Abstract
Mitochondria are critical sources of hydrogen peroxide (H2O2), an important secondary messenger in mammalian cells. Recent work has shown that O2•-/H2O2 emission from individual sites of production in mitochondria is regulated by protein S-glutathionylation. Here, we conducted the first examination of O2•-/H2O2 release rates from cardiac and liver mitochondria isolated from mice deficient for glutaredoxin-2 (GRX2), a matrix-associated thiol oxidoreductase that facilitates the S-glutathionylation and deglutathionylation of proteins. Liver mitochondria isolated from mice heterozygous (GRX2+/-) and homozygous (GRX2-/-) for glutaredoxin-2 displayed a significant decrease in O2•-/H2O2 release when oxidizing pyruvate or 2-oxoglutarate. The genetic deletion of the Grx2 gene was associated with increased protein expression of pyruvate dehydrogenase (PDH) but not 2-oxoglutarate dehydrogenase (OGDH). By contrast, O2•-/H2O2 production was augmented in cardiac mitochondria from GRX2+/- and GRX2-/- mice metabolizing pyruvate or 2-oxoglutarate which was associated with decreased PDH and OGDH protein levels. ROS production was augmented in liver and cardiac mitochondria metabolizing succinate. Inhibitor studies revealed that OGDH and Complex III served as high capacity ROS release sites in liver mitochondria. By contrast, Complex I and Complex III were found to be the chief O2•-/H2O2 emitters in cardiac mitochondria. These findings identify an essential role for GRX2 in regulating O2•-/H2O2 release from mitochondria in liver and cardiac tissue. Our results demonstrate that the GRX2-mediated regulation of O2•-/H2O2 release through the S-glutathionylation of mitochondrial proteins may play an integral role in controlling cellular ROS signaling.
Highlights
-
•
GRX2 regulates ROS release from liver and heart mitochondria.
-
•
Loss of GRX2 decreases ROS release from PDH and OGDH in liver.
-
•
GRX2 deficiency increases PDH and OGDH S-glutathionylation.
-
•
Absence of GRX2 increases ROS release from cardiac mitochondria.
1. Introduction
Mitochondria are quantifiably the most important sites for ROS production and have been documented to contain up to 12 O2•-/H2O2 producing enzymes associated with nutrient metabolism [1]. High capacity sites for ROS production include Complex III and OGDH in liver mitochondria [2]. In skeletal muscle, the most important O2•-/H2O2 sources are OGDH and PDH and Complexes I, II, and III [3]. By contrast, complex I and III seem to be critical sources for ROS release in cardiac tissue [4], [5]. Examination of the ROS forming capacity of individual sites of production is vital since mitochondria utilize H2O2 as a secondary messenger. Indeed, mitochondrial H2O2 is required to stimulate cell division, differentiation, and growth and is required to regulate steroidogenesis, circadian rhythms, T-cell function, and hematopoiesis [2]. Complex III is often regarded as the main source of O2•-/H2O2 in cell signaling [6]. However, recent evidence also indicates that other sites, like OGDH, which accounts for a substantial fraction of the ROS formed in liver mitochondria, can potentially make a major contribution to H2O2 signaling as well [2].
To serve as an effective secondary messenger, H2O2 levels need to be controlled through its production and degradation. This is important since ROS can be damaging at high levels causing oxidative distress and tissue damage [7]. The degradation of O2•- and H2O2 is facilitated by antioxidant systems while production of either molecule is modulated by several mechanisms. Proton leaks represent one possible mechanism for modulating mitochondrial ROS release [8]. Unfortunately, there is still no clear consensus on the physiological importance of proton return in controlling ROS formation. In addition, it has been suggested that the rate of mitochondrial ROS production does not directly depend on protonmotive force or the rate of respiration [9]. Redox signals mediated through protein S-glutathionylation has emerged as another potential mechanism for the regulation of mitochondrial ROS emission. Work on the redox regulation of mitochondrial bioenergetics can be traced back to almost 20 years ago when it was found that the inhibition of Complex I correlates with a depletion of glutathione (GSH) pools and the blocking of cysteine residues [10]. It was later found that S-glutathionylation modifies O2•-/H2O2 release from Complex I and that GRX2 was required to regulate this process [11], [12]. Moreover, it was documented that the conjugation and removal of GSH from Complex I by GRX2 is sensitive to changes in mitochondrial redox buffering capacity [11], [13]. Studies have found that GRX2 and S-glutathionylation are integral for modulating overall ROS emission and phosphorylating respiration in response to changes in redox poise [11], [14]. The importance of GRX2 in mediating S-glutathionylation reactions in mitochondria is underscored by the consequences associated with a loss or deficiency in the enzyme. For example, GRX2 deficiency leads to the development of cataracts, signs of heart disease, hypertension, and perturbs embryonic development [11], [15], [16]. It has also recently been shown to play a role in regulating mitochondrial morphology and low GRX2 transcript levels correlates with development of heart disease in humans [17]. This is associated with impaired mitochondrial ATP production and higher than normal ROS production rates [11]. By contrast, its overexpression protects from doxorubicin-induced cardiac injury and prevents apoptosis [18]. Therefore, protein S-glutathionylation reactions mediated by GRX2 are critical for modulating mitochondrial metabolism and function in response to changes in redox poise.
A number of protein S-glutathionylation targets have been identified in mitochondria [19]. However, only a few GRX2 targets have been documented so far and its function in controlling ROS release from the 12 available O2•-/H2O2 producing sites in mitochondria has not been thoroughly examined. Our group recently found that PDH and OGDH are targeted for S-glutathionylation which modulates O2•-/H2O2 emission from either enzyme complex [14]. Chemical inducers for S-glutathionylation, diamide and disulfiram, induced a ~90% decrease in ROS release from PDH and OGDH, which was associated with conjugation of GSH to either enzyme [14]. In addition, it was found that GRX2 can deglutathionylate purified PDH and OGDH restoring native ROS release rates [14], [20]. In the present study, we profiled the ROS release rates of liver and cardiac mitochondria isolated from GRX2 deficient mice exposed to different substrate and inhibitor combinations. To our knowledge, this is the first study to perform a detailed assessment of the native O2•-/H2O2 release rates from mitochondria deficient in GRX2. In order to examine the effect of GRX2 deficiency on native ROS release rates from different sites, O2•-/H2O2 production was measured using mitochondria oxidizing different substrates and exposed to selective ROS release inhibitors (Fig. 1). To properly measure ROS release rates from the electron transport chain exclusively, mitochondria were supplemented with succinate only, which feeds electrons directly into the ubiquinone pool of respiratory chain (Fig. 1). Assessment of ROS release rates from PDH and OGDH was achieved by measuring O2•-/H2O2 emission from mitochondria oxidizing either pyruvate or 2-oxoglutarate (Fig. 1). Various selective inhibitors for ROS release from OGDH and Complexes I, II, and III were also utilized. It was found that GRX2 deficiency led to the development of different ROS emission profiles in liver and cardiac mitochondria which we attribute to variances in substrate preference for O2•-/H2O2 production between the two tissues. For example, GRX2 deficiency induced a significant decrease in mitochondrial ROS production in liver mitochondria oxidizing pyruvate or 2-oxoglutarate but had the opposite effect in mitochondria from cardiac tissue. Intriguingly, liver mitochondria yielded high amounts of ROS when oxidizing these substrates whereas succinate served as an important source for O2•-/H2O2 production in cardiac mitochondria. The importance of these findings in regard to understanding the importance of GRX2 and protein S-glutathionylation in modulating ROS release in mitochondria oxidizing different nutrient sources is discussed herein.
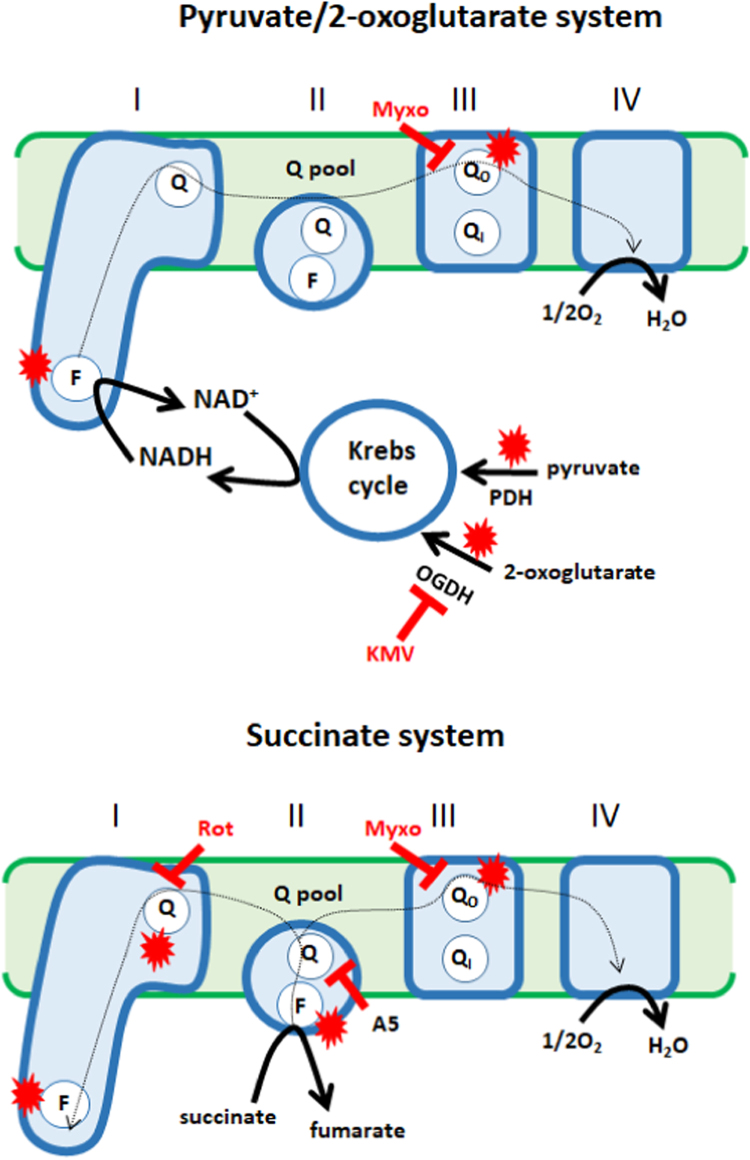
Fig. 1.
