Abstract
Since the discovery of c-di-GMP almost three decades ago, cyclic dinucleotides (CDNs) have emerged as widely used signaling molecules in most kingdoms of life. The family of second messengers now includes c-di-AMP and distinct versions of mixed cyclic GMP-AMP (cGAMP) compounds. Along with these nucleotides, a vast number of proteins for the production and turnover of these molecules have been described, as well as effectors that translate the signals into physiological responses. The latter include but are not limited to mechanisms for adaptation and survival in prokaryotes, persistence and virulence of bacterial pathogens, as well as immune responses to viral and bacterial invasion in eukaryotes. In this review we will focus on recent discoveries and emerging themes that illustrate the ubiquity and versatility of cyclic dinucleotide function at the transcriptional and post-translational levels and, in particular, on insights gained through mechanistic structure-function analyses.
Introduction
In 1987, the discovery of cyclic 3′,5′-diguanylic acid (c-di-GMP) as an allosteric regulator of cellulose biosynthesis in Gluconacetobacter xylinus1 was reported (Figure 1a). The group behind this study also described the first c-di-GMP binding protein associated with the cellulose synthase complex2 and the diguanylate cyclases (DGCs) and phosphodiesterases (PDEs) responsible for c-di-GMP production and degradation, respectively (Figure 1b)1,3. Despite these early discoveries, their impact only became evident later when c-di-GMP and its metabolizing enzymes were identified in multiple other bacteria including Caulobacter crescentus – a popular model for studying asymmetric cell division4,5, Escherichia coli – the workhorse of bacterial genetics6, and pathogenic Salmonella enterica, Pseudomonas aeruginosa, and Vibrio cholerae6–9. These and related works were fueled by comprehensive bioinformatics studies that revealed the conservation of c-di-GMP metabolizing units in the vast majority of bacterial species10. Over the following decade, c-di-GMP transpired as a key bacterial second messenger whose underlying signaling networks control major adaptational and life-style changes, including biofilm formation and pathogenicity (Figure 2).
Figure 1. Cyclic dinucleotide (CDN) signaling.
a. Structural overview of the four prevalent cyclic dinucleotides. b. Regulation of CDN signaling. A generic route for the synthesis and degradation of CDNs is shown. Regulatory feedback loops controlling cellular CDN levels have been described in some instances, e.g. for I-site-containing GGDEF domain proteins or the effect of linear di-GMP (pGpG) on PDE-A activity. Note that the listed receptor/effector classes are common examples and not mutually exclusive. Enzymatic activities arise from stand-alone, single-domain proteins or, more typically, from multi-domain proteins, including proteins with both cyclase (i.e. GGDEF) and PDE (i.e. EAL) domains. Regulatory domains (X) determine the mode of enzyme regulation (e.g. environmental sensing, canonical two-component regulation, etc.). Known stimuli include allosteric ligands, post-translational modifications, light, gases, and mechanotransduction. c. Prevalence, biosynthesis and degradation pathways for the four major CDNs.
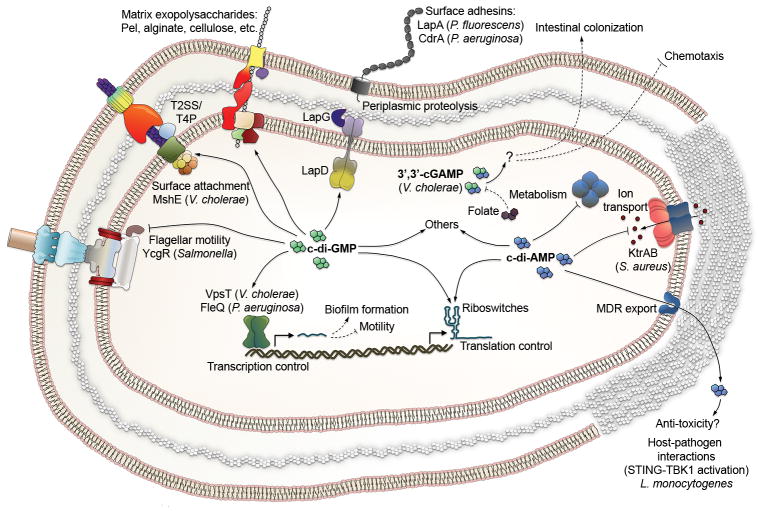
Figure 2. Bacterial CDNs and representative regulatory mechanisms.
Solid lines show direct binding or transport; dashed lines illustrate indirect effects. The prevalent distribution of c-di-GMP/cGAMP and c-di-AMP in Gram-negative and Gram-positive bacteria, respectively, is illustrated by differences in the cell envelope, with a thick peptidoglycan layer illustrating the Gram-positive cell wall. Abbreviations are as follows: T2SS - Type 2 secretion system; T4P - type 4 pili; LmPC - L. monocytogenes pyruvate carboxylase.
Trailing the discovery of c-di-GMP by about two decades, the second CDN, c-di-AMP, surfaced serendipitously during crystallographic analyses of a bacterial checkpoint protein that turned out to harbor diadenylate cyclase (DAC) activity (Figures 1a and 1c)11. C-di-AMP has since been found in many bacterial and archeal species. Interestingly, c-di-AMP can be both essential, which makes it unique among second messengers, and toxic, when overproduced12–14. Similarly to c-di-GMP, c-di-AMP controls a spectrum of cellular processes, including gene expression15, DNA repair11, cell wall synthesis16, metabolism17 and potassium homeostasis (Figure 2)12,18,19.
The latest additions to the family of CDNs are hybrid cGAMP molecules (Figure 1a). 3′,3′-cGAMP was first identified as a second messenger regulating chemotaxis and intestinal colonization in V. cholerae20, but shortly thereafter numerous reports revealed a distinct 2′,3′-cGAMP as key to innate immune signaling and antiviral response in eukaryotes (Figure 2).
Adding to the complexity of CDN signal transduction is the conformational flexibility of the nucleotides themselves, both in solution and bound to proteins and riboswitches (Figures 3a and 3b). This degree of freedom contributes not only to the versatility and vast spectrum of processes controlled by these ubiquitous second messengers, but also to difficulties in the prediction of novel CDN targets.
