Abstract
CD44 is a widely-distributed type I transmembrane glycoprotein that binds hyaluronic acid (HA) in most cell types, including primary tumor cells and cancer-initiating cells and has roles in cell migration, cell-cell, and cell-matrix adhesion. HA-derived conjugates and nanoparticles that target the CD44 receptor on cells have been reported for targeted delivery of therapeutics and imaging agents. Altering crucial interactions of HA with CD44 active sites holds significant importance in modulating targeting ability of hyaluronic acid to other cancer types that do not express the CD44 receptor or minimizing the interaction with CD44+ cells that are not target cells. The approach adopted here was deacetylation of the N-acetyl group and selective sulfation on the C6-OH on the HA polymer, which form critical interactions with the CD44 active site. Major interactions identified by molecular modeling were confirmed to be hydrogen bonding of the C6-OH with Tyr109 and hydrophobic interaction of the N-acetyl group with Tyr46, 83 and Ile 92. Modified HA was synthesized and characterized and its interactions were assessed by in vitro and molecular modeling approaches. In vitro techniques included flow cytometry and fluorescence polarization, while in silico approaches included docking and binding calculations by a MM-PBSA approach. These studies indicated that while both deacetylation and sulfation of HA individually decrease CD44 interaction, both chemical modifications are required to minimize interaction with CD44+ cells. The results of this study represent the first step to effective retargeting of HA-derived NPs for imaging and drug delivery.
Graphical abstract
Introduction
Hyaluronic acid (HA) is a linear, non-sulfated, and negatively charged polysaccharide that is comprised of {β1→3} N-acetylglucosamine (GlcNAc) and {β1→4} glucuronic acid (GlcUA) units. HA, an integral part of the extracellular matrix (ECM),1–3 contributes to lubrication of joints,4 cell migration5 during embryonic morphogenesis,2 cell adhesion, tumor cell proliferation,2,3,6 metastasis, angiogenesis, tissue regeneration, leukocyte trafficking, and progression of inflammation and cancer.2,3,7–13 The native ligand for HA is the transmembrane receptor CD44.14–16 HA binds to the N-terminus of CD44, which functions as the docking site and is lined by a mixture of primarily basic and hydrophobic amino acids.17 The CD44 gene contains 20 exons, 10 of which can be regulated by alternative splicing leading to generation of other splice variants (variant or ‘v’ exons), which are translated to a polypeptide of molecular weight 80–90 kDa depending on the splice variant.18 Biological functions, such as cell migration, adhesion, and structural integrity during anti-inflammatory processes, rely on HA-CD44 interaction.10,11,19–21 The smallest CD44 isoform, CD44 standard (CD44s), is ubiquitously expressed, whereas the variant isoforms are expressed in a few epithelial tissues and in cancers.10,22,23 The expression of variants of CD44 is heterogeneous among progression of various tumors. For example, CD44v10 and v3 are significantly associated with head and neck squamous cell carcinoma (HNSCC) primary tumors, and were shown to stimulate cell growth, proliferation and over-expression of metalloproteinases (MMP), whereas high expression of CD44v4-9 is observed in other tumors.24–33 HA is also known to bind to other receptors such as RHAMM and TSG-6. Major interactions of RHAMM involve association of CD44 for binding34, whereas for TSG-635 the binding events are controlled by a small 45 amino acid binding domain, as compared to large 160 amino acid domain in CD44. Recognition of hyaluronic acid by CD44 regulates various downstream pathways, e.g. activating/inhibiting phosphorylation of tyrosine kinases, activation of Nanog, which leads to overexpression of MDR1/P-gp gene, phosphorylation of c-Jun n-terminal kinases (JNK), and activation of GSK3β.36–44 The downstream events are triggered when CD44 recognizes certain key moieties in the hyaluronic acid polymer chain,38 which include hydrophobic interactions between the N-acetyl group of HA with the phenyl ring of Tyr83, the side chain of Ile92 and the disulfide bond between Cys81 and Cys101 of CD44; water-mediated hydrogen bonding between the carboxylate group of HA with Tyr46 and Arg45 of CD44; hydrogen bonding between the C6-hydroxy group of HA with Tyr109 of CD44, which serves to lock HA to the CD44 active site; and hydrogen bonding between the vicinal diols of HA with guanidine groups of Arg45 and Arg82.17 These interactions are summarized in Figure 1. Desirable features of CD44-HA binding are the absence of ionic interactions and strong prevalence of a hydrophobic core.17 The molecular weight (MW) of HA is also known to play a crucial role in the recognition and binding to the CD44 protein in the ECM.45
Figure 1.
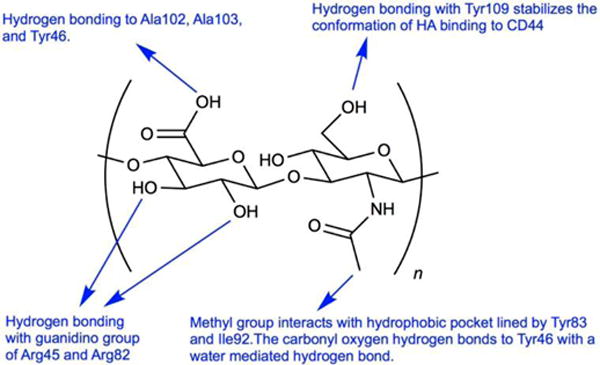
Interactions of {β1→3} N-acetylglucosamine (GlcNAc) and {β1→4} glucuronic acid (GlcUA) repeat unit of hyaluronic acid with crucial amino acids in the murine CD44 active site.
Recently, several groups have attempted to alter the properties of HA by chemical modification. The modified HA target other proteins and enzymes and regulate CD44-independent biological processes. One such chemical modification that has been extensively explored is the sulfation of HA. Sulfated HA showed selective binding, as measured by surface plasmon resonance (SPR), for isoform 165a of vascular endothelial growth factor (VEGF165a)46 and sclerostin, a secreted glycoprotein that has an integral role in bone biology.47 Integration of in silico (molecular docking and dynamics simulations) and in vitro SPR studies showed that binding to sclerostin was dependent on the degree and pattern of HA sulfation. Others showed that increased sulfation of HA resulted in higher binding affinity to TGF-β1 compared with native HA or chondroitin sulfate.48
In the current study, HA was rationally modified to investigate the potential to minimize its interaction with CD44. Our group has previously observed the uptake of HA-derived nanoparticles (NPs) into CD44-expressing tumors and organs, e.g. liver and speen.49–51 Liver cells express stabilin-2 (sub-family of CD44) and spleen cells have high expression of CD44.52 Similar results were observed by Lin et al.53, where HA-IR-780-based nanosystems accumulated largely in liver and spleen. HA-functionalized glycyrrhetinic acid nanoparticles accumulate in major CD44 clearance organs – liver and spleen54. This demands new approaches to design polymers by precisely tuning their structure to achieve maximum tumor accumulation by reducing CD44 interactions nontarget organs. Ultimately, we postulate that NPs derived from structurally-modified HA could have decreased uptake in liver and spleen and/or could be retargeted by conjugation of other targeting ligands to specific receptors with the end goal of increasing accumulation of NPs in tumor. By chemically modifying HA, a wide spectrum of binding partners can be exploited in rationally designing a robust drug/dye delivery system for enhanced tumor recognition. The investigation reported here studies HA-CD44 binding after HA deacetylation, sulfation, and a combination thereof. We hypothesized that deacetylating the HA backbone as one modification, selectively sulfating the C6-OH as a second modification, and incorporation of both deacetylation and sulfation as a third modification, would decrease the binding of these modified derivatives of HA to CD44. Interactions of the modified HA with CD44 were measured using flow cytometry, fluorescence polarization, and in silico approaches.
