Abstract
Mitochondrial respiratory chain (RC) disease is a heterogeneous and highly morbid group of energy deficiency disorders for which no proven effective therapies exist. Robust vertebrate animal models of primary RC dysfunction are needed to explore the effects of variation in RC disease subtypes, tissue-specific manifestations, and major pathogenic factors contributing to each disorder, as well as their pre-clinical response to therapeutic candidates. We have developed a series of zebrafish (Danio rerio) models that inhibit, to variable degrees, distinct aspects of RC function, and enable quantification of animal development, survival, behaviors, and organ-level treatment effects on function as well as mitochondrial biochemistry and physiology. Here, we characterize four pharmacologic inhibitor models of mitochondrial RC dysfunction in early larval zebrafish, including rotenone (complex I inhibitor), azide (complex IV inhibitor), oligomycin (complex V inhibitor), and chloramphenicol (mitochondrial translation inhibitor that leads to multiple RC dysfunction). A range of concentrations and exposure times of each RC inhibitor were systematically evaluated on early larval development, animal survival, integrated behaviors (touch and startle responses), organ physiology (brain death, neurologic tone, heart rate), and fluorescence-based mitochondrial physiology in zebrafish skeletal muscle. Pharmacologic RC inhibitor effects were validated by spectrophotometric analysis of Complex I, II and IV activities, or relative quantitation of ATP levels in larvae. Outcomes were prioritized that utilize in vivo animal imaging and quantitative behavioral assessments, as may optimally inform the translational potential of pre36 clinical drug screens for future clinical study in human mitochondrial disease subjects. The RC complex inhibitors each delayed early embryo development, with short-term exposures of these three agents or chloramphenicol from 5–7 days post fertilization also causing reduced larval survival and organ-specific defects ranging from brain death, behavioral and neurologic alterations, reduced mitochondrial membrane potential in skeletal muscle (rotenone), and/or cardiac edema with visible blood pooling (oligomycin). Remarkably, we demonstrate that treating animals with probucol, a nutrient-sensing signaling network modulating drug that has been shown to yield therapeutic effects in a range of other RC disease cellular and animal models, both prevented acute rotenone-induced brain death in zebrafish larvae, and significantly rescued early embryo developmental delay from either rotenone or oligomycin exposure. Overall, these zebrafish pharmacologic RC function inhibition models offer a unique opportunity to gain novel insights into diverse developmental, survival, organ-level, and behavioral defects of varying severity, as well as their individual response to candidate therapies, in a highly tractable and cost-effective vertebrate animal model system.
Keywords: D. rerio, Complex I, Complex IV, Complex V, mitochondrial translation
CHEMICAL COMPOUNDS: Rotenone (PubChem CID: 6758), sodium azide (PubChem CID: 33557), oligomycin (PubChem CID: 16760598), chloramphenicol (PubChem CID: 5959), mitotracker green FM (PubChem CID: 59705974), TMRE (PubChem CID: 2762682), probucol (PubChem CID: 4912)
1.1 INTRODUCTION
Cellular energy is primarily derived by aerobic metabolism of nutrient-derived reducing equivalents to generate adenosine triphosphate (ATP) through the process of oxidative phosphorylation that is carried out by the integrated function of five enzyme complexes within the inner mitochondrial membrane that together comprise the respiratory chain (RC). RC dysfunction in one or more complexes leads to multi-systemic failure of high-energy demand organs, clinically presenting as severe neurodevelopmental, cardiac, myopathic, renal, hepatic, endocrine, hearing, and/or vision disabilities, as well as global metabolic instability with lactic acidosis. Indeed, nearly 300 unique genes in both the nucleus and mitochondria itself already implicated in primary mitochondrial diseases (McCormick et al., 2013). Recognized as the most common group of inborn metabolic diseases, primary (genetic-based) mitochondrial disease has a combined minimal prevalence of at least 1 in 4,300 (Elliott et al., 2008; Gorman et al., 2015; Haas et al., 2007; Schaefer et al., 2008). In addition, RC dysfunction is broadly implicated in the pathogenesis of a host of modern-day complex diseases ranging from metabolic syndrome (Irving and Nair, 2007) to ischemia-reperfusion injury after stroke (Chouchani et al., 2014) to neurodegenerative diseases (Mandemakers et al., 2007). Regardless of cause, once RC dysfunction occurs, the effective result is the alteration of cellular NADH/NAD+ redox balance that influences a wide range of cellular reactions (McCormack et al., 2015; Zhang et al., 2013), as well as the induction of both oxidative and proteotoxic stress (Segref et al., 2014). In other words, RC dysfunction results in a complex series of cellular adaptations, which, although attempting to maintain cellular function and survival in the face of critical energy deficiency, are themselves major contributors to the clinical pathophysiology of mitochondrial disease (Zhang et al., 2013).
Effective therapies for both primary (genetic) and secondary (acquired) RC diseases are lacking (Avula et al., 2014; Parikh et al., 2009), since little is known about the biochemical and physiologic abnormalities that contribute to their diverse clinical manifestations. Clinical complexity and imprecisely defined and/or understood biochemical phenotypes leave clinicians unable to effectively apply or monitor targeted therapies for RC disease (Avula et al., 2014; Chinnery et al., 2006; Dimauro and Rustin, 2008). Mitochondrial medication ‘cocktails’ variably include vitamins (B1, B2, C), antioxidants (CoQ10, lipoic acid, vitamin E, epi-743)(Martinelli et al., 2012; Parikh et al., 2009), and metabolic modifiers (creatine, L-carnitine, L-arginine (Koga et al., 2005; Parikh et al., 2009), folinic acid (Garcia-Cazorla et al., 2008)). Although efficacy, toxicity or optimal dose of these drugs have not been objectively assessed in human RC disease patients, they are often empirically prescribed in hopes of enhancing residual RC enzymatic function or quenching ‘toxic’ metabolites that are theorized to accumulate in RC dysfunction (Avula et al., 2014; Pfeffer et al., 2013). Provision of these therapies generally adopts a “one size fits all” approach, overtly ignoring inherent variation in RC disease subtypes, tissue-specific manifestations, and major pathogenic factors such as the predominant downstream metabolic and signaling alterations (Zhang et al., 2014; Zhang et al., 2013).
Robust animal models of mitochondrial respiratory chain dysfunction are needed in which to explore these key variables as well as their response to therapy. While simpler model animals such as bacteria, yeast, C. elegans, D. melanogaster (Rea et al., 2010), as well as an increasing array of mouse models of RC disease (Ruzzenente et al., 2016) each have clear experimental value for dissecting disease mechanisms and evaluating therapeutic candidates, there remains a need for efficient, higher-throughput screening of vertebrate animal models in which to cost-effectively quantify mitochondrial physiology and function of individual organs and animal behaviors (Camp et al., 2016). To this end, we have developed a series of zebrafish (Danio rerio) pharmacologic inhibitor models of distinct aspects of mitochondrial RC function in which to efficiently quantify both animal and organ-level treatment effects (Baden et al., 2007; Kari et al., 2007; Steele et al., 2014).
Here, we report the development and characterization of 4 pharmacologic inhibitor models of mitochondrial RC dysfunction in early larval zebrafish animals. These 4 models include rotenone (complex I inhibitor), azide (complex IV inhibitor), oligomycin (complex V inhibitor), and chloramphenicol (mitochondrial translation inhibitor causing multiple RC dysfunction (Abou-Khalil et al., 1980). A range of inhibitor concentrations are discussed at a variety of exposure times, with quantitation of inhibitor effects on early larval development, behaviors (startle response), organ physiology (heart rate, brain death, neurologic tone), and mitochondrial physiology (mitochondrial mass and membrane potential quantitation in muscle with fluorescent dyes, with potential to utilize stable ro-GFP or mito-roGFP lines to quantify cellular or intra-mitochondrial oxidant or antioxidant levels (Albrecht et al., 2014; Gutscher et al., 2008; Seiler et al., 2012). To gain deeper insight into organ-specific treatment effects, zebrafish can also be studied by histological, immunohistochemical, and gene or protein expression studies. However, studies described here are limited to those amenable to in vivo animal imaging, survival, and quantitative behavioral assessment, which are relevant translational outcomes to inform the selection of candidate drugs for study in human clinical trials that aim to improve feeling, function, and/or survival of mitochondrial disease subjects.
1.2 MATERIALS AND METHODS
1.2.1 Generation of zebrafish larvae
All protocols and methods described below have been performed in accordance with IACUC regulations (Protocol ID: IAC 15-001154) for care and use of D. rerio at the Children’s Hospital of Philadelphia Research Institute. Unless otherwise specified, all reagents were obtained from Sigma-Aldrich (St Louis, MO, USA). Embryos and larvae were maintained at 28°C throughout the duration of the experiments. Adult zebrafish (strain AB or TLF) were set pairwise in undivided mating tanks, and resultant embryos were collected and sorted on 0 days post-fertilization (0 dpf) and placed in embryo water (E3) in a 28°C incubator overnight. On 1 dpf, embryos were again sorted to remove non-viable embryos, sanitized with sodium hypochlorite, and treated with pronase by standard methods to promote uniform hatching. On 2 dpf, pronase was removed and larvae were placed in E3/phenylthiourea 0.03ug/l (PTU) for the remainder of the experiment to prevent larval pigment formation, except for those fish in the chronic exposure protocols, which were not maintained in PTU but rather in E3.