Figure 3. Conformational adaptability and mechanism of action of CDNs.
a. Using c-di-GMP as an example, CDN conformations found in protein co-crystal structures are depicted. Several of these conformations have also been shown to be sampled in solution. b. Representative cases of protein regulation via CDNs, exemplified by c-di-GMP binding modules, are shown. Cartoons depict concepts derived from available crystal structures (or modeling in the case CLP). A detailed description of these modes of action are provided in the main text.
Cyclic dinucleotide (CDN) synthesis and degradation
Distinct enzymes are responsible for the biosynthesis and degradation of the different CDNs (Figure 1c). C-di-GMP-producing DGCs contain GGDEF domains with adenylyl cyclase-like folds21,22. The first structure of a DGC, PleD from C. crescentus, also revealed the so-called inhibitory (I−) site as prevalent among GGDEF domain-containing proteins22 (Figure 1b). A conserved I-site RxxD motif partakes in a secondary c-di-GMP binding site, that is distal from the enzyme’s active site and can be involved in negative-feedback regulation and/or protein-protein interactions22,23. For c-di-GMP degradation, β-barrel EAL or the unrelated HD-GYP domains confer specific PDE activity to proteins24,25. While HD-GYP domains typically break the CDN fully to GMP, EAL domains usually lead to the accumulation of linear di-GMP (pGpG), which may suggest additional roles for pGpG in intracellular signaling26. Recently, oligoribonuclease was shown to be the main enzyme to complete the catabolism of pGpG to GMP27,28 (Figure 1c).
Typically, GGDEF and EAL domains are part of multidomain proteins and are flanked by regulatory modules involved in environmental sensing and activity control by altering the intramolecular domain arrangement and/or oligomeric state of the proteins (Figure 1b)10. This includes proteins where the regulatory modules can be catalytically inactive GGDEF or EAL domains5, as well as proteins with dual catalytic activity. In addition, most bacteria encode more than one GGDEF, EAL, or tandem domain-containing protein and their number and complexity of the downstream signaling networks correlates roughly with the adaptability and ‘IQ’ of the organism in changing environmental conditions29.
While c-di-AMP-producing DAC domains are structurally distinct from GGDEF domains, they also occur as modules in larger proteins, where additional domains can impact the enzymes’ quaternary structure and activity30. Another parallel to c-di-GMP signaling is the occurrence of c-di-AMP-specific PDE activity in two different protein folds, DHH/DHHA1 and HD domains, (Figure 1c)13,31, but thus far, c-di-AMP signaling networks are perceived as less complex and widespread than their c-di-GMP counterparts.
V. cholerae DncV and metazoan cGAS are functional homologs producing 3′,3′-cGAMP and 2′,3′-cGAMP, respectively (Figure 1c)32. Interestingly, although they share less than 10% sequence homology and use distinct reaction paths to generate their products, they share striking structural homology32. Reverse engineering of the human cGAS’ active site based on that of DncV produced cGAS variants that synthesized exclusively 3′,3′-cGAMP32. Furthermore, a recent study also identified folate as an unexpected regulator of DncV33. Although the exact purpose of regulation by folate is not well understood, it is intriguing to note that folate binds to DncV in a similar pocket as double-stranded DNA does to cGAS33. Together, these observations paint an evolutionary picture in which metazoan cells could have adopted a bacterial cyclase to create, with relatively modest changes, a cytosolic DNA sensor as defence against intracellular pathogens. Alternatively, the occurance of cGAS-like activity in two kigndoms of life could indicate convergent enzyme evolution as a result of similar environmental pressures or cues.
Adding to the complexity of CDN signaling, a recent study identified GGDEF domain-containing proteins, dubbed hybrid promiscuous (Hypr) GGDEF enzymes, that produced 3′,3′-cGAMP and c-di-AMP, in addition to c-di-GMP, as a function of cellular ATP:GTP ratio (Figure 1c)34. This raises the question whether other enzymes exist that can produce alternative linkages or use distinct substrates, expanding the second messenger chemical space and potential physiological effects.
CDN protein sensors and physiological effects
Today’s wealth of DNA sequencing data and cross-genome comparative studies has allowed the identification of conserved signaling modules implicated in both CDN metabolism and signal transmission10. As discussed, bacteria can encode multiple conserved GGDEF and EAL domain-containing proteins and while a number of these modules lack conserved residues necessary for catalysis, they could nevertheless serve in dinucleotide signal relay. To date, several examples of inactive EAL domains binding c-di-GMP at their degenerate active sites have been described as signal transduction modules35,36. The I-site on GGDEF domains, on the other hand, can not only serve for feedback inhibition in the case of active enzymes, but can also provide a mechanism for c-di-GMP sensing and/or signal transmission in both degenerate and active DGCs23,37,38. Bioinformatics studies have also pinpointed PilZ domains as bona fide c-di-GMP sensors based on a phyletic distribution similar to those of GGDEF and EAL modules and a likely role in c-di-GMP mediated processes39. While such ‘educated guesswork’ has identified a number of other CDN sensors as well40–42, recent advances have offered various unbiased screening approaches. An important example is the development of functionalized (e.g. biotinylated) dinucleotide homologues as capture compounds for the selective pull-down of CDN-binding proteins17,18,43,44. In addition, the Differential Radial Capillary Action of Ligand Assay (DRaCALA) – a method relying on labeled, protein-bound ligand retention upon spotting onto a nitrocellulose membrane – has been used with both purified proteins and expression library lysates for the systematic identification of novel CDN sensors18,45,46.
Transmembrane signaling through CDN turnover domains
Many proteobacteria are expected to utilize a tripartite ‘inside-out’ signaling system for biofilm formation (Figure 4a)36,47. In various Pseudomonads, the core of the system consists of the transmembrane c-di-GMP sensor LapD, a periplasmic protease LapG and a surface adhesion system36,47,48. LapD is a dimeric, transmembrane protein with a catalytically inactive cytosolic GGDEF-EAL tandem that senses intracellular c-di-GMP. At low c-di-GMP levels LapD adopts an autoinhibited conformation. In it the GGDEF module occludes the c-di-GMP-binding pocket on the EAL domain as the latter interacts with a helical extension (signaling or S-helix) of the membrane-proximal HAMP domain36. Increased cellular c-di-GMP has been proposed to induce ‘swing-and-lock’ conformational changes in the cytosolic modules, which relieve the autoinhibition and are stabilized by direct EAL domain dimerization36. Importantly, the HAMP domain transmits these changes to the periplasmic Per/Arnt/Sim (PAS)-like output domain, upon which LapD sequesters its partner protease LapG and prevents proteolytic release of biofilm-associated surface adhesins36,47,49.