Materials and Methods
Materials
Sodium hyaluronate, 10 kDa, was purchased from Lifecore Biomedical (Chaska, MN). Tetrabutylammonium (TBA) hydroxide, DOWEX 50WX8-400 ion exchange resin, sulfur trioxide pyridine complex (SO3-pyridine 98%), hydrazine, hydrazine sulfate, N-(3-Dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS) and fetal bovine serum (FBS) were purchased from Sigma-Aldrich (St. Louis. MO). All water was purified with Barnstead™ Nanopure™ Diamond system (Thermo Scientific; Waltham, MN). Methanol, N,N-dimethylformamide (DMF), 96-well tissue culture plates (Falcon), 12-well tissue culture plates and dialysis tubing (MWCO = 3500) were purchased from Fisher Scientific (Pittsburgh, PA). Ethanol was purchased from the Warner-Graham Company (Cockeysville, MD). Recombinant human CD44-Fc Tag (HPLC–verified) was purchased from Acrobiosystems. Ninhydrin reagent and hydridantin dehydrate 96% and fluoresceinamine isomer I, 99% were purchased from ACROS Organics and 2-methoxyethyl acetate was obtained from TCI America. Anti-CD44 antibody was purchased from BD Pharmigen BD Biosciences. NMR was performed on a 500 MHz Bruker or 600 MHz Varian system using a 5 mm probe at room temperature. Deuterated water (D2O, 99.9% D) was purchased from Cambridge Isotope Laboratories. FTIR measurements were performed on Nicolet IR200 FT-IR instrument using single-reflection ZnSe ATR crystal. Penicillin/streptomycin (100× solution) was purchased from Corning. Cell lines (PC-3, MDA-MB-231, RKO and LNCaP cells) were obtained from American Type Culture Collection (Manassas, VA) and were grown in RPMI-1640 (HyClone, GE Healthcare Life Sciences) with 10% fetal bovine serum and 1% penicillin/streptomycin (P/S).
Preparation of modified HA derivatives
Modifications to HA were synthesized based on methods described in the literature46,55 with slight modifications as described below (Scheme 1).
Scheme 1.
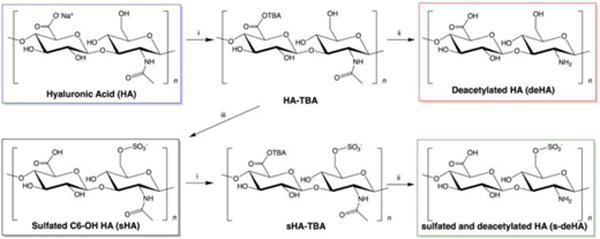
Synthesis of deacetylated, sulfated, and deacetylated and sulfated HA. i: ion exchange with TBA in H2O; ii: hydrazine, hydrazine sulfate, DMF; and iii: sulfur trioxide pyridine, DMF.
Synthesis of TBA salt of hyaluronic acid (HA-TBA)
Sodium hyaluronate (10 kDa, 250 mg) was dissolved in 100 ml ultrapure water, which was mixed with 5 g of DOWEX 50WX8-400 ion- exchange resin to allow the substitution of sodium ions with TBA. This mixture was stirred for 6–8 h for the effective exchange process. The reaction mixture was then filtered through a 0.45 μm membrane filter paper obtained from Millipore. This solution was then lyophilized to yield a white powder with a yield of 90% (HA-TBA).
Synthesis of deacetylated HA-TBA (deHA-TBA)
HA-TBA (200 mg) was dissolved in a three-neck round bottom flask with 20 mL DMF under argon flow at room temperature (rt) to aid the formation of a homogenous mixture. Hydrazine (20 mL) and hydrazine sulfate (200 mg) were added to the mixture and the reaction was allowed to proceed for 8 h under an atmosphere of argon at 100°C. After the reaction was complete, the mixture was cooled to rt and the pH was adjusted to 10 using 4.0 M aqueous NaOH. The mixture was dissolved in 100 mL of 1:1 acetone:water, which was then extensively dialyzed (MWCO = 3500 Da, Spectrum Laboratories) against 1:1 methanol:water for 24 h, followed by 1:1 ethanol:water for 24 h with 4 changes and finally 8 changes of water over 72 h to remove unreacted TBA. The material obtained had a yield of 24% and was then lyophilized and stored at −20°C.
Selective sulfation of hyaluronic acid at C6 position (sHA)
HA-TBA (200 mg) was dissolved in a three-neck round bottom flask with 20 mL DMF under argon for 15 min. Sulfur trioxide pyridine complex (0.325 g) was dissolved in 4 mL of DMF and added dropwise to the reaction mixture. The temperature was maintained at rt for 40 min, and then the reaction was quenched by adding 20 mL of ultrapure water. Acetone was added to the reaction mixture to precipitate the product, which was then filtered and washed with acetone and water. The product was dissolved in water and dialyzed against 1:1 ethanol:water for 24 h with 4 changes followed by 48 h of ultrapure water with 8 changes. This solution was then lyophilized yielding an off-white solid with 82% yield that was stored at −20°C.
Synthesis of deacetylated and sulfated hyaluronic acid (s-deHA)
Sulfated HA-TBA, as prepared above, was utilized for the preparation of deacetylated and sulfated hyaluronic acid. Sulfated hyaluronic acid-TBA compound (200 mg) was dissolved in 20 mL of DMF under argon for 15 min in a three-neck round bottom flask. Hydrazine (20 mL) and hydrazine sulfate (200 mg) were added to the reaction under argon. The reaction mixture was stirred for 8 h at 100°C. It was then allowed to cool to rt and its pH was adjusted to 10 using 4.0 M NaOH. The mixture was then dissolved and dialyzed as described for deHA-TBA above, lyophilized, and stored at −20°C (yield = 34%).
Deacetylation quantification
The colorimetric ninhydrin assay was used to quantify the primary amine produced by deacetylation of the N-acetyl-D-glucosamine monosaccharide. Standard solution (0.1 mg/mL) of glucosamine was used to build the standard curve in acetate buffer (0.5 mL; pH 5.5; 4 M). deHA (0.5 mg) and s-deHA (0.5 mg) were used for the assay. The ninhydrin reagent was prepared by mixing 1 g ninhydrin, 0.12 g hydridantin, 23 mL of 2-methoxyethyl acetate with 12.8 mL of 4 N acetate buffer. The total volume of the sample was 2 mL, and this solution was heated in a boiling water bath for 15 min. The reaction mixture was cooled subsequently to rt followed by measuring the absorbance at 570 nm. Absorption and spectra were recorded on Evolution 220 spectrophotometer (Thermo Scientific) in quartz cells with the optical path length of 10.0 mm at 25°C.