1.2.2 Zebrafish rotenone model of mitochondrial complex I inhibition and probucol treatment
Rotenone was applied in 10 mM Tris pH 7.2, 0.1% DMSO. Larvae were treated from 5 hpf to 36 hpf with 30–100 nM rotenone. On 7 dpf, larvae were treated with 100 nM rotenone for 4 hours, after which their brain phenotype was scored. Control larvae were treated with equal amount of ethanol (vehicle). Rotenone stock solutions (100 mM) were prepared with ethanol. Probucol was applied at 5 uM in 10 mM Tris pH 7.2, 0.1% DMSO to larvae from 3.5 dpf and refreshed daily and on 7 dpf larvae were treated with 100 nM rotenone, as above, in presence of probucol.
1.2.3 Zebrafish azide models of mitochondrial complex IV deficiency
Both acute and chronic models of azide toxicity were evaluated. For the chronic azide model, zebrafish embryos from 3 hpf were treated with sodium azide (50–100 uM) in E3/0.1%DMSO/Tris buffer continuously up to 7 dpf. Control fish were maintained in E3/0.1% DMSO. 2 dpf larvae were manually dechorionated by standard methods to facilitate observation. On subsequent days up to 7 dpf, embryos and larvae were scored for their attainment of microscopic developmental milestones according to the descriptions contained in Kimmel et al. (Kimmel et al., 1995), as well as for mortality, startle and touch response, development of the swim bladder, and heartrate. For the acute azide model, zebrafish larvae at 6 dpf were treated with the RC complex IV inhibitor sodium azide (75–150 uM) in E3/PTU/0.1% DMSO/10mM Tris buffer. Larvae on 7 dpf were observed for toxic effects of the azide from 12 and 24 hours of treatment. Control fish were maintained in E3/PTU/0.1% DMSO/10mM Tris buffer. Larvae were scored for effects of azide exposure compared to buffer-treated controls for physiologic parameters such as development of brain cell death, swimming behavior (touch response) when larvae were manually touched with a probe, startle response when the culture vessel was lightly tapped with a probe, heart rate, and ability to remain upright. Brain cell death indicative of a diseased state was recorded in fish exhibiting gray discoloration in their brain regions, which does not occur in healthy zebrafish larvae. As healthy larvae reliably and swiftly swim away in response to physical contact with a blunt metal probe, the presence or absence of the expected swim response to the probe served as a measure of healthy motor function in the larvae. As healthy larvae exhibit a greater capacity to rest and swim in an upright position similar to adult zebrafish, larval ability to maintain an upright, resting position was measured as an indicator of general health.
1.2.4 Zebrafish oligomycin model of mitochondrial complex V deficiency
At 2 dpf, zebrafish larvae were placed in fresh E3 +10mM Tris, pH 7.4, solution in 6-well dishes (10 fish per well). At this time, up to 4 conditions were tested: (1) Ethanol used at 1:1000 as control, (2) Oligomycin at a final concentration of 300 nm (3) Oligomycin at a final concentration of 1 µM, or (4) Oligomycin at a final concentration of 3 µM. Oligomycin was dissolved in ethanol and administered to each well so that the maximum ethanol concentration of each of the treatment conditions was 1:1000. In some trials, treatment started at 5 hours post fertilization (hpf), rather than 2 dpf. In all cases, E3 and drug were refreshed daily. Fish were monitored each day for survival, structural defects, heart rate and were euthanized on 7 dpf by adding tricane and then placed in ice for at least 20 minutes.
1.2.5 Zebrafish chloramphenicol model of mitochondrial translation deficiency
A zebrafish model of chronic mitochondrial translation deficiency was created using the pharmacologic inhibitor chloramphenicol. At 2 dpf, zebrafish were placed in fresh E3 +10mM Tris, pH 7.4, solution in 6 well dishes (10 fish per well). At this time, 2 conditions were tested, (1) Chloramphenicol alone, and (2) untreated control. Fresh stock of chloramphenicol dissolved in water (6 mM) was administered to each well and buffered in 10 mM Tris pH 7.4. Fish were monitored each day for survival and euthanized on 7 dpf by adding tricane and then placed in ice for at least 20 minutes.
1.2.6 Mitochondrial physiology evaluation in living zebrafish muscle with fluorescence dye imaging
Approximately 900 picoliters of 2 mM tetramethylrhodamine ethyl ester perchlorate (TMRE) together with 4 ug/ul mitotracker green FM (MTG) in DMSO was injected into the yolk of freshly fertilized larvae. Fish were grown to 6 dpf and imaged with a Zeiss LSM 710 microscope. Control (ethanol) larvae were compared to larvae treated for 3 hours with 50 nM rotenone. To analyze relative fluorescence intensity, a frame was drawn around 2 somites in Fiji (imageJ) and the mean brightness of this area measured for both TMRE and MTG. As skin showed much brighter staining with MTG than TMRE, it was excluded from analysis. TMRE was normalized in a given region of interest relative to MTG as marker of mitochondrial content. 6 animals were analyzed per condition.
1.2.7. Probucol rescue of RC inhibitor induced zebrafish embryo developmental delay
Embryos were treated from 6 hpf with 100 nM rotenone, 1 uM oligomycin, 500 uM azide or vehicle control in 10 mM Tris, pH 7.2, 0.1% DMSO. For rescue experiments, 50 uM probucol was concurrently added at 6 hpf. Embryo development was assessed at 30 hpf and embryos were sorted based on head and tail bud appearance relative to normally developing larvae as divided into 4 general developmental stages between 10 and 30 hpf. Average developmental age was calculated for each group in three biological replicate experiment to screen for rescue of each RC inhibitor induced delay.
1.2.8. Electron transport chain (ETC) activity assays
Embryos harvested were homogenized in 50–150 µl (~30 fish per 100 uL, based on fish counts in each tube) buffer (250 mM sucrose, 20 mM Tris-HCl, 3 mM EDTA, pH 7.4) by combination of grinding, 1–2 sec of sonication, and liquid nitrogen freeze-thawing. Mitochondrial200 enriched larval fractions were obtained by differential centrifugation. All spectrophotometric assays were performed at 30°C in 170 µl final volume using a Tecan Infinite 200 PRO plate reader. Complex I and complex II enzyme activities were determined by the reduction of 2,6-dichlorobenzenone-indophenol sodium salt (2,6-DCPIP) at 600 nm (ε = 21 mM−1 cm−1). The assay buffer for the complex I enzyme assay contained 25 mM KH2PO4, pH 7.4, 5 mM MgCl2, 3 mg/ml bovine serum albumin (BSA), 25 µM ubiquinone Q1, 5 µM antimycin A and mitochondrial-enriched zebrafish extract, the reaction was started with 150 µM NADH in the presence and absence of 5 µM rotenone, and rates were calculated after the subtraction of rotenone-insensitive activities. The assay buffer for the complex II enzyme activity assay contained 25 mM KH2PO4 pH 7.4, 5 mM MgCl2, 3 mg/ml BSA, 25 µM ubiquinone Q1, 5 µM antimycin A, 5 µM rotenone, and mitochondrial-enriched zebrafish extract. The reaction was started with 20 mM succinate. Complex IV enzyme activity was measured by following the oxidation of reduced cytochrome c at 550 nm (ε = 21 mM−1 cm−1). The assay buffer contained 25 mM KH2PO4 pH 7.4, 5 mM MgCl2, 0.015% n-dodecyl-β-D-maltoside, 5 µM antimycin A, 5 µM rotenone, and mitochondrial-enriched zebrafish extract. The reaction was started with 15 µM reduced cytochrome c. The rates were calculated as a first order constant. The specific activity was normalized by protein concentration as estimated by Bradford assay (Bradford, 1976).
1.2.9 Measurement of ATP levels in zebrafish
ATP levels were measured in zebrafish using the CellGlo assay (Promega). Treated zebrafish were anesthetized with tricaine, collected in an Eppendorf tube, and E3 was removed completely. Fresh larvae were then resuspended in mitochondrial respiration medium, MiR05 (Fashing and Gnaiger, 2016), at a concentration of 1 larva/50 uL. Anesthetized fish were then homogenized using a benchtop homogenizer set to 30 rpm using a tight-fitting pestle. Homogenate was aliquoted as 50 uL per well onto a 96-well plate with at least two biological replicates performed per condition. 50 uL of Mir05 was added per well. At least 5 wells were also prepared with 100 uL of Mir05 as background control, and a three-point serial dilution of ATP in Mir05 from 1 uM to 10 nM of ATP was used as a standard curve. CellGlo was then performed as per protocol. In brief, the plate was allowed to equilibrate to room temperature for 30 minutes, after which 100 uL of CellGlo reagent was added per well. The plate was placed on an orbital shaker for 2 minutes, incubated for an additional 10 minutes at room temperature, then analyzed by plate reader to record luminescence. Due to variance between runs, likely related to timing and room conditions, ATP amount was normalized to the median ATP in the concurrently analyzed DMSO-treated controls.
1.2.10 Statistical analyses
All experimental analyses were performed in 3–20 animals per condition, with a minimum of 3 biological replicate experiments. Group comparisons were compared using unpaired Student’s T test with statistical significance set at p < 0.05.