Experimental design for the assessment of the effect of GRX2 deficiency on native ROS release rates from different sites of production. Oxidation of Krebs cycle linked metabolites, pyruvate and 2-oxoglutarate, allows the assessment of ROS release rates from pyruvate dehydrogenase (PDH) and 2-oxoglutarate (OGDH) and the electron transport chain. Succinate was used to test ROS release from the electron transport chain since it feeds electrons directly into the ubiquinone pool (Q pool), bypassing the Krebs cycle. The site of action for the selective ROS release inhibitors (KMV; 3-methyl-2-oxo valeric acid, rot; rotenone, A5; atpenin A5, myxo; myxothiazol), which allow the estimation of rates of ROS release production from individual sites, are indicated in the diagram. Sites for ROS production in the different experimental systems are denoted with a red star. Direction of electron flow is indicated by a dotted arrow.
2. Experimental
2.1. Animals and genotyping
Mice heterozygous for GRX2 were a gift from Dr. Mary-Ellen Harper (Faculty of Medicine, University of Ottawa). GRX2 homozygotes and heterozygotes were generated using the C57Bl6N mouse strain. All experiments were approved by Memorial University's Animal Care and Use committee and conducted according to institutional and Federal animal care guidelines. Mice were housed in the animal care unit at room temperature (~23 °C, 12 h (h) dark/12 h light cycle, lights on at 0700 h) and given free access to water and chow (Teklad Global 18% Protein Rodent Diet, 2018). Age-matched male and female mice heterozygous for GRX2 were paired for breeding and the generation of litters. At 3 weeks of age, male pups were ear notched for genotyping. Mice were weighed weekly and euthanized at 8 weeks of age by cervical dislocation under isoflurane anesthesia. Cardiac and liver tissue were extracted for mitochondrial isolation.
DNA extraction for RT-PCR was carried out using the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich) according to the manufacturer's instructions. DNA was amplified using primer sequences for the Grx2 gene [5’-GAC CTA GCC TAC CAG ACT TGG CTG AAA TTT ATT C−3’ (forward), 5’-CAT AGA CAC TCT TCA CTT TCA AGC CCA CCC TC−3’ (reverse)]. Briefly, the DNA extract was mixed with 1 µL of 0.5 µM Grx2 primer, 10 µL of REDExtract-N-Amp Tissue PCR Kit Reaction Mixture, and nuclease-free water providing a final reaction volume of 20 µL. DNA sequences were then amplified using an Eppendorf Mastercycler pro PCR System for 30 cycles. PCR samples were then electrophoresed in a 1.5% w/v agarose gel containing SYBR Safe DNA Gel Stain (1/10000 dilution, Fischer Scientific). DNA fragment size was estimated by electrophoresing Trackit 100 bp DNA Ladder (Fisher Scientific). Nucleotide sequences corresponding to the amplified Grx2 gene were visualized with the Alpha Innotech ChemiImager Ready System. WT mice produced a single nucleotide sequence that was 729 base pairs (bp) in length while GRX2-/- mice produced a fragment 510 bp in size. Samples collected from GRX2+/- mice contained both nucleotide fragments (510 bp and 729 bp).
2.2. Preparation of mitochondria
All steps were performed on ice or 4 °C. After extraction, livers and hearts were immediately placed in freshly prepared mitochondrial isolation buffer containing mannitol (220 mM; to quench hydroxyl radical), EGTA (1 mM), sucrose (70 mM), and Hepes (10 mM) (pH 7.4, abbreviated MESH buffer) supplemented with 0.5% fatty acid free BSA (MESH-B). Note that we found in a previous study that this buffer system does not induce the auto-oxidation of Amplex Ultra Red [21]. Livers were cut into small pieces, rinsed, and then minced on a Teflon watch glass. Minced livers were then homogenized in 15 mL MESH-B using the Potter-Elvejham method. Homogenates were centrifuged at 800×g for 9 min. The supernatant was collected and centrifuged at 10,000×g for 9 min to pellet mitochondria. The supernatant was decanted, excess fat was wiped clean from the sides of the tubes, and then mitochondrial pellets were washed in 15 mL MESH-B and centrifuged at 10,000×g for 9 min. The final mitochondrial pellet was resuspended in 500 µL MESH giving a final concentration of 15–20 mg/mL.
Hearts were initially cut into smaller pieces to wash away any excess blood. After several rinses, heart pieces were minced on a Teflon watch glass. Minced hearts were homogenized in MESH-B containing 1 unit of protease (subtilisin A, to disrupt any myofibers containing mitochondria) using the Potter-Elvejham method. Homogenates were first centrifuged at 10,000×g for 9 min to remove subtilisin A. The pellet was resuspended in 15 mL MESH-B and centrifuged at 800×g for 9 min. The supernatant was collected and centrifuged at 10,000×g for 9 min. The resulting mitochondrial pellet was resuspended in 100 µL MESH providing a final protein concentration of 6–8 mg/mL. Protein content was quantified by Bradford Assay using BSA as a standard.
2.3. Measurement of O2•-/H2O2 release
Mitochondrial O2•-/H2O2 production was examined using the Amplex Ultra Red (AUR) assay as described previously [2]. O2•- is often considered the proximal ROS formed by mitochondria. However, it has been documented that flavin sites (either FAD or FMN) form a mixture of O2•- and H2O2 [22], [23]. In addition, any O2•- formed is quickly dismutated to H2O2 by endogenous SOD. AUR cannot discriminate between H2O2 formed during nutrient oxidation or by the dismutation of O2•-. Therefore, AUR measurements for ROS release by mitochondria are defined as the rate of O2•-/H2O2 release. Liver mitochondria and cardiac mitochondria were first diluted to 3 mg/mL and 1 mg/mL in MESH, respectively, and then stored on ice. Liver and cardiac mitochondria were then diluted to 0.3 mg/mL and 0.1 mg/mL in the wells of a black 96-well plate containing MESH-B. Mitochondria were treated with or without 3-methyl-2-oxo valeric acid (KMV, 10 mM), myxothiazol (4 µM), atpenin A5 (40 µM), or rotenone (4 µM) and incubated for 5 min at 25 °C. Horseradish peroxidase (HRP, 3 U/mL), superoxide dismutase (SOD, 25 U/mL), and AUR reagent (10 µM) were then added to individual wells. Reactions were initiated by the addition of pyruvate (50 µM)/malate (50 µM), 2-oxoglutarate (50 µM)/malate (50 µM), or succinate (50 µM). Note that malate was added to complete the Krebs cycle to ensure the full oxidation of pyruvate and 2-oxoglutarate allowing for the measure of ROS release by PDH, OGDH, and the electron transport chain. Succinate oxidation, on the other hand, by-passes the Krebs cycle completely ensuring measurement of ROS released by the electron transport chain only. Changes in fluorescence were tracked at excitation and emission wavelengths 565 nm/600 nm using a Spectramax M2 microplate reader (Molecular Devices) for 5 min at 25 °C. For genotype comparisons, O2•-/H2O2 release rates by GRX2+/- or GRX2-/- mitochondria were expressed as a percentage of WT production rates. Inhibitor effects were expressed as a percent of control (mitochondria not treated with O2•-/H2O2 release inhibitors).
2.4. Polarographic measurement of mitochondrial respiration
The different states of respiration in mitochondria prepared from liver and cardiac tissue was monitored using an Oxytherm electrode system (Hansatech). Liver and cardiac mitochondria were diluted to 0.5 mg/mL and 0.1 mg/mL, respectively, in the Oxytherm electrode chamber containing respiration buffer (220 mM mannitol, 70 mM sucrose, 2 mM Hepes, 1 mM EGTA, 10 mM KH2PO4, 5 mM MgCl2, pH 7.4) and allowed to equilibrate until a stable oxygen baseline was reached. Pyruvate (10 mM) and malate (2 mM) were injected in the reaction chamber to assess state 2 respiration. State 3 respiration (phosphorylating respiration) was induced by adding ADP (1 mM). Oligomycin (4 µg/mL), an inhibitor for ATP synthase and any contaminating ATPases, was then injected into the reaction mixture to assess state 4 respiration (nonphosphorylating respiration – proton leaks). Oxygen consumption not associated with the electron transport chain (e.g. oxygenases) was measured by adding antimycin A (4 µM), a Complex III inhibitor. Rates of mitochondrial respiration were normalized by subtracting oxygen consumption not associated with the electron transport chain (rate of consumption – oxygen consumed after antimycin A addition). Respiratory control ratios were calculated as the ratio of state 3 to state 4 respiration.
2.5. Immunoblot
Mitochondria were diluted to 1–3 mg/mL in Laemmli buffer and then heated at 100 °C for 10 min. Ten to sixty micrograms of protein were loaded in each well and then samples were electrophoresed through either a 10% isocratic SDS gel [for detection of PDH, OGDH, and protein glutathione mixed disulfides (PSSG)] or a 12% isocratic SDS gel (detection of GRX1 and GRX2). Note that for PSSG detection, 25 mM N-ethylmaleimide (NEM) was include in the isolation buffer, including the homogenizing buffer, to preserve protein glutathione adducts. This is in contrast to previous studies where it was only included in Laemmli buffer after mitochondria had been isolated [24]. Once completed, proteins were transferred from the gels to nitrocellulose membranes by tank transfer. Membranes were stained with Ponceau S and documented to ensure successful protein transfer. In addition, Ponceau S stains served as the loading control for blots used for PSSG detection. Membranes were rinsed with Tris-buffered saline (TBS) containing 0.1% (v/v) Tween-20 (TBS-T) to remove the Ponceau S stain and blocked for 1 h under constant agitation at room temperature with TBS-T containing 5% (w/v) non-fat skim milk. Membranes were then washed twice with TBS-T and probed with anti-OGDH (20 µg for liver, 10 µg for cardiac, 1/3000, Abcam), anti-PDH (20 µg for liver, 10 µg for cardiac, 1/3000, Abcam), anti-GRX1 (30 µg for liver, 60 µg for cardiac, 1/2000, Abcam), anti-GRX2 (40 µg for liver, 1/500, Abcam), anti-PSSG (20 µg for liver, 20 µg for cardiac, 1/500, Abcam), anti-SDHA (1/3000, Santa Cruz), or anti-SOD2 (1/3000, Santa Cruz) diluted in TBS-T containing 5% (w/v) BSA and 0.02% (w/v) NaN3. Membranes were incubated in primary antibody overnight under constant agitation at 4 °C. The specificity of the anti-PSSG antibody for S-glutathionylated proteins was confirmed by treating samples diluted in Laemmli buffer with 2% (v/v) β-mercaptoethanol. Membranes were then washed twice in TBS-T and probed with anti-mouse and anti-rabbit horseradish peroxidase conjugate secondary antibodies (1/3000, Santa Cruz, diluted in blocking solution) for 1 h at room temperature under constant agitation. Bands were visualized using WestPico Super Signal Chemiluminescent substrate and ImageQuant LAS 4000. Band intensities were quantified using ImageJ software.