Synthesis of hyaluronic acid conjugate with fluoresceinamine (HA-FITC)
Sodium hyaluronate (90–95 mg, Mn = 10–20 kDa) was dissolved in 20 mL of ultrapure water in a 50 mL round bottom flask. Fluoresceinamine (8 mol %) was dissolved in 10 mL DMSO under constant stirring. NHS and EDC, 96–132 mmol (10× molar excess to fluoresceinamine) were dissolved into HA solution and stirred for 30 mins to allow activation of carboxylic groups on HA. The fluoresceinamine DMSO solution was then added dropwise to HA solution under constant stirring. The reaction mixture was stirred for 24 h at rt, wrapped in foil to prevent light exposure. The reaction mixture was then dialyzed against 1:1 ethanol:water for 48 h with 4 changes followed by water alone for 72 h with 8 changes. The product was lyophilized and stored at −20°C with a yield of 68%.
Flow cytometry analysis for binding assays
A cellular binding assay was performed using HA-FITC as the competing probe. MDA-MB-231, PC-3, RKO and LNCaP cells, approximately 90,000 cells/well, were seeded in a 12-well plate. The cells were pre-incubated with sHA, deHA, s-deHA and HA (200 μM) for 1 h at rt with constant shaking. HA-FITC (20 μg/mL) was added to the mixture for 1 h under constant shaking in the dark as the competing ligand to allow for displacement of the unlabeled sample. Cells incubated with unlabeled HA served as the negative control, whereas cells incubated with HA-FITC alone served as the positive control. After incubation, the cells were diluted to 300 μL with PBS in FACS tubes. A FACS LSRII-green flow cytometer (BD) was used for all flow cytometry measurements. A total of 10,000 gated events were acquired per sample and the mean fluorescence intensity was plotted in a histogram-based graphical representation. Each data point is representative of the mean of three independent measurements on the flow cytometer. Data were analyzed with FlowJo (Tree Star) software.
Fluorescence polarization assay
Fluorescence polarization measurements were performed in a 384-well low-volume black round-bottom polystyrene NBS microplate (Corning) using a Spectromax M5 plate reader (Molecular Devices, Sunnyvale, CA, USA). Polarization values are reported as millipolarization units (mP). The fluorescent probe, HA-FITC, synthesized above and rhCD44-Fc were dissolved in 1× PBS. For direct binding assay between the fluorescent probe and the CD44 protein, 1 μL of 50 nM fluorescent probe and 10 μL of solution with increasing concentration of recombinant CD44 protein (130 nM-72 μM) in PBS were transferred to the microplate wells. The final volume of the reaction mixture was 11 μL in each well, and all measurements were performed in triplicate. The microplate was shaken for 5 min in the dark at rt before being read by the plate reader (λex = 489 nm; λem = 538 nm). Estimation of binding of modified HA derivatives was carried out using a competitive binding assay. To each well in the 384-well plate, 10 μl of 23.4 μM recombinant CD44 was added along with 1 μl of increasing concentration of unlabeled HA derivatives (64 nM - 1000 μM) and 1 μl of fluorescent probe (HA-FITC, 50 nM). The total volume of the reaction mixture was 12 μL in each well. Fluorescence measurements were made after a 15 min incubation. The data was fitted and IC50 values were determined using a non-linear least squares fit to a single site binding model (Graphpad v7.0).
Molecular docking of hyaluronic acid derivatives
The X-ray diffraction crystal structure of mouse CD44 hyaluronan-binding domain was obtained from Protein Data Bank (PDB ID: 4MRD).17 There is similarity in binding of HA between the murine and human CD44 active site, which is conserved.17,56 Molecular docking was performed to gain deeper insight into the interaction of modified hyaluronic acid derivatives compared to native hyaluronic acid. The protein structure was cleaned by removing water molecules and other inorganic ions/atoms. The HA ligand consisting of 12 units was prepared in Maestro molecular modelling software (Schrodinger). Docking studies were performed using Auto-DockTools (AutoDock Vina).57 The grid box of dimensions 40 Å × 40 Å × 40 Å was generated with a 0.375 Å spacing to perform docking. AutoDock Tools were used to add Gasteiger charges and polar hydrogens to CD44 and modified hyaluronic acid derivatives. The structures were saved in .pdbqt file format. These systems were loaded in the graphic user panel and the grid pane, grid box, and the box dimensions were set accordingly to completely occupy the entire ligand-protein surface to perform docking using AutoDock vina software.58 10 runs were performed for statistical analysis on the obtained docking results. Docking was also performed using SwissDock (http://swissdock.vital-it.ch/),59,60 provided as a free web service. The ligands were docked to the desired protein structures by using suitable docking parameters allowing the online web service to produce docking results using CHARMM energies.60 Similar grid size box dimensions used in AutoDock were used for SwissDock. The docked ligand file was visualized with the protein using UCSF Chimera 1.4.61
Statistical Analysis
Analysis of competitive binding flow cytometry, fluorescence polarization, and molecular docking were performed with one-way ANOVA with Tukey multiple comparisons test, Student’s t test, and non-linear least squares fit to a single site binding model. All statistical analysis was done in Prism 7.0 (GraphPad Software; La Jolla, CA) and Microsoft Excel.
