Abstract
Lyme disease, the most commonly reported vector-borne disease in the United States, results from infection with Borrelia burgdorferi. Early clinical diagnosis of this disease is largely based on the presence of an erythematous skin lesion for individuals in high-risk regions. This, however, can be confused with other illnesses including southern tick-associated rash illness (STARI), an illness that lacks a defined etiological agent or laboratory diagnostic test, and is co-prevalent with Lyme disease in portions of the Eastern United States. By applying an unbiased metabolomics approach with sera retrospectively obtained from well-characterized patients we defined biochemical and diagnostic differences between early Lyme disease and STARI. Specifically, a metabolic biosignature consisting of 261 molecular features (MFs) revealed that altered N-acyl ethanolamine and primary fatty acid amide metabolism discriminated early Lyme disease from STARI. More importantly, development of classification models with the 261 MF biosignature and testing against validation samples differentiated early Lyme disease from STARI with an accuracy of 85 to 98%. These findings revealed metabolic dissimilarity between early Lyme disease and STARI, and provide a powerful and new approach to objectively distinguish early Lyme disease from an illness with nearly identical symptoms.
Introduction
Lyme disease is a multisystem bacterial infection that in the United States is primarily caused by infection with Borrelia burgdorferi sensu stricto. Over 300,000 cases of Lyme disease are estimated to occur annually in the United States, with over 3.4 million laboratory diagnostic tests performed each year (1, 2). Symptoms associated with this infection include fever, chills, headache, fatigue, muscle and joint aches, and swollen lymph nodes; however, the most prominent clinical manifestation in the early stage is the presence of one or more erythema migrans (EM) skin lesions (3). This annular, expanding erythematous skin lesion occurs at the site of the tick bite in 70 to 80% of infected individuals and is typically 5 cm or more in diameter (4, 5). Although an EM lesion is a hallmark for Lyme disease, other types of skin lesions can be confused with EM (3, 5, 6). These include rashes caused by tick-bite hypersensitivity reactions, certain cutaneous fungal infections, bacterial cellulitis and the rash of southern tick-associated rash illness (STARI) (7, 8).
STARI, is associated with a bite from the lone star tick (Amblyomma americanum), and in addition to the development of an EM-like skin lesion, individuals with STARI can present with mild systemic symptoms (including muscle and joint pains, fatigue, fever, chills, and headache) that are similar to those occurring in patients with Lyme disease (7, 9, 10). These characteristics of STARI have led some to postulate that the etiology of this illness is a Borrelia species, including B. burgdorferi (10, 11) or B. lonestari (12–15); however, multiple studies have refuted that STARI is caused by B. burgdorferi (7, 16–19) and additional cases associating B. lonestari with STARI have not emerged (20, 21). Additionally, STARI patients have been screened serologically for reactivity to rickettsial agents, but no evidence was obtained to demonstrate that rickettsia causes this illness (10, 22). Thus, at present no infectious etiology is known for STARI.
STARI cases occur over the geographic region where the lone star tick is present. This includes a region that currently expands from central Texas and Oklahoma upward into the Midwestern states and eastward, including the southern states and along the Atlantic coast into Maine (23). Unlike STARI, Lyme disease is transmitted to humans through the bite of the blacklegged tick (Ixodes scapularis) that is present in the northeastern, mid-Atlantic, and north-central United States, and the western blacklegged tick (I. pacificus), which is present on the Pacific Coast (24). The geographic distribution of human Lyme disease and the vectors for this disease is expanding (24–26), and there is a similar expansion of areas inhabited by the lone star tick (23). Importantly, a strict geographic segregation of Lyme disease and STARI does not exist, as there are regions where STARI and Lyme disease are co-prevalent (25). Thus, there is a growing need for diagnostic methods to differentiate between Lyme disease and STARI, and that facilitate proper treatment, patient management and disease surveillance.
Clinically, the skin lesions of STARI and early Lyme disease are indistinguishable, and no laboratory tool or method exists for the diagnosis of STARI or differentiation of STARI from Lyme disease. The only biomarkers evaluated for differential diagnosis of early Lyme disease and STARI have been serum antibodies to B. burgdorferi (10, 16). However, these tests have poor sensitivity for early stages of Lyme disease, and thus a lack of B. burgdorferi antibodies cannot be used as a reliable differential marker for STARI. We previously demonstrated that metabolic profiling of sera provided a high level of accuracy in differentiating early Lyme disease patients from healthy individuals and those with diseases or conditions that are confounders for existing serological-based laboratory tests of Lyme disease (27). Thus, we now postulate that a metabolomics-driven approach will identify biomarkers that discriminate early Lyme disease from STARI, and provide evidence that these two diseases are biochemically distinct. A retrospective cohort of well-characterized sera from patients with early Lyme disease and STARI was evaluated to identify a differentiating metabolic biosignature. Using statistical modeling, this metabolic biosignature accurately classified test samples that included healthy controls. Additionally, the metabolic biosignature revealed that N-acyl ethanolamine (NAE) and primary fatty acid amide (PFAM) metabolism differed significantly between these two diseases.
Results
Clinical samples
A total of 220 well-characterized retrospective serum samples from three different repositories were used to develop and test a metabolic biosignature that accurately classifies early Lyme disease and STARI (table S1). All samples from Lyme disease patients were culture confirmed and/or PCR positive for B. burgdorferi. The median age for early Lyme disease patients was 45 years and 74% were males. STARI patients had an overall median age of 45 years and 55% were males.
To establish a Lyme disease diagnostic baseline, the recommended two-tiered serology testing for Lyme disease was performed on all samples. First-tier testing was performed using the C6 EIA and was positive for 66% of Lyme disease samples. When STARI and healthy controls were tested by the C6 EIA, two STARI samples (2%) and five healthy controls (9%) tested positive or equivocal. Two-tiered testing using IgM and IgG immunoblots as the second-tier test following a positive or equivocal first-tier assay resulted in a sensitivity of 44% for early Lyme disease samples (duration of illness was not considered for IgM immunoblot testing). The sensitivity of two-tiered testing for early Lyme disease samples included in the Discovery/Training-Sets and the Test-Sets was 40% and 50%, respectively. All STARI and healthy control samples were negative by two-tiered testing (table S1).
Development of a metabolic biosignature for early Lyme disease and STARI differentiation
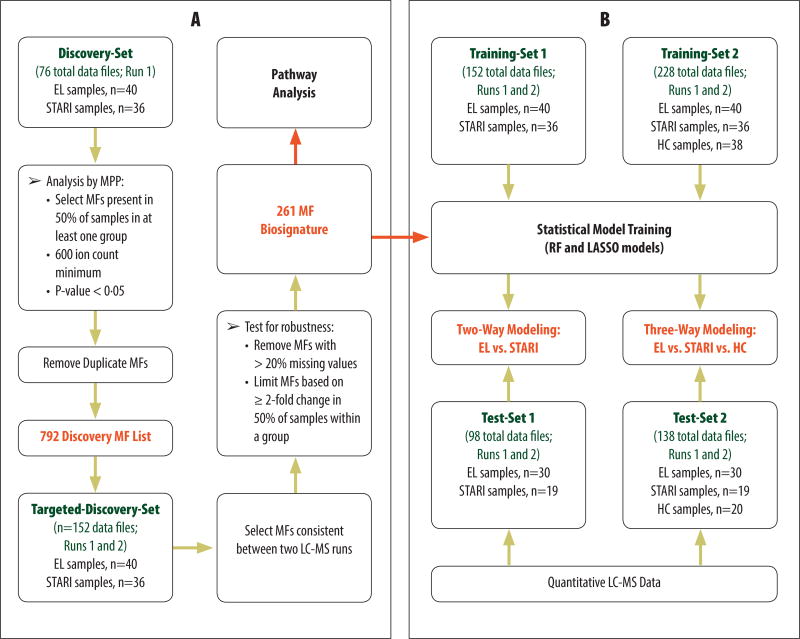
Metabolic profiling by liquid chromatography-mass spectrometry (LC-MS) of a retrospective cohort of well-characterized sera from patients with early Lyme disease (n=40) and STARI (n=36) (table S1 and Fig. 1A) comprising the Discovery-Set (i.e. Test-Set samples were not used in molecular feature selection) resulted in a biosignature of 792 molecular features (MFs) that differed significantly (adjusted-p<0.05) with a ≥ 2 fold change in relative abundance between early Lyme disease and STARI. Down-selection of MFs based on their robustness in replicate analyses of the same sera produced a refined biosignature of 261 MFs (Fig. 1A and table S2). Of these 261 MFs, 60 and 201 displayed an increased and decreased abundance, respectively, in early Lyme disease as compared to STARI. The large number of MFs that differed significantly between early Lyme disease and STARI patients indicated that these two patient groups had distinguishing biochemical profiles. These variances could be applied to define alterations of specific metabolic pathways (Fig. 1A) and used to develop diagnostic classification models (Fig. 1B).
Fig. 1. Metabolic profiling for the identification and application of differentiating molecular features (MFs).
(A) LC-MS data from an initial Discovery-Set of early Lyme disease (EL) and STARI samples was used to identify a list of MFs that were targeted in a second LC-MS run. The data from both LC-MS runs was combined to form the Targeted-Discovery-Set. The MFs were then screened for consistency and robustness and this resulted in a final early Lyme disease-STARI biosignature of 261 MFs. This biosignature was used for downstream pathway analysis and for classification modeling. (B) Two training-data sets along with the 261 MF biosignature list were used to train multiple classification models, random forest (RF) and least absolute shrinkage and selection operator (LASSO). Data from samples of twoTest-Sets (not included for the Discovery/Training-Set data) were blindly tested against the two-way (EL vs STARI) and three-way [EL vs STARI vs healthy controls (HC)] classification models. The regression coeficients used for each MF in the LASSO two-way and three-way classification models are provided in table S4 and S6, resepectively.