1.3 RESULTS
1.3.1 Zebrafish rotenone model of complex I inhibition
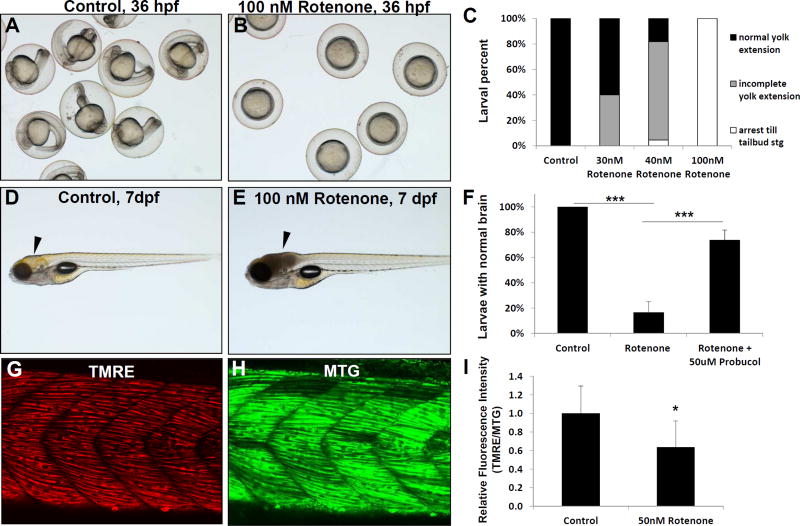
To evaluate the effect of rotenone on development we exposed newly fertilized eggs from 5 hours post-fertilization (5 hpf). Treatment with 100 nM rotenone arrested development between late gastrulation and tail-bud stage, while no cell death was apparent (Fig 1B) and buffer-control treated larvae developed normally (Fig 1A). Rotenone effect was concentration-dependent, with 30–40 nM concentrations causing less strong effects and having a higher fraction of nearly normal larvae (Fig 1C). This effect was similar to oxygen deprivation at this stage (Padilla and Roth, 2001), suggesting that rotenone inhibition of mitochondrial function and cellular bioenergetics stalls development at 5 hpf in a similar fashion to hypoxia-induced suspended animation. We also evaluated whether rotenone could induce organ-specific effects in fully developed animals at 7 dpf. Indeed, we found that treatment with 100 nM rotenone induced a reproducible brain death phenotype within 4 hours of rotenone exposure on 7 dpf (Fig 1D–E), where 86% of rotenone-treated fish had developed grey brain together with an absent response to both touch and sound stimuli, indicating substantial cellular death occurs in their brain (Fig 1F). We also sought to evaluate in vivo mitochondrial physiologic effects of mitochondrial inhibition in zebrafish. Therefore, we co-injected newly fertilized eggs with TMRE and MTG FM to fluorescently label mitochondrial membrane potential and mitochondrial content, respectively, throughout the cells of each animal. Based on the strong fluorescence intensity of these dyes and easy to identify landmarks in skeletal muscle tissue, we quantified the relative trunk muscle fluorescence brightness (Fig 1G–H). As expected, rotenone induced a substantial reduction in TMRE brightness after 3 h hours treatment on 7 dpf (normalized to MTG) (Fig 1I), consistent with reduced mitochondrial membrane potential. Thus, rotenone provides an effective model of complex I inhibition in zebrafish in which to evaluate a variety of effects depending on timing of animal exposure, including early developmental delay, brain death and reduced responsiveness, as well as decreased skeletal muscle mitochondrial membrane potential.
Fig 1. Rotenone inhibition of RC complex I in zebrafish.
Rotenone treatment effects were tested on larval development, brain physiology, and mitochondrial membrane potential. Rotenone inhibits development in a concentration-dependent manner when larvae are treated from 6–36 hours post fertilization (hpf). (A) While control treated larvae developed normally, (B) Rotenone (100 nM) treatment drastically inhibited development, such that embryos did not develop past the tail bud stage. (C) Developmental effects were concentration dependent, as scored both by the larvae arresting before the tail bud stage and their greater size of yolk extension. (D–E) Rotenone induced brain cell death occurred when larvae were treated for 4 hours on 7 days post fertilization (dpf), as indicated by arrowhead in control (D) and rotenone-treated (E) animals. (F) The severe brain phenotype induced by rotenone on 7 dpf was significantly prevented by pre-treating larvae with 50 uM probucol (***, p < 0.001). N=3 biological replicate experiments. (G–I) Rotenone treatment decreased the mitochondrial membrane potential of 7 dpf larval trunk muscle. Relative fluorescence quantitation in a flank skeletal muscle region of interest was performed with TMRE to assess mitochondrial membrane potential (G) compared to Mitotracker green FM (MTG, H) as loading control and indicator of mitochondrial content. (I) Rotenone treatment for 4 hours on 7 dpf induced a significant reduction of mean TMRE brightness normalized to MTG (I). n=6 replicates evaluated. *, p=0.026. Error bars represent (± SD).
Given the robustness of our 7 dpf acute rotenone model, we used it to evaluate effects of probucol, a candidate drug for RC disease therapy. Probucol is a lipophilic, phenolic compound that has antioxidant and lipid-lowering capabilities (Stocker, 2009). Our previous work has shown the benefit of probucol in both treatment and prevention of renal glomerular disease in an RC deficient B6.Pdss2−/− mouse model with coenzyme Q biosynthetic defect (Falk et al., 2011). Probucol has also shown benefit in RC deficient gas-1(fc21) C. elegans with deficient complex I (NDUFS2 orthologue) activity as well as in multiple human cell models of RC diseases, with its mechanism involving inhibition of mTORC1 signaling while also activating AMPK and PPAR pathway signaling that are dysregulated in primary RC disease (Peng et al., 2015). Thus, we decided to test probucol as a therapy to prevent the observed phenotypes of the rotenone induced complex I inhibition zebrafish larval model. Remarkably, we show that pre-treatment of larvae with 5 µM probucol significantly attenuated their brain death upon exposure to rotenone at 7dpf (Fig 1F). Elucidation of the mechanism by which probucol can prevent zebrafish brain necrosis remains under active investigation in our lab. Overall, these data reveal the strong potential to utilize the rotenone-inhibited vertebrate animal model to screen candidate mitochondrial RC complex I disease therapies on individual organs.
1.3.2 Zebrafish azide model of complex IV inhibition
We developed two different models of azide exposure in zebrafish. The first we designate as the “chronic exposure” model, in which 3–5 hpf old embryos were exposed to varying concentrations of sodium azide for 7 days to evaluate the effects on embryonic and larval development. The second model examined a comparatively “acute, short-term exposure” of zebrafish larvae to sodium azide, where larvae were dosed on day 6 post-fertilization (6 dpf), and observations were made 12–24 hours after exposure to the toxicant.
1.3.2.1 Azide chronic exposure zebrafish model
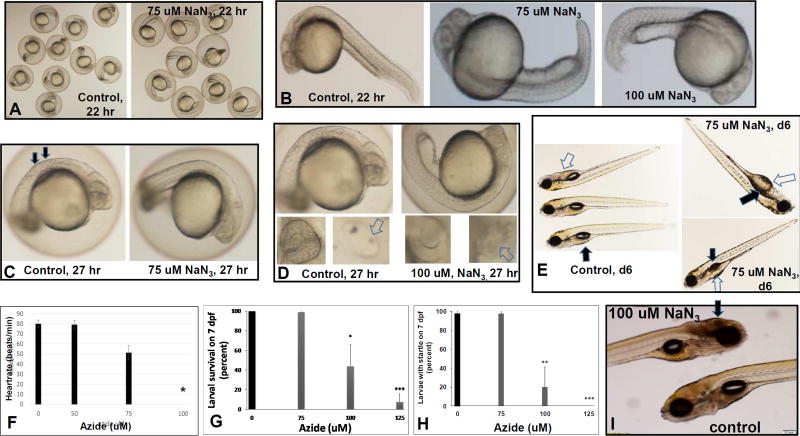
After exposing early embryos to sodium azide, we observed developmental arrest in a dose-specific and time-dependent manner (Fig 2). Developmental delay after 22 hour exposure to 75 uM azide included shorter tails, delayed pigment development and larger yolk sac size compared to controls, suggesting a reduction in nutrient absorption (Fig 2A–B). All larvae continued to develop after 27 hours of exposure to 75 uM or 100 uM azide, although 100 uM azide-treated animals were delayed compared to controls, with absent otoliths within their otic capsules and absence of pigmented cells in their flanks and eyes (Fig 2C–D). By 6 dpf, larvae that had been chronically exposed from the 5 hpf embryo stage to 75 uM azide showed continued developmental delay, with uninflated swim bladders and reduced absorption of yolk (Fig 2E). In addition, cardiac development was delayed as 100uM azide treated fish did not have a heartbeat 26 hours after treatment (31 hpf) (Fig 2F & controls found in Supplementary File S1_Movie 1). Thus, chronic azide exposure can be used to variably delay many aspects of normal larval development, without causing complete larval arrest.
Fig 2. Azide chronic and acute inhibition of RC complex IV.
