Abstract
Esophageal carcinoma is a malignancy that severely threatens human health, with a high incidence rate and a low 5-year survival rate. Resistance to chemotherapy frequently emerges during its treatment, partly due to the induction of autophagy. Therefore, targeting autophagy may be a promising therapeutic approach for the treatment of esophageal carcinoma. In the present study, it was investigated how chloroquine (CQ) can influence the growth ability and biological behaviors of EC109 esophageal squamous carcinoma cells in vitro, as well as the potential molecular mechanisms behind its activity. It was demonstrated that CQ could suppress the growth and proliferation of EC109 cells in a time- and dose-dependent manner; migration and colony formation abilities were also inhibited by CQ. Furthermore, subsequent to the exposure to CQ, the number of autophagosomes was clearly increased in EC109 cells overexpressing green fluorescent protein tagged-light chain (LC)3 when observed by fluorescence microscopy. Protein expression of the endogenous autophagy markers LC3-II and p62 was elevated subsequent to CQ treatment, whereas the expression of proteins from the protein kinase B/mechanistic target of rapamycin target of rapamycin pathway was inhibited. This suggested that CQ could induce the formation of autophagosomes in the initiation of autophagy, but inhibit the degradation of autophagosomes in a later stage of autophagy. The overall effect was that autophagic cell death was activated by CQ, as confirmed by flow cytometry. Overall, the anticancer effect of chloroquine on EC109 was revealed to be mediated through modulating autophagy, and this may produce promising therapeutic benefits for esophageal carcinoma.
Keywords: chloroquine, esophageal carcinoma, autophagy, protein kinase B/mechanistic target of rapamycin signal transduction pathway, p62
Introduction
Esophageal cancer is the eighth most common cancer worldwide, with 456,000 new cases and ~0.4 million mortalities reported in 2012. Due to the geographical variation in the occurrence of esophageal cancer, 49% of new cases occurred in China (1), particularly in two counties renowned for the high occurrence rates of esophageal cancer: Linxian, Henan and Yanting, Sichuan (2). This is due to the tumors being asymptomatic in early stages, meaning >50% of patients present at an advanced stage that is too late for treatment by esophagectomy. Chemotherapy is the main treatment for advanced esophageal cancer (3). However, chemotherapy is associated with drug resistance and side effects. Therefore, the identification of novel and safe treatments for esophageal cancer is necessary.
Autophagy is an intricate and conserved process. Autophagic flux refers to the process by which damaged organelles and unfolded proteins are sequestered into autophagosomes within the cytoplasm, which fuse with lysosomes to form autolysosomes, to induce the degradation of intracellular components (4). The induction of autophagy has been identified as a drug resistance mechanism that promotes cancer cell survival via self-digestion (5,6). Therefore, targeting autophagy may be a promising approach for cancer therapy.
Chloroquine (CQ) has been safely and widely used as an anti-malarial for >60 years (7). A number of studies have suggested that CQ can inhibit autophagy through the prevention of lysosome acidification, and subsequently, the inhibition of autophagosome-lysosome fusion, to block the degradation of autolysosomes at the last step of autophagy (8,9). Previous studies have indicated that CQ exhibited an antitumor effect on several types of cancer, including glioblastoma (10,11), hepatocellular carcinoma (12), breast cancer (13), prostate cancer (14) and pancreatic cancer (15), which prompts us to hypothesize that CQ may be able to influence the growth of esophageal carcinoma through the modulation of autophagy. Therefore, the aim of the present study is to explore the antitumor effect of CQ on the esophageal squamous carcinoma cell line EC109, and the potential mechanism for this effect.
Materials and methods
Cells and reagents
EC109 human esophageal squamous cell carcinoma cells were obtained from the Institute of Rheumatism Immunity, Affiliated Hospital of North Sichuan Medical College (Nanchong, China). EC109 cells were grown in RPMI-1640 supplemented with 10% fetal bovine serum (both from Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), penicillin (100 U/ml) and streptomycin (100 µg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells were incubated in a humidified incubator in 5% CO2 at 37°C.