In silico analysis of metabolic pathways
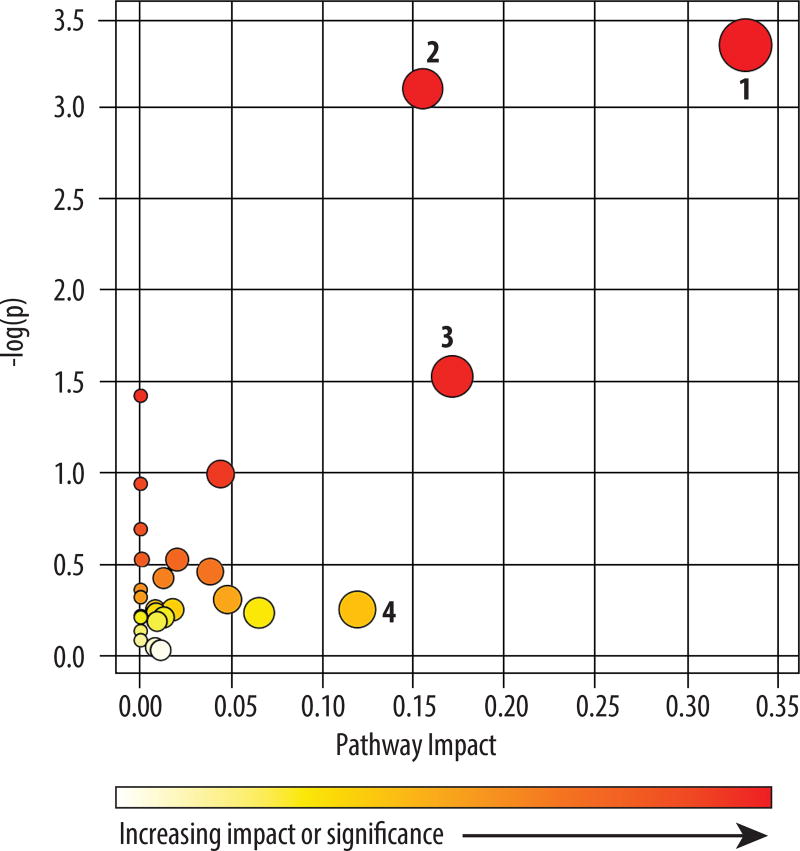
Presumptive chemical identification was applied to the 261 MFs. This yielded predicted chemical formulas for 149 MFs and 122 MFs were assigned a putative chemical structure based on interrogation of each MF’s monoisotopic mass (+ or − 15 ppm) against the Metlin database and the Human Metabolome Database (HMDB) (table S2). An in silico interrogation of potentially altered metabolic pathways was performed using the presumptive identifications for the 122 MFs and MetaboAnalyst (28). Four differentiating pathways were predicted to have the greatest impact, with glycerophospholipid and sphingolipid metabolism having the most number of assigned metabolites (Fig. 2 and table S3). Specifically, the MetaboAnalyst analysis indicated that differences in phosphatidic acid, phosphatidylethanolamine, phosphatidylcholine and lysophosphotidylcholine were the major contributors to altered glycerophospholipid metabolism between STARI and early Lyme disease (table S3). Altered sphingolipid metabolism between these two groups was attributable to changes in the relative abundances of sphingosine, dehydrosphinganine and sulfatide (table S3). Manual interrogation of the predicted structural identifications revealed that 26 and 7 of the 122 MFs assigned a putative structural identification were associated with glycerophospholipid and sphingolipid metabolism, respectively (table S2).
Fig. 2. Pathways differentially regulated in patients with early Lyme disease and STARI.
The 122 presumptively identified MFs were analyzed using MetaboAnalyst to identify perturbed pathways between early Lyme disease and STARI. The color and size of each circle is based on P values and pathway impact values. Pathways with a > 0.1 impact were considered to be perturbed and differentially regulated between patients with early Lyme disease and STARI. A total of four pathways were affected: 1) glycerophospholipid metabolism; 2) sphingolipid metabolism; 3) valine, leucine and isoleucine biosynthesis; and 4) phenylalanine metabolism.
Elucidation of altered NAE metabolism
The prediction of altered metabolic pathways was based on the presumptive structural identification of the early Lyme disease versus STARI differentiating MFs. Thus, to further define the metabolic differences between these two patient groups, structural confirmation of selected MFs was undertaken. Two MFs that displayed relatively large abundance differences (m/z 300.2892, RT 19.66; and m/z 328.3204, RT 20.72) were putatively identified as sphingosine-C18 or 3-ketosphinganine, and sphingosine-C20 or N,N-dimethyl sphingosine, respectively. However, both of these MFs had alternative predicted structures of palmitoyl ethanolamide and stearoyl ethanolamide, respectively. The interrogation of authentic standards against these two serum MFs revealed RTs and MS/MS spectra that identified the m/z 300.2892 and m/z 328.3204 products as palmitoyl ethanolamide (Fig. 3A and 3B) and stearoyl ethanolamide (fig. S1), respectively. These two products, as well as other NAEs, are derived from phosphatidylethanolamine and phosphatidylcholine, and represent a class of structures termed endocannabinoids and endocannabinoid-like (29) (Fig 3C). Further analysis of the 122 MFs identified five additional MFs with a predicted structure that mapped to the NAE pathway. Specifically, MF m/z 286.2737, RT 19.08 was putatively identified as a sphingosine-C17 or pentadecanoyl ethanolamide, and was confirmed to be the latter (fig. S2). MF m/z 356.3517, RT 21.67 was putatively identified and confirmed to be eicosanoyl ethanolamide (fig. S3), and MF m/z 454.2923, RT 18.08 was confirmed to be glycerophospho-N-palmitoyl ethanolamine (fig. S4), an intermediate in the formation of palmitoyl ethanolamide. A second group of lipids, the PFAMs that act as signaling molecules and that are potentially associated with the metabolism of NAEs were also identified as having significant relative abundance differences between the early Lyme disease and STARI patient samples. Specifically, MFs m/z 256.2632, RT 20.08; m/z 284.2943, RT 21.15; and m/z 338.3430, RT 22.14 were confirmed to be palmitamide (Fig. 3D and 3E), stearamide (fig. S5) and erucamide (fig. S6), respectively.
Fig. 3. Metabolite identification and association with NAE and PFAM metabolism.
Structural identification of palmitoyl ethanolamide (A and B) and other NAEs (fig S1–S3) in the 261 MF biosignature indicated alteration of NAE metabolism (C), a pathway that can influence the production of PFAMs. Further MF identification revealed that palmitamide (D and E) and other PFAMs (fig S5 and S6) also differed in abundance between STARI and early Lyme disease patients. Structural identification was achieved by retention-time alignment (A and D) of authentic standard (top panel), authentic standard spiked in pooled patient sera (middle panel), and the targeted metabolite in pooled patient sera (bottom panel), and by comparison of MS/MS spectra (B and E) of the authentic standards (top) and the targeted metabolites in patient sera (bottom). Retention-time alignments for palmitoyl ethanolamide (A) and palmitamide (D) were generated with extracted ion chromatograms for m/z 300.2892 and m/z 256.2632, respectively. The relationship of PFAM formation to NAE metabolism is highlighted in light green in diagram C. PLA, phospholipase A; PLC, phospholipase C; PLD, phospholipase D; ADH, alcohol dehydrogenase; PAM, peptidylglycine α-amidating monooxygenase.
The large number of differentiating MFs associated with NAE metabolism suggested that this is a major biological difference between STARI and early Lyme disease (Fig. 3C and table S2). Four additional MFs of the 261 MF biosignature, and that fit known host biochemical pathways were also structurally confirmed. These included L-phenylalanine (fig. S7), nonanedioic acid (fig. S8), glycocholic acid (fig. S9) and 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) (fig. S10). Additionally, two MFs that provided strong matches to MS/MS spectra in the Metlin databases were putatively identified as arachidonoyl lysophosphatidic acid [Lyso PA (20:4)] (fig. S11) and 3-ketosphingosine (fig. S12).