(A–F), Effects of chronic azide exposure beginning at 5 hpf. (A–B) Developmental delay was induced by early azide treatment after 22 hours of treatment, where (A) low-magnification shows that 75 uM azide treated animals had shorter tails (right panel) (B) Higher magnification of 75 uM and 100 uM azide treated embryos clearly demonstrate their developmental delay compared to buffer treated controls after 22 hours, where azide-treated animals had shorter tails and decreased absorption of their yolk. These embryos were manually dechorionated to facilitate observation. (C) After 27 hours of 75 uM or 100 uM azide treatment, delayed pigment development was observed (arrows, left panel). (D) 100 uM azide-treated animals were delayed compared to controls. Control fish (left panels) observed at this time have developed otoliths (white arrows) within their otic capsules (lower right panel) as well as pigmented cells in their flanks (top panel) and eyes (lower left panel). The 100 uM azide treated (right panels) have not developed pigment in their flanks (top panel) or eyes (lower left panel) nor have they developed otoliths (lower right panel). (E) Developmental delay induced by 75 uM azide chronic treatment from 5 hpf (right panels) relative to control fish on 6 dpf (left panel). Azide treatment at 75 uM concentration delayed normal development, but did not arrest it. Azide-treated animals on 6 dpf had delayed inflation of their swim bladder (black arrows) and reduced absorption of yolk (white arrows). (F) Azide chronic exposure from 5hpf and observed after 26 hours of treatment, revealed that cardiac function was delayed. 100 uM azide treated embryos were sufficiently delayed that they did not yet have a heartbeat after azide treatment for 26 hours. n=3 biological replicates with 3 fish per experiment. *p=0.0072. (G–I), Effects of acute azide exposure beginning on 6 dpf. (G) Survival of 7 dpf zebrafish after 16 hr of sodium azide (NaN3) treatment, n=3 biological replicates with 10 fish per treatment condition. * p=0.0109, **p=0.0001. (H) Effects of acute azide exposure model on zebrafish startle-response after 16 hours azide treatment. Of note, all 125 uM azide treated fish were unresponsive to tapping on the dish. n=4 biological replicates with 10 fish per treatment condition. ** p=0.0033, ***p=0.00001. (I) Treatment with 100 uM azide for 17 hours (acute exposure) resulted in brain necrosis (arrow), which begins at the posterior of the brain and can be observed to move forward (cephalad) as it progresses over time. Error bars represent (±SD).
1.3.2.2 Azide acute exposure zebrafish models
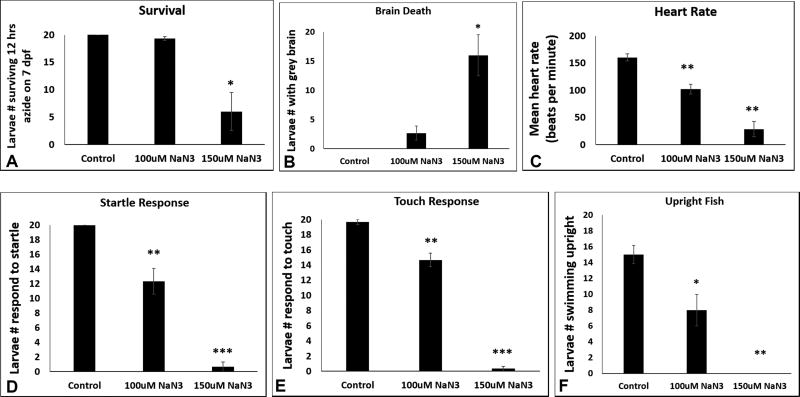
Treating larvae for 12–24 hours with sodium azide beginning on 6 dpf caused a dose- and time-dependent phenotype. By 16 hours of sodium azide exposure, animals displayed a clear-dose dependent reduction in survival, with an LD50 of approximately 100 uM azide (Fig 2G). Similarly, the animals’ startle response at 16 hours exposure (Supp File S2_Movie 2) was severely compromised at 100 uM azide and not present at all in animals exposed to 125 uM azide (Fig 2H and Supp File S3_Movie 3). Most discernibly, 100 uM azide exposure starting at 6 dpf for 17 hours resulted in reproducible and quantifiable brain cell death that visually presented as gray brain (Fig 2I). The gray brain phenotype was first observed at the posterior end of the brain and progressed cephalad (anteriorly) over time. Given these results, we sought to develop a short-term model that would enable simultaneous quantitative profiling of azide effects on zebrafish survival, brain death, heart rate, and neurologic-based behaviors including startle response, touch response, and upright swimming (Fig 3). Specifically, all six of these outcome parameters were assessed in 3 biological replicates of 7 dpf larvae treated for 12 hours with 100 uM and 150 uM sodium azide relative to untreated control. All 6 outcomes showed a clear dose-dependence. Survival was preserved at 12 hours in the 100 uM azide treated larvae (Fig 3A) with few animals developing a gray brain, suggestive of brain death (Fig 3B), although their physiology was already substantially impaired with significant reductions in heart rate (Fig 3C), startle response (Fig 3D), touch response (Fig 3E), and upright swimming ability (Fig 3F). In comparison, larvae exposed to 150 uM azide on 6 dpf for 12 hours had significantly reduced survival (Fig 3A), greater than 80% incidence of gray brain suggestive of brain cell death (Fig 3B), and severely impaired physiology with very few animals having any heart rate (Fig 3C), startle response (Fig 3D) or touch response (Fig 3E) and complete absence of their upright swimming ability (Fig 3F). Thus, short-term azide exposure in fully-formed zebrafish larvae on 6 dpf results in dose- and time-dependent survival as well as physiologic and behavioral effects, including decreased heart rate, loss of motor function, inability to respond to tactile stimulation, as well as evidence of severe neurological brain damage.
Fig 3. Quantitative profiling of azide effects on animal survival, heart rate, and behaviors.
All six outcome parameters were assessed in 3 biological replicates of 7 dpf larvae treated for 12 hours with sodium azide (NaN3, 100 and 150 uM) or untreated control with n=20 larvae per condition. All data are reported as the mean and standard error of number of larvae meeting each parameter across 3 replicate experiments. (A) Survival. Zebrafish number alive on 7 dpf after 12 hours of azide exposure. Relative to untreated control, 100 uM azide p = 0.092 and 150 uM azide p = 0.028. (B) Brain death. Prevalence of larvae with grey brain on 7 dpf after 12 hours of azide exposure. Relative to untreated control, 100 uM azide p=0.078 and 150 uM p=0.022. (C) Heart rate. Average heart rates of larvae on 7 dpf after 12 hours of azide exposure. Within each experimental condition, 6 fish heart rates were manually observed and averaged to represent the group of 20. Relative to untreated control, 100 uM azide p = 0.003 and 150 uM azide p = 0.002. (D) Startle response. Prevalence of larval motor response to dish tapping on 7 dpf after 12 hours of azide exposure. Relative to untreated control, 100 uM azide p=0.004 and 150 uM azide p<0.001. (E) Touch response. Prevalence of larval motor response to touch with metal probe on 7 dpf after 12 hours of azide exposure. Relative to untreated control, 100 uM azide p=0.007 and 150 uM azide p<0.001. (F) Upright behavior. Prevalence of larval in an upright resting state on 7 dpf after 12 hours of azide exposure. Relative to untreated control, 100 uM azide **p=0.028 and 150 uM azide *** p=0.003. Error bars represent (± SEM).
1.3.3 Zebrafish oligomycin model of complex V inhibition had reduced survivorship and abnormal cardiac function
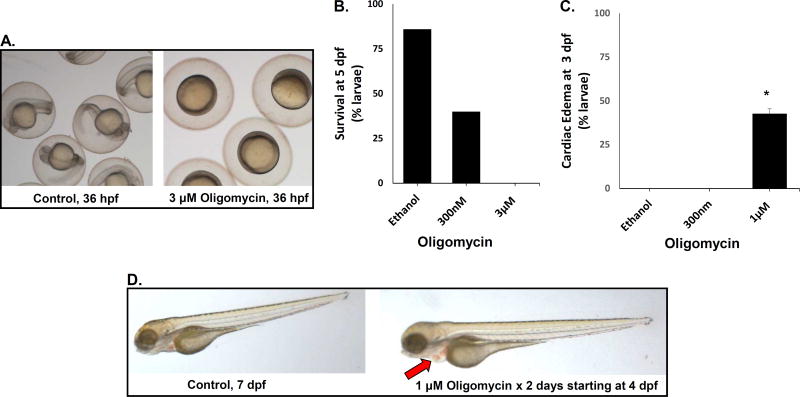
Mitochondrial complex V directly produces cellular energy in the chemical form of ATP by dissipating the proton gradient to convert adenosine diphosphate (ADP) and inorganic phosphate within the mitochondrial matrix. Mutations in several complex V subunits are a well-established cause of human disease, often featuring neurologic disease and occasionally cardiomyopathy (Lopez-Gallardo et al., 2014; Sperl et al., 2006). Oligomycin is a complex V inhibitor (Galante et al., 1979), which we evaluated as a means to produce an animal model of this disease with variable severity depending on concentration used. Wild-type larvae exposed to oligomycin from 2 dpf through 5 dpf were observed closely for survivorship and morphologically characterized compared to fish treated with ethanol as a vehicle control. Our results showed that exposing embryos to oligomycin substantially delayed their development by 36 hpf (Fig 4A). The mean lethal dose (LD50) at 5 dpf was 300 nM oligomycin (Fig 4B). 1 µM had similar lethality at 4–5 dpf (data not shown), but also caused cardiac dilatation with blood pooling in the heart by 3 dpf (Fig 4C–D). In contrast, no visible cardiac edema occurred in fish treated with 300 nM oligomycin, and larvae surviving 300 nM oligomycin exposure were morphologically normal. However, larvae surviving 1 µM oligomycin exposure appeared to be developmentally delayed with less advanced swim bladder development and decreased touch response as compared to controls. At higher doses (3 µM) oligomycin, larval effects were very severe, with 100% fatality seen within 12 hours of exposure to oligomycin started at both 5 hpf (Fig 4A–B) and 2 dpf (data not shown).