2.6. BioGEE switch assays
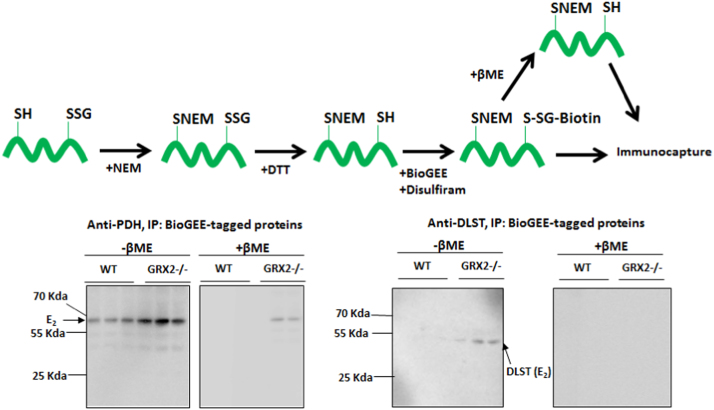
The degree of PDH and OGDH S-glutathionylation in liver mitochondria was assayed using the BioGEE switch assay as described previously with a few modifications (Fig. 6) [14], [25]. Liver tissue from WT and GRX2-/- mice was homogenized and mitochondria were isolated as described above except 25 mM NEM was included in every step. Upon isolation, mitochondria were resuspended in MESH containing 25 mM NEM. For the biotin switch assays, mitochondria were diluted to 4 mg/mL in 100 µL of lysis buffer containing 25 mM NEM and incubated for 10 min at 37 °C. Excess NEM was removed using Zeba micro spin desalting columns (ThermoFisher). Samples were then collected and treated with 5 mL dithiothreitol (DTT) to reduce any protein-glutathione adducts. Samples were then desalted as described above and treated with biotinylated glutathione ethyl ester (BioGEE; 1 mM) and disulfiram (1 mM; an S-glutathionylation catalyst) and incubated for 30 min at 25 °C. For control experiments, BioGEE and disulfiram treated samples were also supplemented with 2% (v/v) β-mercaptoethanol to reduce BioGEE-modified proteins. Samples were supplemented with 50 µL of streptavidin-modified magnetic beads (Dynabeads; Invitrogen) and incubated for 2 h at room temperature under constant agitation. Beads were then magnetically separated from supernatant using a magnetic rack (Biorad), washed twice with PBS, and then heated at 100 °C for 15 min to separate proteins from antibodies. The beads were removed and the resulting supernatant was treated with Laemmli buffer and electrophoresed for immunoblot as described above. The E2 subunit for OGDH and PDH was detected using anti-DLST anti-serum (DLST; dihydrolipoamide succinyl-transferase, E2 subunit for OGDH, 1/2000; Abcam) and anti-PDH cocktail (1/2000; Abcam), respectively. Immunoreactive bands were visualized as described above.
Fig. 6.
E2subunit of PDH and OGDH displays a higher degree of S-glutathionylation in GRX2 deficient liver mitochondria. Liver mitochondria were first treated with NEM to block exposed thiols and then protein glutathione mixed disulfides were reduced with DTT. The newly exposed protein cysteine thiols previously modified by glutathione were then re-glutathionylated with biotinylated glutathione ethyl ester (BioGEE) with chemical S-glutathionylation catalyst disulfiram. Samples were also treated with β-mercaptoethanol to remove the BioGEE group to confirm antibody specificity for the immunocapture and immunoblot. Proteins were then immunoprecipitated using streptavidin beads and probed for the E2 subunit of OGDH and PDH by Western blot. Samples treated with β-mercaptoethanol (βME; reducing conditions) prior to streptavidin immunocapture served as the control. n = 3.
2.7. Data analysis
Amplex Ultra Red assays were performed 5 times and in duplicate. Polarographic measure of mitochondrial respiration was conducted 4–5 times and in duplicate. Immunoblots were conducted in duplicate or triplicate and densitometry normalized to loading controls. ROS release results were normalized to paired control values to estimate the percent change in O2•-/H2O2 formation following inhibitor treatment which can limit power for analysis. All results were analyzed using GraphPad Prism 6 software using two-tailed paired student T-Test or 1-way ANOVA with Fisher's LSD posthoc test. * or #; p ≤ 0.05, ** or ##; p ≤ 0.01, *** or ###; p ≤ 0.001.
3. Results
3.1. GRX2 deficiency does not induce a compensatory increase in Grx1
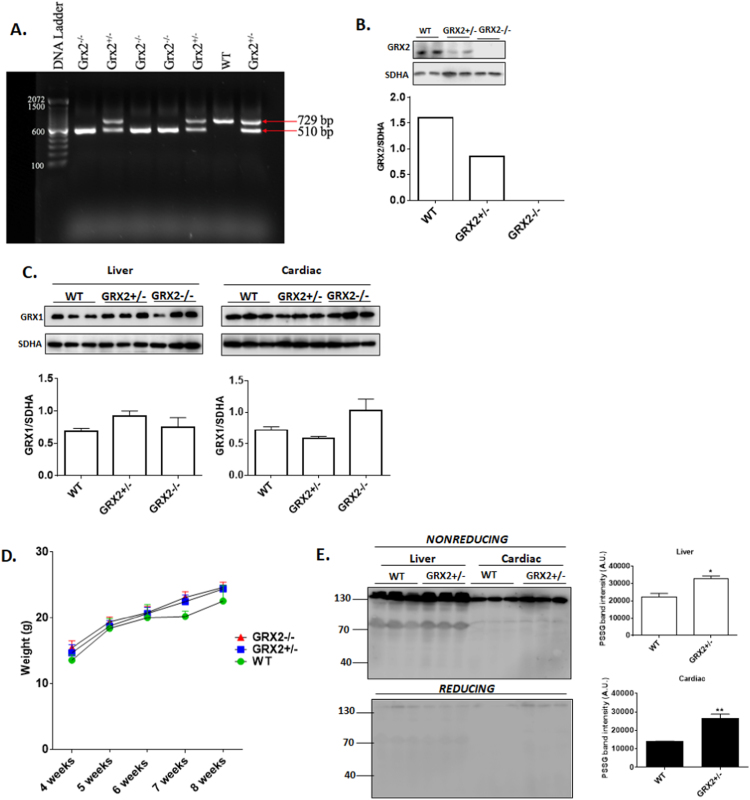
Fig. 2A demonstrates that GRX2 mouse litters contained either the full length and/truncated Grx2 gene, pointing to the successful knockdown (GRX2+/-) or out (GRX2-/-) of the protein. Identification of WT, GRX2+/-, or GRX2-/- mice was conducted by immunoblotting for GRX2 protein in liver mitochondria (Fig. 2B). GRX1 has been reported to localize to the intermembrane space of mitochondria where it regulates protein import and redox signaling [26]. Thus, we decided to test if GRX2 deficiency results in a compensatory increase in GRX1. Immunoblot analysis revealed no significant changes in GRX1 protein levels even though a few samples displayed a slight increase or decrease in band intensity (Fig. 2C). Moreover, previous studies have demonstrated that the ablation or overexpression of the Grx2 gene does not induce a compensatory increase in thioredoxin-2 [11], [18], [27]. The physiological and anatomical impact of the absence of GRX2 was characterized in two previous studies [11], [27]. Knockout of GRX2 does not change feeding behavior or linear growth and weight gain. Fig. 2D confirms that weight gain does not significantly change in mice heterozygous or homozygous for GRX2. It has been demonstrated previously that GRX2 deletion increases the overall number of S-glutathionylated proteins in cardiac mitochondria [11]. Here, we found that the partial deletion of GRX2 also augmented protein S-glutathionylation (Fig. 2E). Performing the electrophoresis in the presence of β-mercaptoethanol (reducing conditions) substantially decreased the intensity of these bands confirming that the protein S-glutathionylation adduct anti-serum is specific for proteins modified by glutathione.
Fig. 2.
Impact of GRX2 deletion on the mitochondrial S-glutathionylated proteome and the expression of GRX1. A. Genotypic characterization of mouse litters. RNA was extracted from ear notches and subjected to PCR analysis for the presence of the full length or truncated forms of the Grx2 gene. B. Immunoblot analysis confirming the partial knockout and complete abolishment of GRX2. Assessment of GRX2 protein levels was conducted on mitochondria isolated from liver tissue. SDH subunit A served as the loading control. C. Depletion or abolishment of GRX2 does not induce a compensatory increase in GRX1 in cardiac or liver tissue. Band intensities were quantified by ImageJ. SDH subunit A served as the loading control. n = 3, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test. D. GRX2+/- and GRX2-/- do not display a significant change in overall body weight when compared to WT littermates. n = 12–21, mean ± SEM, 1-way ANOVA Fisher's LSD post-hoc test. E. Partial ablation of the Grx2 gene induces a significant increase in the overall number of S-glutathionylated proteins in cardiac and liver mitochondria. Samples were electrophoresed under reducing (+ 2% v/v β-mercaptoethanol) or nonreducing (no β-mercaptoethanol) and probed for the presence of S-glutathionylated proteins using protein-glutathione disulfide (PSSG) anti-serum. n = 3, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test.