Figure 4. Tripartite transmembrane signaling through HAMP domain-containing proteins with active or degenerate GGDEF and EAL domains.

a. Inside-out signaling via c-di-GMP. A model of ‘inside-out’ signaling through the LapADG system in P. fluorescens is shown integrating crystallographic snapshots of the LapDG components. Conformational changes upon c-di-GMP recognition via the LapDEAL module lead to release of autoinhibitory intramolecular interactions, transmembrane signal transmission to the periplasmic output domain, and LapG protease sequestration. LapG recruitment to LapD• c-di-GMP prevents the protease reaching its substrate, LapA, at the outer membrane surface. As a result, LapA is retained at the cell surface, constituting an important regulatory step in biofilm formation. b. ‘Outside-in’ signaling via the YfiBNR system in P. aeruginosa. YfiN is a HAMP domain-containing DGC. Cell wall stress can release YfiN inhibition through the sequestration of the inhibitory YfiR partner by the outer membrane component YfiB. Conformational changes are transmitted through the membrane and a membrane-proximal HAMP module to activate intracellular c-di-GMP production.
While this model explains ‘inside-out’ transmission of the c-di-GMP signal, it does not explain CDN access to its binding pockets in the first place. Follow-up studies on the periplasmic domain’s interaction with LapG suggested that LapD acts as a transmembrane coincidence detector: transient, low-affinity LapDG interactions generate ‘outside-in’ signals that destabilize the S-helix-EAL domain autoinhibitory interaction. Coincident increase in c-di-GMP would in turn stabilize the active intracellular conformation and signal out to enhance periplasmic LapG sequestration49.
The ‘outside-in’ signaling component suggests that LapD may have evolved from active transmembrane enzymes that sense periplasmic signals to adjust their intracellular catalytic activity. One such example is the conserved YfiBNR tripartite system that activates upon cell wall and/or periplasmic redox stress in P. aeruginosa (Figure 4b)50,51. YfiN is a dimeric, transmembrane DGC with a periplasmic PAS domain and an intracellular HAMP-GGDEF module. Upon cell envelope stress, the periplasmic protein YfiR gets released from an inhibitory complex with the cyclase’s PAS domain to interact with an outer membrane acceptor, YfiB. Concomitantly, the conformational changes in the freed PAS domain are transmitted intracellularly to activate the DGC via GGDEF domain dimerization50. Thus, the molecular events regulating YfiN are very similar but reversed to those described for the LapDG system, suggesting that the latter could have diverged from active outside-in signaling enzymes.
CDN-binding transcription factors
Gene expression modulation constitutes a recurrent theme in CDN signal transduction. It can be achieved at the translational level through CDN-binding riboswitches or at the transcription initiation level through targeting transcription factors (Figure 2 and Figure 5). While only one protein – the repressor DarR controlling fatty acid metabolism and cold shock protein expression in Mycobacterium smegmatis – has been identified as a direct transcription effector specific for c-di-AMP15, c-di-GMP targets several and diverse transcription regulators (Figure 3). Examples include the master biofilm regulators VpsT (Figure 5a) and VpsR of V. cholerae41,42, the mobility regulator FleQ of P. aeruginosa (Figure 5b) and its homolog FlrA of V. cholerae40,52, the sporulation repressor BldD in filamentous actinomycetes (Figure 5c)53, the CRP/FNR-like virulence gene regulator Clp of Xanthomonas spp.54, and the TetR-like regulator of lipid transport and metabolism genes in M. smegmatis, LtmA55. As opposed to membrane-associated signaling proteins, c-di-GMP–dependent transcription factors typically lack standard recognition modules such as GGDEF, EAL or PilZ domains.
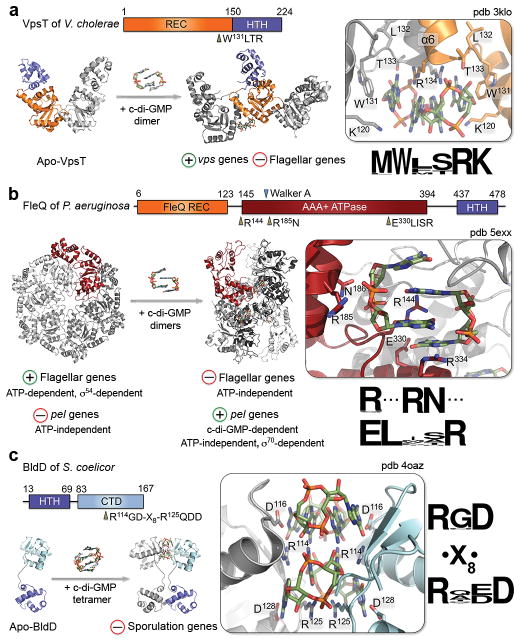
Figure 5. Structures and nucleotide recognition of c-di-GMP-regulated transcription factors.
Domain organizations, crystal structures, c-di-GMP-dependent changes in oligomerization, CDN-binding sites, consensus binding motifs, and observed physiological effects are shown for VpsT of V. cholerae (a), FleQ of P. aeruginosa (b), and BldD of S. coelicor (c). Key binding site side chains, as well as bound c-di-GMP molecules, are shown as sticks. Abbreviations are as introduced in the text: REC, receiver domain; HTH, helix-turn-helix motif; CTD, C-terminal domain.