CQ was kindly provided by Dr Yibin Deng (Hormel Institute, University of Minnesota, Austin, MN, USA). RPMI-1640 was purchased from Hyclone; GE Healthcare Life Sciences. All primary antibodies, including light chain (LC)3 (cat. no. 4108), p62/SQSTM1 (cat. no. 5114), phosphorylated (p)-mechanistic target of rapamycin (mTOR; cat. no. 2971), p-protein kinase B (Akt; cat. no. 9271), p-70S6K (cat. no. 9205), p-4E binding protein 1 (4EBP1; cat. no. 9451)and GAPDH (cat. no. 2118) were obtained from CST Biological Reagents Co., Ltd. (Shanghai, China).
MTT viability assay
The cells were seeded at a density of 2×103 per well in a 96-well plate and incubated for 24 h. CQ was added to each well in a concentration series of 50, 100, 150 and 200 µmol/l and incubated for a further 12, 24 and 36 h. A total of 20 µl MTT (5 mg/ml; Sigma-Aldrich) was added to each well and incubated at 37°C for 4 h, then 100 µl DMSO was added to each well to dissolve crystals and the plate was agitated for 10 min. Absorbance values at 490 nm were detected by a multi-mode detection platform (Molecular Devices Austria GmbH, Wals, Austria). Inhibition rate was calculated as [(Acontrol - Ablank) - (Atreated - Ablank)]/(Acontrol - Ablank)x100. Each experiment was assayed in triplicate.
Scratch assay
Cells were seeded in a 6-well plate at a density of 5×105 cells per well and cultured until the cells formed a confluent monolayer. A 10 µl pipette tip was used to scratch the cells and create a wound. The medium was removed and the cultures were washed twice with PBS to remove non-adherent cells. The cells were then incubated with complete RPMI medium, with and without 200 µmol/l. The migratory ability of the cells was determined according to the extent of closure of the scratch, observed at 0 and 24 h after incubation with CQ. Images were acquired using an inverted microscope (DMI 3000B; Leica microsystems GmbH, Wetzlar, Germany).
Transwell migration assay
Cells (6×104) in RPMI 1640 with 5% FBS were seeded into the upper chamber of polycarbonate membrane Transwell inserts (Corning Incorporated, Corning, NY, USA) with or without 200 µmol/l CQ. RPMI-1640 supplemented with 20% FBS was added to the lower chamber as a chemo-attractant solution. Following a 48-h incubation, the remaining cells on the upper chamber surface were removed, and the cells that had migrated to the underside of the chamber were fixed in methanol and stained with 10% Giemsa for 10 min at room temperature. The cell migration ability was determined by the number of cells that had migrated to the lower side of the filter. These experiments were repeated three times, and the filters were photographed under an inverted microscope.
Soft agar colony-forming assay
To assess the transformation ability of EC109 cells upon CQ treatment, 8×103 cells were suspended in complete RPMI medium containing 0.6% soft agar, seeded in triplicate on 60 mm dishes pre-coated with 3.0% agar in complete growth medium, and incubated at 37°C in a 5% CO2 humidified atmosphere with and without 200 µmol/l CQ. The culture medium was replaced every 4 days. Following an incubation of 18 days, colonies were photographed using an inverted microscope.
Green fluorescent protein (GFP)-LC3 transfection
EC109 cells were transfected with a GFP-LC3 plasmid using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and incubated for 24 h and treated with and without CQ. The images were obtained by fluorescence microscope.