3.2. Effect of GRX2 deficiency on ROS release during the oxidation of pyruvate and 2-oxoglutarate
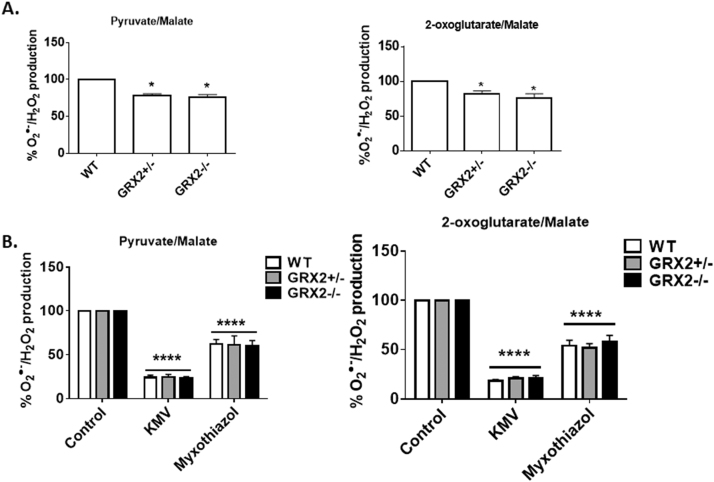
Our group has reported that OGDH and Complex III are high capacity sites for ROS release in liver mitochondria, accounting for almost ~90% of the total O2•-/H2O2 formed during the oxidation of pyruvate or 2-oxoglutarate [2]. Therefore, we decided to test the impact of GRX2 deficiency on production from these sites in liver mitochondria first. As shown in Fig. 3A, ROS release was significantly lower in liver mitochondria isolated form mice heterozygous or homozygous for GRX2. Next, we used KMV and myxothiazol to determine if OGDH and Complex III served as high capacity sites for ROS release. For this, we used a method employed by our group and others to examine the ROS release rates of individual sites of production [2], [28]. It was shown previously that KMV is a strong and specific allosteric inhibitor for O2•-/H2O2 production by OGDH [28]. Our group has found that KMV can induce a ~90% decrease in ROS production by liver mitochondria metabolizing pyruvate or 2-oxoglutarate [2]. KMV (10 mM) induced a ~85% decrease in ROS emission from WT, GRX2+/-, or GRX2-/- liver mitochondria metabolizing either pyruvate or 2-oxoglutarate (Fig. 3B). Since KMV also lowers NADH production, the decrease in ROS release during 2-oxoglutarate and pyruvate oxidation could also be associated with diminished electron flow through the respiratory chain (Fig. 1). We decided to conduct the same assays with mitochondria treated with myxothiazol, which inhibits ROS production from Complex III by binding the QO binding site preventing semiquinone production. Our group reported previously that Complex III accounts for ~45% of the ROS release from liver mitochondria [2]. We had also found in the same study that rotenone elicits a similar effect (decreases ROS release by ~45%), confirming that Complex III is a major ROS producer in liver cells [2]. In addition, in the same previously published study, it was found that treating liver mitochondria with both rotenone and KMV or rotenone and ATP decreased ROS release by ~90%. Myxothiazol induced a ~45% decrease in ROS production in mitochondria oxidizing pyruvate and an almost 50% decrease in samples metabolizing 2-oxoglutarate (Fig. 3B). Therefore, based on the cumulative KMV and myxothiazol effects, OGDH (and to a lesser extent PDH) and Complex III could potentially account for up to ~90% of the ROS release from liver mitochondria. The other 10% is likely contributed by other sites [2].
Fig. 3.
Loss of GRX2 decreases pyruvate and 2-oxoglutarate induced O2•-/H2O2release from liver mitochondria. A. O2•-/H2O2 release was examined in mitochondria prepared from the livers of WT, GRX2+/-, and GRX2-/- mice oxidizing pyruvate or 2-oxoglutarate. Mitochondria (0.3 mg/mL) were diluted in reaction buffer containing either pyruvate (50 µM) or 2-oxoglutarate (50 µM) and malate (50 µM) and the production of H2O2 was measured using AUR reagent. n = 5, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test. B. Examination of the high capacity site(s) for O2•-/H2O2 release in liver mitochondria. Mitochondria (0.3 mg/mL) were preincubated in KMV (10 mM) or myxothiazol (4 µM) in reaction buffer and then H2O2 production was measured using AUR and pyruvate (50 µM) or 2-oxoglutarate (50 µM) and malate (50 µM). n = 5, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test.
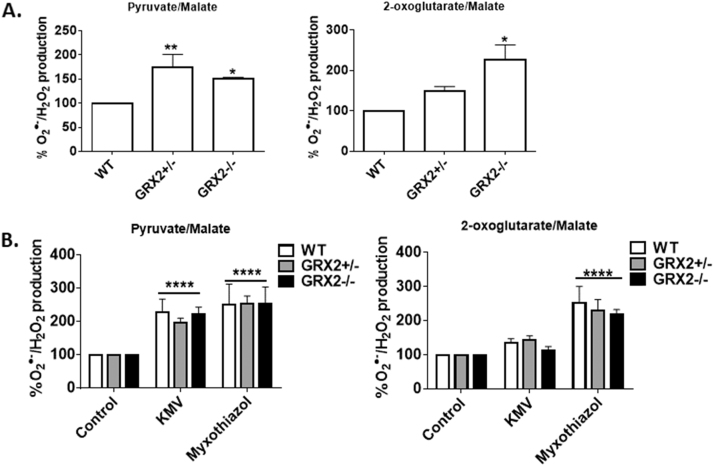
Next, we conducted similar experiments with cardiac mitochondria. To our surprise the ROS release profile in cardiac mitochondria was completely different from the one observed with liver mitochondria. ROS production rates were significantly increased in GRX2+/- and GRX2-/- cardiac mitochondria oxidizing pyruvate (Fig. 4A). Similar observations were made with mitochondria oxidizing 2-oxoglutarate (Fig. 4A). Next, the source of the increase in ROS production during pyruvate and 2-oxoglutarate metabolism was tested. Mitochondria were treated with either KMV or myxothiazol. Surprisingly, KMV exposure induced a significant increase in ROS release from WT, GRX2+/-, and GRX2-/- cardiac mitochondria oxidizing pyruvate (Fig. 4B). This effect is attributed to blockage of Krebs cycle metabolism and the accumulation of NADH and metabolites which can augment ROS release from the electron transport chain. Myxothiazol also induced a significant increase in O2•-/H2O2 release from mitochondria prepared from all three genotypes. However, no differences in ROS production were observed in the different treatment groups during pyruvate oxidation. Similar experiments were conducted with mitochondria metabolizing 2-oxoglutarate. (Fig. 4B). Myxothiazol augmented mitochondrial ROS release while KMV did not have an effect (Fig. 4B). Myxothiazol is known to lower ROS release from Complex III by blocking semiquinone formation. However, some other studies have demonstrated that myxothiazol can actually augment ROS release when substrates that form NADH are metabolized in rat heart and brain mitochondria [29]. This effect is attributed to the blockage of electron flow through the respiratory chain increasing ROS release from sites of formation upstream from Complex III like Complex I, which is typically regarded as a major source of O2•-/H2O2 in cardiac mitochondria [30].
Fig. 4.
Loss of GRX2 increases pyruvate and 2-oxoglutarate induced O2•-/H2O2release from cardiac mitochondria. A. O2•-/H2O2 release was examined in mitochondria prepared from the hearts of WT, GRX2+/-, and GRX2-/- mice oxidizing pyruvate or 2-oxoglutarate. Mitochondria (0.1 mg/mL) were diluted in reaction buffer containing either pyruvate (50 µM) or 2-oxoglutarate (50 µM) and malate (50 µM) and the production of H2O2 was measured using AUR reagent. n = 5, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test. B. Examination of the high capacity site(s) for O2•-/H2O2 release in liver mitochondria. Mitochondria (0.1 mg/mL) were preincubated in KMV (10 mM) or myxothiazol (4 µM) in reaction buffer and then H2O2 production was measured using AUR and pyruvate (50 µM) or 2-oxoglutarate (50 µM) and malate (50 µM). n = 5, mean±SEM. 1-way ANOVA Fisher's LSD post-hoc test.
