Abstract
At the neuronal cell level, long-term memory formation emerges from interactions between initial activity-dependent molecular changes at the synapse and subsequent regulation of gene transcription in the nucleus. This in turn leads to strengthening of the connections back at the synapse that received the initial signal. However, the mechanisms through which this synapse-to-nucleus molecular exchange occurs remain poorly understood. Here we discuss recent studies that delineate nucleocytoplasmic transport of a special class of synaptically-localized transcriptional regulators that upon receiving initial external signal by the synapse move to the nucleus to modulate gene transcription.
Keywords: Memory, Memory enhancement, Nucleocytoplasmic shuttling, Transcription, CREB, CRTC1, HDAC4, NF-κB
INTRODUCTION
Activity-dependent neuronal plasticity allows organisms to adapt and respond to changes in the environment (West and Greenberg, 2011). Neuronal plasticity includes modifications in synaptic structure and function, and, in particular, long-term modifications require gene transcription (Alberini, 2009; Klann and Dever, 2004; Mayford et al., 2012). To initiate neuronal activity-dependent gene transcription, signals must be relayed from active synapses to the nucleus (Ch'ng and Martin, 2011; Greer and Greenberg, 2008; Panayotis et al., 2015). This article summarizes work on synaptically-localized transcriptional regulators that directly transmit information regarding synaptic activity by moving to the nucleus and regulating transcription (Ch'ng and Martin, 2011; Jordan and Kreutz, 2009). Recent publications demonstrate that certain proteins, which function as transcriptional regulators, are initially localized at synapses but can accumulate in the nucleus in response to synaptic plasticity and learning (Ch'ng et al., 2012; Dieterich et al., 2008; Jordan et al., 2007; Lai et al., 2008; Nonaka et al., 2014; Proepper et al., 2007; Uchida et al., 2017).
Neuronal activity-dependent gene transcription typically requires calcium-dependent signaling (Ebert and Greenberg, 2013). Calcium, as a potent activator of intracellular signaling cascade, is normally maintained at very low concentration within the cytoplasm. Neuronal activity leads to calcium influx into the postsynaptic cells either through NMDA receptors or L-type voltage-sensitive calcium channels (Cole et al., 1989; Murphy et al., 1991). Neuronal activity-induced elevation in intracellular calcium activates multiple signaling molecules such as calcium/calmodulin-dependent protein kinases (CaMKII and CaMKIV) and calcium-dependent protein phosphatases (such as calcineurin), which regulate the activity of transcriptional factors by phosphorylating or dephosphorylating them (Takemoto-Kimura et al., 2017).
Neuronal activity not only regulates sequence-specific DNA-binding proteins, but also modifies chromatin structure to control gene transcription (Crosio et al., 2000; Day and Sweatt, 2011; Graff and Tsai, 2013; Kumar et al., 2005; Lopez-Atalaya and Barco, 2014; Peixoto and Abel, 2013; Uchida et al., 2011). In particular, epigenetic processes, such as histone modifications, are important for activity-dependent regulation of transcription (Graff and Tsai, 2013; Lopez-Atalaya and Barco, 2014; Peixoto and Abel, 2013). Moreover, synaptically localized transcription modulators were recently linked to histone modifications that are required for memory enhancement (de la Fuente et al., 2015; Sando et al., 2012; Uchida et al., 2017).
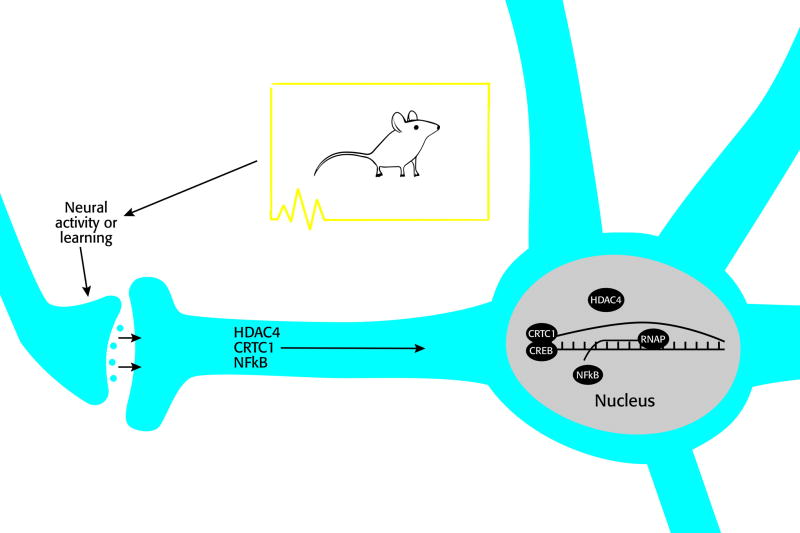
In this review, we will discuss current advances in the field that have shown unique characteristics of the transcription modulators CRTC1, HDAC4 and NF-κB, which are localized at the synapse but, upon neuronal stimulation (such as learning), move to the nucleus to regulate gene transcription, which is essential for synaptic plasticity and memory formation (Fig. 1).
Figure 1. Schematic presentation of synapse-to-nucleus transport of transcription modulators HDAC4, CRTC1 and NF-κB to regulate gene transcription.
ACTIVITY-DEPENDENT NUCLEOCYTOPLASMIC SHUTTLING OF CRTC1 IS REQUIRED FOR MEMORY FORMATION
CREB-dependent gene expression is essential for synaptic plasticity, learning and memory (Bito et al., 1996; Bourtchuladze et al., 1994; Deisseroth et al., 1996; Impey et al., 1998; Josselyn et al., 2004; Kida et al., 2002; Kida and Serita, 2014; Mayford et al., 2012; Silva et al., 1998; Suzuki et al., 2011). Membrane depolarization or an increase in cAMP induces CREB phosphorylation at serine amino acid 133 (Ser133), and the mutation from serine to alanine (Ser133Ala) blocks the induction of CREB-dependent transcription (West et al., 2002). Various kinases phosphorylate CREB at Ser133; however, specific kinases were found to be responsible for its phosphorylation in response to a certain stimulus (Mayr and Montminy, 2001). The nuclear protein kinase CaMKIV is crucial for rapid activity-dependent phosphorylation of CREB at Ser133 in neurons (Ho et al., 2000; Ribar et al., 2000). Eventually, calcium influx leads to CREB dephosphorylation at Ser133 through calcium-dependent activation of the protein phosphatases PP1 and PP2A (Lonze and Ginty, 2002). Although CREB-mediated transcription requires CREB phosphorylation, the latter is not a reliable predictor of target gene activation; additional regulatory processes are required for the engagement of transcriptional promoter elements. For instance, histone acetyltransferase CBP is critical, and the CBP paralog p300 also enhances CREB functionality, resulting in upregulation of plasticity-related genes such as Bdnf and Arc (Impey and Goodman, 2001; Patterson et al., 2001).
Genetic disruption of CREB function leads to a decrease in long-term potentiation (LTP), a form of synaptic plasticity, and long-term memory in mice (Bourtchuladze et al., 1994; Kida et al., 2002; Pittenger et al., 2002). Conversely, genetic enhancement of CREB activity in forebrain increases both LTP and long-term memory through the activation of Bdnf gene expression (Barco et al., 2002; Barco et al., 2005; Suzuki et al., 2011). Moreover, viral-mediated acute increase in CREB function locally in the dentate gyrus leads to memory enhancement (Sekeres et al., 2012). Importantly, neurons overexpressing CREB by viral-mediated gene transfer in the lateral amygdala are recruited selectively into the trace of cued fear memory (Han et al., 2007). Thus, activity-dependent CREB activation plays a key role in memory enhancement by participating in memory trace (Kida and Serita, 2014).
