva M. Liquid chromatography-tandem mass spectrometry for the determination of low-molecular mass aldehydes in human urine. J Chromatogr B. 2010;878:653–658. doi: 10.1016/j.jchromb.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Miller JH, Gardner WP, Gonzalez RR. UHPLC Separation with MS Analysis for Eight Carbonyl Compounds in Mainstream Tobacco Smoke. J Chromatogr Sci. 2010;48:12–17. doi: 10.1093/chromsci/48.1.12. [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Mizukami T, et al. Salivary acetaldehyde concentration according to alcoholic beverage consumed and aldehyde dehydrogenase-2 genotype. Alcohol Clin Exp Res. 2008;32:1607–1614. doi: 10.1111/j.1530-0277.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 21.Brombacher S, Oehme M, Beukes JA. HPLC combined with multiple mass spectrometry (MSn): an alternative for the structure elucidation of compounds and artefacts found in smog chamber samples. J Environ Monitor. 2001;3:311–316. doi: 10.1039/b101204p. [DOI] [PubMed] [Google Scholar]
- 22.Kolliker S, Oehme M. Structure elucidation of nanogram quantities of unknown designer drugs based on phenylalkylamine derivates by ion trap multiple mass spectrometry. Anal Bioanal Chem. 2004;378:1294–1304. doi: 10.1007/s00216-003-2411-2. [DOI] [PubMed] [Google Scholar]
- 23.Olson KL, Swarin SJ. Determination of Aldehydes and Ketones by Derivatization and Liquid-Chromatography Mass-Spectrometry. J Chromatogr. 1985;333:337–347. [Google Scholar]
- 24.Zweiner C, Glauner T, Frimmel FH. Method optimization for the determination of carbonyl compounds in disinfected water by DNPH derivatization and LC-ESI-MS-MS. Anal Bioanal Chem. 2002;372:615–621. doi: 10.1007/s00216-002-1233-y. [DOI] [PubMed] [Google Scholar]
- 25.Kolliker S, Oehme M, Dye C. Structure elucidation of 2,4-dinitrophenylhydrazone derivatives of carbonyl compounds in ambient air by HPLC/MS and multiple MS/MS using atmospheric chemical ionization in the negative ion mode. Anal Chem. 1998;70:1979–1985. doi: 10.1021/ac9709458. [DOI] [PubMed] [Google Scholar]
- 26.Kolliker S, Oehme M, Merz L. Unusual MSn fragmentation patterns of 2,4-dinitrophenylhydrazine and its propanone derivative. Rapid Commun Mass Sp. 2001;15:2117–2126. [Google Scholar]
- 27.Chen JL, Huang YJ, Pan CH, Hu CW, Chao MR. Determination of urinary malondialdehyde by isotope dilution LC-MS/MS with automated solid-phase extraction: a cautionary note on derivatization optimization. Free Radic Biol Med. 2011;51:1823–1829. doi: 10.1016/j.freeradbiomed.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Szarka S, Prokai-Tatrai K, Prokai L. Application of Screening Experimental Designs to Assess Chromatographic Isotope Effect upon Isotope-Coded Derivatization for Quantitative Liquid Chromatography-Mass Spectrometry. Anal Chem. 2014;86:7033–7040. doi: 10.1021/ac501309s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L, Liu P, Wang YL, Yu QW, Yuan BF, Feng YQ. Profiling of aldehyde-containing compounds by stable isotope labelling-assisted mass spectrometry analysis. Analyst. 2015;140:5276–5286. doi: 10.1039/c5an00657k. [DOI] [PubMed] [Google Scholar]
- 30.Tie C, Hu T, Jia ZX, Zhang JI. Novel derivatization strategy for the comprehensive characterization of endogenous fatty aldehydes using HPLC-MRM. Anal Chem. 2016;88:7762–7768. doi: 10.1021/acs.analchem.6b01756. [DOI] [PubMed] [Google Scholar]
- 31.Guan XY, Rubin E, Anni H. An Optimized Method for the Measurement of Acetaldehyde by High-Performance Liquid Chromatography. Alcohol Clin Exp Res. 2012;36:398–405. doi: 10.1111/j.1530-0277.2011.01612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levsen K, Schiebel HM, Terlouw JK, Jobst KJ, Elend M, Preiss A, et al. Even-electron ions: a systematic study of the neutral species lost in the dissociation of quasi-molecular ions. J Mass Spectrom. 2007;42:1024–1044. doi: 10.1002/jms.1234. [DOI] [PubMed] [Google Scholar]
- 33.Lachenmeier DW, Monakhova YB. Short-term salivary acetaldehyde increase due to direct exposure to alcoholic beverages as an additional cancer risk factor beyond ethanol metabolism. J Exp Clin Cancer Res. 2011;30:3. doi: 10.1186/1756-9966-30-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