Fig 4. Oligomycin inhibition of RC complex V.
(A) Representative larvae at 36 hpf after exposure since 5 hpf with ethanol control buffer (left panel) or 3 µM oligomycin (right panel). Oligomycin causes gastrulation arrest. (B) Survival of zebrafish larvae treated with oligomycin from 5 hpf through 5 dpf is reduced. Each concentration of oligomycin was tested on 24 larvae per condition. Percent survival is shown from a single representative experiment to demonstrate dose response, where four biological replicate experiments showed similar trends with varying doses between experiments to yield LD50. (C) Cardiac edema occurred in larvae exposed to 1 uM oligomycin from 2 dpf through 3 dpf. Black bars represent average presence of edema calculated and error bars convey standard error. Each oligomycin concentration was tested on n ≥ 11 larvae per condition. At least 3 biological replicates were performed per condition, except for 1 µM oligomycin that was performed on 2 biological replicates each with n ≥ 20 animals per condition. *p=0.046 (D) Cardiac edema is demonstrated at 4 dpf in larvae exposed to 1 µM oligomycin starting at 2 dpf relative to buffer-only ethanol control larvae (left panel). Oligomycin-treated fish have enlarged heart, with blood pooling (arrow).
1.3.4 Zebrafish chloramphenicol model of mitochondrial translation inhibition has reduced survivorship, morphologic, and behavior defects
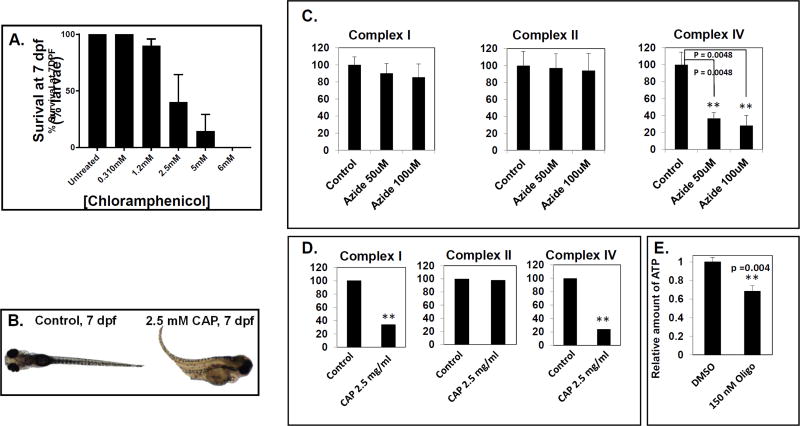
Translation of mRNAs that are transcribed from the mitochondrial genome is critical for the expression of the 13 mtDNA-encoded lipophilic proteins embedded within the inner mitochondrial membrane that are all essential for oxidative phosphorylation (OXPHOS) function by the RC and associated with disease (Boczonadi and Horvath, 2014). Thus, we sought to develop a chemical model to recapitulate the phenotypes of translation defects observed in humans using zebrafish. We exposed wild-type, AB line larvae to the antibiotic chloramphenicol (CAP), a demonstrated inhibitor of mitochondrial translation (Abou-Khalil et al., 1980; Li et al., 2010; McKee et al., 2006), starting at 2 dpf through 7 dpf. Larvae were observed daily for survivorship and characterized morphologically and behaviorally at 5 concentrations of chloramphenicol, in parallel with an untreated control. Our results showed that the lethal dose (LD50) of CAP was approximately 2.5 mM after 5 days of CAP exposure (7 dpf) (Fig 5A). Curiously, some variability was observed between different experimental days as the LD50 fluctuated between 2.5 mM and 5 mM. The lowest concentration of CAP did not appear to have any adverse effects on survival and all larvae remained healthy through 7 dpf, consistent with previously reported data (Ali et al., 2014). No early embryo developmental delay was seen with CAP between 2.5 and 6 mM concentrations. However, severe effects were observed over time at higher concentrations, particularly 6 mM CAP with which larvae did not survive beyond 6 dpf. Although we observed an LD50 at 2.5 mM CAP, the surviving larvae exhibited severe morphological defects when compared to healthy aged-matched controls (Fig 5B). These larvae have underdeveloped swim bladders, distorted tails, severe edema, gray brains and have reduced sensitivity to touch/startle responses (Supplementary File S4_Movie 4).
Fig. 5. Chloramphenicol inhibition of mitochondrial translation, and quantitation of biochemical effects in zebrafish larvae.
(A) Survival of zebrafish larvae treated from 2 dpf through 7 dpf with varying concentrations of chloramphenicol are shown. Black bars represent mean survival and error bars represent standard error of the mean (±SEM). n ≥ 10 larvae per condition. A minimum of 3 biological replicates were performed per condition, except for 5 mM chloramphenicol in which only 2 biological replicates were performed. (B) Representative images are shown of 7 dpf age-matched, untreated control larvae (left panel) and larvae exposed to 2.5 mM chloramphenicol from 2 dpf (right panel). (C–D) Biochemical effects of RC inhibitors on ETC enzyme activities and ATP content in zebrafish larvae. (C) Mitochondrial complex I, complex II, and complex IV activities in zebrafish treated with 50 or 100 µM sodium azide from 5 dpf for 8 hours. (D) Mitochondrial complex I, complex II, and complex IV ETC enzyme activities in zebrafish treated with 2.5 mg/mL chloramphenicol from 2 dpf through 7 dpf. (E) ATP content in zebrafish treated with 150 nM oligomycin from 5 dpf for 24 hours. For panels (C–E), black bars represent mean ETC activity levels (% control) or relative ATP levels and error bars represent standard error. A minimum of 3 biological replicates each with 3 technical replicates were performed per condition (except for (D) where n=1 with 3 technical replicates performed). N = 30–50 total larvae per condition. Control ETC enzyme activity spectrophotometry assay values were: complex I (µmoles of DCPIP reduction/min/mg protein), 0.384 (C) and 0.366 (D); complex II (µmoles of DCPIP reduction/min/mg protein), 0.206 (C) and 0.197 (D); complex IV (µmoles of cytochrome c oxidized/min/mg protein), 15.3 (C) and 11.2 (D). **p < 0.01, student’s T-test.
1.3.5 Modeling drug treatment effects on ETC activities and ATP content
Although the RC inhibitors chosen in this study are well-established and extensively used mitochondrial toxins, the extent of these inhibitors’ efficacy and their impact on overall metabolism have not prior been reported in zebrafish embryos. Therefore, we quantified their inhibitory effects on complex I, II, and IV electron transport chain enzyme specific activities or total larval ATP content. Mitochondrial-enriched larval fractions were prepared from flash-frozen zebrafish larvae populations that had been treated in vivo with each drug, where RC inhibitor time points and/or concentrations evaluated for biochemical assays were less than the lethal (LD50) dose used in the in vivo phenotype assays since dead larvae quickly disintegrate and do not provide suitable tissue for biochemical analysis. Surprisingly, rotenone 100 nM treatment for 4 hours on 7 pdf led to only a 20% decrease in complex I activity (data not shown), whereas the same treatment led to the development of grey brain death in 86% of treated fish (Fig 1F); we suspect this discrepancy was a technical artefact since the larvae possible to collect for further study were the few still alive after 4 hours of rotenone exposure. In contrast, we were able to clearly see the azide effect on complex IV activities (Fig 5C). Even at the lower concentration (50 uM), earlier time point (5 dpf) and shorter duration (8 hours) of azide studied, conditions that did not yield behavioral abnormalities, larval complex IV activity was drastically decreased by more than 60%. The inhibitory effect of azide was dose-dependent and selective for inhibition of complex IV, not complex I or complex II (Fig 5C). Similarly, in vivo treatment with the mitochondrial translation inhibitor, chloramphenicol, also led to a significant decrease in both complex I and complex IV activities, with no change in complex II activity (Fig 5D). The absence of chloramphenicol effect on complex II activity supports its known specificity for inhibition of the mitochondrial translation system, as mtDNA-encoded subunits are part of complexes I and IV but not complex II. Finally, the mitochondrial complex V (ATP synthase) inhibitor, oligomycin, treatment of zebrafish larvae at 10 nM, 100 nM, and 200 nM did not show any significant change in ETC enzyme activities (data not shown), as expected. However, ATP levels in fish homogenate were significantly decreased by 32% (Fig. 5E), consistent with in vivo complex V inhibition of ATP synthesis by oligomycin. Overall, these biochemical analyses demonstrate the anticipated RC inhibitory effects of the zebrafish larval models.