Several of the identified c-di-GMP-regulated transcription factors, including VpsT, FleQ, and FlrA, can be viewed as orphan response regulators (RRs) that contain a phosphorylation-incompetent receiver (REC) domain and/or lack an associated histidine kinase (Figures 5a and 5b). VpsT inversely controls Vibrio exopolysaccharide secretion and flagellar mobility to induce biofilm formation in response to elevated c-di-GMP41. It belongs to the LuxR/FixJ family of RRs and carries a helix-turn-helix (HTH) DNA-binding module C-terminal to its REC domain (Figure 5a)41. The latter utilizes two unusual dimerization interfaces: i) an α1 dimerization interface that is sampled in the absence of dinucleotide and ii) a dimerization interface provided by a C-terminal helical extension of the canonical REC domain fold, α6, that both contributes to and depends on dimer-to-dimer CDN recognition41. Dimeric c-di-GMP binds to an exposed M(W/F/M)(T/S)RK motif in a pocket formed at the N-proximal end of this interface (Figures 5a and 3)41. While structural in vitro analyses suggested the formation of higher-order VpsT oligomers exploiting both dimerization interfaces, fluorescence imaging studies have visualized the c-di-GMP-dependent formation of such macromolecular VpsT clusters in cellulo56. The resultant reorganization of bound promoter DNA causing downstream effects on target gene expression, however, remains to be further examined.
Although FleQ and FlrA contain a divergent REC domain at their N-terminus, c-di-GMP binds to a central AAA+ ATPase σ54-interaction (AAA) domain that precedes the C-terminal HTH module (Figure 5b). By domain organization and sequence homology the proteins are classified as NtrC-like bacterial enhancer binding proteins (bEBPs) but are subject to non-canonical signal regulation. FleQ controls flagellar motility, CdrAB adhesin expression and Pel exopolysaccharide secretion to secure planktonic to sessile transition. Mechanistically, it exhibits remarkable versatility as it can act as an ATP- and σ54-dependent transcription activator, a nucleotide-independent repressor or, finally, an ATP-independent, c-di-GMP- and σ70-dependent transcription activator depending on the target promoters and nucleotide availability57,58.
Recent structural data of the apo–, ADP– and ATP-γ-S–bound AAA domain of FleQ reveal conformations consistent with the activated hexameric rings of classical bEBPs (Figure 5b)59. While at flagellar promoters the hexamers likely activate σ54–dependent transcription through ATP hydrolysis, at the pel promoter they are proposed to act as repressors. As revealed by the crystal structure of the c-di-GMP–complexed AAA domain, binding of c-di-GMP occurs at a composite site distinct from the ATP substrate pocket and leads allosterically to active site obstruction, hexameric ring destabilization and putative reorganization into a discrete dimer-of-trimers hexameric species (Figures 5b and 3b)59. This is proposed to facilitate not only flagellar gene deactivation, but also pel promoter de-repression and the characteristic σ70-dependent transcription activation59.
FleQ binds dimeric c-di-GMP through a trio of conserved motifs (Figures 5b and 3a): a proximal LFR144S motif (R-switch) located at the N-terminus of the bilobal AAA domain, residues R185N186 in helix α7 in the N-terminal lobe (post-Walker A), and a distal E330xxxR334 motif in helix α13 of the C-terminal lobe. These motifs are conserved in FlrA, where earlier modeling and functional studies had helped identify the key role of arginines R135 and R176 (R144 and R185 in FleQ, respectively) in CDN binding52,59. Interestingly, while VpsR shares very similar domain architecture and has been shown to bind c-di-GMP42, it features a phosphorylation-competent REC domain and does not share conservation with FleQ/FlrA’s c-di-GMP binding motifs59,60.
Sporulating actinomycetes’ BldD employs a drastically different mode of c-di-GMP sensing (Figures 5c and 3)53. It binds DNA through an N-terminal XRE HTH motif and acts as a master regulator to inhibit sporulation during vegetative growth61. BldD of Streptomyces venezuelae was recovered during c-di-GMP-affinity purification of cell lysates and c-di-GMP was confirmed to bind and induce dimerization of its C-terminal winged-helix domain (BldDCTD) in vitro53. Strikingly, crystal structures of BldDCTD–c-di-GMP complexes (S. venezuelae and S. coelicor), as well as of full length BldD bound to both c-di-GMP and cognate DNA, demonstrate that c-di-GMP binds as an ion-free tetramer to form a bridge between the two BldDCTD modules that are otherwise ~10 Å apart (Figures 5c and 3)53. The CDN binds to a consensus RxD-x8-RxxD motif as two hydrogen-bonded, intercalated dimers in a sequential and cooperative manner (Figures 5c and 3a). This leads to BldD dimerization stabilized by weak hydrophobic contacts between the HTH motifs and by binding to the pseudo-palindromic cognate DNA53.
Membrane transport and ion homeostasis
While cellular homeostasis requires constant transport of small molecules and ions across the plasma membrane, some cells actively secrete substances as a hallmark of their physiology or differentiation. CDNs have been shown not only to regulate such processes in both pro- and eukaryotic cells, but also in some cases to signal in trans, i.e. by traversing the envelope of their cells of origin.
Bacterial potassium transporters are central to the upkeep of osmotic and pH homeostasis. Based on unbiased c-di-AMP-affinity pull-downs, the KtrAB complex of Staphylococcus aureus was identified as a direct c-di-AMP target (Figure 2) and similar regulatory mechanisms have been since confirmed in additional species12,18. Potassium transport is mediated by the membrane KtrB subunit, which forms a four-repeat pseudosymmetric transporter related to tetrameric cation channels (Figure 6a). A KtrB dimer interacts with a KtrA octamer, which in turn senses cellular ATP to regulate transport62. The KtrA monomers have a bilobal architecture: the ATP-binding N-terminal RCK_N (Regulator of Conductance for K+) lobes form an octameric ring interacting with KtrB, while the C-terminal RCK_C domains form peripheral pairwise contacts (Figure 6a)62. Nucleotide binding to RCK_N has been proposed to induce conformational changes in the KtrA ring that are transmitted to the KtrB module62. C-di-AMP, on the other hand, binds as a U-shaped monomer to the RCK_C dimers and could thus limit the conformational cycle of the active transporter, consistent with its inhibitory effects on potassium transport18,63.
Figure 6. CDN-dependent regulation of ion transport.
a. Crystal structures of the KtrAB duo and mode of CDN recognition. A cartoon representation of the ATP-bound KtrAB crystal structure is shown (left panel). Transparent surface representation is included for the transmembrane KtrB dimer, ATP is shown as spheres, and a dimer of KtrA protomers is shown in color. The right panel depicts an full-length KtrA dimer extracted from the structure shown on the left (top) and the c-di-AMP-bound RCK_C module of a homolog (bottom). Conformational changes in the RKC-C dimer interface upon c-di-AMP recognition could be transmitted to the KtrB-proximal, ATP-binding RCK_N modules. b. A schematic representation of the Hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4) tetramer and its antagonistic regulation by cyclic mono- and dinucleotides.