Results and Discussion
Synthesis and characterization of modified hyaluronic acid derivatives (deHA, sHA and s-deHA)
Deacetylated, sulfated, and deacetylated sulfated HA derivatives were prepared according to Scheme 1 using previously reported methods with slight modifications. Based on their importance for CD44-HA binding,17 the N-acetyl and the C6-OH moieties were chosen for synthetic modifications. We synthesized deacetylated, sulfated, and deacetylated sulfated derivatives of HA of molecular weight 10 kDa by controlling the amount of hydrazine and hydrazine sulfate for deacetylation reaction and molar ratios of sulfur trioxide-pyridine complex per repeat units of HA for selective sulfation reaction. The N-acetyl group was deacetylated using the hydrazine/hydrazine sulfate mixture following the Wolff-Kishner reaction under reflux conditions under an atmosphere of argon, cleaving the acetyl group with the initial evolution of gas to produce the amino group during the reaction. The degree of deacetylation of HA changed with the reaction times from 4 to 8 h. The 8 h reaction time was close to 100% deacetylation with complete absence of N-acetyl peak in the corresponding 1H NMR spectra (2–2.2 ppm) as seen in Figure S5 with reference to 1H and 13C NMR spectra of HA in Figures S6–7. The deacetylation reaction by hydrazinolysis has been reported to cause a reduction in overall molecular weight62 due to C-5 epimerization and β-elimination at the C-4 position leading to breakdown of the glycosidic bonds.63 We observed a lower yield of deacetylated HA presumably due to the above explanation. At the reaction temperature, the deacetylating agents, hydrazine and hydrazine sulfate, convert the free carboxylic group to carboxylic hydrazides. For this reason, the TBA salt was synthesized to avoid generation of side products. Ninhydrin colorimetric assay was used to quantify the degree of deacetylation of HA. The ninhydrin reaction is specific to the primary amino group generated during deacetylation reaction producing a distinct purple violet color which was quantified by absorption spectroscopy. 1H-NMR was also used to assess the degree of deacetylation, where one repeat unit of native HA bears 3 methyl protons for every 2 anomeric protons leading to the theoretical integral ratio between the two of 3:2, respectively. Using this ratio equation,64 the degree of deacetylation was calculated to be 82.6 and 88% for deHA and s-deHA obtained from Ninhydrin assay standard curve. Selective sulfation of HA was controlled by using 3:1 molar ratio of sulfur trioxide-pyridine complex to repeat units of HA. Major sulfation sites on HA are the primary hydroxyl group being the most reactive, while other (secondary) hydroxyl groups are less susceptible to the attack of the sulfating nucleophile moiety due to steric hindrance and spatial orientation of these groups on HA. 1H and 13C NMR analysis was performed on sHA and s-deHA to confirm the sulfation of HA. The reactive center C-6 OH linked to CH2 exhibited a downward shift to 3.55 ppm due to the increased electron withdrawing caused by the sulfate group. Absence of additional peaks between 3.9–4.4 confirmed no side products at positions C-2′, C-3′ and C-4′, indicating selective sulfation of the C-6 OH (Figures S1–S4). The FTIR data in Figure 2 demonstrates the peaks for vibrations of C-O-S and S=O in the range of 800 and 1290 cm−1, indicating the presence of sulfate moieties in sHA and s-deHA. These results were also confirmed by elemental analysis for sulfate content (Table S1). 13C NMR spectra confirmed that HA was sulfated at C6 position by the shift of the C-6 peak from 61.9 to 68 ppm with complete disappearance of the former shown in Figure S3. The surface charge for the modified polymers was assessed using zeta potential to further support the results from colorimetric assay and NMR spectroscopy. Deacetylation was responsible for introducing positive charges in the polymer, thereby shifting the zeta potential of highly negative HA to near neutral values. The s-deHA had a zeta potential of −6 mV, indicating counter balancing effects of both positive and negative charges induced due to deacetylation and sulfation (Figure 3). At the same time, sulfation lead to further negative charge.
Figure 2.
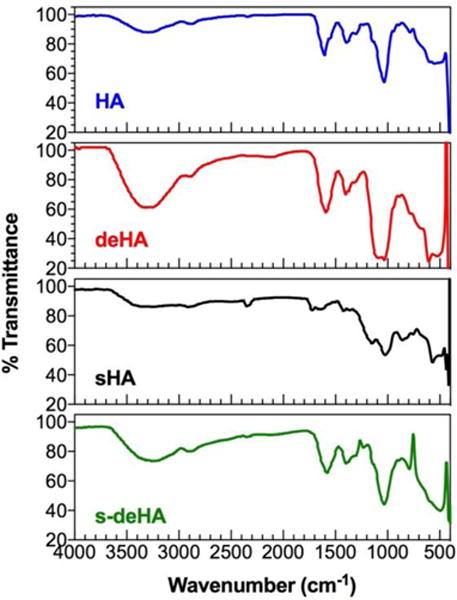
FTIR spectra of native and modified 10 kDa HA.
Figure 3.
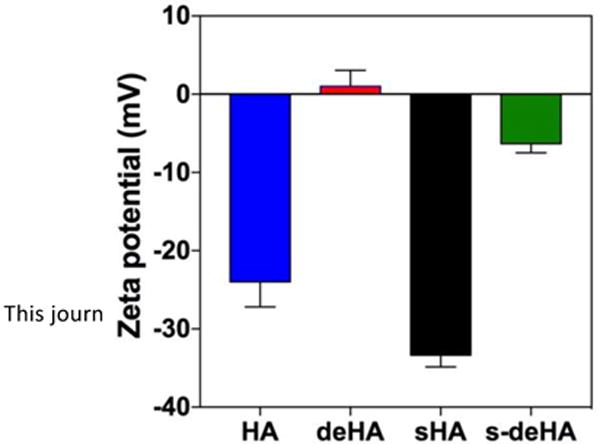
Zeta potential of HA and modified HA.
Flow cytometry analysis of binding of modified HA polymers to CD44
The in vitro binding affinity of modified hyaluronic acid polymers was studied using CD44-expressing cell lines (PC-3, MDA-MB-231 and RKO) and a CD44− cell line (LNCaP). PC-3 prostate, MDA-MB-231 breast, RKO colon cancer cells overexpress CD44 on their surface. The cell lines were first confirmed for CD44 expression using a PE-labelled anti-CD44 antibody. Analysis confirmed that the LNCaP prostate cancer cells did not express the CD44 receptor, while the other cell lines had pronounced CD44 expression (Figure 4).
Figure 4.
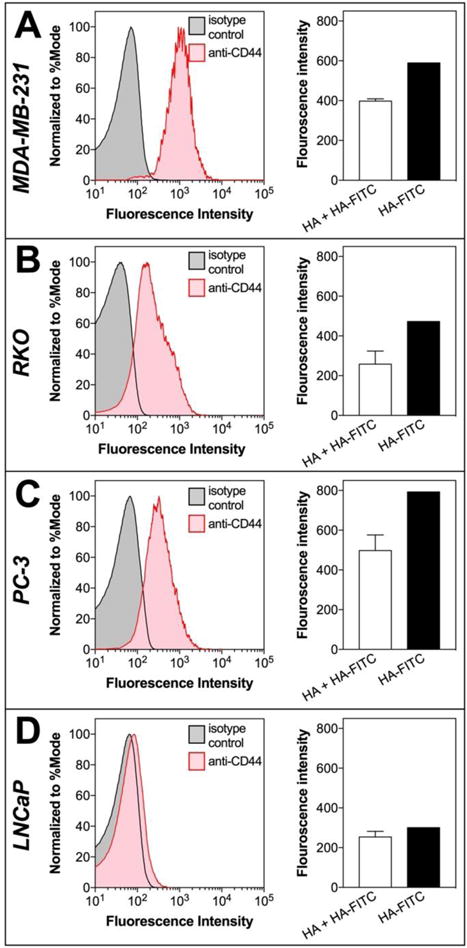
CD44 expression studies. Flow-cytometry histograms showing the expression of CD44 receptor (left plot) and the ability for HA to block HA-FITC binding (right plot) of CD44+ (A) MDA-MB-231, (B) RKO, (C) PC-3 and CD44− (D) LNCaP cell lines. HA competitively inhibited HA-FITC binding in CD44+ cell lines, while CD44- cell lines had lower HA-FITC signal and no difference in signal after competition with HA.
To study the interactions of modified HA with CD44+ and CD44− cell lines we used an assay based on the concept of competitive binding, where the modified HA polymers were initially pre-incubated with the cells for 1 h to allow their binding to the CD44 receptor on the cell surface. This was followed by incubation with HA-FITC for another 1 h to allow competition of the native ligand with the modified HA for the CD44 receptor. Cell staining was then analyzed using the FITC gate to allow acquisition of FITC fluorescence intensity over 10,000 events. In all CD44+ cell lines, HA-FITC effectively displaced the modified polymers, yielding a higher fluorescence intensity compared to the cells incubated with HA. When the cells were incubated with native unlabeled HA, the displacement by the HA-FITC was not effective compared to the modified HA polymers, indicating that unlabeled HA had a higher binding affinity than modified HA. To confirm the findings, the assay was performed using the CD44- cell line, LNCaP. Here, the overall fluorescence intensity for all HA derivatives was significantly lower compared to CD44+ cell lines, demonstrating little or no affinity for CD44− cells. Results are reported as both fluorescence intensity and fluorescence intensity for the modified HA polymers for individual cell lines (Figure 5). Fluorescence index (FI) values were calculated using the following equation, where MFI is mean fluorescence intensity:
Figure 5.