While CREB phosphorylation is essential it is not sufficient for CREB-dependent gene transcription (Impey et al., 1996; Mayr and Montminy, 2001). Thus, neuronal activity-dependent CREB-mediated gene regulation could be controlled by additional mechanisms, such as the coactivators that can modulate CREB activity (Bito et al., 1996; Lonze and Ginty, 2002; Zhang et al., 2005). The CREB-mediated transcriptional coactivators (CRTCs, also known as TORCs; Fig. 2) may potentiate the interaction of CREB with CBP/p300 (Xu et al., 2007) and dramatically increase CREB transcriptional activity independently of Ser133 phosphorylation (Conkright et al., 2003; Iourgenko et al., 2003). Importantly, CRTC1 is translocated from synapses/dendrites to the nucleus in response to neural activity and learning (summarized in Table 1) (Ch'ng et al., 2012; Kovacs et al., 2007; Li et al., 2009; Nonaka et al., 2014; Parra-Damas et al., 2017; Uchida et al., 2017). Nuclear-cytoplasmic redistribution of CRTCs is known to be dependent on its activity-regulated phosphorylation status (Altarejos and Montminy, 2011). Calcium signals promote nuclear translocation of CRTC1 via activation of calcineurin, which directly dephosphorylates CRTC1 at Ser151 (Bittinger et al., 2004; Ch'ng et al., 2012; Screaton et al., 2004). Indeed, a CRTC1 mutant lacking two calcineurin binding sites was confined to the cytoplasm following neuronal stimulation (Nonaka et al., 2014; Uchida et al., 2017). In addition, dephosphorylation of CRTC1 at Ser151 and Ser167 by salt-inducible kinases is required for cytoplasm-to-nucleus translocation of CRTC1 (Bittinger et al., 2004; Ch'ng et al., 2012; Li et al., 2009; Sasaki et al., 2011; Screaton et al., 2004). Both CRTC1 Ser151Ala and CRTC1 Ser151Ala/Ser167Ala mutants were localized to the cytoplasm, but were transported to the nucleus when neurons were stimulated (Ch'ng et al., 2012; Uchida et al., 2017). Thus, dephosphorylation of CRTC1 at Ser151 and Ser167 may not be necessary for the initial translocation of CRTC1 to the nucleus. Indeed, a recent report has shown that two phosphorylation sites (S151 and S245) contribute to nuclear import of CRTC1 (Nonaka et al., 2014).
Figure 2. Neuronal activity-dependent gene expression program required for memory formation.
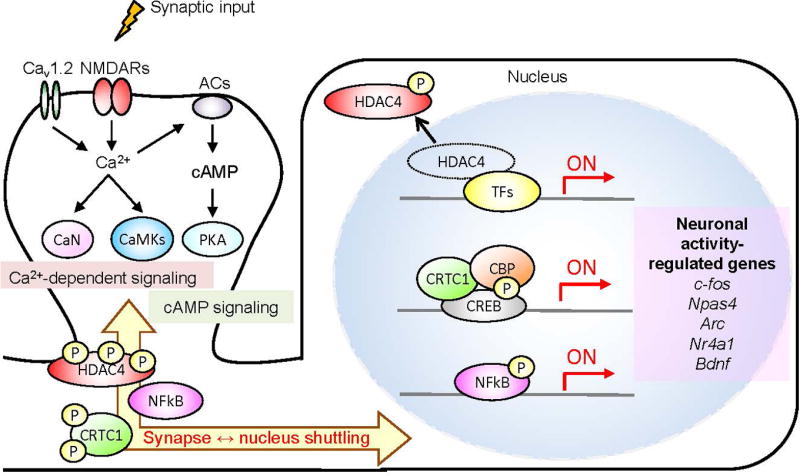
Activation of NMDA receptors (NMDARs) and L-type voltage-sensitive calcium channel (Cav1.2) triggers calcium influx and induces calcium-dependent signaling molecules such as calcineurin (CaN) and Ca2+/calmodulin-dependent protein kinases (CaMKs). Calcium influx also activates members of the cAMP signaling pathway, such as protein kinase (PKA) via Ca2+-sensitive adenylate cyclase (ACs). These signaling molecules modulate the synapse-to-nucleus shuttling of transcription modulators (HDAC4, CRTC1 and NF-κB) via phosphorylation and dephosphorylation. Then, these transcriptional modulators contribute to the control of activity-dependent gene transcription which is required for synaptic plasticity and memory formation. P: phosphorylation; the red arrow marked with “ON” represents gene transcription.
Table 1.
Brief summary of activity-dependent nucleocytoplasmic shuttling of CRTC1 and its role in synaptic plasticity and memory.
| References | System | Findings |
|---|---|---|
| (Zhou et al., 2006) | Cultured hippocampal neurons Hippocampal slices | Neuronal stimulation induces nuclear translocation of CRTC1. |
| CRTC1 regulates neuronal stimulation-induced gene expression of Bdnf. | ||
| TORC1 nuclear translocation correlates with late-phase LTP induction. | ||
| Requirement of TORC1 for late-phase LTP. | ||
| (Kovacs et al., 2007) | Cultured hippocampal or cortical neurons Hippocampal slice cultures | CRTC1 imports to the nucleus upon activation of calcium and cAMP pathways. |
| CRTC1 mediates the synergistic activation of CREB-mediated transcription by calcium and cAMP in neurons. | ||
| CRTC1 is involved in L-LTP maintenance in the Schaffer collateral–CA1 pathway. | ||
| (Li et al., 2009) | Cultured cortical neurons | Increased nuclear translocation of CRTC1 in response to neuronal stimulation. |
| CRTC1 is required for activity-induced dendrite arborization. | ||
| Mouse | CRTC1 is required for dendrite growth in the cortex. | |
| (Espana et al., 2010) | Cultured hippocampal neurons | Amyloid β oligomers impair CRTC1-dependent signaling. impair |
| Amyloid β impairs CRTC1/CREB-dependent transcription by CRTC1/CREB affecting CRTC1 dephosphorylation. | ||
| Mouse | Gene transcription mediated by CRTC1 is impaired in the hippocampus from an animal model of Alzheimer’s disease. | |
| (Ch'ng et al., 2012) | Cultured hippocampal neurons | Activity-dependent synapse-to-nucleus translocation of CRTC1 in excitatory neurons. |
| Nuclear translocation of CRTC1 requires activation of calcineurin. cAMP regulates the persistence of CRTC1 in the nucleus. | ||
| CRTC1 is required for activity-dependent induction of specific CREB target genes (c-fos, Arc, Zif268). | ||
| Hippocampal slice cultures | Chemical LTP induces synapse-to-nucleus translocation of CRTC1. | |
| (Sekeres et al., 2012) | Mouse | Herpes simplex virus-mediated local and acute increase of CRTC1 levels in the dentate gyrus of hippocampus during training enhances consolidation of contextual fear memory. |
| Increasing CRTC1 levels in the dentate gyrus of hippocampus enhances reconsolidation of an established contextual fear memory. | ||
| (Parra-Damas et al., 2014) | Mouse | Learning (Morris water maze)-induces nuclear accumulation of CRTC1 in the CA1 and CA3 subregion of hippocampus. |
| Human | Altered CRTC1 levels and transcriptional changes (ARC, NR4A2) in human brain at intermediate Alzheimer's disease pathological stages. | |
| (Nonaka et al., 2014) | Cultured hippocampal or cortical neurons | Dephosphorylation of CRTC1 at Ser151 and Ser245 is critical for its activity-dependent nuclear import and CRE-dependent transcriptional activity. |
| CRTC1 mutant lacking calcineurin-binding motifs disrupts nuclear translocation of CRTC1 following neuronal stimulation. | ||
| Mouse | Overexpression of the constitutive active form of CRTC1 (Ser151Ala/Ser245/Ala) in the CA suregion of hippocampus enhances activity-dependent gene expression (Arc, c-fos, Bdnf) and long-term fear memory formation. | |
| Learning (contextual fear conditioning) induces nuclear accumulation of CRTC1 in the basolateral amygdala. | ||
| Knockdown of CRTC1 in the basolateral amygdala reduces long-term fear memory. | ||