The 261 MF biosignature list revealed metabolic dissimilarity between Lyme disease and STARI
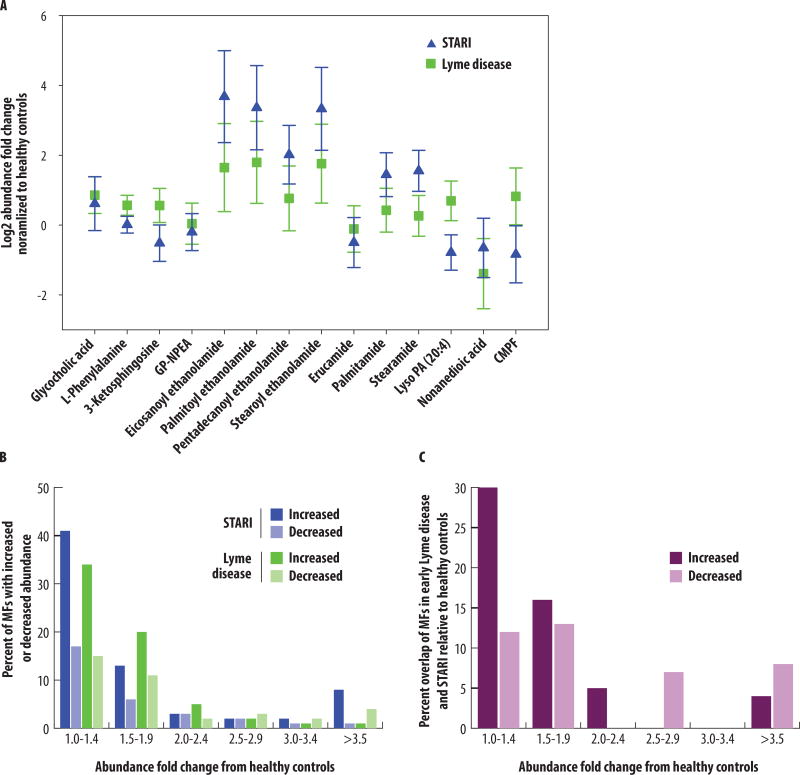
We hypothesized that early Lyme disease and STARI represent distinct metabolic states that would be reflected in a comparison of MFs’ relative abundances in these two disease states to those of healthy controls. The majority of MFs in early Lyme disease and STARI sera maintained fold change differences with respect to healthy controls. This provided evidence for metabolic separation of early Lyme disease and STARI patient samples (Fig. 4A). For three MFs (3-ketosphingosine, CMPF, and Lyso PA 20:4), the levels in early Lyme disease were increased as compared to the healthy controls while the levels in STARI were decreased. Additionally, all of the NAEs and PFAMs had relative abundances in early Lyme disease patients that were closer to those of healthy controls, whereas the relative abundances in STARI were greatly increased. This analysis was expanded to all 261 MFs of the early Lyme disease-STARI biosignature. The total number of MFs with increased and decreased abundances relative to healthy controls was similar for both early Lyme disease and STARI within the different abundance fold change ranges (Fig. 4B). However, when individual MFs with increased or decreased abundances relative to healthy controls were compared between early Lyme disease and STARI a strong difference emerged, with only 0 to 30% of MFs shared between early Lyme disease and STARI yielding the same directional and similar abundance fold change (Fig. 4C). This indicated that the metabolic changes in early Lyme disease and STARI as compared to healthy controls differed.
Fig. 4. Comparison of MF abundances from the Lyme disease-STARI biosignature against healthy controls.
(A) Fourteen of the metabolites with level 1 or level 2 structural identifications were evaluated for abundance differences between early Lyme disease (green squares) and STARI (blue triangles) normalized to the metabolite abundance in healthy controls. Included are metabolites identified for NAE and PFAM metabolism. GP-NAE: glycerophospho-N-palmitoyl ethanolamine; Lyso PA (20:4): arachidonoyl lysophosphatidic acid; CMPF: 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid. The relative mean abundance and 95% confidence intervals are shown for each metabolite. (B) Abundance fold change ranges (x-axis) plotted against the percent of MFs from the 261 MF early Lyme disease-STARI biosignature that have increased (dark blue) or decreased (light blue) abundances in STARI relative to healthy controls, and increased (dark green) or decreased (light green) abundances in early Lyme disease relative to healthy controls. (C) The percentage of identical MFs in STARI and early Lyme disease that had the same directional and similar abundance fold change difference relative to healthy controls (y-axis). MFs were grouped based on abundance fold change ranges:1.0–1.4, 1.5–1.9, 2.0–2.4, 2.5–2.9, 3.0–3.4, and ≥3.5 (x-axis). MFs with increased fold changes relative to healthy controls are indicated in dark purple or and those with a decreased fold changes are indicated in light purple.
Diagnostic classification of early Lyme disease vs STARI
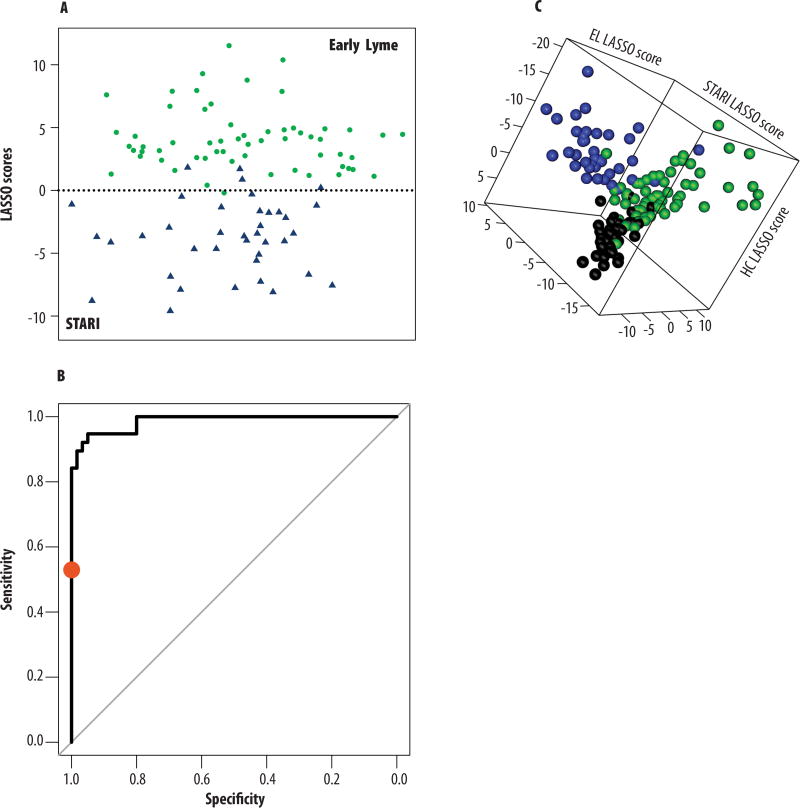
Classification models were used to determine whether the 261 MF biosignature could be applied to discriminate early Lyme disease from STARI (Table 1 and Fig. 1B). Specifically, two classification models, least absolute shrinkage and selection operator (LASSO) and random forest (RF) were trained with the 261 MF biosignature using abundance data from the Training-Set samples only (Fig. 1B). Test-Set samples were not used for molecular feature selection or to train the classification models. The LASSO model selected 38 MFs, and RF by default does not perform feature selection and thus used all 261 MFs for classification of the STARI and early Lyme disease patient populations (table S2). Note, table S4 provides the regression coefficients for the 38 MFs selected by LASSO. When Test-Set samples (Fig. 1B) (i.e. those not included in the Discovery/Training-Set) were tested in duplicate, early Lyme disease samples were classified by RF and LASSO with an accuracy of 97% and 98%, respectively. The STARI samples had a classification accuracy of 89% with both models (Table 1 and table S5). A depiction of the LASSO scores for the Test-Set data showed segregation of the early Lyme disease and STARI patient samples, and demonstrated the discriminating power of the 38 MFs selected by the LASSO model (Fig. 5A). A receiver operating characteristic (ROC) curve was plotted to demonstrate the performance of the LASSO model for differentiating early Lyme disease from STARI patients. The area under the curve (AUC) was calculated to be 0.986 (Fig. 5B). The 38 MFs of the LASSO model encompassed four of the 14 structurally confirmed metabolites: CMPF, L-phenylalanine, palmitoyl ethanolamide, and arachidonoyl lysophosphatidic acid (table S2).
Table 1. Classification modeling using the 261 MF biosignature list.
RF, random forest; LASSO, least absolute shrinkage and selection operator; MF; molecular feature.
| Classification Model |
Test-Set Sample Group |
Number of Data Files* |
RF (261 MFs) | LASSO (38/82 MFs±) | ||
|---|---|---|---|---|---|---|
| Number Correctly Predicted |
% Classification Accuracy |
Number Correctly Predicted |
% Classification Accuracy |
|||
| 1: Two-Way Model | Early Lyme Disease | 60 | 58 | 97 | 59 | 98 |
| STARI | 38 | 34 | 89 | 34 | 89 | |
| 2: Three-Way Model | Early Lyme Disease | 60 | 51 | 85 | 51 | 85 |
| STARI | 38 | 35 | 92 | 35 | 92 | |
| Healthy Controls | 40 | 38 | 95 | 37 | 93 | |
Samples were analyzed in duplicate by LC-MS.
A total of 38 MFs were selected by the LASSO model for two-way modeling and 82 MFs were selected by the LASSO for three-way modeling.
Fig. 5. Evaluation of classification models’ performance.
(A) LASSO scores (Xβ; i.e. the linear portion of the regression model) were calculated for Test-Set data of early Lyme disease (green dots) and STARI (blue triangles) serum samples by multiplying the transformed abundances of the 38 MFs identified in the two-way LASSO model by the LASSO coefficients of the model and summing for each sample. Scores are plotted along the y-axis; serum samples are plotted randomly along the x-axis for easier viewing. (B) An ROC curve demonstrates the level of discrimination that is achieved between early Lyme disease and STARI using the 38 MFs of the two-way LASSO classification model by depicting a true positive rate (sensitivity; early Lyme disease) versus a false positive rate (specificity; STARI) for the Test-Set samples (table S6). The AUC was calculated to be 0.986. The diagonal line represents an AUC value of 0.5. The performance of two-tiered testing (red dot) on the same sample set (Test-Set 1) was included as a reference for the sensitivity and specificity of the current clinical laboratory test for Lyme disease. (C) LASSO scores (Xβi) were calculated for the Test-Set data of early Lyme disease (green spheres), STARI (blue spheres), and healthy control (black spheres) serum samples by multiplying the transformed abundances of the 82 MFs identified in the three-way LASSO model by each of three LASSO coefficients used in the model. Each axis represents the sample score in the direction of one of the three sample groups. Scores are used in calculation of probabilities of class membership, with highest probability determining the predicted class. Although there is overlap, the three groups predominantly occupy distinct areas of the plot.