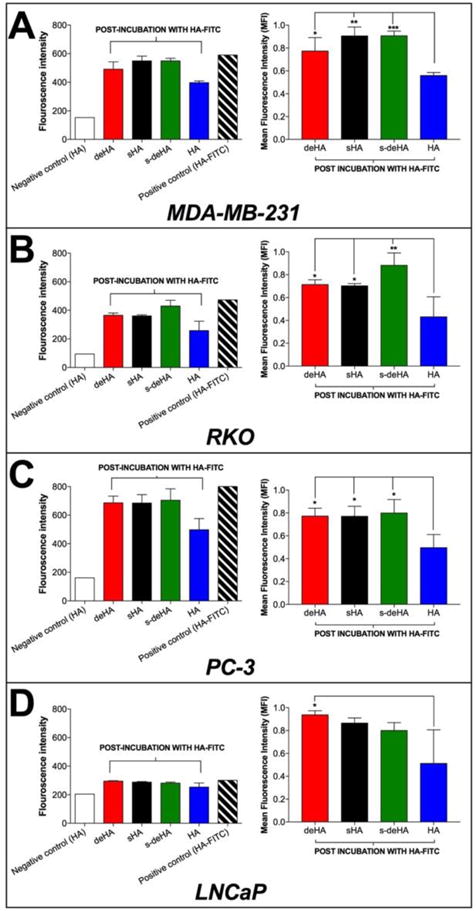
Competition binding assay using flow cytometry analysis. Binding of HA-FITC (20 μg/ml) for 1 hr in the presence of modified HA and native HA determined in (A) MDA-MB-231, (B) RKO, (C) PC-3, and (D) LNCaP cancer cell lines. Negative control comprised of incubation of cells with unlabeled HA while positive control cells were incubated only with HA-FITC. The left column indicates fluorescence intensities obtained after competition binding of modified HA polymers during flow analysis, while the right column indicates the normalized fluorescence intensities based on the obtained negative and positive fluorescence intensities. Data are shown as mean ± S.D., * p<0.05, ** p<0.01, *** p<0.001; One-way ANOVA with Tukey multiple comparisons test.
Interaction of modified HA polymers with CD44 recombinant protein studied by fluorescence polarization
The interactions between CD44 and modified HA polymers were studied using fluorescence polarization via a competitive binding assay method. Fluorescence polarization (FP) is a powerful technique used to study biomolecular interactions. FP assays require relatively small amounts of expensive reagents as they have been miniaturized to 384 and 1536 well formats. FP values in an assay are dependent on the rotational rate which correlates with the molecular weight of the fluorescent species. A low FP value indicates the presence of unbound fluorescent probe, which rotates rapidly, resulting in depolarization of light, whereas binding of the fluorescent probe to a high molecular weight protein or bio-macromolecule rotates the complex slowly, thereby yielding higher FP values. Fluorescence polarization is usually recorded in mP (millipolarization units). The advantage of using FP is that the protein and the fluorescent probe are not immobilized on the surface, therefore allowing no interference between the probe and the binding site domain of the protein. The binding affinity of HA for CD44 was determined by titrating HA-FITC probe (50 nM) with CD44 (130 nM - 72 μM) (Figure 6A). A dose-dependent increase in the FP values was observed in HA-CD44 binding while no such effect was observed with fluorescein as a probe, which is indicative of CD44-HA binding. Binding affinity (Kd) of HA-FITC was determined to be 21 μM using nonlinear least square fitting to a single-site binding model. This is consistent with studies reported in literature, where the binding affinity (Kd) of 24.6 μM was determined by isothermal titration calorimetry16 and SPR.65,66
Figure 6.
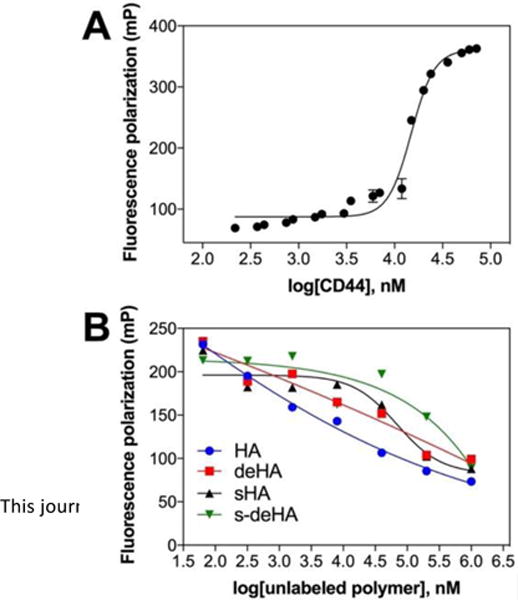
(A) Binding curve isotherm of HA-FITC (50 nM) to CD44-Fc protein (130 nM-72 μM). Kd value obtained for the conjugate = 21 μM. Values are expressed as mean ± s.d of n=3 (B) Effect of increasing concentration of unlabeled derivatives of HA (64nM- 1000μM) and constant CD44 concentration of 23.4 μM competing with pre-incubated HA-FITC (50 nM).
To evaluate the effect of HA modifications on CD44 binding, we conducted a series of competition assays. In each assay, a constant mixture of the HA-FITC and CD44 was titrated with modified HA. The IC50 values of unlabeled modified HA were determined using nonlinear least square fitting to a single-site binding model. The data shows that individual modifications (deacetlylation and sulfation) indeed reduce binding to CD44 (Figure 6B). Incorporating both modifications to HA results in reduced binding to CD44 compared to single modifications; specific values could not be obtained as the slopes were very broad for double modified HA. Based on these results we suggest that the secondary amino and sulfo groups on modified HA reduce the affinity of HA for CD44.