| Overexpression of the constitutive active form of CRTC1 induces a significant enlargement of spine heads. | ||
| (Uchida et al., 2017) | Primary hippocampal neurons | Neuronal stimulation induces nuclear translocation of CRTC1. |
| Two phosphorylation sites of CRTC1 at Ser151 and Ser167 are important for its nuclear retention rather than nuclear import following neuronal stimulation. | ||
| Mouse | Learning (contextual fear conditioning and object location task) induces synapse-to nucleus translocation of CRTC1 in the CA subregion of hippocampus and these are dependent on the strength of learning. | |
| Learning-induced nuclear translocation of CRTC1 is dependent on microtubule stability. | ||
| Loss-of-function CRTC1 in the CA subregion of hippocampus disrupts LTP and long-term memory. | ||
| Gain-of-function CRTC1 in the CA subregion of hippocampus leads to memory enhancement. | ||
| Calcineurin-mediated CRTC1 nuclear translocation is required for memory enhancement. | ||
| CRTC1/CREB/KAT5-mediated epigenetic regulation of Fgf1b expression is required for memory enhancement. | ||
| (Parra-Damas et al., 2017) | Mouse | Contextual fear learning induces CRTC1 dephosphorylation in the hippocampus. |
| Contextual fear learning induces CRTC1 nuclear translocation in the CA3 subregion of hippocampus. |
Please note that the findings listed here do not cover all the contents of the reference papers and that we do not cite all the related articles here, due to the space limitation,
Several lines of evidence have shown recently that CRTC1 plays a key role in synaptic plasticity and memory formation in rodents (Nonaka et al., 2014; Sekeres et al., 2012; Uchida et al., 2017; Zhou et al., 2006). The phosphorylation of CRTC1 in the hippocampus is decreased following contextual fear conditioning (Parra-Damas et al., 2017) and nuclear transport of CRTC1 is observed in the CA1 and CA3 pyramidal neurons, but not in the dentate gyrus granule neurons, of the hippocampus following contextual fear conditioning, novel object location task and Morris water maze task (Parra-Damas et al., 2017; Uchida et al., 2017). This learning-dependent nuclear translocation of CRTC1 also occurs in the basolateral amygdala following contextual fear conditioning (Nonaka et al., 2014). Viral-mediated enhancement of CRTC1 activity in the dentate gyrus (Sekeres et al., 2012) and CA area of the hippocampus (Nonaka et al., 2014; Uchida et al., 2017) increases contextual fear. Conversely, shRNA-mediated knockdown of CRTC1 in the CA region of the hippocampus (Uchida et al., 2017) or basolateral amygdala (Nonaka et al., 2014) leads to decreased contextual fear along with decreased CREB-mediated gene transcription. Furthermore, increased CRTC1 function promoted spine enlargement in the CA1 pyramidal cells (Nonaka et al., 2014) and the loss-of-function CRTC1 disrupted LTP in CA1 (Uchida et al., 2017; Zhou et al., 2006), strongly suggesting that CRTC1 is involved in structural and synaptic plasticity.
EMERGING ROLE OF LEARNING-DEPENDENT NUCLEAR TRANSLOCATION OF CRTC1 IN MEMORY ENHANCEMENT
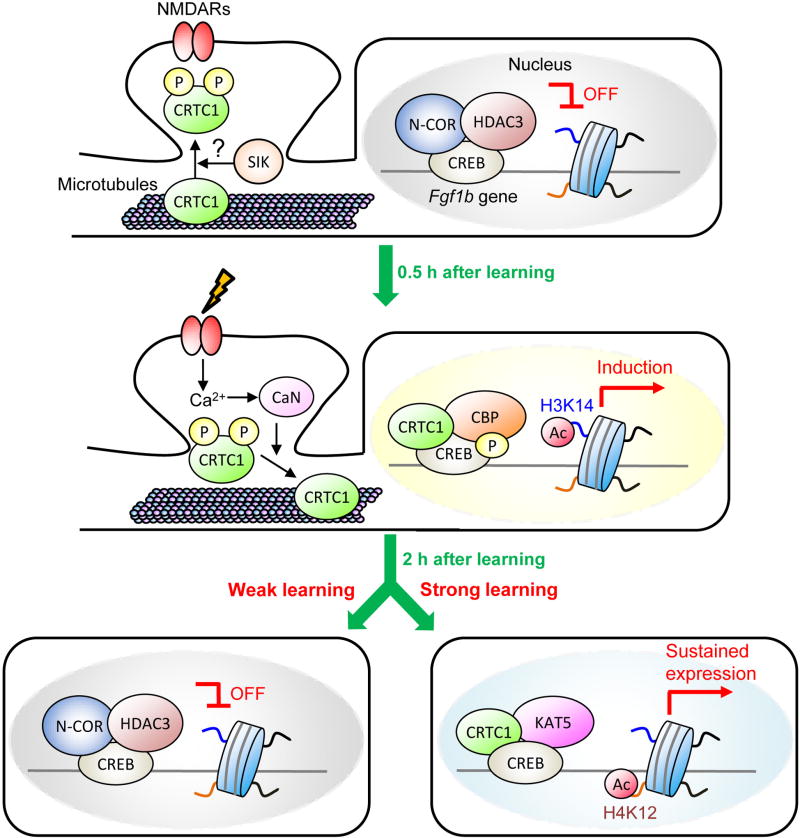
Activity-dependent synapse-to-nucleus translocation of transcription regulators is believed to be an important regulation of synaptic function and memory, but the exact mechanisms of this translocation remain unclear. As mentioned above, the studies of CRTC1 have largely focused on the role of its nuclear translocation in memory processing. CRTC1 is transported from the synapses/dendrites to the nucleus following learning (Fig. 2), but how the strength of activity that reaches the synapse can be transmitted to the nucleus and be involved in epigenetic regulation of gene transcription is unknown. To this end, a recent study has found that hippocampus-dependent learning induces calcineurin-dependent transport of CRTC1 into the nucleus of excitatory neurons of the hippocampal CA subregion (Uchida et al., 2017). CRTC1 nuclear translocation in the CA1 and CA3 areas is more prominent in mice that received strong training (3 footshocks) compared to weak training (1 footshock) in contextual fear conditioning (Fig. 3). Moreover, this translocation is dependent on microtubules, as it is disrupted by microtubule destabilizer drug nocodazole, in agreement with the observations showing key role of microtubule dynamics in synaptic plasticity and memory (Dent, 2017; Jaworski et al., 2009; Martel et al., 2016; Uchida et al., 2014; Uchida and Shumyatsky, 2015). Genetic enhancement of CRTC1 activity in the CA area strengthened both contextual fear memory and object location memory even following weak training. Conversely, viral-mediated knockdown of CRTC1 with shRNA against Crtc1 in the CA area weakened long-term memory following strong training and this impairment was reversed by overexpression of shRNA-resistant CRTC1, but not by mutant CRTC1 that has constitutive cytosolic localization, suggesting that nuclear transport of CRTC1 plays a key role in memory enhancement. The phosphorylated form of CREB at Ser133 occupied the Fgf1b promoter following both weak and strong training in contextual fear and the recruitment of CRTC1 and CBP enhanced the acetylation of H3K14, which is one of the substrates of CBP (Peixoto and Abel, 2013), leading to Fgf1b gene transcription. This molecular events were transient, because the occupancies of the phosphorylated CREB and CBP on the Fgf1b gene promoter were no longer observed 1 h following weak training (Fig. 3). Interestingly, sustained CRTC1 recruitment on Fgf1b gene promoter (up to 2 hours) following strong training was associated with long-lasting upregulation of H4K12 acetylation, which was not observed in weak training condition. This sustained transcription following strong training is mediated at least in part by substitution of histone acetyltransferase KAT5 for CBP in a CRTC1-dependent manner. Strong training maintains upregulation of Fgf1b transcription 2 h after learning by recruiting KAT5 to the promoter region independently of CREB phosphorylation, enhancing H4K12 acetylation (Fig. 3). Given that increased H4K12 acetylation might be associated with memory enhancement (Guan et al., 2009; Peleg et al., 2010), nuclear translocation of CRTC1 and subsequent initiation of KAT5-dependent enhancement of H4K12 are critical for enduring synaptic plasticity and memory enhancement. More importantly, this study suggests that synaptic CRTC1 may be a “molecular sensor” that transmits data to the nucleus regarding the strength of synaptic input.