Western blot analysis
Cells treated with or without 200 µmol/l CQ were lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) containing proteinase inhibitors, and lysates were collected. The protein samples were detected using SDS-PAGE (8–15% gel). A total of 40 µg of protein was loaded into each lane of the SDS-PAGE gel and transferred onto a polyvinyl difluoride membrane. Protein was quantified using a BCA assay. Membranes were blocked in 5% fat-free milk for 1 h at room temperature, and probed with the previously specified primary antibodies (dilution 1:1,000) overnight at 4°C, followed by horseradish peroxidase-conjugated secondary antibodies (1:5,000; Boster Biological Technology, Pleasanton, CA, USA) and incubated for 1 h at room temperature. Bands were visualized using an enhanced chemiluminscence kit (Merck KGaA).
Cell death assessment by flow cytometry
Cells were seeded at 5×105 cells per well in a 6-well plate and incubated with or without 200 µmol/l CQ for 24 h. Cells were harvested by trypsinization and washed with PBS. Subsequent to staining with Annexin V-fluorescein isothiocyanate for 15 min at 4°C and propidium iodide for 2 min at room temperature (Nanjing KeyGen BioTech, Co., Ltd., Nanjing, China), cells were detected using flow cytometry using FACSComp (version 4.1; Beckman Coulter, Inc., Brea, CA, USA).
Statistical analysis
The results are expressed as the mean ± standard deviation and t-tests or one-way analysis of variance followed by a least significant difference t-test were performed using SPSS (version 13.0; SPSS, Inc., Chicago, IL, USA) statistical software. P<0.05 was considered to indicate a statistically significant difference.
Results
Proliferation, migration and colony formation abilities of EC109 are inhibited by CQ
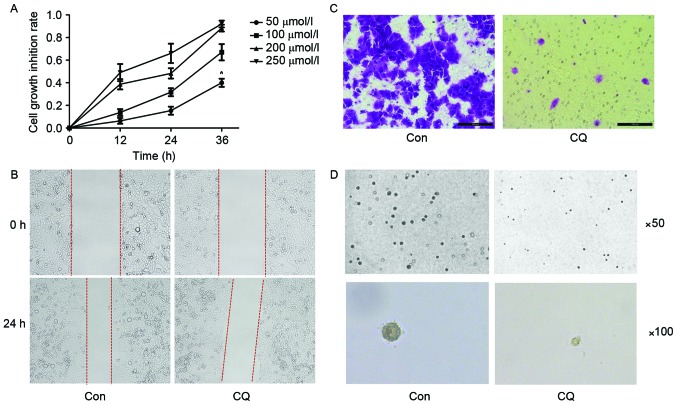
An increasing number of studies have described the anti-tumor effect of CQ. Thus, the first aim of the present study was to explore whether CQ affected the growth, migration or colony-forming ability of EC109 cells. According to the results of the MTT assay, CQ suppressed the viability of EC109 cells in a time and dose-dependent manner (Fig. 1A). The scratch assay indicated that the migration ability of EC109 cells was inhibited after a 24 h CQ treatment (Fig. 1B). A Transwell migration assay was performed for the same purpose, and a similar result was obtained (Fig. 1C). The colony formation ability of EC109 cells was evaluated using a soft agar assay; it was revealed that the number and size of colonies were reduced in the CQ group compared with the control group (Fig. 1D).
Figure 1.
CQ treatment induced the inhibition of the proliferation, migration and colony-formation ability of EC109. (A) Cell viability detected by an MTT assay, displaying the dose- and time-dependent effect of CQ treatment on EC109 cells. Effect of CQ treatment on cell migration illustrated by (B) a scratch assay (magnification, ×100) and (C) a Transwell assay (magnification, ×200). (D) The colony-formation ability of EC109 cells as evaluated on soft agar. Each experiment was performed three times, independently. *P<0.05 vs. CON (determined by unpaired t-test). CQ, chloroquine; CON, control.
Cell death is induced by CQ in EC109 cells
The results of flow cytometry demonstrated that the cell death ratio in the CQ group was significantly increased compared with the control group (P<0.05; Fig. 2).
Figure 2.