Diagnostic classification of early Lyme disease vs STARI vs healthy controls
Separate three-way classification models using LASSO and RF were developed by including LC-MS data collected for healthy controls in the Training-Set samples (Fig. 1B). For model training LASSO selected 82 MFs (table S2). The regression coefficients for the 82 MFs selected by LASSO are provided in table S6. Evaluation of the RF and LASSO three-way classification models with Test-Set samples (those not used in the Discovery/Training-Sets) reveled classification accuracies of 85% and 92% for early Lyme disease and STARI, respectively. Surprisingly, healthy controls were classified with accuracies of 95% and 93% with the RF and LASSO models, respectively (Table 1 and table S7). Plotting of LASSO scores calculated for Test-Set data revealed three groupings that corresponded with early Lyme disease, STARI and healthy controls (Fig. 5C). Of the early Lyme disease samples that were misclassified with the RF model (n=9), all were predicted to be healthy controls; and those misclassified by the LASSO model (n=9), three were classified as STARI and six as healthy controls. Of the STARI samples that were misclassified by the RF and LASSO models (n=3 for both models), all samples were misclassified as early Lyme disease. When healthy controls were misclassified using the RF model (n=2) and LASSO model (n=3), all were misclassified as early Lyme disease.
Of the 38 MFs selected by LASSO for the two-way classification model, 33 were included in the 82 MFs of the LASSO three-way classification model (table S2). The 82 MFs of the LASSO three-way classification included seven of the 14 structurally confirmed metabolites: 3-ketosphingosine, glycocholic acid and pentadecanoyl ethanolamide, as well as the four included in the LASSO two-way classification model (table S2).
Biosignature was not influenced by geographic variability
Since retrospective samples collected by multiple laboratories were used in these studies, we assessed whether a geographic bias was introduced. Pair-wise comparisons, by sample source, of mean LASSO scores and mean RF probabilities of STARI and healthy control Test-Set data were performed with ANOVA followed by computation of simultaneous confidence intervals using Tukey’s method (table S8 and S9). These results demonstrated that all STARI samples grouped together regardless of source, and differ from healthy controls. Likewise, the healthy controls from Colorado and New York were considered a single group that differed from STARI based on the LASSO healthy control or STARI scores, and the RF classification probabilities for healthy controls. The only outlier noted in theses analyses was the New York healthy controls, but this was only when the ANOVA was performed with RF classification probabilities for STARI.
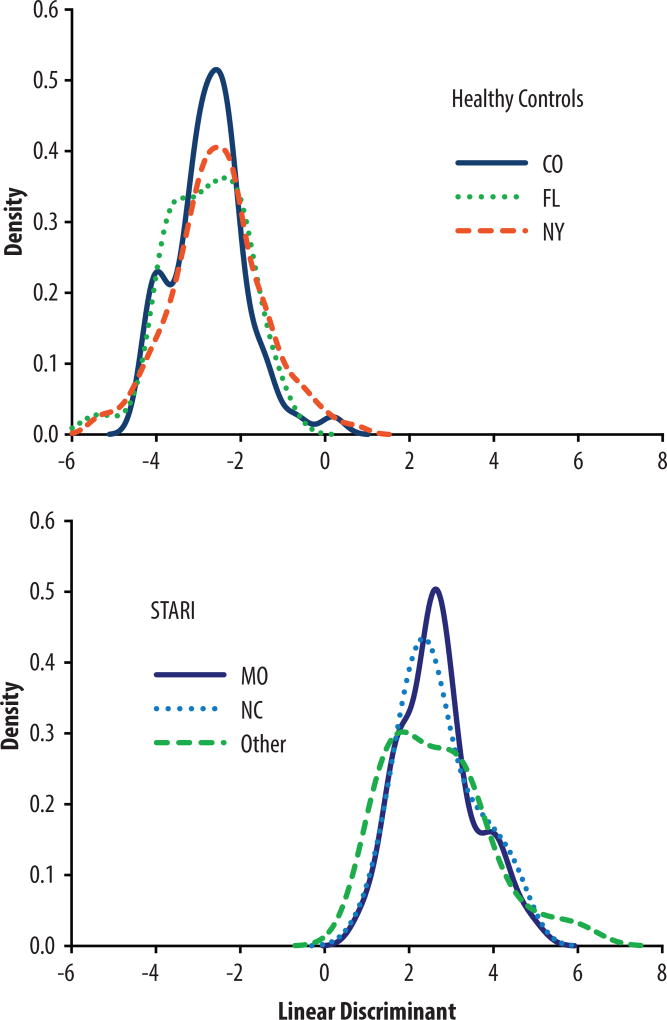
The grouping of the samples based on disease state and not geographic distribution was also evaluated by linear discriminant analysis using the 82 MFs of the LASSO three-way classification model (Fig. 6). For this analysis healthy controls from Florida, a region with low prevalence for Lyme disease and reported to have STARI cases, were included to evaluate whether samples collected in the southern United States would differ from those collected in New York or Colorado. For STARI, three patient samples groups collected in Missouri, NC and other states (included VA, GA, KY, TN, AL, IA and NE) were compared. The linear discriminant analysis demonstrated that although slight variation exists between the three healthy control groups (NY, CO and FL), there is greater variability between all healthy controls and all STARI samples than within healthy controls or STARI samples based on geographic location of collection (Fig. 6).
Fig. 6. Evaluation of intra-and inter-group variability.
Linear discriminant analysis was performed using the 82 MFs picked by LASSO in the three-way classification model to assess the intra-group variability based on the geographical region or laboratory from which STARI (CO-blue, solid; FL-green, dotted; and NY-red, dashed) and healthy control (MO-dark blue, solid; NC-light blue, dotted; and Other-green, dashed) sera were obtained. Only slight intra-group variation was observed. This analysis also compared and showed clear differentiation of all healthy control from STARI samples regardless of geographical region or laboratory origin. Healthy controls from FL were included in this analysis to demonstrate that healthy controls from an area with low incidence of Lyme disease and where STARI cases occur do not differ from the healthy controls obtained from other regions and used in the classification modeling.
Discussion
The inability to detect B. burgdorferi by PCR or culture, no serological response to B. burgdorferi antigens in STARI patients, and transmission by different tick species are accepted as evidence that the etiologies of STARI and Lyme disease differ (7–8,16, 25). Nevertheless, an overlap in clinical symptoms, including the development of an EM-like skin lesion, creates confusion and controversy for the clinical differentiation of STARI and Lyme disease (30). The data reported here demonstrated marked differences between the metabolic profiles of early Lyme disease and STARI patients, and thus provide compelling positive data to support the concept that these two illnesses are distinct entities. Metabolic pathway analyses and the structural identification of several MFs with significant abundance differences between early Lyme disease and STARI identified multiple NAEs. These endogenous lipid mediators are derived from phosphatidylcholine and phospahtidylethanolamine via the endocannabinoid system (Fig. 3C) (29). Arachidonoylethanolamide (AEA), a widely studied endocannabinoid, is an endogenous agonist of the cannabinoid receptors; however, it is a minor component of animal tissues. In contrast, congeners of AEA, such as the NAEs identified in the early Lyme disease-STARI biosignature, are significant products of animal tissues, including the skin (29, 31). The serum levels of NAEs possessing long-chain saturated fatty acids were significantly increased in the serum of STARI patients. These are produced in response to inflammation, and act in an anti-inflammatory manner as agonists of PPAR-α or by enhancing AEA activity (32, 33). The NAEs can be converted to N-acylglycine structures via an alcohol dehydrogenase, and further degraded to PFAMs (34). Interestingly, our data not only demonstrated a STARI-associated increase in NAEs with saturated fatty acids, but also an increase in the corresponding PFAMs. Although the mechanism for the increased NAE and PFAM levels in STARI patients is unknown, decreased fatty acid amide hydrolase activity that liberates fatty acids from both NAEs and PFAMs (35) would result in the observed increase in abundance of these metabolites. The anti-inflammatory activity of the NAEs also raises the possibility that these metabolites are partially responsible for the milder symptoms associated with STARI (9). As the enzymes involved with the genesis and degradation of NAEs and PFAMs are known (29, 36), studies can be constructed to further elucidate the mechanism(s) by which NAEs and PFAMs accumulate in the sera of STARI patients.
We previously demonstrated proof-of-concept for a discriminating metabolic biosignature of early Lyme disease (27). This current work expands on the utility of this approach by demonstrating the ability to distinguish early Lyme disease from an illness with nearly identical symptoms or what would be considered a Lyme disease-like illness (37). The existing diagnostic algorithm for Lyme disease is a two-tiered serologic approach that utilizes an EIA or IFA as a first-tier test followed by IgM and IgG immunoblotting as the second-tier test (38). For early Lyme disease, the sensitivity of this diagnostic is 29–40% and the specificity is 95–100% (39). The current antibody-based approaches do not distinguish between active and previous infections, an important limitation. In the current study all of the STARI samples were negative by two-tiered testing, and only 2% were positive by the first-tier EIA. Early Lyme disease samples were 44% positive (40% positivity for the early Lyme disease samples used in the Discovery and Training Sets and 50% positivity for early Lyme disease samples used in the Test Sets) by two-tiered testing. In contrast, when classification modeling was applied to the 261 MFs of the early Lyme disease-STARI biosignature, diagnostic accuracy for early Lyme disease was dramatically increased (85 to 98% accuracy depending on the model) as compared to serology. Classification by RF or LASSO was overall highly accurate for early Lyme disease and STARI, in particular when using the two-way classification models. Interestingly, when healthy controls were introduced and used to develop a three-way classification model there was a slight increase in the accuracy for STARI and decrease in the accuracy for early Lyme disease, but healthy controls were classified with a 93–95% accuracy. This was surprising as healthy controls were not used to create the initial 261 MF biosignature, and furthers supported that STARI and early Lyme disease are metabolically distinct from healthy controls, but in different ways.