Figure 3. Proposed model how CRTC1 controls Fgf1b transcription and enhances memory.
Under basal conditions, CRTC1 is phosphorylated and anchored to the synapses and dendrites. In the nucleus, HDAC3–N-CoR complex represses Fgf1b transcription. Upon learning (both weak and strong learning), Ca2+ signal potentiate CRTC1 dephosphorylation via activation of calcineurin (CaN). Dephosphorylated CRTC1 translocates to the nucleus, where it binds to phosphorylated CREB (pCREB) and histone acetyltransferases (CBP) and enhances Fgf1b gene transcriptional activity by increasing the acetylation of H3K14 on its promoter. These molecular events are terminated at 2h following weak training (e.g., one foot-shock contextual fear conditioning). Conversely, strong training (e.g., three foot-shock contextual fear conditioning) maintains nuclear localization of CRTC1 and upregulates Fgf1b transcription independently of pCREB even 2 h after learning by enhancing H4K12 acetylation via KAT5 recruitment to its promoter region. Learning-induced KAT5 recruitment acetylates H4K12 on the Fgf1b promoter, thereby enhancing synaptic plasticity and memory formation. P: phosphorylation; Ac: acetylation.
IMPLICATIONS OF CRTC1 FOR ALZHEIMER’S DISEASE
Alzheimer’s disease is a neurodegenerative disorder, characterized by progressive decline in memory, cognitive functions, and changes in behavior and personality (Mattson, 2004; Reddy and Beal, 2008). Alzheimer’s disease is also associated with the loss of synapses, synaptic function, and neuronal loss. Many molecules that are involved in the synaptically-localized proteins that influence activity-dependent regulation of gene transcription are implicated in Alzheimer’s disease. Among them, CRTC1 has been suggested to be associated with Alzheimer’s disease in both animal and human studies. Nuclear translocation of CRTC1 following learning was impaired in a mouse model of Alzheimer’s disease (Parra-Damas et al., 2017; Parra-Damas et al., 2014). It is interesting to note that CREB/CRTC1-mediated regulation of gene expression in the hippocampus of a mouse model of Alzheimer’s disease was changed not only in naïve conditions but also in response to learning (Parra-Damas et al., 2014). Moreover, disrupted memory and decreased induction of activity-dependent genes (Nr4a1 and Nr4a2) following learning in mice lacking the Alzheimer’s disease-linked presenilin genes (presenilin conditional double knockout) were ameliorated by CRTC1 overexpression into the dorsal hippocampus (Parra-Damas et al., 2017), suggesting an impairment in activity-dependent gene regulation in Alzheimer’s disease. A study in humans found a reduction of both total and phosphorylated CRTC1 in human hippocampus at Braak IV and V–VI pathological stages of Alzheimer’s disease (Parra-Damas et al., 2014). Taken together, aberrant activity-dependent nucleocytoplasmic shuttling of CRTC1 and subsequent dysregulation of gene transcription are implicated in the pathophysiology of Alzheimer’s disease.
NUCLEOCYTOPLASMIC SHUTTLING OF HDAC4 REGULATES MEMORY
In addition to CRTC1, a recent study has shown that activity-dependent nuclear export of HDAC4 is important for gene transcription and memory formation (Fig. 2). HDAC4 is abundant in neurons, where it is predominantly localized to cytoplasm and dendritic spines (Darcy et al., 2010; Grozinger et al., 1999; Wang et al., 1999). Phosphorylation of HDAC4 by CaMKs promotes its localization to the cytoplasm. Phosphorylation of HDAC4 at Ser246, Ser467, and Ser632 through calcium signaling facilitates its nuclear export (Chawla et al., 2003; McKinsey et al., 2001; Schlumm et al., 2013) and derepression of transcription factor MEF2. Conversely, dephosphorylation of HDAC4 by calcineurin allows its nuclear translocation (Paroni et al., 2008). Nuclear HDAC4 binds to chromatin, as well as to MEF2A and CREB, leading to histone deacetylation and repression of neuronal gene expression (Bolger and Yao, 2005; Chen and Cepko, 2009; Li et al., 2012; Youn et al., 2000). In the dentate gyrus, the majority of HDAC4 is localized in the cytoplasm, but it is localized in the cell nuclei in a proportion of CA1 and CA3 areas (Darcy et al., 2010). Interestingly, a subset of puncta is associated with dendritic shafts and dendritic spines. Studies in primary cultures have shown that neuronal activity induces the nuclear export of HDAC4. The subcellular localization of HDAC4 is regulated by NMDA receptors, as application of the NMDA receptor antagonist AP5 induces nuclear accumulation of HDAC4 (Sando et al., 2012). Mutation of HDAC4 Ser246, Ser467, and Ser632 (HDAC4-3SA mutant) results in nuclear retention (Chawla et al., 2003; Sando et al., 2012; Schlumm et al., 2013). A recent report showed that HDAC4 regulates a transcription program that is essential for synaptic transmission and information processing in the brain (Sando et al., 2012). This pathway involves an activity-dependent association of HDAC4 with transcription factors and neuronal chromatin. In addition, transgenic mice expressing HDAC4-3SA mutant, a constitutive nuclear localization, showed reduced gene expression essential for synaptic function, including Camk2a, Homer1, and Snap25 (Sando et al., 2012). Moreover, transgenic mice expressing a truncated form of HDAC4, which accumulates in the nucleus and thus acts as a gain-of-function transcriptional repressor, showed impaired acquisition and retention of memory in Barnes maze (Sando et al., 2012), indicating that nuclear HDAC4 is a negative regulator of memory formation. It should be noted that HDAC4-mediated repression of gene expression and reduced memory formation are independent of HDAC activity, as the truncated form of HDAC4 does not have the catalytic domain necessary for HDAC activity (Sando et al., 2012). This is likely mediated through repression of transcription factors such as MEF2 (Sando et al., 2012). However, it is still unclear whether nuclear import/export of HDAC4 occurs following learning. In addition, the nature of the interaction between HDAC4 and MEF2 or CREB during memory formation is largely unknown. Further examination will be necessary to uncover roles of nucleocytoplasmic shuttling of HDAC4 and its transcription machinery during memory formation.