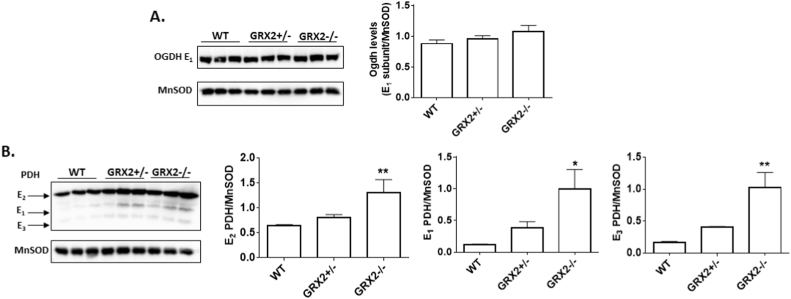
3.3. GRX2 deficiency alters PDH and OGDH protein levels in liver and cardiac mitochondria
The rate of ROS genesis is heavily influenced by the concentration of the electron donating site [31]. Thus, we tested the impact of GRX2 deficiency on the protein levels of PDH and OGDH. Notable differences in the expression of the different subunits for PDH were observed. Fig. 5A shows that there no significant change in the level of the E1 subunit for OGDH in liver mitochondria from GRX2+/- or GRX2-/- mice relative to control. The different subunits for PDH displayed a more drastic shift in expression. Indeed, the E1, E2, and E3 subunits for PDH displayed significant increases in GRX2-/- mitochondria when compared to WT samples (Fig. 5B). An almost 2-fold increase in expression of the E2 subunit (top band) was observed in GRX2-/- mitochondria and ~5-fold increase was recorded for the E1 and E3 subunits. No significant changes in the expression of all three subunits was observed in the GRX2+/-. OGDH and PDH have been documented to undergo S-glutathionylation in different tissues [10], [14], [32]. Therefore, we decided to test if the decrease in ROS release in liver mitochondria from GRX2 deficient mice was associated with an increase in the S-glutathionylation of either enzyme. To test this, we conducted a BioGEE switch assay where sites for S-glutathionylation are modified with a glutathione analog conjugated to biotin allowing for enzyme immunocapture (Fig. 6). As shown in Fig. 6, stronger immunoreactive bands corresponding to the E2 subunit of PDH were detected in mitochondrial samples collected from GRX2-/- mice. Notably, we were also able to successfully modify the E2 subunit of PDH with BioGEE in WT liver samples indicating that a small fraction of the total PDH pool may be S-glutathionylated in isolated mitochondria (Fig. 6). Similar observations were made for OGDH (Fig. 6). However, in this particular case we only detected BioGEE-modified E2 subunit corresponding to OGDH in mitochondria enriched from GRX2-/- liver. Reduction of the disulfide bridge between BioGEE and the E2 subunit with β-mercaptoethanol before immunoprecipitation decreased or abolished the presence of immunoreactive bands corresponding to the E2 subunit of PDH or OGDH (Fig. 6). Collectively, these results demonstrate that deletion of the Grx2 gene augments the protein levels of PDH. Furthermore, our findings show that PDH and OGDH are targets for S-glutathionylation in vitro and that the absence of GRX2 augments the protein S-glutathionylation of either protein complex in isolated liver mitochondria.
Fig. 5.
Ablation of GRX2 induces a significant increase in PDH protein levels in liver mitochondria. A. Immunoblot analysis of the levels of OGDH using E1 subunit anti-serum. Band intensities were quantified by ImageJ. MnSOD served as the loading control. n = 3, mean±SEM. 1-way ANOVA Fisher's LSD post-hoc test. B. Immunoblot analysis of the levels of E1, E2, and E3 subunits using PDH antibody cocktail. MnSOD served as the loading control. Band intensities were quantified by ImageJ. n = 3, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test.
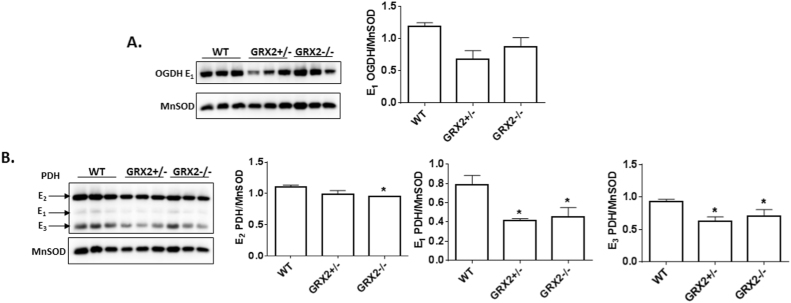
Next, the protein levels for PDH and OGDH were examined in cardiac mitochondria. A trend for a decrease in the protein levels of the E1 subunit for OGDH was observed in GRX2+/- and GRX2-/- mitochondria (Fig. 7A). Significant decreases in the E1 and E3 subunits for PDH cardiac mitochondria from GRX2+/- and GRX2-/- were also recorded (Fig. 7B). In addition, it was found that the E2 subunit for PDH displayed a small but significant decrease but only in GRX2-/- cardiac mitochondria (Fig. 7B). Thus, despite the decrease in PDH and OGDH, ROS production rates are still significantly higher in GRX2+/- and GRX2-/- mitochondria. Collectively, these findings demonstrate that the partial absence or complete loss of GRX2 has opposing effects in liver and cardiac mitochondria.
Fig. 7.
Loss of GRX2 induces a significant decrease in PDH protein levels in cardiac mitochondria. A. Immunoblot analysis of the levels of OGDH using E1 subunit anti-serum. Band intensities were quantified by ImageJ. MnSOD served as the loading control. n = 3, mean±SEM. 1-way ANOVA Fisher's LSD post-hoc test. B. Immunoblot analysis of the levels of E1, E2, and E3 subunits using PDH antibody cocktail. MnSOD served as the loading control. Band intensities were quantified by ImageJ. n = 3, mean±SEM. 1-way ANOVA Fisher's LSD post-hoc test.
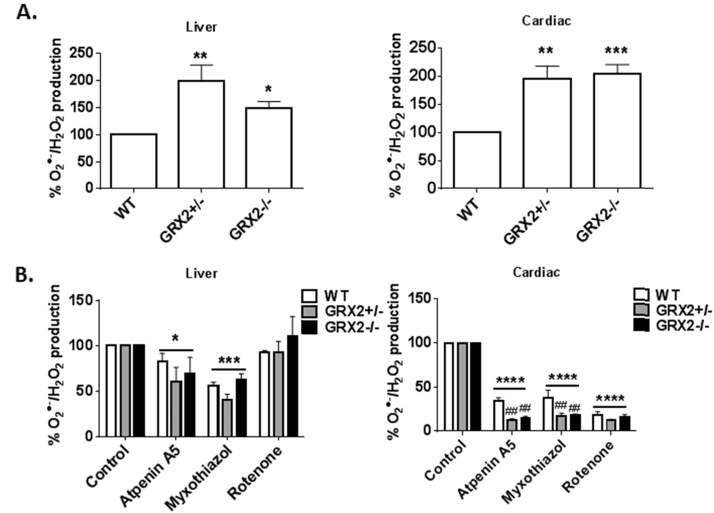
3.4. GRX2 deficiency augments succinate-driven O2•-/H2O2 production
Next, the impact of GRX2 deficiency on O2•-/H2O2 formation by the respiratory chain was examined. For this, succinate served as the sole carbon source for respiration which bypasses the Krebs cycle donating electrons to the ubiquinone pool, negating ROS release from PDH and OGDH (Fig. 1). Fig. 8A demonstrates that succinate-driven ROS production was significantly higher in GRX2+/- and GRX2-/- mitochondria from liver and cardiac tissue. Next, ROS production by different sites in the electron transport chain was examined. Liver mitochondria were treated with atpenin A5, myxothiazol, or rotenone to determine which site of production was affected by GRX2 depletion. Atpenin A5 binds the ubiquinone binding pocket in Complex II (succinate dehydrogenase) blocking electron flow to Complex I or III during succinate oxidation (Fig. 1). Rotenone binds the ubiquinone binding pocket in Complex I and thus serves as a means to determine how much ROS is formed during reverse electron flow from succinate (Fig. 1). As shown in Fig. 8B, inclusion of atpenin A5 induced a small but significant decrease in O2•-/H2O2 formation in liver mitochondria from all three genotypes. Inclusion of myxothiazol induced a more significant decrease in ROS production by liver mitochondria (Fig. 8B). Finally, inclusion of rotenone did not alter O2•-/H2O2 formation indicating Complex I is not a significant source of ROS in liver mitochondria. These results confirm that complex III is an important ROS source and complex I is not a producer in liver mitochondria. Next, the effect of the different inhibitors on O2•-/H2O2 production in WT and GRX2 deficient cardiac mitochondria was tested. Inclusion of atpenin A5 induced a robust decrease in ROS production in all three genotypes (Fig. 8B). Intriguingly, the effect was much larger in cardiac mitochondria isolated from GRX2+/- and GRX2-/- mice. In GRX2+/- and GRX2-/- mitochondria, atpenin A5 decreased ROS production by ~85% whereas in WT samples it was lowered by ~70% (Fig. 8B). A similar effect was observed in samples treated with myxothiazol (Fig. 8B). Finally, rotenone decreased ROS production by cardiac mitochondria isolated from WT, GRX2+/-, and GRX2-/- mice by ~85% but no differences were observed between the genotypes. These results indicate Complex I and potentially Complex III are important ROS sources in cardiac mitochondria, an observation consistent with previous findings [5].
Fig. 8.
Partial loss or ablation of GRX2 induces a significant increase in O2•-/H2O2 release from liver and cardiac mitochondria oxidizing succinate. A. Liver (0.3 mg/mL) and cardiac (0.1 mg/mL) mitochondria were diluted in reaction buffer containing succinate (50 µM) and the production of H2O2 was measured using AUR reagent. n = 5, mean±SEM. 1-way ANOVA Fisher's LSD post-hoc test. B. Examination of the high capacity site(s) for O2•-/H2O2 release in liver mitochondria oxidizing succinate. Mitochondria (0.3 mg/mL) were preincubated in KMV (10 mM) or myxothiazol (4 µM) in reaction buffer and then H2O2 production was measured using AUR and pyruvate (50 µM) or 2-oxoglutarate (50 µM) and malate (50 µM). n = 5, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test. n = 4, mean ± SEM, 2-way ANOVA Fisher's LSD post-hoc test.