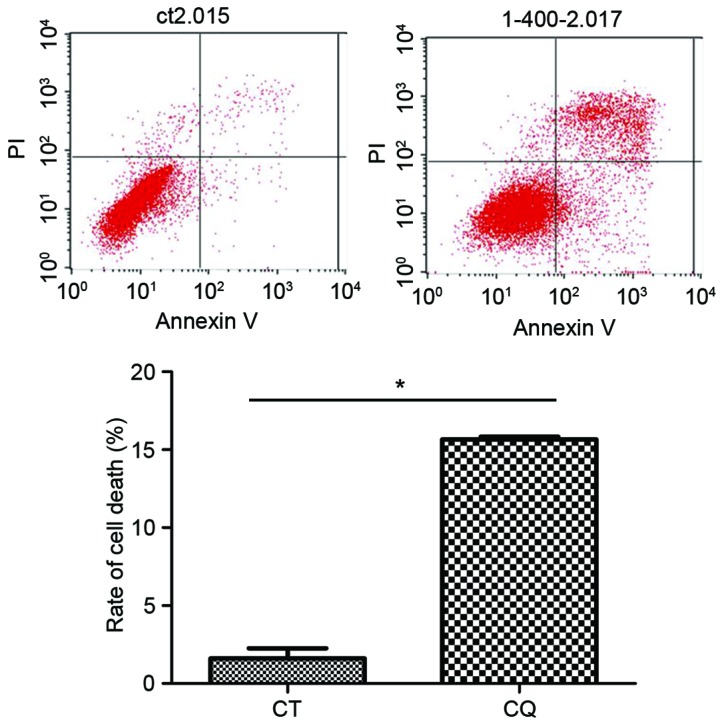
CQ initiation of autophagic cell death in EC109 cells as analyzed by Annexin V-fluorescein isothiocyanate/PI staining. *P<0.05 vs. CON (determined by one-way analysis of variance). CQ, chloroquine; PI, propidium iodide; CT, control.
CQ modulates autophagy through inducing the formation of autophagosomes and preventing the degradation of autophagosomes in EC109 cells
A number of studies have indicated that the induction of autophagy promoted tumor cell survival, whereas others have achieved the opposite result (16). Therefore, the present study aimed to explore whether CQ could modulate autophagy and influence the survival of EC109 cells.
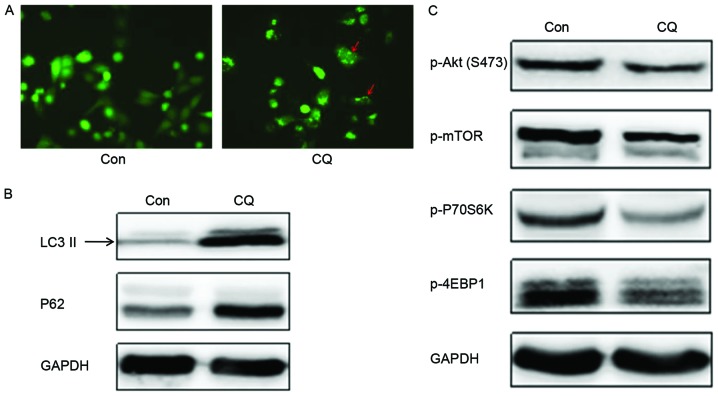
The GFP-LC3 fused protein translocates rapidly to nascent autophagosomes in a punctate distribution during the initiation of autophagy. Subsequent to the transfection of a GFP-LC3 plasmid into EC109 cells to monitor autophagosome formation by fluorescence microscopy, it was observed that the number of autophagosomes was evidently increased following CQ treatment (Fig. 3A). LC3 has two isoforms, LC3-I and LC3-II; LC3-I is cleaved by autophagy associated protein ATG4 and lipidated to form LC3-II during the initiation of autophagy. Thus, the expression of LC3-II is often used as a marker to evaluate the progression of autophagy (17). Western blot analysis revealed that CQ treatment markedly enhanced LC3-II expression (Fig. 3B), which indicated that CQ could induce the formation of autophagosomes in EC109 cells; this result corroborated the conclusion drawn from the GFP-LC3 transfection experiment.