The effects of c-di-AMP on ion homeostasis are not limited to the KtrAB duo. C-di-AMP has been shown to bind to the RCK_C domain of another S. aureus transporter, the cation-proton antiporter CpaA, and to the KdpD sensor histidine kinase, which typically controls the expression of another potassium uptake system, as well as several virulence factors in the pathogen18,19.
The most surprising role of CDNs in ion homeostasis, however, is the recent discovery that c-di-GMP, c-di-AMP and both cGAMP variants can bind and modulate the activity of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels in mammalian cardiac pacemaker myocytes64. HCN channels are tetrameric ion channels with an N-terminal voltage sensor extension to their membrane pore module and a C-terminal CRP/FNR-like cyclic nucleotide-binding domain (Figure 6b). cGMP (or cAMP) binding to the latter is transmitted via an α-helical linker (‘C-linker’) to the pore to enhance HCN channel opening in response to voltage, thus leading to increased heart rate. Strikingly, the C-linker was shown to bind a variety of CDNs both in silico and in isolated myocytes ex vivo, with net inhibitory effect on action potential firing (Figure 6b)64. This raises interesting questions such as whether myocytes produce endogenous cGAMP to regulate HCN activity and heart rate, whether pathogen-secreted CDNs can affect heart rate to cause morbidity (e.g. by heart-affecting intracellular Listeria) or whether such modulation is coincidental but could be exploited pharmacologically64.
C-di-GMP control of bacterial secretion systems (SS)
Bacterial SS are macromolecular nanomachines securing the physical conduit, protection and energetics for the export of a versatile arsenal of biopolymers across the double bacterial envelope in Gram-negative and mycobacterial species65. Secretion is often key for bacterial adaptation, competitiveness, and virulence, and in many cases is regulated by c-di-GMP either at gene expression or by targeting proteins that regulate or directly partake in secretion at the cell envelope. For example, the LapDG system regulates surface maintenance of the Type I SS-transported adhesin LapA in P. fluorescens, while it controls the adhesin CdrA, cargo of a two-partner Type Vb secretion pair, in P. aeruginosa (Figure 4a)36,47,48. In parallel, the expression of these adhesins is regulated by the c-di-GMP-sensing FleQ, thus providing an additional regulatory boost to the system (Figure 5b)48.
Among identified c-di-GMP sensors are also various SS-powering ATPases. Examples include Type IV pili/Type II SS-associated MshE homologues from V. cholerae and P. aeruginosa, the flagellar export ATPase FliL from diverse bacteria and rotary ATPases HrcN and ClpB2 associated with pseudomonad Type III and Type VI SS, respectively46,66 (Figure 2). Residues key for c-di-GMP recognition have been identified for some of these targets and the recognition mechanisms appear to differ significantly from that of the ATPase FleQ59. For example, recent work demonstrated that MshE binds c-di-GMP at its N-terminal domain primarily through hydrophobic interactions with a 24-residue-long RLGxx(L/V/I)(L/V/I)xxG(L/V/I)(L/V/I)xxxxLxxxLxxQ sequence67.
Perhaps the archetypal SS under c-di-GMP control are the ones for exopolysaccharide secretion, which build up the biofilm matrix in many species and where c-di-GMP was originally discovered as a regulatory effector (Figure 7a)1. Again, c-di-GMP can act at gene expression by binding non-canonical transcription factors (e.g. VpsT, VpsR or FleQ41,42,58), or by directly binding regulatory modules associated with the secretion machineries. Examples include synthase-dependent systems for cellulose secretion in various species, the Pel and alginate systems in P. aeruginosa, and the poly-N-acetylglucosamine (PNAG) secretion system in E. coli (Figure 7a). Interestingly, while these systems likely share functional and architectural similarities, the mechanisms of c-di-GMP activation are strikingly diverse68.
Figure 7. C-di-GMP dependent regulation of exopolysaccharide secretion in biofilms.
a. Examples of synthase-dependent systems subject to direct c-di-GMP regulation. Color-coding for functionally homologous proteins is shown on the right and c-di-GMP binding is indicated by yellow stars. b. Cartoon representation of the crystal structure and topology of a catalytic BcsAB complex from R. sphaeroides. C. Crystallographic snapshots of the active site pocket in different CDN-free and bound states. Gating loop residues (dark blue), the cellulose product (light grey), and bound c-di-GMP (olive green) are shown as sticks, the UDP-glucose substrate as spheres, and inhibitory salt bridge interactions are indicated by dotted lines.
Mechanistically best-studied is the BcsA-BcsB cellulose biosynthetic complex of Rhodobacter sphaeroides (Figure 7b)69. BcsB is an accessory protein essential for cellulose secretion, whereas BcsA integrates glycosyl-transferase (GT), inner-membrane transporter, and c-di-GMP sensing activities69. Nucleotide-free BcsA rests in an autoinhibited conformation where the GT active site is capped by a conserved ‘gating loop’, which protrudes from the lipid headgroup layer to block substrate entry69. C-di-GMP binds to the C-terminal PilZ domain as a dimer, interacting with a conserved D609xS611xxG614 motif on the β-barrel surface and π-stacking with two arginines (R580xxxR584 motif) from the membrane-proximal PilZ domain linker (Figure 7c). Nucleotide coordination by R580 thus breaks a gating loop-tethering salt bridge to relieve the autoinhibition and allow substrate coordination and catalysis69. Interestingly, cellulose secretion in E. coli, Salmonella, and other enterobacteria is controlled by an additional c-di-GMP-sensing module. The cytosolic BcsE protein binds c-di-GMP via a conserved I-site-like motif (RxGD) in its so-called GIL domain and contributes to maximal cellulose production through an unresolved mechanism70.