To date the development of a diagnostic tool for STARI or for differentiation of early Lyme disease and STARI has received little attention. As the geographic distribution of Lyme disease continues to expand (25, 26), so will the geographic range where there is overlap of Lyme disease and STARI. Thus, a diagnostic tool that accurately differentiates these two diseases could have a major impact on patient management. Lyme disease is treated with antibiotics, and although there is no defined infectious etiology for STARI, this illness is also commonly treated in a similar manner (7, 20, 40). Establishment of a robust diagnostic tool would not only facilitate antibiotic stewardship, it would also allow for proper studies to assess the true impact of therapies for STARI. Lyme disease is also a reportable disease and in order to maintain accurate disease surveillance in low incidence areas, it is essential that diseases such as STARI be excluded (30). Additionally, vaccines are currently being developed for Lyme disease (41–44) and as these are tested, it will be important to identify STARI patients in order to properly assess vaccine efficacy.
To apply the discoveries of this work towards the development of an assay that can be used for the clinical differentiation of early Lyme disease and STARI, it must first be determined whether an emphasis should be placed on the diagnosis of Lyme disease or STARI. As there is no defined etiology of STARI, and Lyme disease is not necessarily self-limiting without antibiotics and can have subsequent complications if untreated, we envision that the final assay would focus on being highly sensitive for early Lyme disease and be primarily applied in regions where Lyme disease and STARI overlap. Although existing laboratory tests for Lyme disease emphasize specificity, this strategy needs to be reconsidered for a differential diagnostic test of STARI and early Lyme disease, since any illness presenting with an EM in a region with a known incidence of Lyme disease would likely be treated with antibiotics (7, 20, 40). As with all diagnostic tests, use of a metabolic biosignature for differentiation of early Lyme disease and STARI would need to be performed in conjunction with clinical evaluation of the patient, and consideration of their medical history and epidemiologic risk for these two diseases.
The approach outlined in this study applies semi-quantitative mass spectrometry and the use of biochemical signatures for the classification of patients. Clinical application of such an approach would likely occur in a specialized clinical diagnostic laboratory. At this point it is difficult to accurately assess the cost benefits of a MS based assay; however, it should be noted that the second-tier immunoblot assays for the serological diagnosis of Lyme disease are already performed in specialized laboratories (1, 45, 46). Further a LC-MS assay for a single sample performed as described in this work takes about 1.5 h from sample processing to data collection. MS assays are currently used in clinical laboratories for the analyses of small molecule metabolites. The majority of these tests are under Clinical Laboratory Improvement Amendments (CLIA) guidelines, but an FDA cleared mass spectrometry-based test for inborn metabolic errors is in use (47). The most accurate quantification of metabolites by mass spectrometry is achieved by Multiple Reaction Monitoring (MRM) assays (48). Such assays are developed with the knowledge of a MF’s chemical structure. To this end, we have identified the chemical structure of 14 MFs and are continuing to identify those that have the greatest fold change difference and reproducibility. It should be noted that the NAEs and PFAMs that were revealed via our pathway analyses are amenable to MRM assays (49). These metabolites are now being investigated for their ability to accurately classify STARI and early Lyme disease.
The data reported here were generated from the analysis of retrospectively collected serum samples from various repositories that have been archived for different lengths of time. To reduce the impact of the potential variability associated with these samples, stringent criteria were applied to the data analysis. In addition to the requirement of a significant fold change, those MFs selected for the final early Lyme disease-STARI biosignature were required to be present in at least 80% of samples within a sample group and maintain the median fold-change difference in at least 50% of samples within a group. While the STARI and healthy control sera were collected by multiple laboratories and from multiple geographic locations, the early Lyme disease sera was obtained from a single laboratory. This is a potential limitation of the study. However, linear discriminate analysis was applied to assess the variability within the healthy control and STARI samples collected by different laboratories. This analysis demonstrated little to no variability among the STARI or healthy control samples indicating that the criteria used for MF selection effectively reduced non-biological variability. As we have noted, data were collected by non-absolute semi-quantitative mass spectrometry. Nevertheless, this is a common practice applied in the development of differentiating biosignatures for infectious diseases (27, 50–53), and our workflow ensured that the most robust MFs were selected and used for classification modeling. This included the use of full technical replicates that controlled for potential variability introduced during sample processing and LC-MS analyses. Although the use of replicate samples could have biased the discriminating power of the classification models for the Training-Set data, the use of an independent hold out Test-Set prevents this from being a problem for model testing.
Without knowledge of a known etiologic agent, we recognize that STARI simply encompasses a clinical syndrome. The STARI samples used in this current work included those collected in studies used to define this illness (9), as well as samples collected outside those original studies. Ongoing efforts are directed at prospective collection of additional samples that will be useful to assess the applicability of our current metabolic biosignature in a real-world scenario. Future sample collection will also target patient populations with non-Lyme EM-like lesions, including tick-bite hypersensitivity reactions, certain cutaneous fungal infections and bacterial cellulitis. Additionally, other factors such as confections with other vector-borne pathogens will need to be addressed with prospective studies. In the Southeastern United States, there is evidence for enzootic transmission of B. burgdorferi; however, it is debatable whether Lyme disease occurs in this region (11, 30, 54, 55). The current study was not designed to provide evidence for or against the presence of Lyme disease in the southern United States. Nevertheless, metabolic profiling offers a novel approach that is orthogonal to the methods currently employed to address this issue.
Materials and Methods
Study design
STARI is an illness that has received little attention over the years, but is a confounding factor in diagnosing early Lyme disease in areas where both illnesses overlap and contributes to the debate surrounding the presence of Lyme disease in the southern United States. No diagnostic tool exists for STARI or for differentiating early Lyme disease from STARI. Based on documented differences between early Lyme disease and STARI (9, 16, 56), we hypothesized that metabolic profiling of serum would permit development of a biochemical biosignature that when applied could accurately classify early Lyme disease and STARI patients. For this reason, we designed an unbiased-metabolomics study to directly compare the metabolic host responses between these two illnesses, and subsequently evaluate how this metabolic biosignature distinguishes these two illnesses. The use of unbiased metabolomics for biosignature discovery does not lend itself to power calculations to determine sample size. Thus, sample sizes were selected based on our previous studies (27, 50, 51). To obtain a sufficient number of well-characterized STARI sera, retrospectively collected samples from two separate studies were used. Specifically, the first set of STARI serum samples (n=33) was obtained from the CDC repository. These samples were collected through a prospective study performed between 2007 and 2009 (57). Patients were enrolled through CDC outreach efforts (n=17) or by contract with the University of North Carolina at Chapel Hill (n=16). The states where patients were recruited included NC, 18; VA, 4; TN, 3; KY, 2; GA, 2; IA, 2; AL, 1; and NE, 1. All samples were collected pre-treatment with the exception of one patient who was treated with doxycycline 1–2 days before the serum sample was obtained. The second set of STARI samples (n=22) was obtained from the New York Medical College serum repository (20). These samples were collected between 2001 and 2004 from patients living in Missouri.
Sufficient numbers of well-characterized early Lyme disease serum samples were acquired from New York, an area of high incidence for Lyme disease and low incidence of STARI (9). Specifically, all early Lyme disease samples (n=70) were culture and/or PCR positive for B. burgdorferi and were collected pre-treatment. To ensure appropriate representation of both non-disseminated and disseminated forms of early EM Lyme disease, samples from patients with a single EM that were skin culture and/or PCR positive for B. burgdorferi and blood culture negative (n=35), and patients with multiple EMs or a single EM that were blood culture positive (n=35) were used. Early Lyme disease samples were collected between 1992 and 2007, and 1 to 33 days post-onset of symptoms. To understand the relationship of our findings to a healthy control population serum samples from healthy donors were also included in the study. These were procured from repositories at New York Medical College, the CDC and the University of Central Florida. A detailed description of inclusion and exclusion criteria for each patient and donor population is provided in table S1. All participating institutions obtained institutional review board (IRB) approval for this study. IRB review and approval for this study ensured that the retrospective samples used had been collected under informed consent.
All samples were analyzed in duplicate and were randomized prior to processing for LC-MS analyses. Healthy control sera were used as quality control samples for each LC-MS experiment. The serum samples and respective LC-MS data files of each patient group and healthy controls were randomly separated into a Discovery-Set,/Training-Sets 1 and 2, and Test-Sets 1 and 2. Specifically, 40 of the 70 early Lyme disease and 36 of the 55 STARI samples were randomly selected as the Discovery-Set samples. This sample set was used for molecular feature selection. To train the classification models, two training-sets were used. The first, Training-Set 1, was identical to the Discovery-Set (i.e. contained the same early Lyme disease and STARI samples) and the second, Training-Set 2, consisted of the same samples as Training-Set 1 with the addition of 38 of the 58 healthy control samples. Lastly, Test-Sets 1 and 2 were created. Test-Set 1 was comprised of 30 early Lyme disease and 19 STARI samples that were not included in the Discovery/Training samples sets. Test-Set 2 consisted of the same samples as those used in Test-Set 1 with the addition of 20 healthy control samples that were not included in the Training-Set 2 samples. Test-Sets 1 and 2 were exclusively used for blinded testing of the classification models.