1.3.6 Modeling drug treatment effects on embryo development in zebrafish RC inhibitor models
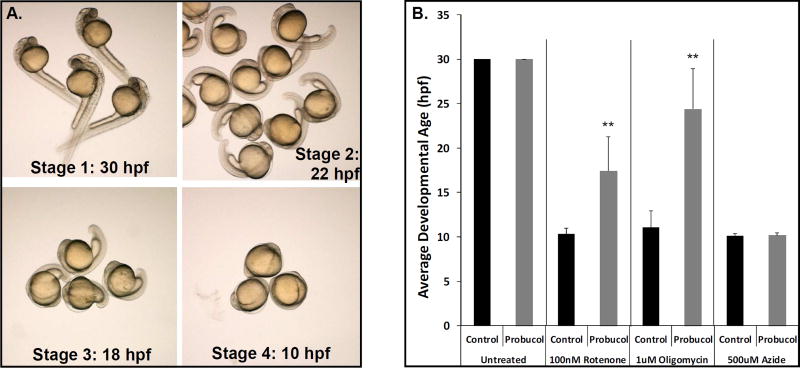
Given the common early developmental delay observed with several the RC inhibitors tested, we sought to use these models to evaluate the comparative therapeutic effectiveness in different RC complex models of a single drug treatment. Specifically, we tested whether probucol, an agent showing therapeutic benefit in diverse RC disease models as discussed above, can rescue developmental delay in the early treatment embryo models of rotenone, azide, and oligomycin that inhibit complex I, IV, and V, respectively. We created a grading scale to describe developmental stages when treatment was started at 4–6 hpf for the embryo developmental stage assessed at approximately 30 hpf: (stage 1) 28–30 hpf stage with normal head and tail bud, (stage 2) 22 hpf stage with smaller head and bent tail, (stage 3) 18 hpf stage with small head and minimal tail extension, (stage 4) 10 hpf stage with no distinguishable head or tail extension (Fig 6A). Using this scoring system, we calculated the average developmental stage of embryos exposed to each RC inhibitor alone or with concurrent 50 uM probucol treatment from 6 hpf through 30 hpf (Fig 6B). Interestingly, probucol significantly rescued embryo develop on exposure to rotenone (p < 0.002) or oligomycin (p < 0.004), but not azide. These data demonstrate the utility of using the early developmental delay RC disease inhibitors model to screen therapeutic compounds and potentially identify therapeutic candidates for distinct RC complex disorders.
Fig 6. Probucol rescues embryo developmental delay in pharmacologic inhibition models of RC complex I (rotenone) and complex V (oligomycin) but not complex IV (azide).
(A) Larvae treated with RC complex I, IV, and V inhibitors from 6 hpf show marked developmental delay. The extent of developmental delay upon evaluation at 30 hpf was classified into four stages based on approximate timing of developmental progress typical of normally developing embryos, including 30 hpf (normal, stage 1), 22 hpf (mild delay, stage 2), 18 hpf (moderate delay, stage 3), and 10 hpf (severe delay, stage 4). (B) Treatment with 100 nM rotenone, 500 uM azide, or 1 uM oligomycin from 6 hpf to 30 hpf caused severely delayed embryo development (stage 4). Developmental inhibition was significantly reversed with concurrent 50 uM probucol treatment for rotenone (p < 0.002) and oligomycin (p < 0.004), but no developmental rescue was seen for azide. Bars represent average and standard deviation of 3 biological replicate experiments per condition, with at least 10 embryos studied per condition in each technical replicate.
1.4 DISCUSSION
1.4.1. Zebrafish rotenone model of complex I inhibition
Rotenone, a potent mitochondrial complex I inhibitor, when exposed to zebrafish within several hours of fertilization, inhibited their early embryonic and larval development. Thus, the early embryo rotenone exposure assay provides an easily scorable early developmental phenotype, quantifiable within a 36 hour timeframe. These results are consistent with previous studies in which rotenone induced developmental delay, although a similar concentration to what was used in our studies appeared to have a less severe effect (Pinho et al., 2013). In addition, once larval development was complete, brief rotenone exposure for only 4 hours on 7 dpf reproducibly caused brain cell death and loss of behavioral responses, as well as a significant reduction of mitochondrial membrane potential quantifiable in skeletal muscle. Rotenone has previously been attempted to establish a zebrafish model for Parkinson’s disease in 7 dpf larvae, although a four-fold lower dose of rotenone used in that study (25 nM) reportedly failed to cause detectable problems in larval behavior, brain physiology or dopaminergic neurons (Bretaud et al., 2004). Interestingly, another group showed that 4 week chronic exposure of adult zebrafish to rotenone does induce symptoms comparable to those seen in Parkinson Disease (Wang et al., 2016). Remarkably, when we used the 7 dpf acute rotenone exposure model to test probucol, a candidate mitochondrial disease therapy, we found that probucol pre-treatment substantially prevented zebrafish larval brain death upon subsequent rotenone exposure. This therapeutic effect in a mitochondrial RC disorder is consistent with our prior reported study of probucol in other animal and cell models, where probucol both prevented and reversed an otherwise lethal renal focal segmental glomerulosclerosis-like disease in a Pdss2 genetic mouse model of coenzyme Q10 deficiency (Falk et al., 2011), rescued lifespan in a C. elegans gas-1(fc21) model of genetic-based complex I (NDUFS2 homologue), and rescued cellular viability in RC deficient patient cells (Peng et al., 2015). Thus, finding physiologic benefit of probucol in an acute rotenone exposure model of brain death demonstrates the potential utility of cross-species investigations to evaluate a candidate compound’s therapeutic efficacy, and clearly demonstrates the feasibility and utility of the acute rotenone pharmacologic zebrafish larvae model for effectively screening organ-specific outcomes of potential candidate therapies for RC disease. In addition, we demonstrated the utility of using the early developmental delay rotenone model for screening for candidate drug treatment effects upon exposure to rotenone as well as other RC inhibitors (azide and oligomycin), where probucol significantly rescued embryo development in both rotenone and oligomycin models.
1.4.2 Zebrafish azide model of complex IV inhibition
Sodium azide (NaN3) has been shown to inhibit cellular respiration by preventing electron transport through complex IV (CIV) of the mitochondrial electron transport chain. The azide moiety binds to the heme iron of cytochrome a3 when it is in its oxidized (ferric, Fe3+) state (Stannard and Horecker, 1948), and thus inhibits the redox cycling of cytochrome c through the activity of cytochrome c oxidase. The basic toxicology of sodium azide in the larval zebrafish model has only been investigated in the last several years (Ali et al., 2014; Ali et al., 2011), and the effects of azide are dependent on the age and developmental stage of the zebrafish larvae. D. rerio embryos can be developmentally arrested within the first 24 hrs post-fertilization by subjecting the embryos to anoxia (Padilla and Roth, 2001). Further, the arrest can be maintained for up to 25 hours without damage to the embryos, which will continue to develop normally if normoxia is reestablished (Padilla and Roth, 2001). Anoxia inhibits the normal activity of the mitochondrial RC, as oxygen is not available to act as the final electron acceptor at the end of the electron transport system of the RC. We postulated that chronic exposure of young zebrafish embryos to compounds which inhibit the mitochondrial RC may show a similar effect, and that sodium azide inhibition of complex IV will result in developmental arrest of the embryos in a dose-specific manner. The degree of developmental arrest (complete arrest versus developmental delay) was indeed found to be proportional to the degree that RC function is disrupted by toxicant treatment in our “chronic azide exposure” model, in which embryos from 3–5 hpf were exposed to varying concentrations of sodium azide for 7 days to evaluate the effects on embryonic and larval development. In addition, fully-developed larval zebrafish exposed to toxicants that inhibit electron transport, impede oxidative phosphorylation capacity, and limit ATP production may display deleterious effects in a dose-responsive manner. Indeed, we describe a highly informative “acute, short-term exposure” model of fully formed zebrafish larvae exposed to varying concentrations of sodium azide model starting on 6 dpf for 12–24 hrs. Zebrafish at this age are no longer able to be reversibly arrested in their development, as are the early embryos and instead display effects such as decreased heartrate, loss of motor function, inability to respond to tactile stimulation, severe neurological damage, and, at high doses, increased mortality. These phenotypes are quantifiable and offer a robust series of survival, physiologic and behavioral outcomes that may potentially aide in the evaluation of therapeutic candidates for varying severity of complex IV disease.
1.4.3 Zebrafish oligomycin model of complex V inhibition
Oligomycin, a potent inhibitor of mitochondrial complex V, which is the ATP synthase that directly dissipates the proton gradient to generate ATP, recapitulated relevant human phenotypes of mitochondrial complex V deficiency. Oligomycin-treated larval phenotypes included cardiomyopathy, decreased survival, and abnormal neurologic response that occur in patients with mutations in genes encoding mitochondrial complex V subunits, such as MT-ATP6. Phenotypes seen in the oligomycin chemical model also recapitulated complex V subunit gene morpholino-based knock460 down phenotypes (data not shown). Future biochemical characterization of oligomycin-treated zebrafish tissues will likely advance the understanding of the precise relationship between complex V inhibition magnitude and specific pathophysiology with the observed organ-level and behavioral phenotypes. The close resemblance of this chemical inhibition-based model of complex V deficiency with genetic models in zebrafish and humans is encouraging, and suggests that the oligomycin model may enable deeper understanding of the currently poorly understood biochemical processes that become altered in primary complex V deficiency. The oligomycin zebrafish model will also enable screening of potential treatments to evaluate their efficacy and toxicity on representative phenotypes that occur in complex V deficiency, where we demonstrated that probucol treatment significantly rescues early developmental delay caused by oligomycin treatment.