Molecular docking
Docking studies and binding energy calculations were used to predict the recognition of modified HA derivatives to CD44 receptor. Docking was carried out using modified HA derivatives and native HA with CD44 protein. Docking energies were obtained using Swissdock and Autodock docking software packages and compared to observe a trend in decrease of binding energies based on the modifications in the HA structure. The modified HA derivatives, deacetylated and sulphated HA, had lower binding energies of −8.7 kcal/mol and −7.6 kcal/mol, respectively, compared to −13.2 kcal/mol for native HA. This implies that the lower binding energies after the N-acetyl and the C6-OH group were chemically modified caused the differences in docking energies due to repulsive interactions of the amino group interactions and sulfo group. The repulsive interaction hindered locking of the HA structure on the active site of the protein, as indicated in Figure 7 and Table 1, consistent with AutoDock and SwissDock calculations. s-deHA had the lowest binding energy compared to single modifications. This implies that both modifications combined to make interaction with CD44 least favorable. Findings from FP indicate the order of affinity HA > deHA > sHA > s-deHA, whereas from molecular docking study we observe the order of HA > sHA > deHA > s-deHA. Both modifications decrease binding to CD44. It was observed that only one disaccharide unit is responsible for interaction with the active site, as illustrated in Figure 1. The described modifications on the disaccharide were observed to interfere with the major interactions which are key determining factors for HA and CD44 binding. Such structural changes, being introduced with the same substitution ratio, should have similar effect regardless of the MW of the polymer, as only its small fragment is responsible for the interaction. This provided the rationale of exploring only one lower molecular weight (10 kDa) of hyaluronic acid. In contrast, different substitutions ratios will have different effect on the binding affinity even with the polymer backbone of the same molecular weight. However, an important consideration should be kept in mind that hyaluronic acid belongs to family of natural carbohydrates67, where controlling the exact molecular weight and chemical substitution ratio is extremely challenging. Even with precise stoichiometric substitution ratio under control, it will be virtually impossible to control such modifications site-by-site. This will lead to the possibility for the less (or completely unmodified) fragments of the polymer to determine the interaction with CD44.
Figure 7.
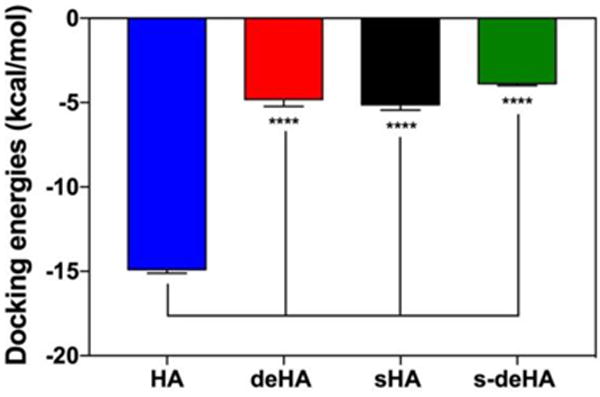
Docking energy calculated by Autodock software of HA and modified HA derivatives with CD44 indicate that each of the modifications had a lower binding energy compared to native HA with CD44. Graphs show mean ± SD (n=10), **** p < 0.0001; One-way ANOVA with Tukey multiple comparisons test.
Table 1.
Docking scores from Autodock software plotted indicating; One-way ANOVA with Tukey multiple comparisons test. (B) Docking energies of modified HA polymers using SwissDock and Autodock software.
| HA Derivative | Docking Energy (kcal/mol)
|
|
|---|---|---|
| SwissDock energies | Autodock energies | |
| HA | −11.87 | −13.2 |
| s-HA | −7.6 | −5.11 |
| deHA | −8.7 | −4.81 |
| s-deHA | −5.2 | −2.57 |
Conclusions
The reported study was focused on evaluating the effects of chemical modifications of hyaluronic acid on its binding affinity to standard CD44 glycoprotein receptor. Chemically modified HA was previously shown to selectively bind various proteins, e.g. TIMP-3, VEGF165a, FGF-2, sclerotin, and hBMP-4. Modifying HA structure, which alters CD44 interactions, could also potentially affect other receptors such as RHAMM and TSG-6, since these interactions are exquisitely sensitive to compromising the selectivity by altering the overall chemical structure. However, modifications such as deacetylation and sulfation of HA have not been thoroughly evaluated for their influence on CD44 binding. Here we have described a novel methodology for tuning HA properties to control its binding to CD44. Our in vitro and theoretical modeling studies have revealed good correlation of reduced binding with chemical modifications on HA. Future studies will evaluate higher molecular weight HA, since recognition of different molecular weight of HA and downstream activation pathways are different for the polymers of various MW.68,45,69,70 Further, we will investigate in vivo the role of HA modification on overall biodistribution, tumor accumulation, and pharmacokinetic profiles by developing nanosystems based on polymers designed to increase payload of imaging agents/drugs to tumors.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health grants R01 EB019449, P20 GM103480, Shared Instrumentation Grant 1S10RR17846 and 1S10RR027940, and the Fred and Pamela Buffett Cancer Center at UNMC (P30 CA036727). We thank the High-performance computing (Holland Computing Center) at University of Nebraska, Lincoln for docking studies. We thank Ms. Megan Holmes for technical assistance on cell experiments.
Footnotes
References
- 1.Lapcík L, Jr, Lapcík L, De Smedt S, Demeester J, Chabrecek P. Hyaluronan: Preparation, Structure, Properties, and Applications. Chem Rev. 1998;98(8):2663–2684. doi: 10.1021/cr941199z. [DOI] [PubMed] [Google Scholar]
- 2.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4(7):528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 3.Toole BP. Hyaluronan in morphogenesis. Semin Cell Dev Biol. 2001;12(2):79–87. doi: 10.1006/scdb.2000.0244. [DOI] [PubMed] [Google Scholar]
- 4.Gotoh S, Onaya J, Abe M, Miyazaki K, Hamai A, Horie K, Tokuyasu K. Effects of the molecular weight of hyaluronic acid and its action mechanisms on experimental joint pain in rats. Ann Rheum Dis. 1993;52(11):817–822. doi: 10.1136/ard.52.11.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2(11):793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 6.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine M Lou, Calabro A, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106(3):349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorokin L. The impact of the extracellular matrix on inflammation. Nat Rev Immunol. 2010;10(10):712–723. doi: 10.1038/nri2852. [DOI] [PubMed] [Google Scholar]
- 8.Joyce Ja, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lennon FE, Singleton Pa. Hyaluronan regulation of vascular integrity. Am J Cardiovasc Dis. 2011;1(3):200–213. [PMC free article] [PubMed] [Google Scholar]
- 10.Ponta H, Sherman L, Herrlich Pa. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 11.Toole BP, Wight TN, Tammi MI. Hyaluronan-cell interactions in cancer and vascular disease. Journal of Biological Chemistry. 2002:4593–4596. doi: 10.1074/jbc.R100039200. [DOI] [PubMed] [Google Scholar]