Randomization into Discovery/Training-Sets or Test-Sets was done in a manner that ensured bias was not introduced based on the repository from which STARI samples were obtained or on whether the early Lyme disease samples were from a non-disseminated or disseminated case. Biosignature development was performed by screening MFs based on stringent criteria outlined in Fig. 1A and detailed in the Biosignature development section (below).
Lyme disease serologic testing of all serum samples
Standard two-tiered testing was performed on all samples (38). The C6 B. burgdorferi (Lyme) ELISA (Immunetics, Boston, MA) was used as a first-tier test, and any positive or equivocal samples were reflexed to Marblot IgM and IgG immunoblots (MarDx Diagnostics, Inc., Carlsbad, CA) as the second-tier test. Serologic assays were performed according to the manufacturer’s instructions, and the data were interpreted according to established CDC guidelines (38). Duration of illness, however, was not considered for test interpretation.
Liquid chromatography-mass spectrometry (LC-MS)
Serum samples were randomized prior to extraction of small molecule metabolites and LC-MS analyses. Small molecule metabolites were extracted from sera as previously reported (27). An aliquot (10 µl) of the serum metabolite extract was applied to a Poroshell 120, EC-C8, 2.1 × 100 mm, 2.7 µm LC Column (Agilent Technologies, Palo Alto, CA). The metabolites were eluted with a 2–98% nonlinear gradient of acetonitrile in 0.1% formic acid at a flow rate of 250 µl/min with an Agilent 1200 series LC system. The eluent was introduced directly into an Agilent 6520 quadrapole time of flight mass (Q-TOF) spectrometer and MS was performed as previously described (27, 50). LC-MS and LC-MS/MS data were collected under the following parameters: gas temperature, 310°C; drying gas at 10 liters per min; nebulizer at 45 lb per in2; capillary voltage, 4,000 V; fragmentation energy, 120 V; skimmer, 65 V; and octapole RF setting, 750 V. The positive-ion MS data for the mass range of 75 to 1,700 Da were acquired at a rate of 2 scans per sec. Data were collected in both centroid and profile modes in 4-GHz high-resolution mode. Positive-ion reference masses of 121.050873 m/z and 922.009798 m/z were introduced to ensure mass accuracy. To monitor instrument performance, quality control samples consisting of a metabolite extract of healthy control serum (BioreclamationIVT, Westbury, NY) was analyzed in duplicate at the beginning of each analysis day and every 20 samples during the analysis day.
Biosignature development
LC-MS data from an initial Discovery-Set of samples comprised of randomly selected early Lyme disease (n=40) and randomly selected STARI patients (n=36) that were exclusively used for molecular feature selection and classification model training were processed with the Molecular Feature Extractor algorithm tool of the Agilent MassHunter Qualitative Analysis software version B.05.00 (Agilent Technologies, Santa Clara, CA). The MFs were aligned between data files with a 0.25 min retention time window and 15 ppm mass tolerance. Comparative analyses of differentiating MFs between patient groups were performed using the workflow presented in Fig.1A. Specifically, the Discovery-Set data was analyzed using Mass Profiler Pro (MPP) software version B.12.05 (Agilent Technologies). Using MPP a univariate, unpaired t-test was performed on each metabolite to test for a difference in mean (standardized) abundance between early Lyme disease and STARI groups. Multiple testing was accounted for by computing false-discovery rate (FDR)-adjusted p-values (Benjamin and Hochberg, 1995). To prevent selection of MFs biased by uncontrolled variables (diet, other undisclosed illnesses, etc.), only MFs present in 50% or more of samples in at least one group and that differed between the groups with a significance of adjusted-p<0.05 were selected. Quantitative Analysis software version B.05.01 (Agilent Technologies) was used to extract area abundance values for all differentially selected MFs from the MS data files. Duplicate MFs were removed by assessing adduct ions, as well as mass, retention time and abundance similarities; this resulted in the Discovery MF List. A duplicate LC-MS analysis of the Discovery-Set samples was performed and the area abundance for MFs of the discovery MF List were extracted using the Quantitative Analysis software. These data with those from the first LC-MS analysis formed the Targeted-Discovery-Set.
Abundance data from the Targeted-Discovery-Set data files were normalized using a two-step method. First, abundances (area under the peak for the monoisotopic mass) of each Discovery MF were normalized by the median intensity of the stable MFs detected in each individual sample (58). Stable MFs were those identified in the original extraction of LC-MS data files with the Agilent MassHunter Qualitative Analysis software and present in at least 50% of all sample data files. Secondly, median fold changes of stable MFs between the initial quality control sample (applied at the beginning of the LC-MS analysis) and each of the subsequent quality control samples (applied every 20 clinical samples throughout the LC-MS analysis) were calculated. The median fold change calculated for the quality control sample that directly followed each series of 20 clinical samples was multiplied against the normalized Discovery-MF abundances in the clinical samples of that series. This second normalization step was performed to correct for instrument variability. To apply stringency to the development of a final early Lyme disease-STARI biosignature, MFs were filtered based on consistency in the duplicate LC-MS data sets by requiring the same directional abundance change between the patient groups. Specifically, MFs with at least a ≥ 2-fold abundance difference and a 1.5-fold abundance difference between the medians of the two groups (early Lyme disease and STARI) for LC-MS analysis-1 and LC-MS analysis-2, respectively, were selected. Further criteria applied to ensure that the most robust MFs were being selected included: removing MFs with > 20% missing values in both groups, and selecting only MFs where at least 50% of the samples within a patient group produced a fold change of ≥ 2 in comparison to the mean of the other patient group. This selection process resulted in the MFs included in the early Lyme disease-STARI biosignature.
Prediction and verification of MF chemical structure
Confirmation of the chemical structures of selected MFs was performed by LC-MS-MS to provide level-1 or level-2 identifications (59). Commercial standards palmitoyl ethanolamide, stearoyl ethanolamide, eicosanoyl ethanolamide, glycerophospho-N-palmitoyl ethanolamine, pentadecanoyl ethanolamide, and erucamide were obtained from Cayman Chemical (Ann Arbor, MI, USA). Commercial standards piperine and nonanedioic acid were obtained from Sigma Aldrich (Saint Louis, MO, USA). Commercial standards methyl oleate, stearamide, palmitamide, CMPF, and glycocholic acid were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The LC conditions used were the same as those used for the LC-MS analyses of serum metabolites. MS/MS spectra of the targeted MFs and commercial standards were obtained with an Agilent 6520 Q-TOF mass spectrometer. Electrospray ionization was performed in the positive ion mode as described for MS analyses, except the mass spectrometer was operated in the 2 GHz extended dynamic range mode. The positive ion MS/MS data (50 to 1,700 Da) were acquired at a rate of 1 scan per sec. Precursor ions were selected by the quadrupole and fragmented via collision-induced dissociation (CID) with nitrogen at collision energies of 10, 20, or 40 eV. To provide a level-1 identification, the MS/MS spectra of the targeted metabolites were compared to spectra of commercial standards. Additionally, LC retention time comparisons between the targeted MF and the respective standard were made. A retention time window of ± 5 sec was applied as a cutoff for identification. The MS/MS spectra of selected serum metabolites were compared to spectra in the Metlin database for a level-2 identification.
Metabolic pathway analysis by MetaboAnalyst
The experimentally obtained monoisotopic masses corresponding to the MFs of the 261 biosignature list were searched against HMDB using a 15 ppm window. The resulting list of potential metabolite structures were applied to the MetaboAnalyst pathway analysis tool (28) Settings for pathway analysis consisted of applying Homo sapiens pathway library; the Hypergeometric Test for the over-representation analysis and Relative-betweenness centrality to estimate node importance in the pathway topology.
Statistical analyses
Methods to filter the list of MFs and to normalize abundances are described in the section on biosignature development. Prior to analysis, the normalized abundances were log2 transformed and each MF was scaled to have a mean of zero and standard deviation of 1. Statistical analyses were performed using R software (60).
Classification Modeling
For classification modeling, Training- and Test-Set samples were used as previously described (27, 50) and as shown in Fig. 1B. Separate classification analyses were performed for comparison of two groups (early Lyme disease and STARI) and three groups (early Lyme disease, STARI and healthy controls). For each scenario, two classification approaches were applied: random forest (RF) using the RandomForest package (61), with 16 features randomly selected for each clade and a total of 500 trees; and LASSO logistic (two-way) and multinomial (three-way) regression analysis using the glmnet package (62), with the tuning parameter chosen for minimum misclassification error over a 10-fold cross-validation. The ROC curve and AUC were generated for predicted responses on the Test-Set samples only using the pROC package (63). For the purpose of visualization, LASSO scores for individual patient samples were calculated by multiplying the respective regression coefficients (tables S4 and S6) resulting from LASSO analysis by the transformed abundance of each MF in the biosignature (38 MFs in the case of two-way classification and 82 MFs in the case of three-way classification) and summing for each sample. The rgl package was used to generate the 3-dimensional scatterplot of LASSO scores (64).
Linear Discriminant Analysis
A linear discriminant analysis was performed with the 82 MFs selected by the three-way LASSO model using linear discriminant analysis function in R. MF abundance data included in the linear discriminant analysis were from healthy controls from Colorado, Florida, and New York, and from STARI patients from North Carolina, Missouri, and other states. Before linear discriminant analysis data were transformed by taking the log2 value and standardizing to the mean 0 and variance 1 within each MF. Samples were differentiated by healthy and STARI.
Supplementary Material
Acknowledgments
We thank Drs. Travis Jewett and Micah Halpern for assistance in the collection of healthy control sera at UCF, and Doug Garcia at CSU for graphics.