1.4.4 Zebrafish chloramphenicol model of mitochondrial translation inhibition with multiple RC complex dysfunction
Chloramphenicol, a potent inhibitor of mitochondrial translation, exposure in zebrafish recapitulates relevant human phenotypes of mitochondrial translation disorders known to cause multiple RC deficiencies. For example, mitochondrial translation deficient patients may clinically present with Leigh syndrome, a classic mitochondrial disease phenotype that can involve any RC complex and cause brainstem dysfunction, dystonia, ataxia, seizures and other multi-systemic phenotypes (Lake et al., 2016). In our chloramphenicol model, we recapitulate these neurodegenerative phenotypes through the presence of gray brain and reduced touch and startle responses. Interestingly, we have also observed on several occasions an apparent seizure phenotype in larvae exposed to chloramphenicol [data not shown], which requires further characterization and may serve as an additional scoring factor to be evaluated during therapeutic testing. Overall, our data provides a clear, highly efficacious and highly reproducible in vivo zebrafish model to test therapies that improve phenotypes and ultimately lead to treatments for humans with mitochondrial translation diseases. Future studies for the development of this model will include measurement and characterization of the mitochondrial mass, membrane potential and reactive oxygen species levels in order to precisely evaluate the efficacy of therapies that improve mitochondrial characteristics. In addition, testing of more specific and clinically utilized compounds, such as oxazolidinone class antibiotics should be evaluated to determine their effects on mitochondrial translation and provide several robust models to screen therapies (McKee et al., 2006).
1.5 CONCLUSION
Four well-characterized pharmacologic inhibitors of complexes I (rotenone), IV (azide), V (oligomycin), and mitochondrial translation (chloramphenicol) that are well-established RC toxins caused reproducible phenotypic both in early embryos and fully formed larval fish. Specifically, rotenone, azide, and oligomycin each delayed early embryo development and/or arrest. Further, later short-term exposures of these three RC complex inhibitors, as well as chloramphenicol that is known to inhibit mitochondrial translation and lead over time to multiple RC inhibition, between 5–7 dpf significantly reduced larval survival and induced organ-specific defects including brain death, behavioral and neurologic alterations, reduced mitochondrial membrane potential in skeletal muscle (rotenone), and/or cardiac edema with visible blood pooling (oligomycin). Remarkably, we demonstrate that acute rotenone-induced brain death could be significantly prevented by pre-treating zebrafish larvae for several days with probucol, as could early embryo developmental delay from either rotenone or oligomycin. Finally, we have shown that rotenone, sodium azide and chloramphenicol do inhibit zebrafish larval ETC enzyme activities whereas oligomycin reduces overall ATP levels in zebrafish larvae, providing biochemical evidence to support the in vivo specificity and robustness of these inhibitor models in zebrafish larvae. Overall, these zebrafish pharmacologic RC function inhibitor models offer a unique opportunity to gain novel insights into diverse developmental, organ-level, and behavioral defects of varying severity, as well as their individual response to candidate therapies, in a highly tractable and cost-effective vertebrate animal model.
Supplementary Material
File S1. Movie_1: Heartbeat in untreated (control) larval zebrafish are shown. Measuring heartrate in zebrafish is straightforward as a result of their transparent body, which make the heart easily discernible.
File S2. Movie_2: Control. Normal startle response in 7 dpf larval zebrafish. Note the vigorous response to tap stimulus.
File S3_Movie_3: Azide startle response. Video shows startle response of 7 dpf larvae after exposure to 100 uM sodium azide for 18 hours. Note the lack of movement, although beating hearts are visible.
File S4. Movie_4: Chloramphenicol startle response. Video shows startle response to larvae at 6 dpf treated with 2.5 mM chloramphenicol from 2 dpf. Unlike controls, most chloramphenicol-exposed larvae exhibit no tap response.
HIGHLIGHTS.
Rotenone causes delayed development, mitochondrial dysfunction & brain death in larvae
Probucol treatment prevents acute rotenone-induced brain death in zebrafish larvae
Azide causes dose/time-dependent delay, brain death, & behavioral changes in larvae
Oligomycin causes dose/time dependent delay, death, behavioral changes & cardiac edema
Chloramphenicol causes decreased zebrafish survival, brain death, & behavioral changes
Acknowledgments
We are grateful to Tara Gallagher for providing technical assistance in the Children’s Hospital of Philadelphia Laboratory Aquatics Core Facility. This work was funded by in part by the National Institutes of Health (R01-HD065858 and R01-GM120762 to M.J.F. and T32-GM008638 to R.G.), the Will Woleben Family Research Fund; the Jaxson Flynt Family Research Fund; and the Children’s Hospital of Philadelphia Research Institute Genes, Genomes, and Pediatric Disease (GGPD) research affinity group. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABREVIATIONS
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- CHL
chloramphenicol D. rerio, Danio rerio
- dpf
days post fertilization
- hpf
hours post fertilization
- MTG
mitotracker green
- NaN3
sodium azide
- PTU
phenylthiourea
- RC
respiratory chain
- TMRE
Tetramethylrhodamine ethyl ester perchlorate
- ETC
Electron Transport Chain.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abou-Khalil S, Salem Z, Yunis AA. Mitochondrial metabolism in normal, myeloid, and erythroid hyperplastic rabbit bone marrow: effect of chloramphenicol. Am J Hematol. 1980;8:71–79. doi: 10.1002/ajh.2830080109. [DOI] [PubMed] [Google Scholar]
- Albrecht SC, Sobotta MC, Bausewein D, Aller I, Hell R, Dick TP, Meyer AJ. Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J Biomol Screen. 2014;19:379–386. doi: 10.1177/1087057113499634. [DOI] [PubMed] [Google Scholar]
- Ali S, Aalders J, Richardson MK. Teratological effects of a panel of sixty water-soluble toxicants on zebrafish development. Zebrafish. 2014;11:129–141. doi: 10.1089/zeb.2013.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, van Mil HG, Richardson MK. Large-scale assessment of the zebrafish embryo as a possible predictive model in toxicity testing. PLoS One. 2011;6:e21076. doi: 10.1371/journal.pone.0021076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avula S, Parikh S, Demarest S, Kurz J, Gropman A. Treatment of mitochondrial disorders. Curr Treat Options Neurol. 2014;16:292. doi: 10.1007/s11940-014-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden KN, Murray J, Capaldi RA, Guillemin K. Early developmental pathology due to cytochrome c oxidase deficiency is revealed by a new zebrafish model. J Biol Chem. 2007;282:34839–34849. doi: 10.1074/jbc.M703528200. [DOI] [PubMed] [Google Scholar]
- Boczonadi V, Horvath R. Mitochondria: impaired mitochondrial translation in human disease. Int J Biochem Cell Biol. 2014;48:77–84. doi: 10.1016/j.biocel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bretaud S, Lee S, Guo S. Sensitivity of zebrafish to environmental toxins implicated in Parkinson's disease. Neurotoxicol Teratol. 2004;26:857–864. doi: 10.1016/j.ntt.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Camp KM, Krotoski D, Parisi MA, Gwinn KA, Cohen BH, Cox CS, Enns GM, Falk MJ, Goldstein AC, Gopal-Srivastava R, Gorman GS, Hersh SP, Hirano M, Hoffman FA, Karaa A, MacLeod EL, McFarland R, Mohan C, Mulberg AE, Odenkirchen JC, Parikh S, Rutherford PJ, Suggs-Anderson SK, Tang WH, Vockley J, Wolfe LA, Yannicelli S, Yeske PE, Coates PM. Nutritional interventions in primary mitochondrial disorders: Developing an evidence base. Mol Genet Metab. 2016;119:187–206. doi: 10.1016/j.ymgme.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P, Majamaa K, Turnbull D, Thorburn D. Treatment for mitochondrial disorders. Cochrane Database Syst Rev. 2006:CD004426. doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord EN, Smith AC, Eyassu F, Shirley R, Hu CH, Dare AJ, James AM, Rogatti S, Hartley RC, Eaton S, Costa AS, Brookes PS, Davidson SM, Duchen MR, Saeb-Parsy K, Shattock MJ, Robinson AJ, Work LM, Frezza C, Krieg T, Murphy MP. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimauro S, Rustin P. A critical approach to the therapy of mitochondrial respiratory chain and oxidative phosphorylation diseases. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbadis.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk MJ, Polyak E, Zhang Z, Peng M, King R, Maltzman JS, Okwuego E, Horyn O, Nakamaru-Ogiso E, Ostrovsky J, Xie LX, Chen JY, Marbois B, Nissim I, Clarke CF, Gasser DL. Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO Mol Med. 2011;3:410–427. doi: 10.1002/emmm.201100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fashing M, Gnaiger E. OROBOROS O2k-Protocols Chemicals Mitochondrial respiration medium - MiR06. Vol. 1413,Mitochondrial Physiol. Netw. 2016:1–4. [Google Scholar]