Figure 3.
CQ modulation of autophagy through inducing the formation of autophagosomes and preventing the degradation of autophagosomes in EC109 cells. (A) Fluorescence microscopy analysis of the GFP-LC3 translocation in EC109 cells with and without CQ (magnification, ×200). Red arrows indicated autophagosomes. Western blot analysis for (B) LC3-II and p62 and (C) phosphorylated Akt/mTOR signaling pathway protein members. GAPDH was used as a loading control. CQ, chloroquine; GFP, green fluorescent protein; LC3, light chain 3; mTOR, mechanistic target of rapamycin; Con, control; p-, phosphorylated-; 4EBP1, 4E binding protein 1.
Another autophagy marker, p62, is sequestered within autophagosomes and then degraded by lysosomes during the last stage of autophagy. The present study demonstrates that the expression of p62 was upregulated following CQ treatment compared with the control (Fig. 3B). Collectively, these results demonstrate that CQ inhibited autophagy by preventing autophagosome degradation at the last stage of autophagy, despite inducing autophagosome formation during the initiation of autophagy.
Based on the results outlined in the previous section, it is hypothesized that CQ could suppress the growth and proliferation of EC109 cells in a time- and dose-dependent manner and reduce their migration and colony formation ability by preventing autophagosome degradation. In order to elucidate the mechanism for this process as induced by CQ, the expression of key proteins in the Akt/mTOR signaling pathway, which serves an important role in promoting cell growth and proliferation and is associated with the regulation of autophagy, was assessed. Western blotting demonstrated that the expression of autophagic negative regulatory factors, p-Akt and p-mTOR, decreased on CQ treatment. This suggests that autophagosome formation was initiated. The phosphorylation levels of the downstream proteins, including the cell translation regulation factors, p70S6K and 4EBP1, also decreased with CQ treatment (Fig. 3C), and therefore, CQ may initiate the formation of autophagosomes, but inhibit the degradation of the autophagosome in the final stage of autophagy.
Discussion
Esophageal carcinoma is one of the most common types of malignant neoplasm and a leading cause for cancer-associated mortality worldwide (1). Radiotherapy and chemotherapy are the most common treatments for patients with advanced esophageal carcinoma, with cisplatin, fluorouracil, oxaliplatin and capecitabine being the most commonly used chemotherapy agents (18). These drugs gradually induce chemoresistance and side effects due to the higher and longer dosages required. Developing an effective drug may cost $800 million and 15 years (19), therefore, a more effective way to solve the problem may be to develop new chemotherapeutics from existing drugs.
Over the past ten years, autophagy inhibition has been a popular topic in the field of cancer therapy. Autophagy is an important regulatory mechanism involved in cell growth, maturation and death, functions which are associated with a number of diseases, including cancer. Autophagic activity in the context of cancer is a double-edged sword: It degrades damaged organelles and recycles macromolecules to maintain a stable cellular environment, preventing tumor formation (20), but autophagy may also contribute to tumorigenesis by helping cancer cells to survive in response to growth-limiting conditions, including nutrient deficiency, absence of growth factors, hypoxia and exposure to cytotoxic drugs (6). A recent study revealed that in an increasing proportion of cases, chemotherapy failure is due to drug-initiated autophagy, which eventually induces chemoresistance (21).
As an inhibitor of autophagy, a number of studies have suggested CQ as a promising approach for cancer therapy as it is relatively safe and inexpensive (7,22,23). However, the specific anti-tumor mechanisms of CQ remain uncharacterized. In order to explore whether CQ could be used as a potential chemotherapeutic agent for the treatment of esophageal carcinoma, it was used to treat EC109 esophageal squamous cell carcinoma cells. The MTT assay results indicated that CQ was able to suppress the proliferation of EC109 cells in a time and dose-dependent manner, and the migration ability of the cells was also inhibited, as demonstrated using scratch and Transwell chamber tests. Furthermore, CQ suppressed colony formation on soft agar, which indicated that the transformation ability of EC109 cells was impaired. Collectively, these findings demonstrated that CQ may have an antitumor effect on EC109 cells.