Proteomics analysis highlighted HDAC4 as a global regulator associated with the onset of memory deficits in both normal age-associated cognitive decline and Alzheimer’s disease (Neuner et al., 2016). A recent paper showed that ApoE4, a major genetic risk factor for Alzheimer’s disease, increased the nuclear import of HDAC4, which leads to histone deacetylation and epigenetic changes in Bdnf gene transcription (Sen et al., 2015).
THE ROLE OF SYNAPSE-TO-NUCLEUS NF-κB SHUTTLING IN SYNAPTIC PLASTICITY AND MEMORY
NF-κB is a transcription factor that is ubiquitously expressed and responds to diverse stimuli including cytokines and growth factors (Hayden and Ghosh, 2012). In mammals, NF-κB consists of five subunits (RelA/p65, RelB, c-Rel, p105/50, and p100/52), possessing either transcriptional activator (Rel proteins) or repressor (p50, p52) activity. NF-κB signaling encompasses activation of mainly p65, c-Rel, and p50 containing heterodimers within the central nervous system (Engelmann and Haenold, 2016). NF-κB is normally found in the nucleus, but inactive NF-κB dimers are retained in the cytoplasm by binding IκB inhibitory protein. Upon stimulation, the IκB kinase complex becomes activated and phosphorylates IκB, leading to its degradation. This degradation, in turn, allows NF-κB to translocate to the nucleus and regulate gene expression of its target genes (Fig. 2). In mouse hippocampal cultures, synaptosomal NF-κB activity is enhanced by depolarization and exposure to the calcium ionophoreionomicin (Meffert et al., 2003). Some reports show synaptic localization of NF-κB in neurons (Kaltschmidt et al., 1993; Meberg et al., 1996; Meffert et al., 2003; Schmeisser et al., 2012), suggesting a function of NF-κB as a retrograde messenger. Indeed, p65-GFP fusion protein is travelling to the nucleus upon glutamate stimulation in neurons (Meffert et al., 2003; Wellmann et al., 2001). This nuclear accumulation of NF-κB is mediated by CaMKII, and followed by an increase in gene transcription (Meffert et al., 2003). Thus, NF-κB is activated at synapses following synaptic activity and is transported back to the nucleus to regulate transcription of its target genes such as those encoding BDNF, Fos, CaMKIIδ, IGF2, Zif268, C/EBP, PKA, NMDAR, and PKC (de la Fuente et al., 2015; Engelmann and Haenold, 2016; Kaltschmidt et al., 1999).
Several lines of evidence have shown a key role of NF-κB in synaptic and structural plasticity, and memory formation. Genetic ablation of NF-κB in mice resulted in severe defects in the dentate gyrus (Imielski et al., 2012). These mice showed degenerating neurites, hampered axogenesis, synaptogenesis and memory impairment. These structural and behavioral deficits were reversed by the re-activation of NF-κB, suggesting that NF-κB is a crucial regulator of the homeostasis in the dentate gyrus. Another study indicated that NF-κB controls excitatory synapses and dendritic spine formation and morphology in mouse hippocampal neurons (Boersma et al., 2011). NF-κB is activated during LTP in the mouse hippocampus (Freudenthal et al., 2004). A growing body of evidence indicates the involvement of NF-κB in memory processes in rodents, in different learning tasks such as inhibitory avoidance (Freudenthal et al., 2005), radial arm maze (Meffert et al., 2003), contextual fear conditioning (Lubin and Sweatt, 2007) and novel object recognition (Federman et al., 2013).
Hippocampal NF-κB activity increases 45 min following training in inhibitory avoidance and returns to basal levels within 2 h after initial training (Freudenthal et al., 2005). Contextual fear conditioning also increases the activity of hippocampal NF-κB 45 min following training (de la Fuente et al., 2014). The intra-hippocampal injection of sulfasalazine, an NF-κB inhibitor, following contextual fear conditioning leads to deficits in long-term memory in mice (de la Fuente et al., 2014). Moreover, in the novel object recognition task, hippocampal NF-κB is active during memory consolidation, and the inhibition of hippocampal NF-κB signaling after training drastically impairs long-term memory in object recognition (Federman et al., 2013). These results suggest that NF-κB is involved in memory consolidation. Synaptic NF-κB is also activated by learning. Synaptosomal NF-κB activation occurs 5 min following inhibitory avoidance training during consolidation (Salles et al., 2015), suggesting a potential role of retrograde transport of NF-κB in memory consolidation.
In addition to the role of NF-κB in memory consolidation, several reports have shown the role of NF-κB in memory reconsolidation. Hippocampal NF-κB activity is increased after memory retrieval (Boccia et al., 2007; de la Fuente et al., 2011). Moreover, retrieval of contextual fear memory activates the NF-κB pathway to regulate histone H3 phosphorylation and acetylation at specific gene promoters in hippocampus (Lubin and Sweatt, 2007). Importantly, the inhibition of NF-κB DNA-binding complex activity leads to impairment in fear memory reconsolidation, and this deficit can be ameliorated by treatment with HDAC inhibitor, suggesting that histone acetylation is a mechanism by which NF-κB signaling exerts its effects in the process of memory reconsolidation (Lubin and Sweatt, 2007).
Taken together, NF-κB plays an important role in memory consolidation and reconsolidation and has dual function as a synaptic activity detector and as a transcription regulator. However, the role of synapse-to-nucleus shuttling of NF-κB remains unclear. Future studies should clarify direct contribution of synaptic NF-κB to regulation of gene transcription and subsequent memory formation.
CONCLUSION
Recent work has delineated some of the signaling pathways regulating activity-dependent gene transcription by transcriptional cofactors that are transported from synapses to the nucleus following neuronal activity. However, more research has to be done to fully understand the fascinating aspect of these unusual signaling pathways directly connecting synaptic activity with gene transcription, including but not limited to early development, ageing and gender differences. Considering the significant role these pathways play in organism’s responses to changes in the environment, unraveling the molecular interactions of these transcriptional cofactors are critical for understanding of their involvement in the regulation of both health and disease.
Table 2.
Brief summary of activity-dependent transcriptional regulators implicated in neuropsychiatric disorders.