Two of the three P. aeruginosa exopolysaccharide SS employ a c-di-GMP-activatable, synthase-dependent mechanism similar to that for cellulose secretion (Figure 7a)68. Interestingly, while the alginate system employs a PilZ domain-containing c-di-GMP sensor, Alg44, Pel exopolysaccharide secretion is regulated by the degenerate GGDEF domain of the inner membrane protein PelD38,39,68. To date, only structures of PelD’s cytosolic C-terminal GAF-GGDEF domain module have been resolved and show one or two U-shaped c-di-GMP molecules bound to the conserved I-site (R367GLD) of the protein71,72. Further investigation is necessary to uncover how the activating signal is transmitted to the partner GT PelF and the putative inner-membrane transporter PelG.
Finally, PNAG biogenesis relies on a yet different c-di-GMP regulatory mechanism (Figure 7a). The CDN is sensed by a two-partner module comprising the GT-transporter protein PgaC and the small membrane protein PgaD, essential for PNAG production73. At low c-di-GMP, PgaD fails to interact with PgaC and is rapidly degraded, which effectively inhibits secretion. C-di-GMP is proposed to coordinate between membrane-proximal arginine residues from the two proteins, thus stabilizing the PgaCD interaction, protecting PgaD from degradation and securing active PNAG polymerization and export73.
Extra- and intercellular CDN signaling
C-di-AMP is the first CDN for which transmembrane export has been demonstrated experimentally (Figure 2). Intracellular L. monocytogenes secretes c-di-AMP in a multidrug efflux pump (MDR)-dependent manner and thus activates the STING-dependent Type I interferon response in host cells74. The main contributors to the export – MdrM and MdrT – belong to the major facilitator superfamily of MDRs, which are polysubstrate-specific multipass membrane proteins typically operating through rocker-switch alternating-access mechanism75. An MDR-deletion mutant exhibits increased sensitivity to sub-lethal concentrations of vancomycin – an antibiotic that blocks peptidoglycan crosslinking and induces cell wall stress16. As both addition of c-di-AMP to the growth medium and intracellular overexpression of a DAC mitigate the vancomycin effects on the MDR-less mutant16, it is possible that c-di-AMP has both extracellular and intracellular roles in cell wall homeostasis. Furthermore, as intracellular accumulation of c-di-AMP can be toxic, transmembrane export might provide an alternative to enzymatic reduction of its levels13,14.
In the dawn of c-di-GMP signaling research, the CDN was generally defined as unique for bacteria. Nevertheless, it is now known that ‘social’ amoebae such as Dictyostelium discoideum can express conserved GGDEF domain-containing DGCs (Figure 1c), whose c-di-GMP product likely acts as an intercellular, secreted signal to trigger multicellular stalk differentiation upon nutrient depletion76. How the dinucleotide is exported and sensed by the cells, however, remains unknown.
While no extracellular role for prokaryotic cGAMP has been reported, the 2′-3′ eukaryotic version employs various mechanisms for horizontal signal propagation. For example, cGAMP produced by virus-infected cells in response to pathogen DNA can be efficiently packaged into new virions and thus transferred to activate anti-viral signaling in secondary infected cells77,78. In addition, 2′-3′ cGAMP can spread horizontally into neighboring host cells via low-selectivity connexin gap junctions. Interestingly, while in viral infections this can prime bystander cells for antiviral response, metastasizing brain carcinoma can hijack the pathway to drive astrocytes into secreting tumor-protective paracrine signals79,80. Finally, the only PDE known to break eukaryotic cGAMP, ENPP1, is a single-pass membrane protein with extracellular catalytic C-terminal domain81. It is therefore possible that – similarly to c-di-AMP in Listeria and c-di-GMP in Dictyostelium – eukaryotic cGAMP has evolved a mechanism for membrane export of its own.
Eukaryotic sensing through STING
STING (stimulator of interferon genes) is a dimeric, membrane-bound immune system protein that was identified as key to TANK-binding kinase 1 (TBK1)-dependent, CDN-induced Type I interferon response by a forward mutagenesis genetic screen in mice82. STING was confirmed as a direct c-di-GMP/c-di-AMP target83 and, with the discovery of the cytosolic DNA-sensing cGAS, as an even more potent receptor for mammalian cGAMP84–87. Structural studies have been limited to the soluble, C-terminal CDN-binding domain and numerous crystal structures of its CDN-free and -bound forms from various species have been reported (e.g. Figure 8)88–96.
Figure 8. CDN recognition by STING.
Crystal structures of human STING bound to c-di-GMP (a) and cGAMP (b) are shown. The bottom panels depict different views of the nucleotide binding pocket showing key CDN-coordinating side chains and bound ligands as sticks.
The STING CTD protomers adopt a characteristic α/β fold with a central twisted β-sheet surrounded by four α-helices (Figure 8). Of the latter, helix α1 (α5 in the full-length protein) forms an extended, ~40 Å-long secondary structure element, which carries characteristic kinks introduced by a π-helical insertion (Ser162-Arg169 for human STING) and a conserved proline residue (Pro173)88–96. In all structures, as well as in solution, apo-STING adopts a homodimeric V-shaped fold. It harbors a deep central cleft, which is lined by the kinked α1 helices and accommodates the long and mostly unstructured β2-β3 loops of each subunit88,91,93–95. While dimerization is mainly mediated by conserved hydrophobic interactions involving N-proximal residues of α1 and part of α3, the dimer exhibits significant conformational variations among solved apo-structures. In particular, structures of the human protein show a relatively splayed dimer88,91,95, whereas mouse STING crystallizes in a closed conformation with the two protomers rotated and brought in together relative to the 2-fold symmetry axis93. Interestingly, an ancient anemone STING homolog was crystallized in both an ‘open’ and a ‘closed’ state suggesting that the two likely represent alternatively sampled conformations rather than species-specific differences among homologs94.
The two states appear to discriminate among activating ligands and thus can explain differences in the physiological response to pathogen-derived versus endogenous CDNs. All CDN ligands bind in monomeric, U-shaped conformations at the central cleft of the dimer (Figure 8). In all but one c-di-GMP-bound structures, STING exhibits minor conformational changes relative to the apo-protein, with an overall splayed conformation and only subtle rearrangements in the β2-β3 loops (Figure 8a)88,89,91,95. The interaction between STING and c-di-GMP is mediated by direct and solvent-mediated hydrogen bonds, as well as by unique stacking interactions between the purine rings of the ligand and the phenolic group of Tyr167 from the α1 π-helix88,89,91,95.
