Abstract
Iron (Fe) is an essential element for plants; however, its availability is limited as it forms insoluble complexes in the soil. Consequently, plants have developed mechanisms to adapt to low Fe conditions. We demonstrate that ethylene is involved in Fe deficiency-induced physiological responses in Malus xiaojinensis, and describe the identification of MxERF4 as a protein-protein interaction partner with the MxFIT transcription factor, which is involved in the iron deficiency response. Furthermore, we demonstrate that MxERF4 acts as an MxFIT interaction partner to suppresses the expression of the Fe transporter MxIRT1, by binding directly to its promoter, requiring the EAR motif of the MxERF4 protein. Suppression of MxERF4 expression in M. xiaojinensis, using virus induced gene silencing resulted in an increase in MxIRT1 expression. Taken together, the results suggest a repression mechanism, where ethylene initiates the Fe deficiency response, and the response is then dampened, which may require a transient inhibition of Fe acquisition via the action of MxERF4.
Introduction
Iron (Fe) is a critical element for a number of metalloenzymes involved in photosynthesis and respiration, and so imbalances in Fe levels can profoundly affect cellular metabolism. Plants have developed two Fe uptake strategies, Strategies I (non-graminaceous plants) and II (graminaceous plants)1,2, respectively. The Strategy I response includes two main processes: i) the ferric chelates reduction at the root surface; and ii) the absorption of the ferrous Fe across the root plasma membrane3. The Strategy II response relies on the mugineic acids (Mas) biosynthesis and secretion, which are Fe(III)-solubilizing molecules that take up chelated Fe, and so is referred to as the Chelation Strategy1.
It has been demonstrated that plant hormones are involved in signaling associated with the Fe deficiency responses, inducing adaptive responses4,5. As an example, ethylene synthesis is induced during Fe starvation in roots, and may be involved in transducing the Fe deficiency signal to induce adaptive changes6,7. Indeed, exogenous application of the ethylene precursor, was reported to mimic iron deficiency induced morphological responses in tomato (Solanum lycopersicum)8,9 and pea (Pisum sativum). Elevated ethylene production, which has been observed as a consequence with Fe deficiency, has been further confirmed by studies showing the upregulation of ethylene synthesis related genes, such as SAMS, ACS, and ACO, under such conditions10–13. In addition to ethylene synthesis related genes, ethylene signaling genes also show altered expression during Fe starvation10,14,15. Notably, FIT (FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR) acts a central role in up-regulating the expression of key genes involved in Fe uptake16–21. FIT expression is upregulated by Fe deficiency17,18 and in response to ethylene10,22,23. In addition, EIN3 (ETHYLENE-INSENSITIVE3) and EIL1 (EIN3-LIKE1) can interact with FIT23 directly, and the resulting complex also interacts with the Mediator subunits (MED16 and MED25), which positively regulate iron homeostasis23–25. FIT may also interact with gene repressors21. The relative proportions of active and inactive FIT pools may be affected by differential FIT protein-protein interactions21. Recently, a transcription factor ZAT12 was identified which served as a negative regulator of Fe acquisition21.
For keeping the cellular ethylene homeostasis and avoiding toxic effect, it has not yet been determined whether there is repressors function as a negative regulator which can modulate ethylene under prolonged Fe deficient conditions. It is possible that ethylene sensitivity is promoted at the earlier stages of Fe deficiency and then slows the ethylene response once it has been initiated via a dampening mechanism. ERFs (Ethylene-responsive element binding factors), also known as EREBPs (ethylene-responsive element binding proteins), that interact with the GCC-box sequence26. Most of the ERFs were identified as transcriptional activators27–33. Recent studies indicate that some ERF proteins can act as transcriptional repressors, in tobacco and Arabidopsis, ERF3 and ERF4 were shown to repress the expression of a GCC-box-containing reporter gene27,28. ERF factors also play a role in a variety of developmental processes34, abiotic and biotic stress responses27,35,36.