- Galante YM, Wong SY, Hatefi Y. Composition of complex V of the mitochondrial oxidative phosphorylation system. J Biol Chem. 1979;254:12372–12378. [PubMed] [Google Scholar]
- Garcia-Cazorla A, Quadros EV, Nascimento A, Garcia-Silva MT, Briones P, Montoya J, Ormazabal A, Artuch R, Sequeira JM, Blau N, Arenas J, Pineda M, Ramaekers VT. Mitochondrial diseases associated with cerebral folate deficiency. Neurology. 2008;70:1360–1362. doi: 10.1212/01.wnl.0000309223.98616.e4. [DOI] [PubMed] [Google Scholar]
- Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, Feeney C, Horvath R, Yu-Wai-Man P, Chinnery PF, Taylor RW, Turnbull DM, McFarland R. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77:753–759. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Cohen BH. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- Irving BA, Nair KS. Aging and diabetes: mitochondrial dysfunction. Curr Diab Rep. 2007;7:249–251. doi: 10.1007/s11892-007-0039-x. [DOI] [PubMed] [Google Scholar]
- Kari G, Rodeck U, Dicker AP. Zebrafish: an emerging model system for human disease and drug discovery. Clin Pharmacol Ther. 2007;82:70–80. doi: 10.1038/sj.clpt.6100223. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koga Y, Akita Y, Nishioka J, Yatsuga S, Povalko N, Tanabe Y, Fujimoto S, Matsuishi T. L-arginine improves the symptoms of strokelike episodes in MELAS. Neurology. 2005;64:710–712. doi: 10.1212/01.WNL.0000151976.60624.01. [DOI] [PubMed] [Google Scholar]
- Lake NJ, Compton AG, Rahman S, Thorburn DR. Leigh syndrome: One disorder, more than 75 monogenic causes. Ann Neurol. 2016;79:190–203. doi: 10.1002/ana.24551. [DOI] [PubMed] [Google Scholar]
- Li CH, Cheng YW, Liao PL, Yang YT, Kang JJ. Chloramphenicol causes mitochondrial stress, decreases ATP biosynthesis, induces matrix metalloproteinase-13 expression, and solid-tumor cell invasion. Toxicol Sci. 2010;116:140–150. doi: 10.1093/toxsci/kfq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gallardo E, Emperador S, Solano A, Llobet L, Martin-Navarro A, Lopez-Perez MJ, Briones P, Pineda M, Artuch R, Barraquer E, Jerico I, Ruiz-Pesini E, Montoya J. Expanding the clinical phenotypes of MT-ATP6 mutations. Hum Mol Genet. 2014;23:6191–6200. doi: 10.1093/hmg/ddu339. [DOI] [PubMed] [Google Scholar]
- Mandemakers W, Morais VA, De Strooper B. A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J Cell Sci. 2007;120:1707–1716. doi: 10.1242/jcs.03443. [DOI] [PubMed] [Google Scholar]
- Martinelli D, Catteruccia M, Piemonte F, Pastore A, Tozzi G, Dionisi-Vici C, Pontrelli G, Corsetti T, Livadiotti S, Kheifets V, Hinman A, Shrader WD, Thoolen M, Klein MB, Bertini E, Miller G. EPI-743 reverses the progression of the pediatric mitochondrial disease--genetically defined Leigh Syndrome. Mol Genet Metab. 2012;107:383–388. doi: 10.1016/j.ymgme.2012.09.007. [DOI] [PubMed] [Google Scholar]
- McCormack S, Polyak E, Ostrovsky J, Dingley SD, Rao M, Kwon YJ, Xiao R, Zhang Z, Nakamaru-Ogiso E, Falk MJ. Pharmacologic targeting of sirtuin and PPAR signaling improves longevity and mitochondrial physiology in respiratory chain complex I mutant Caenorhabditis elegans. Mitochondrion. 2015;22:45–59. doi: 10.1016/j.mito.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick E, Place E, Falk MJ. Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics. 2013;10:251–261. doi: 10.1007/s13311-012-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother. 2006;50:2042–2049. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla PA, Roth MB. Oxygen deprivation causes suspended animation in the zebrafish embryo. Proc Natl Acad Sci U S A. 2001;98:7331–7335. doi: 10.1073/pnas.131213198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S, Saneto R, Falk MJ, Anselm I, Cohen BH, Haas R, Medicine Society TM. A modern approach to the treatment of mitochondrial disease. Curr Treat Options Neurol. 2009;11:414–430. doi: 10.1007/s11940-009-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Ostrovsky J, Kwon YJ, Polyak E, Licata J, Tsukikawa M, Marty E, Thomas J, Felix CA, Xiao R, Zhang Z, Gasser DL, Argon Y, Falk MJ. Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Hum Mol Genet. 2015;24:4829–4847. doi: 10.1093/hmg/ddv207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G, Horvath R, Klopstock T, Mootha VK, Suomalainen A, Koene S, Hirano M, Zeviani M, Bindoff LA, Yu-Wai-Man P, Hanna M, Carelli V, McFarland R, Majamaa K, Turnbull DM, Smeitink J, Chinnery PF. New treatments for mitochondrial disease-no time to drop our standards. Nat Rev Neurol. 2013;9:474–481. doi: 10.1038/nrneurol.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho BR, Santos MM, Fonseca-Silva A, Valentao P, Andrade PB, Oliveira JM. How mitochondrial dysfunction affects zebrafish development and cardiovascular function: an in vivo model for testing mitochondria-targeted drugs. Br J Pharmacol. 2013;169:1072–1090. doi: 10.1111/bph.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Graham BH, Nakamaru-Ogiso E, Kar A, Falk MJ. Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev Disabil Res Rev. 2010;16:200–218. doi: 10.1002/ddrr.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzenente B, Rotig A, Metodiev MD. Mouse models for mitochondrial diseases. Hum Mol Genet. 2016;25:R115–R122. doi: 10.1093/hmg/ddw176. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, Chinnery PF, Turnbull DM. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Segref A, Kevei E, Pokrzywa W, Schmeisser K, Mansfeld J, Livnat-Levanon N, Ensenauer R, Glickman MH, Ristow M, Hoppe T. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab. 2014;19:642–652. doi: 10.1016/j.cmet.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Seiler C, Davuluri G, Abrams J, Byfield FJ, Janmey PA, Pack M. Smooth muscle tension induces invasive remodeling of the zebrafish intestine. PLoS Biol. 2012;10:e1001386. doi: 10.1371/journal.pbio.1001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl W, Jesina P, Zeman J, Mayr JA, Demeirleir L, VanCoster R, Pickova A, Hansikova H, Houst'kova H, Krejcik Z, Koch J, Smet J, Muss W, Holme E, Houstek J. Deficiency of mitochondrial ATP synthase of nuclear genetic origin. Neuromuscul Disord. 2006;16:821–829. doi: 10.1016/j.nmd.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Stannard JN, Horecker BL. The in vitro inhibition of cytochrome oxidase by azide and cyanide. J Biol Chem. 1948;172:599–608. [PubMed] [Google Scholar]
- Steele SL, Prykhozhij SV, Berman JN. Zebrafish as a model system for mitochondrial biology and diseases. Transl Res. 2014;163:79–98. doi: 10.1016/j.trsl.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Stocker R. Molecular mechanisms underlying the antiatherosclerotic and antidiabetic effects of probucol, succinobucol, and other probucol analogues. Curr Opin Lipidol. 2009;20:227–235. doi: 10.1097/MOL.0b013e32832aee68. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu W, Yang J, Wang F, Sima Y, Zhong ZM, Wang H, Hu LF, Liu CF. Parkinson's disease-like motor and non-motor symptoms in rotenone-treated zebrafish. Neurotoxicology. 2016;58:103–109. doi: 10.1016/j.neuro.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hailat Z, Falk MJ, Chen XW. Integrative analysis of independent transcriptome data for rare diseases. Methods. 2014 doi: 10.1016/j.ymeth.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tsukikawa M, Peng M, Polyak E, Nakamaru-Ogiso E, Ostrovsky J, McCormack S, Place E, Clarke C, Reiner G, McCormick E, Rappaport E, Haas R, Baur JA, Falk MJ. Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PLoS One. 2013;8:e69282. doi: 10.1371/journal.pone.0069282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. Movie_1: Heartbeat in untreated (control) larval zebrafish are shown. Measuring heartrate in zebrafish is straightforward as a result of their transparent body, which make the heart easily discernible.
File S2. Movie_2: Control. Normal startle response in 7 dpf larval zebrafish. Note the vigorous response to tap stimulus.
File S3_Movie_3: Azide startle response. Video shows startle response of 7 dpf larvae after exposure to 100 uM sodium azide for 18 hours. Note the lack of movement, although beating hearts are visible.
File S4. Movie_4: Chloramphenicol startle response. Video shows startle response to larvae at 6 dpf treated with 2.5 mM chloramphenicol from 2 dpf. Unlike controls, most chloramphenicol-exposed larvae exhibit no tap response.