| Disease or model | Targets | Findings | References | |
|---|---|---|---|---|
| Alzheimer's disease | Human study | CREB, CBP/p300 | Reduced CREB, pCREB (Ser133), and CBP/p300 levels in prefrontal cortex in Alzheimer's disease. | (Bartolotti et al., 2016) |
| MEF2 | Association of SNP rs190982 with Alzheimer's disease. | (Lambert et al., 2013) (Ruiz et al., 2014) | ||
| Animal model | HDAC4, MeCP2 | HDAC4 and MeCP2 are identified as top canonical pathways by proteomics analyses. | (Neuner et al., 2016) | |
| HDAC4 | Increased nuclear HDAC4 levels in the hippocampus. | (Sen et al., 2015) | ||
| CRTC1 | Impaired learning-induced nuclear translocation of CRTC1. | (Parra-Damas et al., 2017; Parra-Damas et al., 2014) | ||
| The changes in CRTC1/CREB target genes. | ||||
| Reduced memory in model mice was restored by hippocampal overexpression of CRTC1. | ||||
| Autism/Rett syndrome | Human study | MECP2 | Mutations in the MECP2 gene in approximately 90% of patients with Rett syndrome. | (Amir et al., 1999) |
| MEF2 | MEF2C haploinsufficiency in autism. Small deletion encompassing the MEF2C gene in autism. | (Novara et al., 2010) | ||
| (Le Meur et al., 2010) | ||||
| HDAC4 | HDAC4 haploinsufficiency in autism. | (Pinto et al., 2014) | ||
| Animal model | MeCP2 | Cognitive and social abnormalities. | (Ebert et al., 2013; Li et al., 2008; Moretti et al., 2006) | |
| Deficit in motor behavior. | ||||
| MEF2 | Altered spine number and impaired memory. | (Barbosa et al., 2008) | ||
| Depression | Human study | CREB, HDAC4 | Decreased levels of CREB and increased levels of HDAC4 in patients with depression. | (Hobara et al., 2010; Ren et al., 2011) |
| Animal model | CREB, CRTC1, MEF2, MeCP2, HDAC4, | CREB overexpression in the dentate gyrus induces antidepressant-like behavior. | (Chen et al., 2001; Higuchi et al., 2016; Hutchinson et al., 2012; Lepack et al., 2016; Meylan et al., 2016a; Meylan et al., 2016b; Sailaja et al., 2012; Sarkar et al., 2014; Uchida et al., 2011) | |
| CRTC1 knockout mice showed increased depression-like behavior. | ||||
| Decreased Mef2 mRNA in depressed mice. | ||||
| Increased MeCP2-HDAC2 binding in depressed mice. | ||||
| MeCP2 mutant mice showed increased depression-like behavior. | ||||
| Hippocampal overexpression of HDAC4 induced depression-like behaviors. | ||||
| Increased HDAC4 in the hippocampus or medial prefrontal cortex of depressive mice. | ||||
Please note that the findings listed here do not cover all the contents of the reference papers and that we do not cite all the related articles here, due to the space limitation,
Synaptically-localized transcriptional regulators, CRTC1, HDAC4 and NF-κB, can move to the nucleus upon neuronal stimulation
Their phosphorylation status regulates their synaptic or nuclear localization
These transcriptional regulators are unique, providing means of direct link between synaptic activity and gene transcription
Acknowledgments
We would like to thank Ian Defalco for the artwork in Figure 1. This work was supported by the NIH Grant R01MH107555, Whitehall Foundation (#2008-12-104), March of Dimes (FY13-168), NARSAD Independent Investigator Award (24661) and the New Jersey Commission on Brain Injury Research Grant CBIR15IRG006 (to G.P.S); CREST-JST, KAKENHI (15K09807 and 15H04895), the Takeda Science Foundation and the NAITO Foundation (to S.U.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, Kandel ER. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/s0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- Barco A, Patterson SL, Alarcon JM, Gromova P, Mata-Roig M, Morozov A, Kandel ER. Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron. 2005;48:123–137. doi: 10.1016/j.neuron.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Boccia M, Freudenthal R, Blake M, de la Fuente V, Acosta G, Baratti C, Romano A. Activation of hippocampal nuclear factor-kappa B by retrieval is required for memory reconsolidation. J Neurosci. 2007;27:13436–13445. doi: 10.1523/JNEUROSCI.4430-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma MC, Dresselhaus EC, De Biase LM, Mihalas AB, Bergles DE, Meffert MK. A requirement for nuclear factor-kappaB in developmental and plasticity-associated synaptogenesis. J Neurosci. 2011;31:5414–5425. doi: 10.1523/JNEUROSCI.2456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Ch'ng TH, Martin KC. Synapse-to-nucleus signaling. Curr Opin Neurobiol. 2011;21:345–352. doi: 10.1016/j.conb.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng TH, Uzgil B, Lin P, Avliyakulov NK, O'Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Darcy MJ, Calvin K, Cavnar K, Ouimet CC. Regional and subcellular distribution of HDAC4 in mouse brain. J Comp Neurol. 2010;518:722–740. doi: 10.1002/cne.22241. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente V, Federman N, Fustinana MS, Zalcman G, Romano A. Calcineurin phosphatase as a negative regulator of fear memory in hippocampus: control on nuclear factor-kappaB signaling in consolidation and reconsolidation. Hippocampus. 2014;24:1549–1561. doi: 10.1002/hipo.22334. [DOI] [PubMed] [Google Scholar]
- de la Fuente V, Federman N, Zalcman G, Salles A, Freudenthal R, Romano A. NF-kappaB transcription factor role in consolidation and reconsolidation of persistent memories. Front Mol Neurosci. 2015;8:50. doi: 10.3389/fnmol.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente V, Freudenthal R, Romano A. Reconsolidation or extinction: transcription factor switch in the determination of memory course after retrieval. J Neurosci. 2011;31:5562–5573. doi: 10.1523/JNEUROSCI.6066-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- Dent EW. Of microtubules and memory: implications for microtubule dynamics in dendrites and spines. Mol Biol Cell. 2017;28:1–8. doi: 10.1091/mbc.E15-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, et al. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6:e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann C, Haenold R. Transcriptional Control of Synaptic Plasticity by Transcription Factor NF-kappaB. Neural Plast. 2016;2016:7027949. doi: 10.1155/2016/7027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, Romano A. Nuclear factor kappaB-dependent histone acetylation is specifically involved in persistent forms of memory. J Neurosci. 2013;33:7603–7614. doi: 10.1523/JNEUROSCI.4181-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenthal R, Boccia MM, Acosta GB, Blake MG, Merlo E, Baratti CM, Romano A. NF-kappaB transcription factor is required for inhibitory avoidance long-term memory in mice. Eur J Neurosci. 2005;21:2845–2852. doi: 10.1111/j.1460-9568.2005.04126.x. [DOI] [PubMed] [Google Scholar]
- Freudenthal R, Romano A, Routtenberg A. Transcription factor NF-kappaB activation after in vivo perforant path LTP in mouse hippocampus. Hippocampus. 2004;14:677–683. doi: 10.1002/hipo.20020. [DOI] [PubMed] [Google Scholar]
- Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N, Liauw JA, Blaeser F, Wei F, Hanissian S, Muglia LM, Wozniak DF, Nardi A, Arvin KL, Holtzman DM, et al. Impaired synaptic plasticity and cAMP response element-binding protein activation in Ca2+/calmodulin-dependent protein kinase type IV/Gr-deficient mice. J Neurosci. 2000;20:6459–6472. doi: 10.1523/JNEUROSCI.20-17-06459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielski Y, Schwamborn JC, Luningschror P, Heimann P, Holzberg M, Werner H, Leske O, Puschel AW, Memet S, Heumann R, et al. Regrowing the adult brain: NF-kappaB controls functional circuit formation and tissue homeostasis in the dentate gyrus. PLoS One. 2012;7:e30838. doi: 10.1371/journal.pone.0030838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Goodman RH. CREB signaling--timing is everything. Sci STKE. 2001;2001:pe1. doi: 10.1126/stke.2001.82.pe1. [DOI] [PubMed] [Google Scholar]
- Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nature neuroscience. 1998;1:595–601. doi: 10.1038/2830. [DOI] [PubMed] [Google Scholar]
- Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Kreutz MR. Nucleocytoplasmic protein shuttling: the direct route in synapse-to-nucleus signaling. Trends Neurosci. 2009;32:392–401. doi: 10.1016/j.tins.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Josselyn SA, Kida S, Silva AJ. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol Learn Mem. 2004;82:159–163. doi: 10.1016/j.nlm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B, Uherek M, Wellmann H, Volk B, Kaltschmidt C. Inhibition of NF-kappaB potentiates amyloid beta-mediated neuronal apoptosis. Proc Natl Acad Sci U S A. 1999;96:9409–9414. doi: 10.1073/pnas.96.16.9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt C, Kaltschmidt B, Baeuerle PA. Brain synapses contain inducible forms of the transcription factor NF-kappa B. Mech Dev. 1993;43:135–147. doi: 10.1016/0925-4773(93)90031-r. [DOI] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, Pena de Ortiz S, Kogan JH, Chevere I, Masushige S, Silva AJ. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 2002;5:348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- Kida S, Serita T. Functional roles of CREB as a positive regulator in the formation and enhancement of memory. Brain Res Bull. 2014;105:17–24. doi: 10.1016/j.brainresbull.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lai KO, Zhao Y, Ch'ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci U S A. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, Herrup K. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med. 2012;18:783–790. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Barco A. Can changes in histone acetylation contribute to memory formation? Trends Genet. 2014;30:529–539. doi: 10.1016/j.tig.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel G, Uchida S, Hevi C, Chevere-Torres I, Fuentes I, Park YJ, Hafeez H, Yamagata H, Watanabe Y, Shumyatsky GP. Genetic Demonstration of a Role for Stathmin in Adult Hippocampal Neurogenesis, Spinogenesis, and NMDA Receptor-Dependent Memory. J Neurosci. 2016;36:1185–1202. doi: 10.1523/JNEUROSCI.4541-14.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Siegelbaum SA, Kandel ER. Synapses and memory storage. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg PJ, Kinney WR, Valcourt EG, Routtenberg A. Gene expression of the transcription factor NF-kappa B in hippocampus: regulation by synaptic activity. Brain Res Mol Brain Res. 1996;38:179–190. doi: 10.1016/0169-328x(95)00229-l. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Murphy TH, Worley PF, Baraban JM. L-type voltage-sensitive calcium channels mediate synaptic activation of immediate early genes. Neuron. 1991;7:625–635. doi: 10.1016/0896-6273(91)90375-a. [DOI] [PubMed] [Google Scholar]
- Neuner SM, Wilmott LA, Hoffmann BR, Mozhui K, Kaczorowski CC. Hippocampal proteomics defines pathways associated with memory decline and resilience in normal aging and Alzheimer's disease mouse models. Behav Brain Res. 2016 doi: 10.1016/j.bbr.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka M, Kim R, Fukushima H, Sasaki K, Suzuki K, Okamura M, Ishii Y, Kawashima T, Kamijo S, Takemoto-Kimura S, et al. Region-specific activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron. 2014;84:92–106. doi: 10.1016/j.neuron.2014.08.049. [DOI] [PubMed] [Google Scholar]
- Panayotis N, Karpova A, Kreutz MR, Fainzilber M. Macromolecular transport in synapse to nucleus communication. Trends Neurosci. 2015;38:108–116. doi: 10.1016/j.tins.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Paroni G, Cernotta N, Dello Russo C, Gallinari P, Pallaoro M, Foti C, Talamo F, Orsatti L, Steinkuhler C, Brancolini C. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008;19:655–667. doi: 10.1091/mbc.E07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Damas A, Chen M, Enriquez-Barreto L, Ortega L, Acosta S, Perna JC, Fullana MN, Aguilera J, Rodriguez-Alvarez J, Saura CA. CRTC1 Function During Memory Encoding Is Disrupted in Neurodegeneration. Biol Psychiatry. 2017;81:111–123. doi: 10.1016/j.biopsych.2016.06.025. [DOI] [PubMed] [Google Scholar]
- Parra-Damas A, Valero J, Chen M, Espana J, Martin E, Ferrer I, Rodriguez-Alvarez J, Saura CA. Crtc1 activates a transcriptional program deregulated at early Alzheimer's disease-related stages. J Neurosci. 2014;34:5776–5787. doi: 10.1523/JNEUROSCI.5288-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SL, Pittenger C, Morozov A, Martin KC, Scanlin H, Drake C, Kandel ER. Some forms of cAMP-mediated long-lasting potentiation are associated with release of BDNF and nuclear translocation of phospho-MAP kinase. Neuron. 2001;32:123–140. doi: 10.1016/s0896-6273(01)00443-3. [DOI] [PubMed] [Google Scholar]
- Peixoto L, Abel T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology. 2013;38:62–76. doi: 10.1038/npp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34:447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- Proepper C, Johannsen S, Liebau S, Dahl J, Vaida B, Bockmann J, Kreutz MR, Gundelfinger ED, Boeckers TM. Abelson interacting protein 1 (Abi-1) is essential for dendrite morphogenesis and synapse formation. EMBO J. 2007;26:1397–1409. doi: 10.1038/sj.emboj.7601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Beal MF. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer's disease. Trends Mol Med. 2008;14:45–53. doi: 10.1016/j.molmed.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar TJ, Rodriguiz RM, Khiroug L, Wetsel WC, Augustine GJ, Means AR. Cerebellar defects in Ca2+/calmodulin kinase IV-deficient mice. J Neurosci. 2000;20:RC107. doi: 10.1523/JNEUROSCI.20-22-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles A, Boccia M, Blake M, Corbi N, Passananti C, Baratti CM, Romano A, Freudenthal R. Hippocampal dynamics of synaptic NF-kappa B during inhibitory avoidance long-term memory consolidation in mice. Neuroscience. 2015;291:70–80. doi: 10.1016/j.neuroscience.2015.01.063. [DOI] [PubMed] [Google Scholar]
- Sando R, 3rd, Gounko N, Pieraut S, Liao L, Yates J, 3rd, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151:821–834. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takemori H, Yagita Y, Terasaki Y, Uebi T, Horike N, Takagi H, Susumu T, Teraoka H, Kusano K, et al. SIK2 is a key regulator for neuronal survival after ischemia via TORC1-CREB. Neuron. 2011;69:106–119. doi: 10.1016/j.neuron.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Schlumm F, Mauceri D, Freitag HE, Bading H. Nuclear calcium signaling regulates nuclear export of a subset of class IIa histone deacetylases following synaptic activity. J Biol Chem. 2013;288:8074–8084. doi: 10.1074/jbc.M112.432773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser MJ, Baumann B, Johannsen S, Vindedal GF, Jensen V, Hvalby OC, Sprengel R, Seither J, Maqbool A, Magnutzki A, et al. IkappaB kinase/nuclear factor kappaB-dependent insulin-like growth factor 2 (Igf2) expression regulates synapse formation and spine maturation via Igf2 receptor signaling. J Neurosci. 2012;32:5688–5703. doi: 10.1523/JNEUROSCI.0111-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Sekeres MJ, Mercaldo V, Richards B, Sargin D, Mahadevan V, Woodin MA, Frankland PW, Josselyn SA. Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising memory quality. J Neurosci. 2012;32:17857–17868. doi: 10.1523/JNEUROSCI.1419-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Nelson TJ, Alkon DL. ApoE4 and Abeta Oligomers Reduce BDNF Expression via HDAC Nuclear Translocation. J Neurosci. 2015;35:7538–7551. doi: 10.1523/JNEUROSCI.0260-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Fukushima H, Mukawa T, Toyoda H, Wu LJ, Zhao MG, Xu H, Shang Y, Endoh K, Iwamoto T, et al. Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J Neurosci. 2011;31:8786–8802. doi: 10.1523/JNEUROSCI.3257-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto-Kimura S, Suzuki K, Horigane SI, Kamijo S, Inoue M, Sakamoto M, Fujii H, Bito H. Calmodulin kinases: essential regulators in health and disease. J Neurochem. 