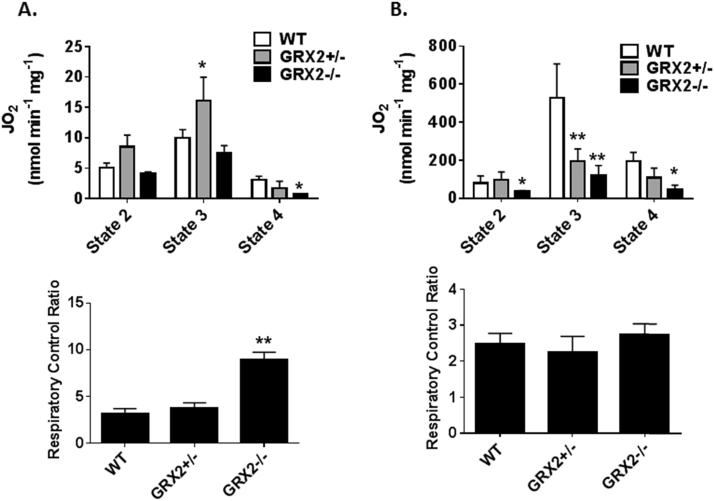
3.5. GRX2 deficiency alters respiration and oxidative phosphorylation
Mitochondrial O2•-/H2O2 production and release relies on the same electron transfer pathways that are used to make ATP. Therefore, the impact of a deficiency or absence in GRX2 on the bioenergetics of liver and cardiac mitochondria was examined. Liver mitochondria from mice heterozygous for GRX2 displayed a significant increase in state 3 (phosphorylating) respiration (Fig. 9A). Liver mitochondria prepared from GRX2-/- mice did not display any change phosphorylating respiration when compared to WT samples (Fig. 9A). However, GRX2-/- liver mitochondria did show small but significant change in respiration associated with proton leaks (state 4 respiration). Next, we calculated the respiratory control ratio (RCR), a proxy measure for the efficiency of ATP production in mitochondria. The significant decrease in proton leaks augmented the coupling efficiency for GRX2-/- mitochondria, increasing its ATP forming capacity (Fig. 9A). By contrast, GRX2+/- mitochondria displayed no significant change in RCR relative to WT mitochondria. We then conducted similar experiments with cardiac mitochondria prepared from WT and GRX2 deficient mice. Interestingly, the absence of GRX2 had the opposite effect on these mitochondria. Mitochondria collected from the cardiac tissue of GRX2+/- and GRX2-/- mice displayed ~50% decrease in state 3 respiration (Fig. 9B). State 2 and 4 respiration was also significantly decreased in GRX2-/- cardiac mitochondria (Fig. 9B). Overall, these results demonstrate that GRX2 has opposing regulatory effects on mitochondrial respiration and oxidative phosphorylation in liver and cardiac tissue.
Fig. 9.
Impact of GRX2 deletion on mitochondrial respiration and oxidative phosphorylation capacity. A. Liver mitochondria (0.5 mg/mL) were diluted in 1 mL of respiration buffer in the Oxytherm electrode reaction chamber. Once a stable baseline was reached, the different states of respiration were measured. State 2 respiration was induced by adding pyruvate (10 mM) and malate (2 mM) followed by the induction of state 3 respiration (injection of ADP to a final concentration of 1 mM). State 4 respiration was induced by adding oligomycin (4 µg/mL). Finally, respiration was stopped by the addition of antimycin A (4 µM). n = 3–5, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test. B. Cardiac mitochondria (0.1 mg/mL) were diluted in 1 mL of respiration buffer in the Oxytherm electrode reaction chamber. Once a stable baseline was reached, the different states of respiration were measured. State 2 respiration was induced by adding pyruvate (10 mM) and malate (2 mM) followed by the induction of state 3 respiration (injection of ADP to a final concentration of 1 mM). State 4 respiration was induced by adding oligomycin (4 µg/mL). Finally, respiration was stopped by the addition of antimycin A (4 µM). n = 4–5, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test.
3.6. Absolute ROS release rates for liver and cardiac mitochondria oxidizing different substrates
The results collected above indicate that the loss of GRX2 has differential effects on ROS production in liver and cardiac mitochondria. In liver mitochondria, loss of GRX2 lowered pyruvate and 2-oxoglutarate driven O2•-/H2O2 production whereas it had the opposite effect in samples isolated from cardiac tissue (Fig. 3, Fig. 4). By contrast, the absence of GRX2 augmented ROS production by succinate oxidation in liver and cardiac mitochondria (Fig. 8). Unfortunately, we were unable to identify the site responsible for enhancing succinate driven ROS production in liver mitochondria. In addition, we were unable to identify which site(s) were responsible for ROS formation in cardiac mitochondria oxidizing Krebs cycle linked sustrates. We reasoned that these discrepancies can be partially attributed to differences in ROS producing capacity of individual sites in liver and cardiac mitochondria. In this study, we used physiological substrate concentrations (50 µM) to stimulate ROS production which may allow for differences in the rate of formation in cardiac and liver mitochondria to be measured. First, we found that succinate is a poor substrate for ROS production in liver mitochondria when compared to cardiac tissue (Table 1). Indeed, cardiac mitochondria from WT or GRX2 deficient mice produced ~4-fold more O2•-/H2O2 than liver mitochondria when oxidizing succinate (Table 1). Also, the absolute rate for ROS release was ~2-fold higher in cardiac or liver mitochondria from GRX2 deficient mice (Table 1). The rate for pyruvate driven ROS production was ~5-fold higher in WT liver mitochondria when compared to cardiac and ~2-fold higher in mitochondria from GRX2 deficient mice (Table 2). The absence of GRX2 also significantly lowered ROS release from liver mitochondria oxidizing pyruvate and had the opposite effect in cardiac mitochondria. Similar results were observed with liver and cardiac mitochondria metabolizing 2-oxoglutarate. Indeed, O2•-/H2O2 emission in liver mitochondria isolated from WT mice oxidizing 2-oxoglutarate was ~4.5-fold higher than cardiac mitochondria (Table 3). In addition, rates of production in GRX2+/- and GRX2-/- were ~2.5 fold higher in liver mitochondria when compared to cardiac (Table 3). Finally, the loss of GRX2 increased ROS release in cardiac mitochondria oxidizing 2-oxoglutarate. By contrast, GRX2 deficiency had the opposite effect on liver mitochondria, lowering ROS release during 2-oxoglutarate metabolism. Overall, this demonstrates that liver and cardiac tissue display a certain level of specificity in terms of which sites will serve as the major sources of ROS.
Table 1.
Comparison of the O2•-/H2O2 emission rates from cardiac and liver mitochondria oxidizing succinate. 2-way ANOVA with a Fisher's LSD post-hoc test. n = 5, mean ± SEM. pmol min−1 mg of protein−1.
| WT | GRX2+/- | GRX2-/- | |
|---|---|---|---|
| Liver | 107.27 ± 14.98 | 195.27 ± 28.15* | 173.83 ± 31.41* |
| Cardiac | 389.11 ± 46.84# | 759.37 ± 133.37**, # | 699.88 ± 134.51**, # |
* corresponds to comparison between genotype, # corresponds to comparison between tissues within genotype. p ≤ 0.05 = * or #, p ≤ 0.01 = **.
Table 2.
Comparison of the O2•-/H2O2 emission rates from cardiac and liver mitochondria oxidizing pyruvate. 2-way ANOVA with a Fisher's LSD post-hoc test. n = 4, mean ± SEM. pmol min-1 mg of protein-1.
| WT | GRX2+/- | GRX2-/- | |
|---|---|---|---|
| Liver | 308.72 ± 43.75 | 203.64 ± 29.71* | 193.61 ± 18.04* |
| Cardiac | 44.96 ± 21.05## | 107.05 ± 20.02*, # | 81.20 ± 1.51*, # |
* corresponds to comparison between genotype, # corresponds to comparison between tissues within genotype. p ≤ 0.05 = * or #, p ≤ 0.01 = ##.
Table 3.
Comparison of the O2•-/H2O2 emission rates from cardiac and liver mitochondria oxidizing 2-oxoglutarate. 2-way ANOVA with a Fisher's LSD post-hoc test. n = 4–5, mean ± SEM. pmol min-1 mg of protein-1.
| WT | GRX2+/- | GRX2-/- | |
|---|---|---|---|
| Liver | 464.72 ± 11.20 | 365.50 ± 45.63* | 368.11 ± 34.26* |
| Cardiac | 65.71± 14.44### | 150.17 ± 14.58*, ## | 194.24 ± 11.23*, ## |
* corresponds to comparison between genotype, # corresponds to comparison between tissues within genotype. p ≤ 0.05 = * or #, p ≤ 0.01 = ##, p ≤ 0.001.
4. Discussion
The present findings show that GRX2 regulates O2•-/H2O2 release from liver and cardiac mitochondria. This is the first study of its kind to demonstrate that GRX2 is required to control the production of H2O2, an observation that has strong implications for how the formation of this secondary messenger for mitochondria-to-cell communication is regulated. It has been put forth that cellular redox switches serve as an interface between changes in the environment (the exposome – e.g. nutrient availability) and the genome/transcriptome/metabolome [33]. In mitochondria, these redox relays are influenced by nutrient oxidation which alters mitochondrial redox poise through H2O2 and NADPH production [34]. Reversible oxidation of cysteine switches on mitochondrial sources of ROS in response to changes in the exposome has been proposed to regulate mitochondria-to-cell signaling [35]. Our findings indicate that GRX2 forms part of this redox interface, modulating how much O2•-/H2O2 is formed by high capacity ROS release sites in liver and cardiac mitochondria in response to changes in nutrient availability.
4.1. GRX2 deficiency has differential effects on pyruvate and 2-oxoglutarate driven ROS production in liver and cardiac mitochondria
O2•-/H2O2 emission is influenced by mitochondrial redox poise and changes in glutathione buffering capacity. PDH and OGDH have been identified as mitochondrial redox sensors which adjust their enzymatic activity and ROS output in response to changes in mitochondrial redox buffering capacity [14], [36], [37]. Our group demonstrated recently that this redox sensing capacity is related to the protein S-glutathionylation of PDH and OGDH in liver mitochondria and that conjugation of glutathione to either enzyme complex alters ROS release during forward and reverse electron flow [14], [20]. Protein S-glutathionylation decreased O2•-/H2O2 emission from both enzyme complexes and purified GRX2 can reverse these effects [14], [20]. In the present study, it was found that a deficiency in GRX2 significantly decreased ROS release in liver mitochondria oxidizing pyruvate and 2-oxoglutarate. BioGEE switch assays revealed that the E2 subunit of OGDH and PDH exhibited increased protein S-glutathionylation in liver mitochondria isolated from GRX2 deficient mice. This confirms observations from previous studies demonstrating that the E2 subunit of OGDH and PDH is targeted for S-glutathionylation [14], [38]. Here, we have extended on these observations by demonstrating that endogenously expressed GRX2 is required to control ROS release from PDH and OGDH in isolated mitochondria. It should be noted though that the results collected in this study were done with isolated mitochondria which may not be a direct reflection of mitochondrial function in vivo. However, this perceived limitation does not negate the novelty of our findings which clearly demonstrate that endogenously expressed GRX2 is required to control ROS release from these enzymes through protein S-glutathionylation. Taken together, OGDH and PDH are targets for protein S-glutathionylation and GRX2 may be required to modulate how much ROS is released from these two enzymes for mitochondria-to-cell signaling in liver.