Funding: Supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant R21/R33 AI100228 (J.T.B and G.P.W), R01 AI099094 (M.W.J), the UCF College of Medicine (M.W.J) and the Centers for Disease Control and Prevention.
Footnotes
Fig. S1. Level 1 identification of stearoyl ethanolamide.
Fig. S2. Level 1 identification of pentadecanoyl ethanolamide.
Fig. S3. Level 1 identification of eicosanoyl ethanolamide.
Fig S4. Level 1 identification of glycerophospho-N-palmitoyl ethanolamine.
Fig. S5. Level 1 identification of stearamide.
Fig. S6. Level 1 identification of erucamide.
Fig. S7. Level 1 identification of L-phenylalanine.
Fig. S8. Level 1 identification of nonanedioic acid.
Fig. S9. Level 1 identification of glycocholic acid.
Fig. S10. Level 1 identification of 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF).
Fig S11. Level 2 identification of Lyso PA (20:4) by MS/MS spectral matching.
Fig. S12. Level 2 identification of 3-ketosphingosine by MS/MS.
Table S1. Serum samples used in this study.
Table S2. 261 MF biosignature list.
Table S3. MetaboAnalyst results.
Table S4. Regression coefficients (β) of the LASSO two-way statistical model.
Table S5. LASSO and RF two-way model classification probability scores.
Table S6. Regression coefficients (β) of the LASSO three-way statistical model.
Table S7. LASSO and RF three-way model classification probability scores.
Table S8. ANOVA results on Lasso and Random Forest scores with sample source as the explanatory variable.
Table S9. Grouping indicated by the ANOVA results.
Author contributions: C.R.M., J.T.B., G.P.W., B.J.J., L.V.A., and M.N.I. designed the experiments. A.M.H., M.J.D. and B.G.A. performed statistical analyses that included linear discriminant analyses, ROC generation and statistical modeling. L.V.A., K.W., and A.P. performed the metabolomics analyses and data processing. M.A.P., B.J.J., G.P.W., I.M. and M.W.J. helped with serum sample collections, clinical data extraction and compiling of patient data.
Competing interests: Dr. Wormser reports receiving research grants from Immunetics, Inc., Institute for Systems Biology, Rarecyte, Inc., and Quidel Corporation. He owns equity in Abbott; has been an expert witness in malpractice cases involving Lyme disease; and is an unpaid board member of the American Lyme Disease Foundation.
References and Notes
- 1.Hinckley AF, Connally NP, Meek JI, Johnson BJ, Kemperman MM, Feldman KA, White JL, Mead PS. Lyme disease testing by large commercial laboratories in the United States. Clin. Infect. Dis. 2014;59:676–681. doi: 10.1093/cid/ciu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson CA, Saha S, Kugeler KJ, Delorey MJ, Shankar MB, Hinckley AF, Mead PS. Incidence of Clinician-Diagnosed Lyme Disease, United States, 2005–2010. Emerg. Infect. Dis. 2015;21:1625–1631. doi: 10.3201/eid2109.150417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet. 2012;379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- 4.Steere AC, Malawista SE, Hardin JA, Ruddy S, Askenase W, Andiman WA. Erythema chronicum migrans and Lyme arthritis. The enlarging clinical spectrum. Ann. Intern. Med. 1977;86:685–698. doi: 10.7326/0003-4819-86-6-685. [DOI] [PubMed] [Google Scholar]
- 5.Wormser GP. Clinical practice. Early Lyme disease. N. Engl. J. Med. 2006;354:2794–2801. doi: 10.1056/NEJMcp061181. [DOI] [PubMed] [Google Scholar]
- 6.Nadelman RB. Erythema migrans. Infect. Dis. Clin. North Am. 2015;29:211–239. doi: 10.1016/j.idc.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Masters EJ, Donnell HD. Epidemiologic and diagnostic studies of patients with suspected early Lyme disease Missouri, 1990–1993. J. Infect. Dis. 1996;173:1527–1528. doi: 10.1093/infdis/173.6.1527. [DOI] [PubMed] [Google Scholar]
- 8.Masters E, Granter S, Duray P, Cordes P. Physician-diagnosed erythema migrans and erythema migrans-like rashes following Lone Star tick bites. Arch. Dermatol. 1998;134:955–960. doi: 10.1001/archderm.134.8.955. [DOI] [PubMed] [Google Scholar]
- 9.Wormser GP, Masters E, Nowakowski J, McKenna D, Holmgren D, Ma K, Ihde L, Cavaliere LF, Nadelman RB. Prospective clinical evaluation of patients from Missouri and New York with erythema migrans-like skin lesions. Clin. Infect. Dis. 2005;41:958–965. doi: 10.1086/432935. [DOI] [PubMed] [Google Scholar]
- 10.Campbell GL, Paul WS, Schriefer ME, Craven RB, Robbins KE, Dennis DT. Epidemiologic and diagnostic studies of patients with suspected early Lyme disease, Missouri, 1990–1993. J. Infect. Dis. 1995;172:470–480. doi: 10.1093/infdis/172.2.470. [DOI] [PubMed] [Google Scholar]
- 11.Clark KL, Leydet B, Hartman S. Lyme borreliosis in human patients in Florida and Georgia, USA. Int. J. Med. Sci. 2013;10:915–931. doi: 10.7150/ijms.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bacon RM, Gilmore RD, Jr, Quintana M, Piesman J, Johnson BJ. DNA evidence of Borrelia lonestari in Amblyomma americanum (Acari: Ixodidae) in southeast Missouri. J. Med. Entomol. 2003;40:590–592. doi: 10.1603/0022-2585-40.4.590. [DOI] [PubMed] [Google Scholar]
- 13.Varela AS, Luttrell MP, Howerth EW, Moore VA, Davidson WR, Stallknecht DE, Little SE. First culture isolation of Borrelia lonestari, putative agent of southern tick-associated rash illness. J. Clin. Microbiol. 2004;42:1163–1169. doi: 10.1128/JCM.42.3.1163-1169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James AM, Liveris D, Wormser GP, Schwartz I, Montecalvo MA, Johnson BJ. Borrelia lonestari infection after a bite by an Amblyomma americanum tick. J. Infect. Dis. 2001;183:1810–1814. doi: 10.1086/320721. [DOI] [PubMed] [Google Scholar]
- 15.Grigery CN, Moyer P, Little SE, Masters EJ. Bacteriocidal activity of lizard and mouse serum for Borrelia lonestari, putative agent of a Lyme-like illness (AKA STARI or Masters disease) in Missouri. Mo. Med. 2005;102:442–446. [PubMed] [Google Scholar]
- 16.Philipp MT, Masters E, Wormser GP, Hogrefe W, Martin D. Serologic evaluation of patients from Missouri with erythema migrans-like skin lesions with the C6 Lyme test. Clin. Vaccine Immunol. 2006;13:1170–1171. doi: 10.1128/CVI.00238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felz MW, Chandler FW, Jr, Oliver JH, Jr, Rahn DW, Schriefer ME. Solitary erythema migrans in Georgia and South Carolina. Arch. Dermatol. 1999;135:1317–1326. doi: 10.1001/archderm.135.11.1317. [DOI] [PubMed] [Google Scholar]
- 18.Kirkland KB, Klimko TB, Meriwether RA, Schriefer M, Levin M, Levine J, Mac Kenzie WR, Dennis DT. Erythema migrans-like rash illness at a camp in North Carolina: a new tick-borne disease? Arch. Intern. Med. 1997;157:2635–2641. [PubMed] [Google Scholar]
- 19.Rich SM, Armstrong PM, Smith RD, Telford SR., 3rd Lone star tick-infecting borreliae are most closely related to the agent of bovine borreliosis. J. Clin. Microbiol. 2001;39:494–497. doi: 10.1128/JCM.39.2.494-497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wormser GP, Masters E, Liveris D, Nowakowski J, Nadelman RB, Holmgren D, Bittker S, Cooper D, Wang G, Schwartz I. Microbiologic evaluation of patients from Missouri with erythema migrans. Clin. Infect. Dis. 2005;40:423–428. doi: 10.1086/427289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters EJ, Donnell HD. Lyme and/or Lyme-like disease in Missouri. Mo. Med. 1995;92:346–353. [PubMed] [Google Scholar]
- 22.Nicholson WL, Masters E, Wormser GP. Preliminary serologic investigation of 'Rickettsia amblyommii' in the aetiology of Southern tick associated rash illness (STARI) Clin. Microbiol. Infect. 2009;15(Suppl 2):235–236. doi: 10.1111/j.1469-0691.2008.02155.x. [DOI] [PubMed] [Google Scholar]