In order to determine whether autophagy was involved in the antitumor activity of CQ, autophagosome formation was monitored by transfecting EC109 cells with a GFP-LC3 plasmid, and observing the expression of LC3 by fluorescence, and LC3-II and p62 by western blot analysis. As the substrates of autophagic flux, LC3 and p62 are involved in the process of autophagosome-lysosome fusion, which can be used to evaluate the formation and degradation of the autophagosome (24). Results demonstrate that CQ treatment increased the expression of LC3 and p62, indicating that autophagosome formation was increased, and autophagosome degradation was markedly inhibited, subsequent to CQ treatment.
To investigate the signaling mechanism of CQ-mediated autophagy in EC109 cells, the Akt/mTOR pathway, which serves a central role in regulating cell growth, was studied. The over-activation of this pathway contributes to cancer development and progression; Akt positively regulates the phosphorylation and activation of mTOR, which is a sensor of nutrient levels to promote cell growth (25). Furthermore, the Akt/mTOR pathway has been demonstrated to be involved in the regulation of autophagy; the inhibition of mTOR induces autophagy via the phosphorylation and inactivation of Unc-51-like autophagy activating kinase 1, a serine/threonine kinase that initiates autophagy (26). mTOR exists in two cellular complexes, mTORC1 and mTORC2 (27). mTORC1 phosphorylates S6K1 and 4E-BP1, which contribute to translation, protein synthesis and cell growth. mTORC2 regulates the phosphorylation of Akt. In the present study, the expression of p-mTOR, p-Akt, p-p70S6K1 and p-4EBP1 were markedly downregulated following CQ treatment.
The increased LC3-II expression and the inhibition of the Akt/mTOR pathway indicated that CQ induced autophagic flux and facilitated the formation of autophagosomes. However, the upregulation of p62 indicated that CQ also inhibited the degradation of the autophagosomal contents. Taken together, these results demonstrate that, as an inhibitor of autophagy, CQ induced the formation of the autophagosome, but inhibited the degradation of autophagosome in the last stage of autophagy. The overall effect was autophagic cell death activation by CQ, as confirmed by flow cytometry. Liu et al came to a similar conclusion regarding cervical cancer by incubating SiHa cells with hydroxychloroquine, the hydroxylated analog of CQ (28); another study suggested that CQ suppressed pancreatic cancer growth independent of TP53/TRP53 status by inhibiting autophagy (15).
Based on the data of the present study, CQ exerted an antitumor effect on EC109 through the modulation of autophagy, suggesting that CQ may exhibit promising therapeutic benefits for the treatment of esophageal squamous cell carcinoma.
Acknowledgements
The authors would like to thank Professor Yibing Deng (Hormel Institute, University of Minnesota, Austin, MN, USA) and Professor Minghui Yang (the Institute of Rheumatism Immunity of Affiliated Hospital of North Sichuan Medical College, Nanchong, China). The present study was funded by the Natural Science Foundation of the Education Department of Sichuan Province (grant no. 16ZA0224) and the Sichuan Province Special Project for the Youth Team of Science and Technology Innovation (grant no. 2015TD0029).