2017 doi: 10.1111/jnc.14020. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Uchida S, Martel G, Pavlowsky A, Takizawa S, Hevi C, Watanabe Y, Kandel ER, Alarcon JM, Shumyatsky GP. Learning-induced and stathmin-dependent changes in microtubule stability are critical for memory and disrupted in ageing. Nat Commun. 2014;5:4389. doi: 10.1038/ncomms5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Shumyatsky GP. Deceivingly dynamic: Learning-dependent changes in stathmin and microtubules. Neurobiol Learn Mem. 2015;124:52–61. doi: 10.1016/j.nlm.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Teubner BJ, Hevi C, Hara K, Kobayashi A, Dave RM, Shintaku T, Jaikhan P, Yamagata H, Suzuki T, et al. CRTC1 Nuclear Translocation Following Learning Modulates Memory Strength via Exchange of Chromatin Remodeling Complexes on the Fgf1 Gene. Cell Rep. 2017;18:352–366. doi: 10.1016/j.celrep.2016.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Bertos NR, Vezmar M, Pelletier N, Crosato M, Heng HH, Th'ng J, Han J, Yang XJ. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann H, Kaltschmidt B, Kaltschmidt C. Retrograde transport of transcription factor NF-kappa B in living neurons. J Biol Chem. 2001;276:11821–11829. doi: 10.1074/jbc.M009253200. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Xu W, Kasper LH, Lerach S, Jeevan T, Brindle PK. Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. EMBO J. 2007;26:2890–2903. doi: 10.1038/sj.emboj.7601734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HD, Grozinger CM, Liu JO. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J Biol Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolotti N, Bennett DA, Lazarov O. Reduced pCREB in Alzheimer's disease prefrontal cortex is reflected in peripheral blood mononuclear cells. Mol Psychiatry. 2016;21:1158–1166. doi: 10.1038/mp.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng TH, Uzgil B, Lin P, Avliyakulov NK, O'Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Shirayama Y, Shin KH, Neve RL, Duman RS. Expression of the cAMP response element binding protein (CREB) in hippocampus produces an antidepressant effect. Biol Psychiatry. 2001;49:753–762. doi: 10.1016/s0006-3223(00)01114-8. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. doi: 10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espana J, Valero J, Minano-Molina AJ, Masgrau R, Martin E, Guardia-Laguarta C, Lleo A, Gimenez-Llort L, Rodriguez-Alvarez J, Saura CA. beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J Neurosci. 2010;30:9402–9410. doi: 10.1523/JNEUROSCI.2154-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi F, Uchida S, Yamagata H, Abe-Higuchi N, Hobara T, Hara K, Kobayashi A, Shintaku T, Itoh Y, Suzuki T, et al. Hippocampal MicroRNA-124 Enhances Chronic Stress Resilience in Mice. J Neurosci. 2016;36:7253–7267. doi: 10.1523/JNEUROSCI.0319-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobara T, Uchida S, Otsuki K, Matsubara T, Funato H, Matsuo K, Suetsugi M, Watanabe Y. Altered gene expression of histone deacetylases in mood disorder patients. J Psychiatr Res. 2010;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Hutchinson AN, Deng JV, Cohen S, West AE. Phosphorylation of MeCP2 at Ser421 contributes to chronic antidepressant action. J Neurosci. 2012;32:14355–14363. doi: 10.1523/JNEUROSCI.2156-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur N, Holder-Espinasse M, Jaillard S, Goldenberg A, Joriot S, Amati-Bonneau P, Guichet A, Barth M, Charollais A, Journel H, et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet. 2010;47:22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepack AE, Bagot RC, Pena CJ, Loh YE, Farrelly LA, Lu Y, Powell SK, Lorsch ZS, Issler O, Cates HM, et al. Aberrant H3.3 dynamics in NAc promote vulnerability to depressive-like behavior. Proc Natl Acad Sci U S A. 2016;113:12562–12567. doi: 10.1073/pnas.1608270113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan EM, Breuillaud L, Seredenina T, Magistretti PJ, Halfon O, Luthi-Carter R, Cardinaux JR. Involvement of the agmatinergic system in the depressive-like phenotype of the Crtc1 knockout mouse model of depression. Transl Psychiatry. 2016a;6:e852. doi: 10.1038/tp.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan EM, Halfon O, Magistretti PJ, Cardinaux JR. The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: Possible relevance for treatment-resistant depression. Neuropharmacology. 2016b;107:111–121. doi: 10.1016/j.neuropharm.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner SM, Wilmott LA, Hoffmann BR, Mozhui K, Kaczorowski CC. Hippocampal proteomics defines pathways associated with memory decline and resilience in normal aging and Alzheimer's disease mouse models. Behav Brain Res. 2016 doi: 10.1016/j.bbr.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka M, Kim R, Fukushima H, Sasaki K, Suzuki K, Okamura M, Ishii Y, Kawashima T, Kamijo S, Takemoto-Kimura S, et al. Region-specific activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron. 2014;84:92–106. doi: 10.1016/j.neuron.2014.08.049. [DOI] [PubMed] [Google Scholar]
- Novara F, Beri S, Giorda R, Ortibus E, Nageshappa S, Darra F, Dalla Bernardina B, Zuffardi O, Van Esch H. Refining the phenotype associated with MEF2C haploinsufficiency. Clin Genet. 2010;78:471–477. doi: 10.1111/j.1399-0004.2010.01413.x. [DOI] [PubMed] [Google Scholar]
- Parra-Damas A, Chen M, Enriquez-Barreto L, Ortega L, Acosta S, Perna JC, Fullana MN, Aguilera J, Rodriguez-Alvarez J, Saura CA. CRTC1 Function During Memory Encoding Is Disrupted in Neurodegeneration. Biol Psychiatry. 2017;81:111–123. doi: 10.1016/j.biopsych.2016.06.025. [DOI] [PubMed] [Google Scholar]
- Parra-Damas A, Valero J, Chen M, Espana J, Martin E, Ferrer I, Rodriguez-Alvarez J, Saura CA. Crtc1 activates a transcriptional program deregulated at early Alzheimer's disease-related stages. J Neurosci. 2014;34:5776–5787. doi: 10.1523/JNEUROSCI.5288-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Dwivedi Y, Mondal AC, Pandey GN. Cyclic-AMP response element binding protein (CREB) in the neutrophils of depressed patients. Psychiatry Res. 2011;185:108–112. doi: 10.1016/j.psychres.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Heilmann S, Becker T, Hernandez I, Wagner H, Thelen M, Mauleon A, Rosende-Roca M, Bellenguez C, Bis JC, et al. Follow-up of loci from the International Genomics of Alzheimer's Disease Project identifies TRIP4 as a novel susceptibility gene. Transl Psychiatry. 2014;4:e358. doi: 10.1038/tp.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja BS, Cohen-Carmon D, Zimmerman G, Soreq H, Meshorer E. Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc Natl Acad Sci U S A. 2012;109:E3687–3695. doi: 10.1073/pnas.1209990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Chachra P, Kennedy P, Pena CJ, Desouza LA, Nestler EJ, Vaidya VA. Hippocampal HDAC4 contributes to postnatal fluoxetine-evoked depression-like behavior. Neuropsychopharmacology. 2014;39:2221–2232. doi: 10.1038/npp.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekeres MJ, Mercaldo V, Richards B, Sargin D, Mahadevan V, Woodin MA, Frankland PW, Josselyn SA. Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising memory quality. J Neurosci. 2012;32:17857–17868. doi: 10.1523/JNEUROSCI.1419-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Nelson TJ, Alkon DL. ApoE4 and Abeta Oligomers Reduce BDNF Expression via HDAC Nuclear Translocation. J Neurosci. 2015;35:7538–7551. doi: 10.1523/JNEUROSCI.0260-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Uchida S, Teubner BJ, Hevi C, Hara K, Kobayashi A, Dave RM, Shintaku T, Jaikhan P, Yamagata H, Suzuki T, et al. CRTC1 Nuclear Translocation Following Learning Modulates Memory Strength via Exchange of Chromatin Remodeling Complexes on the Fgf1 Gene. Cell Rep. 2017;18:352–366. doi: 10.1016/j.celrep.2016.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]