- 23.Springer YP, Jarnevich CS, Barnett DT, Monaghan AJ, Eisen RJ. Modeling the Present and Future Geographic Distribution of the Lone Star Tick, Amblyomma americanum (Ixodida: Ixodidae), in the Continental United States. Am. J. Trop. Med. Hyg. 2015;93:875–890. doi: 10.4269/ajtmh.15-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn MB, Jarnevich CS, Monaghan AJ, Eisen RJ. Modeling the Geographic Distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the Contiguous United States. J. Med. Entomol. 2016 doi: 10.1093/jme/tjw076. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kugeler KJ, Farley GM, Forrester JD, Mead PS. Geographic Distribution and Expansion of Human Lyme Disease, United States. Emerg. Infect. Dis. 2015;21:1455–1457. doi: 10.3201/eid2108.141878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lantos PM, Nigrovic LE, Auwaerter PG, Fowler VG, Jr, Ruffin F, Brinkerhoff RJ, Reber J, Williams C, Broyhill J, Pan WK, Gaines DN. Geographic Expansion of Lyme Disease in the Southeastern United States, 2000–2014. Open Forum Infect. Dis. 2015;2 doi: 10.1093/ofid/ofv143. ofv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molins CR, Ashton LV, Wormser GP, Hess AM, Delorey MJ, Mahapatra S, Schriefer ME, Belisle JT. Development of a metabolic biosignature for detection of early Lyme disease. Clin. Infect. Dis. 2015;60:1767–1775. doi: 10.1093/cid/civ185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iannotti FA, Di Marzo V, Petrosino S. Endocannabinoids and endocannabinoid-related mediators: Targets, metabolism and role in neurological disorders. Prog. Lipid Res. 2016;62:107–128. doi: 10.1016/j.plipres.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Forrester JD, Brett M, Matthias J, Stanek D, Springs CB, Marsden-Haug N, Oltean H, Baker JS, Kugeler KJ, Mead PS, Hinckley A. Epidemiology of Lyme disease in low-incidence states. Ticks Tick Borne Dis. 2015;6:721–723. doi: 10.1016/j.ttbdis.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Dalle Carbonare M, Del Giudice E, Stecca A, Colavito D, Fabris M, D'Arrigo A, Bernardini D, Dam M, Leon A. A saturated N-acylethanolamine other than N-palmitoyl ethanolamine with anti-inflammatory properties: a neglected story. J. Neuroendocrinol. 2008;20(Suppl 1):26–34. doi: 10.1111/j.1365-2826.2008.01689.x. [DOI] [PubMed] [Google Scholar]
- 32.Di Marzo V, Melck D, Orlando P, Bisogno T, Zagoory O, Bifulco M, Vogel Z, De Petrocellis L. Palmitoylethanolamide inhibits the expression of fatty acid amide hydrolase and enhances the anti-proliferative effect of anandamide in human breast cancer cells. Biochem. J. 2001;358:249–255. doi: 10.1042/0264-6021:3580249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo Verme J, Fu J, Astarita G, La Rana G, Russo R, Calignano A, Piomelli D. The nuclear receptor peroxisome proliferator-activated receptor-alpha mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 2005;67:15–19. doi: 10.1124/mol.104.006353. [DOI] [PubMed] [Google Scholar]
- 34.Bradshaw HB, Rimmerman N, Hu SS, Benton VM, Stuart JM, Masuda K, Cravatt BF, O'Dell DK, Walker JM. The endocannabinoid anandamide is a precursor for the signaling lipid N-arachidonoyl glycine by two distinct pathways. BMC Biochem. 2009;10:14. doi: 10.1186/1471-2091-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 36.Divito EB, Cascio M. Metabolism, physiology, and analyses of primary fatty acid amides. Chem. Rev. 2013;113:7343–7353. doi: 10.1021/cr300363b. [DOI] [PubMed] [Google Scholar]
- 37.Masters EJ, Grigery CN, Masters RW. STARI, or Masters disease: Lone Star tick-vectored Lyme-like illness. Infect. Dis. Clin. North Am. 2008;22:361–376. doi: 10.1016/j.idc.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 38.C. f. D. C. a. Prevention. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. Morb. Mortal. Wkly. Rep. 1995;44:590–591. [PubMed] [Google Scholar]
- 39.Aguero-Rosenfeld ME, Wang G, Schwartz I, Wormser GP. Diagnosis of lyme borreliosis. Clin. Microbiol. Rev. 2005;18:484–509. doi: 10.1128/CMR.18.3.484-509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masters EJ. Lyme-like illness currently deserves Lyme-like treatment. Clin. Infect. Dis. 2006;42:580–581. doi: 10.1086/500018. author reply 581–582. [DOI] [PubMed] [Google Scholar]
- 41.Comstedt P, Hanner M, Schuler W, Meinke A, Schlegl R, Lundberg U. Characterization and optimization of a novel vaccine for protection against Lyme borreliosis. Vaccine. 2015;33:5982–5988. doi: 10.1016/j.vaccine.2015.07.095. [DOI] [PubMed] [Google Scholar]
- 42.Wressnigg N, Barrett PN, Pollabauer EM, O'Rourke M, Portsmouth D, Schwendinger MG, Crowe BA, Livey I, Dvorak T, Schmitt B, Zeitlinger M, Kollaritsch H, Esen M, Kremsner PG, Jelinek T, Aschoff R, Weisser R, Naudts IF, Aichinger G. A Novel multivalent OspA vaccine against Lyme borreliosis is safe and immunogenic in an adult population previously infected with Borrelia burgdorferi sensu lato. Clin. Vaccine Immunol. 2014;21:1490–1499. doi: 10.1128/CVI.00406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomes-Solecki M. Blocking pathogen transmission at the source: reservoir targeted OspA-based vaccines against Borrelia burgdorferi. Front Cell Infect. Microbiol. 2014;4:136. doi: 10.3389/fcimb.2014.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richer LM, Brisson D, Melo R, Ostfeld RS, Zeidner N, Gomes-Solecki M. Reservoir targeted vaccine against Borrelia burgdorferi: a new strategy to prevent Lyme disease transmission. J. Infect. Dis. 2014;209:1972–1980. doi: 10.1093/infdis/jiu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson BJB. Lyme Disease: An Evidence-based Approach. chap. 4. CAB International; Wallingford, Oxfordshire, UK; Cambridge, MA: 2011. pp. 73–87. [Google Scholar]
- 46.Wormser GP, Levin A, Soman S, Adenikinju O, Longo MV, Branda JA. Comparative cost-effectiveness of two-tiered testing strategies for serodiagnosis of lyme disease with noncutaneous manifestations. J. Clin. Microbiol. 2013;51:4045–4049. doi: 10.1128/JCM.01853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strathmann FG, Hoofnagle AN. Current and future applications of mass spectrometry to the clinical laboratory. Am. J. Clin. Pathol. 2011;136:609–616. doi: 10.1309/AJCPW0TA8OBBNGCK. [DOI] [PubMed] [Google Scholar]
- 48.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 49.Dumlao DS, Buczynski MW, Norris PC, Harkewicz R, Dennis EA. High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines. Biochim. Biophys. Acta. 2011;1811:724–736. doi: 10.1016/j.bbalip.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahapatra S, Hess AM, Johnson JL, Eisenach KD, DeGroote MA, Gitta P, Joloba ML, Kaplan G, Walzl G, Boom WH, Belisle JT. A metabolic biosignature of early response to anti-tuberculosis treatment. BMC Infect. Dis. 2014;14:53. doi: 10.1186/1471-2334-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voge NV, Perera R, Mahapatra S, Gresh L, Balmaseda A, Lorono-Pino MA, Hopf-Jannasch AS, Belisle JT, Harris E, Blair CD, Beaty BJ. Metabolomics-Based Discovery of Small Molecule Biomarkers in Serum Associated with Dengue Virus Infections and Disease Outcomes. PLoS Negl. Trop. Dis. 2016;10:e0004449. doi: 10.1371/journal.pntd.0004449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tritten L, Keiser J, Godejohann M, Utzinger J, Vargas M, Beckonert O, Holmes E, Saric J. Metabolic profiling framework for discovery of candidate diagnostic markers of malaria. Sci. Rep. 2013;3:2769. doi: 10.1038/srep02769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinayavekhin N, Mahipant G, Vangnai AS, Sangvanich P. Untargeted metabolomics analysis revealed changes in the composition of glycerolipids and phospholipids in Bacillus subtilis under 1-butanol stress. Appl. Microbiol. Biotechnol. 2015;99:5971–5983. doi: 10.1007/s00253-015-6692-0. [DOI] [PubMed] [Google Scholar]
- 54.Clark KL, Leydet BF, Threlkeld C. Geographical and genospecies distribution of Borrelia burgdorferi sensu lato DNA detected in humans in the USA. J. Med. Microbiol. 2014;63:674–684. doi: 10.1099/jmm.0.073122-0. [DOI] [PubMed] [Google Scholar]
- 55.Oliver JH, Jr, Lin T, Gao L, Clark KL, Banks CW, Durden LA, James AM, Chandler FW., Jr An enzootic transmission cycle of Lyme borreliosis spirochetes in the southeastern United States. Proc. Natl. Acad. Sci. U S A. 2003;100:11642–11645. doi: 10.1073/pnas.1434553100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feder HM, Jr, Hoss DM, Zemel L, Telford SR, 3rd, Dias F, Wormser GP. Southern Tick-Associated Rash Illness (STARI) in the North: STARI following a tick bite in Long Island, New York. Clin. Infect. Dis. 2011;53:e142–146. doi: 10.1093/cid/cir553. [DOI] [PubMed] [Google Scholar]
- 57.Vaughn MF, Sloane PD, Knierim K, Varkey D, Pilgard MA, Johnson BJ. Practice-Based Research Network Partnership with CDC to acquire clinical specimens to study the etiology of southern tick-associated rash illness (STARI) J. Am. Board Fam. Med. 2010;23:720–727. doi: 10.3122/jabfm.2010.06.100098. [DOI] [PubMed] [Google Scholar]
- 58.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 59.Salek RM, Steinbeck C, Viant MR, Goodacre R, Dunn WB. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience. 2013;2:13. doi: 10.1186/2047-217X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R. C. Team. R Foundation for Statistical Computing. Vienna, Austria: 2016. [Google Scholar]
- 61.Wiener ALaM. Classification and Regression by randomForest. R News. 2002;3:18–22. [Google Scholar]
- 62.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 63.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adler DMD. rgl: 3D Visualization Using OpenGL. R package version 0.96.0. 2016 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.