Most studies invested in deciphering the impact of protein S-glutathionylation on mitochondrial function and total ROS release have been carried out in cardiac tissue. Complex I in cardiac mitochondria was identified as one of the first sites in mitochondria to undergo protein S-glutathionylation [39]. Several studies have demonstrated that S-glutathionylation alters native ROS release rates from Complex I [12], [39], [40], [41]. In addition, GRX2 is required to conjugate and remove glutathione from Complex I in response to changes in mitochondrial redox poise [13]. In this study, it was found that a deficiency in GRX2 augmented O2•-/H2O2 release from cardiac mitochondria metabolizing pyruvate or 2-oxoglutarate, which is consistent with results collected in other studies [11], [12], [41]. What is notable in our study is that the same effects were not observed in liver mitochondria. In liver mitochondria, it was found that loss of GRX2 induced a significant decrease in ROS release during the oxidation of pyruvate or 2-oxoglutarate due to the maintenance of OGDH and PDH in an S-glutathionylated state. These tissue specific effects may be related to which enzyme(s) serve as the high capacity site(s) for ROS release in different tissues. For instance, our group identified OGDH and Complex III as the chief sites for O2•-/H2O2 release in liver mitochondria, an observation that was confirmed in the present study [2]. Furthermore, we found in this study that pyruvate and 2-oxoglutarate produced ~5 times more O2•-/H2O2 in liver mitochondria when compared to cardiac mitochondria. This falls in line with the conclusion that OGDH and Complex III are high capacity sites for ROS release in liver mitochondria. By contrast, Complex I and III are known as critical ROS sources in cardiac mitochondria [5]. Here, using succinate as a substrate and selective ROS production inhibitors, we found that Complex I and III are integral sources of ROS in cardiac mitochondria, which generated ~4-fold more ROS when compared to liver mitochondria. Furthermore, the observation that myxothiazol augmented ROS release from cardiac mitochondria during pyruvate or 2-oxoglutarate metabolism confirms that Complex I is a major site for production. Myxothiazol has been documented to augment ROS release from cardiac mitochondria due to the over reduction of electron donating centers in Complex I [29], [42]. Finally, atpenin A5 decreased ROS release from cardiac mitochondria oxidizing succinate by ~90%. If Complex II was an important source then ROS release would have increased due to electron accumulation in the FAD prosthetic group of subunit A. Thus, aptenin A5 is inhibiting ROS release by blocking reverse and forward electron flow to Complexes I and III, respectively (Fig. 1). Collectively, these observations reflect the idea that Complex I is one the major sites for ROS release in cardiac mitochondria and that GRX2 regulates ROS release from this respiratory complex. Therefore, these findings show that 1) GRX2 plays an integral role in modulating ROS release from major sites of production through reversible S-glutathionylation reactions and 2) there are tissue specific differences in regard to which ROS forming sites serve as high capacity O2•-/H2O2 producers and GRX2 controls ROS release from these specific sites.
4.2. GRX2 is required to modulate succinate-induced ROS production
Mitochondria isolated from liver and cardiac tissue of GRX2+/- and GRX2-/- mice both displayed a significant increase in ROS production when succinate was being oxidized. However, as indicated above succinate was a more potent driver for ROS release in cardiac tissue, which is attributed to Complex I and III serving as the major sites for O2•-/H2O2 release in heart mitochondria. Our findings have important implications for heart function and disease. As mentioned, this is the first time the impact of GRX2 deficiency on mitochondrial O2•-/H2O2 release from individual sites of production was ever quantified. Previous reports had found that partial or full deletion of the Grx2 results in the over S-glutathionylation of the mitochondrial proteome, increased S-glutathionylation of complex I, and the development of signs of heart disease including left ventricular hypertrophy, mitochondrial fragmentation, fibrosis, and hypertension [11], [43]. Intriguingly, these effects are not associated with oxidative distress or damage [11]. Hydrogen peroxide does play an integral role in modulating myocardial contraction and relaxation but the over-oxidation of protein cysteine switches due to high ROS has also been implicated in heart and cardiovascular disease [44]. Based on our findings, we propose that GRX2 plays a vital role in controlling how much H2O2 is released from Complex I for myocardial signaling and that disabling GRX2 activity compromises redox signaling in cardiomyocytes inducing contractile dysfunction and metabolic inflexibility. It may also be required to modulate how much is released from Complex III, which was recently identified as a target for protein S-glutathionylation [19]. Finally, we cannot discount that the absence of GRX2 will also inhibit ATP synthase, a known target for S-glutathionylation. In this case, the S-glutathionylation of ATP synthase could augment ROS production by decreasing proton return and electron flow through the respiratory chain [45].
4.3. GRX2 has opposing effects on respiration in liver and cardiac mitochondria
Mitochondrial ROS release also relies on the same electron transfer pathways utilized for respiration and ATP production. Therefore, we also examined respiratory capacity in GRX2 deficient mitochondria. Cardiac mitochondria displayed a ~50% drop in state 3 respiration, consistent with findings in a previous study [11]. This is also in line with the observation that loss of GRX2 augments ROS release due to the S-glutathionylation and inhibition of Complex I [11]. Therefore, the lack of control over S-glutathionylation reactions in cardiac mitochondria compromises electron flow through the respiratory chain resulting in the over reduction of electron donating centers increasing O2•-/H2O2 production while simultaneously compromising ATP production. Deletion of the Grx2 gene had the opposite effect on liver mitochondria. In fact, it was found that mitochondria isolated from the liver of mice heterozygous for GRX2 displayed increased state 3 respiration. We are unsure as to why the partial loss of GRX2 would augment phosphorylating respiration. However, it is possible that a 50% decrease in the availability of this protein may force mitochondria to adopt a hyperfused state increasing respiration [46]. In addition, in contrast to cardiac mitochondria, our results indicate that GRX2 is not integral for regulating phosphorylating respiration in the liver. The complete knockout of the Grx2 gene did not alter state 3 respiration relative to WT mitochondria. This could be due to an unidentified system that compensates for the disabling of GRX2 function in liver mitochondria. Indeed, liver mitochondria are enriched with a number of systems that are required to maintain redox poise and balance since hepatocytes are naturally exposed to high levels of ROS. Therefore, it may be that another antioxidant system involved in maintaining redox poise, like the glutathione S-transferase system, may compensate for the loss of GRX2 in liver mitochondria. Unfortunately, the function of glutathione S-transferase systems in modulating the S-glutathionylation of proteins in mitochondria still has not been examined.
5. Conclusions
Our results demonstrate for the first time that GRX2 is required to regulate O2•-/H2O2 release in liver and cardiac mitochondria. In liver, major sites for GRX2 action were found to be OGDH and PDH, observations that were consistent with previous findings showing that both enzyme complexes are targeted for S-glutathionylation. By contrast, different inhibitor and substrate combinations identified Complex I was the major GRX2 regulatory target in cardiac tissue. In aggregate, our findings indicate that GRX2 is integral for controlling ROS release from mitochondria in different tissues.
Acknowledgements
Funding was provided by the Natural Sciences and Engineering Research Council of Canada (NSERC # RGPIN-2016-04829). JC was funded by NSERC. NK was funded by the Canadian Queen Elizabeth II Diamond Jubilee Scholarship Program.