To better understand this ethylene regulatory system in iron stress responses, previously we used Malus xiaojinensis, a woody plant that is used as a rootstock for apple trees and that exhibits highly efficient Fe uptake37–39. Our previous reciprocal grafting experiments showed that iron deficiency responses induction only occurs when M. xiaojinensis was used as the rootstock, compared with the iron inefficiency rootstock38. Here, we identified a new protein-protein interaction partner of M. xiaojinensis FIT, the ethylene response factor (ERF), MxERF4. We show that MxERF4 serves as a repressor of Fe uptake, thereby acting as a regulator in a signal dampening mechanism.
Results
Ethylene is involved in Fe deficiency-induced physiological responses
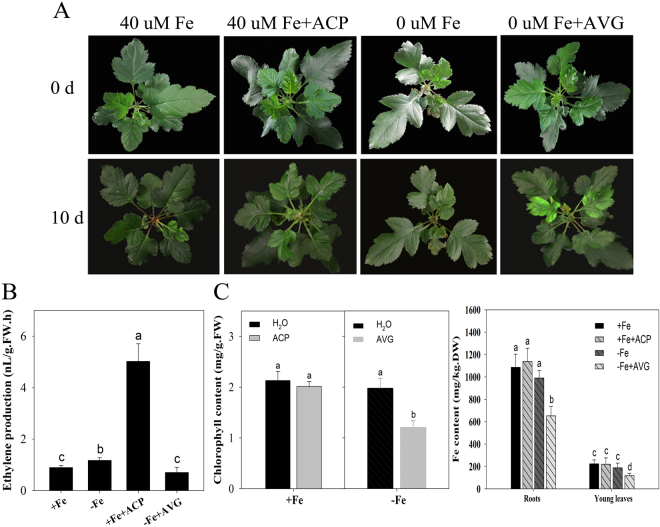
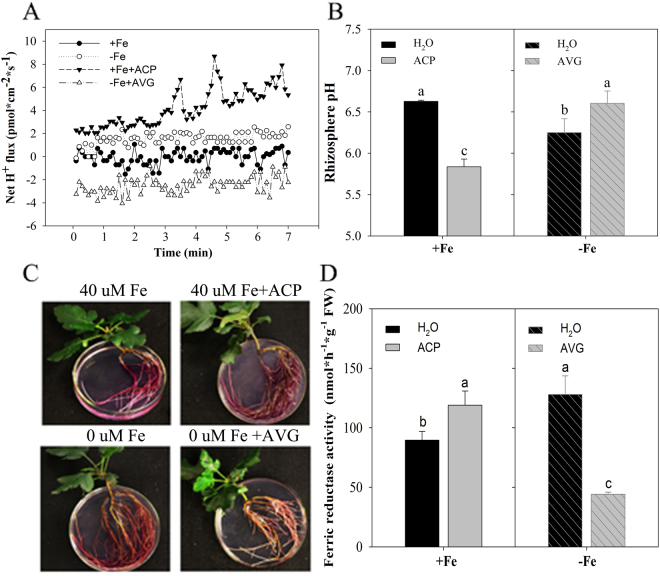
After low Fe treatment of the Fe uptake efficient species, M. xiaojinensiss, chlorosis, a typical Fe-deficiency symptom, was not obvious (Fig. 1A). Figure 1B showed Fe deficiency induced the ethylene production and exogenous application of an ethylene inhibitor, aminoethoxyvinylglycine (AVG) decreased ethylene production. Accordingly, AVG application resulted in Fe deficient chlorosis and reduced chlorophyll content under Fe deficient conditions (Fig. 1A,C). Similarly, exogenous application of AVG induced a significant decrease in the Fe content of roots and young leaves but not significant in ACP treatment (Fig. 1C), indicating that ethylene may play a role in the tolerance of Fe deficiency in M. xiaojinensis. The regulation of root proton (H+) extrusion and Fe(III) reductase (FCR) activity is known to be an important component of the response to Fe deficiency40–43, so we assessed whether the Fe deficiency response M. xiaojinensis involving ethylene was associated with H+ flux and FCR activity. As shown in Fig. 2, Fe deficiency induced a substantial increase in H+ flux and FCR activity, but application of AVG weakened this response. Interestingly, application of ethephon, 2-Chloroethylphosphonic acid (ACP) mimicked Fe deficiency in that it induced both increased H+ flux and FCR activity under normal Fe conditions. These results suggest that ethylene positively regulates Fe deficiency induced physiological responses in M. xiaojinensis.