- 12.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6(7):508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 13.Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y, et al. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J Biol Chem. 1999;274(35):25085–25092. doi: 10.1074/jbc.274.35.25085. [DOI] [PubMed] [Google Scholar]
- 14.Gallatin WM, Weissman IL, Butcher EC. A cell-surface molecule involved in organ-specific homing of lymphocytes. Nature. 1983;304(5921):30–34. doi: 10.1038/304030a0. [DOI] [PubMed] [Google Scholar]
- 15.Lesley J, Hyman R, Kincade PW. CD44 and its interaction with extracellular matrix. Adv Immunol. 1993;54:271–335. doi: 10.1016/s0065-2776(08)60537-4. (Section V) [DOI] [PubMed] [Google Scholar]
- 16.Peach RJ, Hollenbaugh D, Stamenkovic I, Aruffo A. Identification of hyaluronic acid binding sites in the extracellular domain of CD44. J Cell Biol. 1993;122(1):257–264. doi: 10.1083/jcb.122.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerji S, Wright AJ, Noble M, Mahoney DJ, Campbell ID, Day AJ, Jackson DG. Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat Struct Mol Biol. 2007;14(3):234–239. doi: 10.1038/nsmb1201. [DOI] [PubMed] [Google Scholar]
- 18.Nagano O, Okazaki S, Saya H. Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene. 2013;32(44):5191–5198. doi: 10.1038/onc.2012.638. [DOI] [PubMed] [Google Scholar]
- 19.Lesley J, Hascall VC, Tammi M, Hyman R. Hyaluronan binding by cell surface CD44. J Biol Chem. 2000;275(35):26967–26975. doi: 10.1074/jbc.M002527200. [DOI] [PubMed] [Google Scholar]
- 20.Misra S, Heldin P, Hascall VC, Karamanos NK, Skandalis SS, Markwald RR, Ghatak S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011;278(9):1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misra S, Ghatak S, Zoltan-Jones A, Toole BP. Regulation of multidrug resistance in cancer cells by hyaluronan. J Biol Chem. 2003;278(28):25285–25288. doi: 10.1074/jbc.C300173200. [DOI] [PubMed] [Google Scholar]
- 22.He Q, Lesley J, Hyman R, Ishihara K, Kincade PW. Molecular isoforms of murine CD44 and evidence that the membrane proximal domain is not critical for hyaluronate recognition. J Cell Biol. 1992;119(6):1711–1720. doi: 10.1083/jcb.119.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleeman J, Rudy W, Hofmann M, Moll J, Herrlich P, Ponta H. Regulated clustering of variant CD 44 proteins increases their hyaluronate binding capacity. J Cell Biol. 1996;135(4):1139–1150. doi: 10.1083/jcb.135.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wielenga VJ, Heider KH, Offerhaus GJ, Adolf GR, van den Berg FM, Ponta H, Herrlich P, Pals ST. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53(20):4754–4756. [PubMed] [Google Scholar]
- 25.Marangoni E, Lecomte N, Durand L, de Pinieux G, Decaudin D, Chomienne C, Smadja-Joffe F, Poupon MF. CD44 targeting reduces tumour growth and prevents post-chemotherapy relapse of human breast cancers xenografts. Br J Cancer. 2009;100(6):918–922. doi: 10.1038/sj.bjc.6604953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang SJ, Wreesmann VB, Bourguignon LY. Association of CD44 V3-containing isoforms with tumor cell growth, migration, matrix metalloproteinase expression, and lymph node metastasis in head and neck cancer. Head Neck. 2007;29(6):550–558. doi: 10.1002/hed.20544. [DOI] [PubMed] [Google Scholar]
- 27.Günthert U, Hofmann M, Rudy W, Reber S, Zöller M, Hauβmann I, Matzku S, Wenzel A, Ponta H, Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65(1):13–24. doi: 10.1016/0092-8674(91)90403-L. [DOI] [PubMed] [Google Scholar]
- 28.Naor D, Vogt Sionov R, Zahalka M, Rochman M, Holzmann B, Ish-Shalom D. Organ-Specific Requirements for Cell Adhesion Molecules During Lymphoma Cell Dissemination. Springer; Berlin Heidelberg: 1998. pp. 143–166. [DOI] [PubMed] [Google Scholar]
- 29.Ochiai S, Nakanishi Y, Mizuno K, Hashimoto S, Inutsuka S, Kawasaki M, Yatsunami J, Hara N. Expression of CD44 standard and CD44 variant 6 in human lung cancer. Nihon Kyobu Shikkan Gakkai Zasshi. 1997;35(11):1179–1185. [PubMed] [Google Scholar]
- 30.Kurozumi K, Nishida T, Nakao K, Nakahara M, Tsujimoto M. Expression of CD44 variant 6 and lymphatic invasion: Importance to lymph node metastasis in gastric cancer. World Journal of Surgery. 1998;22:853–858. doi: 10.1007/s002689900481. [DOI] [PubMed] [Google Scholar]
- 31.Foekens Ja, Dall P, Klijn JG, Skroch-Angel P, Claassen CJ, Look MP, Ponta H, Van Putten WL, Herrlich P, Henzen-Logmans SC. Prognostic value of CD44 variant expression in primary breast cancer. Int J Cancer. 1999;84(3):209–215. doi: 10.1002/(sici)1097-0215(19990621)84:3<209::aid-ijc2>3.0.co;2-9. DOI: 10.1002/(SICI)1097-0215(19990621)84:3<209::AID-IJC2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Ayhan A, Tok EC, Bildirici I, Ayhan A. Overexpression of CD44 variant 6 in human endometrial cancer and its prognostic significance. Gynecol Oncol. 2001;80(3):355–358. doi: 10.1006/gyno.2000.6014. [DOI] [PubMed] [Google Scholar]
- 33.Ishida T. Immunohistochemical expression of the CD44 variant 6 in colorectal adenocarcinoma. Surg Today. 2000;30(1):28–32. doi: 10.1007/PL00010042. [DOI] [PubMed] [Google Scholar]
- 34.Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Frontiers in Immunology. 2015 doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahoney DJ, Blundell CD, Day AJ. Mapping the Hyaluronan-binding Site on the Link Module from Human Tumor Necrosis Factor-stimulated Gene-6 by Site-directed Mutagenesis. J Biol Chem. 2001;276(25):22764–22771. doi: 10.1074/jbc.M100666200. [DOI] [PubMed] [Google Scholar]
- 36.Bourguignon LYW, Zhu H, Chu A, Iida N, Zhang L, Hung MC. Interaction between the adhesion receptor, CD44, and the oncogene product, p185(HER2), promotes human ovarian tumor cell activation. J Biol Chem. 1997;272(44):27913–27918. doi: 10.1074/jbc.272.44.27913. [DOI] [PubMed] [Google Scholar]
- 37.Ghatak S, Misra S, Toole BP. Hyaluronan constitutively regulates ErbB2 phosphorylation and signaling complex formation in carcinoma cells. J Biol Chem. 2005;280(10):8875–8883. doi: 10.1074/jbc.M410882200. [DOI] [PubMed] [Google Scholar]
- 38.Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, Lowe E, Mahoney DJ, Tammi MI, Kahmann JD, et al. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol Cell. 2004;13(4):483–496. doi: 10.1016/S1097-2765(04)00080-2. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Heldin CH, Heldin P. Inhibition of platelet-derived growth factor-BB-induced receptor activation and fibroblast migration by hyaluronan activation of CD44. J Biol Chem. 2006;281(36):26512–26519. doi: 10.1074/jbc.M605607200. [DOI] [PubMed] [Google Scholar]