References
- 1.Bolt M.J., Mailloux R.J., Rasenick M.M., Wali R.K., Skarosi S., Bissonnette M., Brasitus T.A., Sitrin M.D. Expression of G protein alpha subunits in normal rat colon and in azoxymethane-induced colonic neoplasms. Gastroenterology. 1998;115(6):1494–1503. doi: 10.1016/s0016-5085(98)70028-9. [DOI] [PubMed] [Google Scholar]
- 2.Slade L., Chalker J., Kuksal N., Young A., Gardiner D., Mailloux R.J. Examination of the superoxide/hydrogen peroxide forming and quenching potential of mouse liver mitochondria. Biochim Biophys. Acta. 2017;1861(8):1960–1969. doi: 10.1016/j.bbagen.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Martin W.F., Sies H. Physiological evolution: genomic redox footprints. Nat. Plants. 2017;3:17071. doi: 10.1038/nplants.2017.71. [DOI] [PubMed] [Google Scholar]
- 4.Truong J., Mailloux R.J., Chan H.M. Impact of methylmercury exposure on mitochondrial energetics in AC16 and H9C2 cardiomyocytes. Toxicol. Vitr.: Int. J. Publ. Assoc. BIBRA. 2015;29(5):953–961. doi: 10.1016/j.tiv.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Lesnefsky E.J., Chen Q., Tandler B., Hoppel C.L. Mitochondrial dysfunction and myocardial ischemia-reperfusion: implications for novel therapies. Annu Rev. Pharmacol. Toxicol. 2017;57:535–565. doi: 10.1146/annurev-pharmtox-010715-103335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mailloux R.J. Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol. 2015;4:381–398. doi: 10.1016/j.redox.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailloux R.J., Beriault R., Lemire J., Singh R., Chenier D.R., Hamel R.D., Appanna V.D. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PloS One. 2007;2(8):e690. doi: 10.1371/journal.pone.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beriault R., Hamel R., Chenier D., Mailloux R.J., Joly H., Appanna V.D. The overexpression of NADPH-producing enzymes counters the oxidative stress evoked by gallium, an iron mimetic, Biometals: an international journal on the role of metal ions in biology. Biochem. Med. 2007;20(2):165–176. doi: 10.1007/s10534-006-9024-0. [DOI] [PubMed] [Google Scholar]
- 10.Applegate M.A., Humphries K.M., Szweda L.I. Reversible inhibition of alpha-ketoglutarate dehydrogenase by hydrogen peroxide: glutathionylation and protection of lipoic acid. Biochemistry. 2008;47(1):473–478. doi: 10.1021/bi7017464. [DOI] [PubMed] [Google Scholar]
- 11.Mailloux R.J., Xuan J.Y., McBride S., Maharsy W., Thorn S., Holterman C.E., Kennedy C.R., Rippstein P., deKemp R., da Silva J., Nemer M., Lou M., Harper M.E. Glutaredoxin-2 is required to control oxidative phosphorylation in cardiac muscle by mediating deglutathionylation reactions. J. Biol. Chem. 2014;289(21):14812–14828. doi: 10.1074/jbc.M114.550574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurd T.R., Requejo R., Filipovska A., Brown S., Prime T.A., Robinson A.J., Fearnley I.M., Murphy M.P. Complex I within oxidatively stressed bovine heart mitochondria is glutathionylated on Cys-531 and Cys-704 of the 75-kDa subunit: potential role of CYS residues in decreasing oxidative damage. J. Biol. Chem. 2008;283(36):24801–24815. doi: 10.1074/jbc.M803432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer S.M., Taylor E.R., Brown S.E., Dahm C.C., Costa N.J., Runswick M.J., Murphy M.P. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J. Biol. Chem. 2004;279(46):47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien M., Chalker J., Slade L., Gardiner D., Mailloux R.J. Protein S-glutathionylation alters superoxide/hydrogen peroxide emission from pyruvate dehydrogenase complex. Free Radic. Biol. Med. 2017;106:302–314. doi: 10.1016/j.freeradbiomed.2017.02.046. [DOI] [PubMed] [Google Scholar]
- 15.Wu H., Yu Y., David L., Ho Y.S., Lou M.F. Glutaredoxin 2 (Grx2) gene deletion induces early onset of age-dependent cataracts in mice. J. Biol. Chem. 2014;289(52):36125–36139. doi: 10.1074/jbc.M114.620047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brautigam L., Jensen L.D., Poschmann G., Nystrom S., Bannenberg S., Dreij K., Lepka K., Prozorovski T., Montano S.J., Aktas O., Uhlen P., Stuhler K., Cao Y., Holmgren A., Berndt C. Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1. Proc. Natl. Acad. Sci. USA. 2013;110(50):20057–20062. doi: 10.1073/pnas.1313753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanaan G.N., Ichim B., Gharibeh L., Maharsy W., Patten D.A., Xuan J.Y., Reunov A., Marshall P., Veinot J., Menzies K., Nemer M., Harper M.E. Glutaredoxin-2 controls cardiac mitochondrial dynamics and energetics in mice, and protects against human cardiac pathologies. Redox Biol. 2017;14:509–521. doi: 10.1016/j.redox.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diotte N.M., Xiong Y., Gao J., Chua B.H., Ho Y.S. Attenuation of doxorubicin-induced cardiac injury by mitochondrial glutaredoxin 2. Biochim. Biophys. Acta. 2009;1793(2):427–438. doi: 10.1016/j.bbamcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Gergondey R., Garcia C., Marchand C.H., Lemaire S.D., Camadro J.M., Auchere F. Modulation of the specific glutathionylation of mitochondrial proteins in the yeast Saccharomyces cerevisiae under basal and stress conditions. Biochem. J. 2017;474(7):1175–1193. doi: 10.1042/BCJ20160927. [DOI] [PubMed] [Google Scholar]
- 20.Mailloux R.J., Craig Ayre D., Christian S.L. Induction of mitochondrial reactive oxygen species production by GSH mediated S-glutathionylation of 2-oxoglutarate dehydrogenase. Redox Biol. 2016;8:285–297. doi: 10.1016/j.redox.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young A., Gardiner D., Brosnan M.E., Brosnan J.T., Mailloux R.J. Physiological levels of formate activate mitochondrial superoxide/hydrogen peroxide release from mouse liver mitochondria. FEBS Lett. 2017;591(16):2426–2438. doi: 10.1002/1873-3468.12777. [DOI] [PubMed] [Google Scholar]
- 22.Mailloux R.J., Gardiner D., O'Brien M. 2-Oxoglutarate dehydrogenase is a more significant source of O2(.-)/H2O2 than pyruvate dehydrogenase in cardiac and liver tissue. Free Radic. Biol. Med. 2016;97:501–512. doi: 10.1016/j.freeradbiomed.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Starkov A.A., Fiskum G., Chinopoulos C., Lorenzo B.J., Browne S.E., Patel M.S., Beal M.F. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 2004;24(36):7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailloux R.J., Adjeitey C.N., Xuan J.Y., Harper M.E. Crucial yet divergent roles of mitochondrial redox state in skeletal muscle vs. brown adipose tissue energetics. Faseb J. 2012;26(1):363–375. doi: 10.1096/fj.11-189639. [DOI] [PubMed] [Google Scholar]
- 25.Hurd T.R., Prime T.A., Harbour M.E., Lilley K.S., Murphy M.P. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J. Biol. Chem. 2007;282(30):22040–22051. doi: 10.1074/jbc.M703591200. [DOI] [PubMed] [Google Scholar]
- 26.Pai H.V., Starke D.W., Lesnefsky E.J., Hoppel C.L., Mieyal J.J. What is the functional significance of the unique location of glutaredoxin 1 (GRx1) in the intermembrane space of mitochondria? Antioxid. Redox Signal. 2007;9(11):2027–2033. doi: 10.1089/ars.2007.1642. [DOI] [PubMed] [Google Scholar]
- 27.Mailloux R.J., Xuan J.Y., Beauchamp B., Jui L., Lou M., Harper M.E. Glutaredoxin-2 is required to control proton leak through uncoupling protein-3. J. Biol. Chem. 2013;288(12):8365–8379. doi: 10.1074/jbc.M112.442905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinlan C.L., Goncalves R.L., Hey-Mogensen M., Yadava N., Bunik V.I., Brand M.D. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 2014;289(12):8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starkov A.A., Fiskum G. Myxothiazol induces H(2)O(2) production from mitochondrial respiratory chain. Biochem Biophys. Res. Commun. 2001;281(3):645–650. doi: 10.1006/bbrc.2001.4409. [DOI] [PubMed] [Google Scholar]
- 30.Chouchani E.T., Pell V.R., Gaude E., Aksentijevic D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N.J., Smith A.C., Eyassu F., Shirley R., Hu C.H., Dare A.J., James A.M., Rogatti S., Hartley R.C., Eaton S., Costa A.S.H., Brookes P.S., Davidson S.M., Duchen M.R., Saeb-Parsy K., Shattock M.J., Robinson A.J., Work L.M., Frezza C., Krieg T., Murphy M.P. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H., Denton T.T., Xu H., Calingasan N., Beal M.F., Gibson G.E. Reductions in the mitochondrial enzyme alpha-ketoglutarate dehydrogenase complex in neurodegenerative disease - beneficial or detrimental? J. Neurochem. 2016;139(5):823–838. doi: 10.1111/jnc.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Go Y.M., Jones D.P. Redox biology: interface of the exposome with the proteome, epigenome and genome. Redox Biol. 2014;2:358–360. doi: 10.1016/j.redox.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones D.P., Sies H. The redox code. Antioxid. Redox Signal. 2015;23(9):734–746. doi: 10.1089/ars.2015.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 36.Fisher-Wellman K.H., Gilliam L.A., Lin C.T., Cathey B.L., Lark D.S., Neufer P.D. Mitochondrial glutathione depletion reveals a novel role for the pyruvate dehydrogenase complex as a key H2O2-emitting source under conditions of nutrient overload. Free Radic. Biol. Med. 2013;65:1201–1208. doi: 10.1016/j.freeradbiomed.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLain A.L., Szweda P.A., Szweda L.I. alpha-Ketoglutarate dehydrogenase: a mitochondrial redox sensor. Free Radic. Res. 2011;45(1):29–36. doi: 10.3109/10715762.2010.534163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLain A.L., Cormier P.J., Kinter M., Szweda L.I. Glutathionylation of alpha-ketoglutarate dehydrogenase: the chemical nature and relative susceptibility of the cofactor lipoic acid to modification. Free Radic. Biol. Med. 2013;61:161–169. doi: 10.1016/j.freeradbiomed.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passarelli C., Tozzi G., Pastore A., Bertini E., Piemonte F. GSSG-mediated Complex I defect in isolated cardiac mitochondria. Int. J. Mol. Med. 2010;26(1):95–99. doi: 10.3892/ijmm_00000439. [DOI] [PubMed] [Google Scholar]
- 40.Wu H., Lin L., Giblin F., Ho Y.S., Lou M.F. Glutaredoxin 2 knockout increases sensitivity to oxidative stress in mouse lens epithelial cells. Free Radic. Biol. Med. 2011;51(11):2108–2117. doi: 10.1016/j.freeradbiomed.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor E.R., Hurrell F., Shannon R.J., Lin T.K., Hirst J., Murphy M.P. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 2003;278(22):19603–19610. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 42.Hansford R.G., Hogue B.A., Mildaziene V. Dependence of H2O2 formation by rat heart mitochondria on substrate availability and donor age. J. Bioenerg. Biomembr. 1997;29(1):89–95. doi: 10.1023/a:1022420007908. [DOI] [PubMed] [Google Scholar]
- 43.Kanaan G.N., Ichim B., Gharibeh L., Maharsy W., Patten D.A., Xuan J.Y., Reunov A., Marshall P., Veinot J., Menzies K., Nemer M., Harper M.E. Glutaredoxin-2 controls cardiac mitochondrial dynamics and energetics in mice, and protects against human cardiac pathologies. Redox Biol. 2018;14:509–521. doi: 10.1016/j.redox.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pastore A., Piemonte F. Protein glutathionylation in cardiovascular diseases. Int J. Mol. Sci. 2013;14(10):20845–20876. doi: 10.3390/ijms141020845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S.B., Foster D.B., Rucker J., O'Rourke B., Kass D.A., Van Eyk J.E. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circ. Res. 2011;109(7):750–757. doi: 10.1161/CIRCRESAHA.111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shutt T., Geoffrion M., Milne R., McBride H.M. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 2012;13(10):909–915. doi: 10.1038/embor.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]