References
- 1.Stewart BW, Wild CP. World cancer report 2014. lyon cedex. France: The international agency for research on cancer; [Google Scholar]
- 2.Lin Y, Totsuka Y, Shan B, Wang C, Wei W, Qiao Y, Kikuchi S, Inoue M, Tanaka H, He Y. Esophageal cancer in high-risk areas of China: Research progress and challenges. Ann Epidemiol. 2017;27:215–221. doi: 10.1016/j.annepidem.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Yu L, Gu C, Zhong D, Shi L, Kong Y, Zhou Z, Liu S. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014;355:34–45. doi: 10.1016/j.canlet.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XJ, Chen S, Huang KX, Le WD. Why should autophagic flux be assessed? Acta Pharmacol Sin. 2013;34:595–599. doi: 10.1038/aps.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M, Zhang S, Wang M, Xiao G, Liao H. Autophagy promotes paclitaxel resistance of cervical cancer cells: Involvement of Warburg effect activated hypoxia-induced factor 1-α-mediated signaling. Cell Death Dis. 2014;5:e1367. doi: 10.1038/cddis.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ODonovan TR, OSullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011;7:509–524. doi: 10.4161/auto.7.5.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon VR, Lee H. Chloroquine and its analogs: A new promise of an old drug for effective and safe cancertherapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 8.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanotto-Filho A, Braganhol E, Klafke K, Figueiró F, Terra SR, Paludo FJ, Morrone M, Bristot IJ, Battastini AM, Forcelini CM, et al. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Lett. 2015;358:220–231. doi: 10.1016/j.canlet.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 11.Golden EB, Cho HY, Jahanian A, Hofman FM, Louie SG, Schönthal AH, Chen TC. Chloroquine enhances temozolomide cytotoxicity in malignant gliomas by blocking autophagy. Neurosurg Focus. 2014;37:E12. doi: 10.3171/2014.9.FOCUS14504. [DOI] [PubMed] [Google Scholar]
- 12.Mei L, Chen Y, Wang Z, Wang J, Wan J, Yu C, Liu X, Li W. Synergistic anti-tumour effects of tetrandrine and chloroquine combination therapy in human cancer: A potential antagonistic role for p21. Br J Pharmacol. 2015;172:2232–2245. doi: 10.1111/bph.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefort S, Joffre C, Kieffer Y, Givel AM, Bourachot B, Zago G, Bieche I, Dubois T, Meseure D, Vincent-Salomon A, et al. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy. 2014;10:2122–2142. doi: 10.4161/15548627.2014.981788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrow JM, Yang JC, Evans CP. Autophagy as a modulator and target in prostate cancer. Nat Rev Urol. 2014;11:508–516. doi: 10.1038/nrurol.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang A, Kimmelman AC. Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy. 2014;10:1683–1684. doi: 10.4161/auto.29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White E, Mehnert JM, Chan CS. Autophagy, metabolism, and cancer. Clin Cancer Res. 2015;21:5037–5046. doi: 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 19.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: New estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Li X. Autophagy inhibition enhances sensitivity of endometrial carcinoma cells to paclitaxel. Int J Oncol. 2015;46:2399–2408. doi: 10.3892/ijo.2015.2937. [DOI] [PubMed] [Google Scholar]
- 22.Balic A, Sørensen MD, Trabulo SM, Sainz B, Jr, Cioffi M, Vieira CR, Miranda-Lorenzo I, Hidalgo M, Kleeff J, Erkan M, Heeschen C. Chloroquine targets pancreatic cancer stem cells via inhibition of CXCR4 and hedgehog signaling. Mol Cancer Ther. 2014;13:1758–1771. doi: 10.1158/1535-7163.MCT-13-0948. [DOI] [PubMed] [Google Scholar]
- 23.Fan C, Wang W, Zhao B, Zhang S, Miao J. Chloroquine inhibits cell growth and induces cell death in A549 lung cancer cells. Bioorg Med Chem. 2006;14:3218–3222. doi: 10.1016/j.bmc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Gao C, Yao S, Xie B. Blocking autophagic flux enhances matrine-induced apoptosis in human hepatoma cells. Int J Mol Sci. 2013;14:23212–23230. doi: 10.3390/ijms141223212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8:e55096. doi: 10.1371/journal.pone.0055096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Luo XY, Jiang H, Yang MH, Yuan GH, Tang Z, Wang H. Hydroxychloroquine facilitates autophagosome formation but not degradation to suppress the proliferation of cervical cancer SiHa cells. Oncol Lett. 2014;7:1057–1062. doi: 10.3892/ol.2014.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]