By contrast, all 2′,3′-cGAMP-bound mouse, human and even anemone variants not only adopt a ‘closed’ conformation, but also reveal restructuring of the β2-β3 loops into a four-stranded antiparallel β-sheet (Figure 8b). The latter forms a ‘lid’ that caps the top of the CDN binding pocket and further restricts solvent access92,94,96. Apart from the pivotal role of Tyr167, the dinucleotide is further stabilized by stacking interactions with Arg238, which is itself buttressed by another lid’ residue, Tyr240 92,96. While one of the c-di-GMP-bound structures features similar lid rearrangements and closed conformation90, it features the hSTING-R232 variant that is both more responsive to c-di-GMP activation and binds 2′,3′-cGAMP with much higher affinity than the human reference variant hSTING-H232 84,96. This can explain the particular crystallographic capture of hSTING-R232 in a closed conformation even in the presence of c-di-GMP, as observed in one of the two such structures reported90,91.
Importantly, although anemone STING recognizes a 3′-3′-linked cGAMP signal naturally and binds the dinucleotide in yet a different splayed conformation, its mode of 2′,3′-cGAMP coordination is virtually identical to those of the human and mouse variants94. These data suggest that potent metazoan 2′,3′-cGAMP-STING activation possibly evolved through the exploitation of a highly conserved STING conformational intermediate that is not typically sampled by prokaryotic CDNs94.
Many unanswered questions remain in the field. It is still unclear how CDN binding to STING leads to downstream TBK1 activation or how reported covalent and non-covalent modifications of the receptor – phosphorylation, Ca2+ coordination, or intracellular trafficking89,97 – regulate its function. Importantly, its nearly 30 residue-long C-terminal tail (CTT) are not resolved in the crystal structures but are both necessary and sufficient for downstream signal activation. This, together with the identification of STING mutants that lead to constitutive interferon response, suggests that the protein might maintain its activating CTT in an autoinhibited conformation that is only relieved upon CDN signal recognition95.
Outlook
Recent decades of research have led to the recognition of CDNs as a new, widely used class of second messengers and have identified the functional units controling their biosynthesis, turn-over and signal transduction. The presented signaling modules are only a part of the ever-growing and diverse panel of CDN metabolizing enzymes or signaling targets. That said, researchers have only scratched the surface regarding the physiological triggers that modulate CDN levels, or their spatiotemporal regulation within the complex signaling networks that are beginning to emerge. On the other hand, the available knowledge has already been proven useful in many different ways. Interfering with bacterial signaling can aid the development of novel anti-infectives, as well as the production of natural nanomaterials of biotechnological use98,99. The mammalian pathways, on the other hand, can be exploited in immunization approaches or the pharmacological targeting of infections and cancer80,100. Hopefully, future studies on the adaptational and virulence strategies of bacteria, as well as their evolutionary patterns, can be further harnessed to humankind’s advantage.
Acknowledgments
Work in the Sondermann laboratory is supported by NIH grant R01-AI097307 (H.S.). P.V.K. is supported by the Centre National de la Recherche Scientifique (CNRS) and the Institute for Integrative Biology of the Cell (I2BC) and was previously recipient of a Roux-Cantarini postdoctoral fellowship from the Institut Pasteur. We thank Fitnat Yildiz for critically reading a draft of the review, and Nicolas Reyes, Rémi Fronzes, and Richard Cooley for useful discussions. We apologize to all researchers that have contributed to the field, whose work we could not cover due to space constraints.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1•.Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. Seminal work on the discovery of c-di-GMP as an allosteric regulator of bacterial cellulose synthesis that laid the foundation of CDN signaling research. [DOI] [PubMed] [Google Scholar]
- 2.Weinhouse H, et al. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 3.Tal R, et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul R, et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 6.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 7.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hisert KB, et al. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol Microbiol. 2005;56:1234–1245. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- 9.Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. A comprehensive bioinformatics study highlighting the prevalence of c-di-GMP signaling across the bacterial domain, as well as the multi-component, multiple pathway nature of c-di-GMP signal transduction within highly adaptable species. [DOI] [PubMed] [Google Scholar]
- 11•.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. A structure-function study of B. subtilis DNA damage checkpoint protein DisA, which led to the serendipitous discovery of c-di-AMP and its dedicated diadenylate cyclases. [DOI] [PubMed] [Google Scholar]
- 12.Bai Y, et al. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol. 2014;196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynh TN, et al. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci USA. 2015;112:E747–756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehne FM, et al. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Li W, He ZG. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan Zeevi M, et al. Listeria monocytogenes multidrug resistance transporters and cyclic di-AMP, which contribute to type I interferon induction, play a role in cell wall stress. J Bacteriol. 2013;195:5250–5261. doi: 10.1128/JB.00794-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sureka K, et al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014;158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moscoso JA, et al. Binding of Cyclic Di-AMP to the Staphylococcus aureus Sensor Kinase KdpD Occurs via the Universal Stress Protein Domain and Downregulates the Expression of the Kdp Potassium Transporter. J Bacteriol. 2016;198:98–110. doi: 10.1128/JB.00480-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. Identification of V. cholerae DncV as a virulence factor and a cyclase for previously undescribed hybrid cGAMP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei J, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 22•.Chan C, et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. First structural study of a diguanylate cyclase that led to the characterization of the allosteric I-site for feedback regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahlstrom KM, Giglio KM, Sondermann H, O’Toole GA. The Inhibitory Site of a Diguanylate Cyclase Is a Necessary Element for Interaction and Signaling with an Effector Protein. J Bacteriol. 2016;198:1595–1603. doi: 10.1128/JB.00090-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Barends TR, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. A structure-function study revealing the regulation of c-di-GMP specific phosphodiesterases. [DOI] [PubMed] [Google Scholar]
- 25.Bellini D, et al. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol. 2014;91:26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stelitano V, et al. C-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: analysis of the reaction mechanism and novel roles for pGpG. PLoS One. 2013;8:e74920. doi: 10.1371/journal.pone.0074920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen D, et al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2015;112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orr MW, et al. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci USA. 2015;112:E5048–5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corrigan RM, Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 31.Rao F, et al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kranzusch PJ, et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell. 2014;158:1011–1021. doi: 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu D, et al. Structural biochemistry of a Vibrio cholerae dinucleotide cyclase reveals cyclase activity regulation by folates. Mol Cell. 2014;55:931–937. doi: 10.1016/j.molcel.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 34•.Hallberg ZF, et al. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3′, 3′-cGAMP) Proc Natl Acad Sci USA. 2016;113:1790–1795. doi: 10.1073/pnas.1515287113. Identification of a subclass of GGDEF domain-containing dinucleotide cyclases with broad catalytic spectrum and substrate specificity reflecting cellular AMP:GMP ratios. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro MV, et al. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 2011;9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozaki S, et al. Activation and polar sequestration of PopA, a c-di-GMP effector protein involved in Caulobacter crescentus cell cycle control. Mol Microbiol. 2014;94 doi: 10.1111/mmi.12777. [DOI] [PubMed] [Google Scholar]