Figure 1.
Effect of ethylene on the phenotype of Fe-deficient Malus xiaojinensis. (A) The phenotype of M. xiaojinensis seedlings grown in media supplied with 40 µM Fe, 40 µM Fe + 100 mg/L 2-Chloroethylphosphonic acid (ACP), 0 µM Fe, 0 uM Fe + 10 µM aminoethoxyvinylglycine (AVG) for 10 d respectively. (B) The ethylene production of M. xiaojinensis seedlings grown in media supplied with 40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 µM Fe + 10 µM AVG for 2 d. Bars represent means ± SE of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test). (C) The chlorophyll content and Fe content of M. xiaojinensis seedlings grown in media supplied with 40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 µM Fe + 10 µM AVG for 2 d. Bars represent means ± SE of of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test).
Figure 2.
Applying Ethylene treatments effect on Fe-deficient responses in Malus xiaojinensis. (A) Transient H+ flux in the plant root hair zone during different treatments (40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 µM Fe + 10 µM AVG for 2 d. (B) Rhizosphere pH of plants grown under different treatments (40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 uM Fe + 10 µM AVG) for 2 d. Bars represent means ± SE of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test). (C,D) Rhizosphere ferric reductase activity of plants grown under different treatments (40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 µM Fe + 10 µM AVG) for 2 d. Bars represent means ± SE of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test).
Inverse expression of MxERF4 and Fe deficiency responsive genes during Fe deficiency
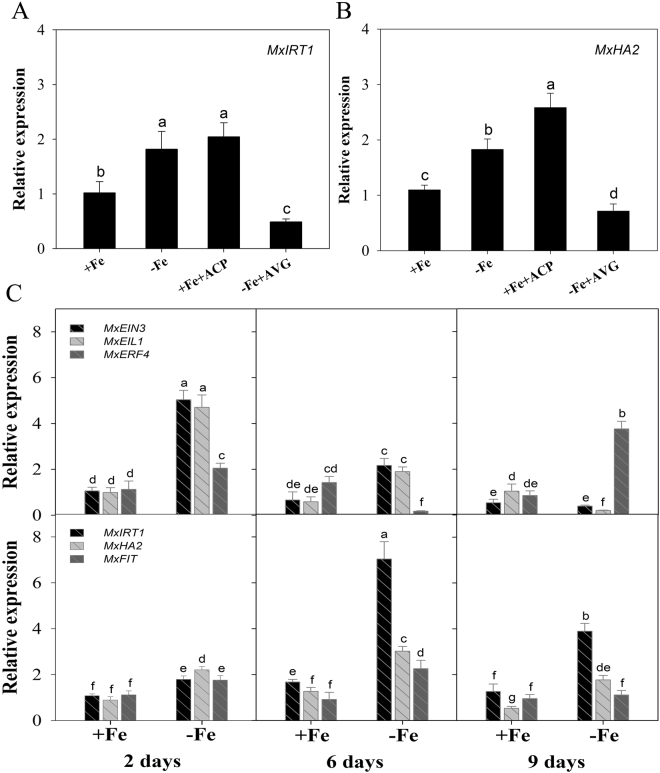
We first investigated whether Fe deficiency responsive genes were regulated by ethylene application. ACP was included during the normal Fe level treatments and AVG was added when the plants were treated to induce deficiency. We found that the expression levels of MxHA2 which is responsible for the major acidification activity were increase by the ethylene treatment (Fig. 3A,B). To identify genes potentially involved in ethylene signaling that might control these Fe deficient responsive genes, we queried transcriptome data that was generated from an earlier study of M. xiaojinensis exposed to Fe-deficient media44. We identified MDP0000324718 as a candidate hub gene for modulating the deficiency response (Fig. S1), which we named MxERF4. Expression of MxERF4 significantly increased in response to Fe deficiency, compared with Fe sufficiency on the ninth day. To determine which biological activities might be coordinated with MxERF4 expression, we targeted genes that positively regulate Fe homeostasis, and that the expression patterns of MxFIT, MxIRT1 and MxHA2 were opposite to that of MxERF4. As expected, upon Fe deficiency, MxFIT, MxIRT1 and MxHA2 expression levels were induced during the first 6 days, but were lower at day 9. EIN3 and EIL1, two transcription factors involved in ethylene signaling, have been associated with regulation of iron homeostasis24 and, as expected, the expression of both genes was induced during the first 6 days of treatment, but was down-regulated at day 9 (Fig. 3C). Taken together, these results indicated that MxERF4 may play a role with MxEIN3 and MxEIL1 together in the dampening mechanism.