- 40.Yu Q, Stamenkovic I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev. 1999;13(1):35–48. doi: 10.1101/gad.13.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourguignon LYW, Peyrollier K, Xia W, Gilad E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283(25):17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohashi R, Takahashi F, Cui R, Yoshioka M, Gu T, Sasaki S, Tominaga S, Nishio K, Tanabe KK, Takahashi K. Interaction between CD44 and hyaluronate induces chemoresistance in non-small cell lung cancer cell. Cancer Lett. 2007;252(2):225–234. doi: 10.1016/j.canlet.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 43.Shigeishi H, Biddle A, Gammon L, Emich H, Rodini CO, Gemenetzidis E, Fazil B, Sugiyama M, Kamata N, MaCkenzie IC. Maintenance of stem cell self-renewal in head and neck cancers requires actions of GSK3b influenced by CD44 and RHAMM. Stem Cells. 2013;31(10):2073–2083. doi: 10.1002/stem.1418. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Qi L, Liang Z, Song W, Liu Y, Wang Y, Sun B, Zhang B, Cao W. Transforming growth factor-β1 induces EMT by the transactivation of epidermal growth factor signaling through HA/CD44 in lung and breast cancer cells. Int J Mol Med. 2015 doi: 10.3892/ijmm.2015.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cyphert JM, Trempus CS, Garantziotis S. Size Matters: Molecular Weight Specificity of Hyaluronan Effects in Cell Biology. International Journal of Cell Biology. 2015 doi: 10.1155/2015/563818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim D-K, Wylie RG, Langer R, Kohane DS. Selective binding of C-6 OH sulfated hyaluronic acid to the angiogenic isoform of VEGF165. Biomaterials. 2016;77:130–138. doi: 10.1016/j.biomaterials.2015.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salbach-Hirsch J, Samsonov SA, Hintze V, Hofbauer C, Picke A-K, Rauner M, Gehrcke J-P, Moeller S, Schnabelrauch M, Scharnweber D, et al. Structural and functional insights into sclerostin-glycosaminoglycan interactions in bone. Biomaterials. 2015;67:335–345. doi: 10.1016/j.biomaterials.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Hintze V, Miron A, Moeller S, Schnabelrauch M, Wiesmann HP, Worch H, Scharnweber D. Sulfated hyaluronan and chondroitin sulfate derivatives interact differently with human transforming growth factor-??1 (TGF-??1) Acta Biomater. 2012;8(6):2144–2152. doi: 10.1016/j.actbio.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 49.Hill TK, Kelkar SS, Wojtynek NE, Souchek JJ, Payne WM, Stumpf K, Marini FC, Mohs AM. Near infrared fluorescent nanoparticles derived from hyaluronic acid improve tumor contrast for image-guided surgery. Theranostics. 2016;6(13):2314–2328. doi: 10.7150/thno.16514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelkar SS, Hill TK, Marini FC, Mohs AM. Near infrared fluorescent nanoparticles based on hyaluronic acid: Self-assembly, optical properties, and cell interaction. Acta Biomater. 2016;36:112–121. doi: 10.1016/j.actbio.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill TK, Abdulahad A, Kelkar SS, Marini FC, Long TE, Provenzale JM, Mohs AM. Indocyanine green-loaded nanoparticles for image-guided tumor surgery. Bioconjug Chem. 2015;26(2):294–303. doi: 10.1021/bc5005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kennel SJ, Lankford TK, Foote LJ, Shinpock SG, Stringer C. CD44 expression on murine tissues. J Cell Sci. 1993;104(Pt 2):373–382. doi: 10.1242/jcs.104.2.373. 1993. [DOI] [PubMed] [Google Scholar]
- 53.Lin T, Yuan A, Zhao X, Lian H, Zhuang J, Chen W, Zhang Q, Liu G, Zhang S, Chen W, et al. Self-assembled tumor-targeting hyaluronic acid nanoparticles for photothermal ablation in orthotopic bladder cancer. Acta Biomater. 2017;53:427–438. doi: 10.1016/j.actbio.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Gu X, Wang H, Sun Y, Wu H, Mao S. Synthesis, characterization and liver targeting evaluation of self-assembled hyaluronic acid nanoparticles functionalized with glycyrrhetinic acid. Eur J Pharm Sci. 2017;96:255–262. doi: 10.1016/j.ejps.2016.09.036. [DOI] [PubMed] [Google Scholar]
- 55.Becher J, Möller S, Riemer T, Schiller J, Hintze V, Bierbaum S, Scharnweber D, Worch H, Schnabelrauch M. Sulfated Glycosaminoglycan Building Blocks for the Design of Artificial Extracellular Matrices. 2012:315–328. [Google Scholar]
- 56.Plazinski W, Knys-Dzieciuch A, Milas M, Rinaudo M, Pérez S, Imberty A, Cañada FJ, Parrilli M, Jiménez-Barbero J, De Castro C, et al. Interactions between CD44 protein and hyaluronan: insights from the computational study. Mol BioSyst. 2012;8(2):543–547. doi: 10.1039/C2MB05399C. [DOI] [PubMed] [Google Scholar]
- 57.Morris G, Huey R. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J …. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256.AutoDock4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trott O, Olson AJ. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39(SUPPL 2) doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011;32(10):2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Babasola O, Rees-Milton KJ, Bebe S, Wang J, Anastassiades TP. Chemically modified N-acylated hyaluronan fragments modulate proinflammatory cytokine production by stimulated human macrophages. J Biol Chem. 2014;289(36):24779–24791. doi: 10.1074/jbc.M113.515783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crescenzi V, Francescangeli A, Renier D, Bellini D. New cross-linked and sulfated derivatives of partially deacetylated hyaluronan: Synthesis and preliminary characterization. Biopolymers. 2002;64(2):86–94. doi: 10.1002/bip.10131. [DOI] [PubMed] [Google Scholar]
- 64.Tan SC, Khor E, Tan TK, Wong SM. The degree of deacetylation of chitosan: advocating the first derivative UV-spectrophotometry method of determination. Talanta. 1998;45(4):713–719. doi: 10.1016/S0039-9140(97)00288-9. [DOI] [PubMed] [Google Scholar]
- 65.Liu LK, Finzel BC. Fragment-based identification of an inducible binding site on cell surface receptor CD44 for the design of protein-carbohydrate interaction inhibitors. J Med Chem. 2014;57(6):2714–2725. doi: 10.1021/jm5000276. [DOI] [PubMed] [Google Scholar]
- 66.Ogino S, Nishida N, Umemoto R, Suzuki M, Takeda M, Terasawa H, Kitayama J, Matsumoto M, Hayasaka H, Miyasaka M, et al. Two-state conformations in the hyaluronan-binding domain regulate CD44 adhesiveness under flow condition. Structure. 2010;18(5):649–656. doi: 10.1016/j.str.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 67.Kogan G, Šoltés L, Stern R, Gemeiner P. Hyaluronic acid: A natural biopolymer with a broad range of biomedical and industrial applications. Biotechnology Letters. 2007:17–25. doi: 10.1007/s10529-006-9219-z. [DOI] [PubMed] [Google Scholar]
- 68.Wolny PM, Banerji S, Gounou C, Brisson AR, Day AJ, Jackson DG, Richter RP. Analysis of CD44-hyaluronan interactions in an artificial membrane system: Insights into the distinct binding properties of high and low molecular weight hyaluronan. J Biol Chem. 2010;285(39):30170–30180. doi: 10.1074/jbc.M110.137562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang C, Cao M, Liu H, He Y, Xu J, Du Y, Liu Y, Wang W, Cui L, Hu J, et al. The high and low molecular weight forms of hyaluronan have distinct effects on CD44 clustering. J Biol Chem. 2012;287(51):43094–43107. doi: 10.1074/jbc.M112.349209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maharjan AS, Pilling D, Gomer RH. High and low molecular weight hyaluronic acid differentially regulate human fibrocyte differentiation. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