- 38.Lee VT, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 40•.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. The first work to demonstrate c-di-GMP control at the transcription initiation level through the regulation of biofilm-related genes by direct c-di-GMP binding to P. aeruginosa FleQ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193 doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duvel J, et al. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods. 2012;88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics. 2012;75:4874–4878. doi: 10.1016/j.jprot.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 45•.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. A methodological work on a quantitative high-throughput assay for the unbiased discovery and characterization of CDN-binding proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roelofs KG, et al. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog. 2015;11:e1005232. doi: 10.1371/journal.ppat.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newell PD, Boyd CD, Sondermann H, O’Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011;9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooley RB, et al. Cyclic Di-GMP-Regulated Periplasmic Proteolysis of a Pseudomonas aeruginosa Type Vb Secretion System Substrate. J Bacteriol. 2016;198:66–76. doi: 10.1128/JB.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterjee D, et al. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. Elife. 2014;3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giardina G, et al. Investigating the allosteric regulation of YfiN from Pseudomonas aeruginosa: clues from the structure of the catalytic domain. PLoS One. 2013;8:e81324. doi: 10.1371/journal.pone.0081324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malone JG, et al. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 2012;8:e1002760. doi: 10.1371/journal.ppat.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol. 2013;90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tschowri N, et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014;158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin KH, et al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 55.Li W, He ZG. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012;40:11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shikuma NJ, Fong JC, Yildiz FH. Cellular levels and binding of c-di-GMP control subcellular localization and activity of the Vibrio cholerae transcriptional regulator VpsT. PLoS Pathog. 2012;8:e1002719. doi: 10.1371/journal.ppat.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baraquet C, Harwood CS. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci USA. 2013;110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuyama BY, et al. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2016;113:E209–218. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauriano CM, Ghosh C, Correa NE, Klose KE. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J Bacteriol. 2004;186:4864–4874. doi: 10.1128/JB.186.15.4864-4874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.den Hengst CD, et al. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol. 2010;78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 62.Vieira-Pires RS, Szollosi A, Morais-Cabral JH. The structure of the KtrAB potassium transporter. Nature. 2013;496:323–328. doi: 10.1038/nature12055. [DOI] [PubMed] [Google Scholar]
- 63.Kim H, et al. Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP) J Biol Chem. 2015;290:16393–16402. doi: 10.1074/jbc.M115.641340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lolicato M, et al. Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness. Nat Chem Biol. 2014;10:457–462. doi: 10.1038/nchembio.1521. [DOI] [PubMed] [Google Scholar]
- 65.Costa TR, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 66.Trampari E, et al. Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger cyclic-di-GMP. J Biol Chem. 2015;290:24470–24483. doi: 10.1074/jbc.M115.661439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang YC, et al. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun. 2016;7:12481. doi: 10.1038/ncomms12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McNamara JT, Morgan JL, Zimmer J. A molecular description of cellulose biosynthesis. Annu Rev Biochem. 2015;84:895–921. doi: 10.1146/annurev-biochem-060614-033930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang X, et al. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol. 2014;93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z, Chen JH, Hao Y, Nair SK. Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J Biol Chem. 2012;287:30191–30204. doi: 10.1074/jbc.M112.378273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitney JC, et al. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J Biol Chem. 2012;287:23582–23593. doi: 10.1074/jbc.M112.375378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steiner S, Lori C, Boehm A, Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2013;32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woodward JJ, Iavarone AT, Portnoy D. A c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453:254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 76•.Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. The first report of c-di-GMP biosynthesis and signal transduction in eukarotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gentili M, et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 78.Bridgeman A, et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349:1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Q, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533:493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li L, et al. Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat Chem Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sauer JD, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83•.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. Identification of STING as a eukaryotic CDN sensor-effector. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. Identification of cGAMP as an endogenous CDN product in mammalian cells, as well as its direct role in type I interferon response through direct STING activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ouyang S, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 91.Shang G, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 92.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chin KH, et al. Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING. Acta Crystallogr D Biol Crystallogr. 2013;69:352–366. doi: 10.1107/S0907444912047269. [DOI] [PubMed] [Google Scholar]
- 94•.Kranzusch PJ, et al. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol Cell. 2015;59:891–903. doi: 10.1016/j.molcel.2015.07.022. Structural studies of an anemone STING homolog revealing the likely ancient origins of eukaryotic 2′-3′ cGAMP recognition through the exploitation of a unique conformational intermediate not sampled by 3′-3′ CDNs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yin Q, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang L, et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mohite BV, Patil SV. A novel biomaterial: bacterial cellulose and its new era applications. Biotechnol Appl Biochem. 2014;61:101–110. doi: 10.1002/bab.1148. [DOI] [PubMed] [Google Scholar]
- 99.Opoku-Temeng C, Zhou J, Zheng Y, Su J, Sintim HO. Cyclic dinucleotide (c-di-GMP, c-di-AMP, and cGAMP) signalings have come of age to be inhibited by small molecules. Chem Commun (Camb) 2016;52:9327–9342. doi: 10.1039/c6cc03439j. [DOI] [PubMed] [Google Scholar]
- 100.Gravekamp C, Chandra D. Targeting STING pathways for the treatment of cancer. Oncoimmunology. 2015;4:e988463. doi: 10.4161/2162402X.2014.988463. [DOI] [PMC free article] [PubMed] [Google Scholar]