Figure 3.
Expression of Fe deficiency responsive genes during ethylene treatments. (A) Expression of the MxIRT1 gene in plants grown under different conditions (40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 µM Fe + 10 µM AVG) for 2 d. Values are the means ± SE of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test). (B) Expression of the MxHA2 gene in plants grown under different conditions (40 µM Fe, 40 µM Fe + 100 mg/L ACP, 0 µM Fe, 0 µM Fe + 10 µM AVG) for 2 d. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test). (C) Expression of the genes (MxEIN3, MxEIL1, MxERF4, MxIRT1, MxHA2 and MxFIT) in plants grown under different conditions (40 µM Fe and 0 µM Fe) for 2 d, 6 d and 9 d. Values are the means ± SE of three replicates.
MxERF4 acts as a repressor and contains a repression domain
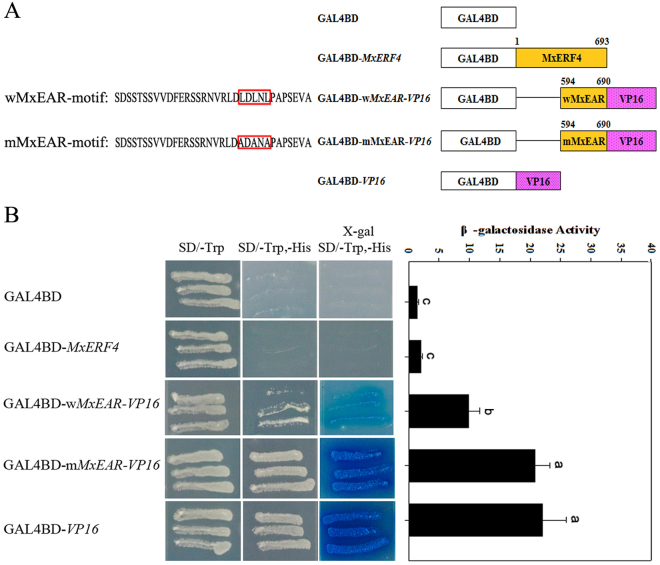
Some ERF-associated amphiphilic repression (EAR) motif containing ERF proteins can act as transcriptional repressors. It has been demonstrated that some ERFs can repress gene expression27,28; however, the repressor activity of MxERF4 proteins containing EAR motifs need further to be demonstrated. To evaluate potential MxERF4 repressor function, we performed a transactivaiton analysis. We used the VP16 transcriptional activation domain as a positive control and a fusion protein of the EAR motif with VP16 at the N-terminus as the effector (Fig. 4). The results showed that inclusion of the EAR motif with the VP16 resulted in a >50% inhibition of the original β-gal activity (Fig. 4). By contrast, mutation of EAR motif did not affect the activity of the β-gal, indicating that MxERF4 can indeed function as a repressor.
Figure 4.
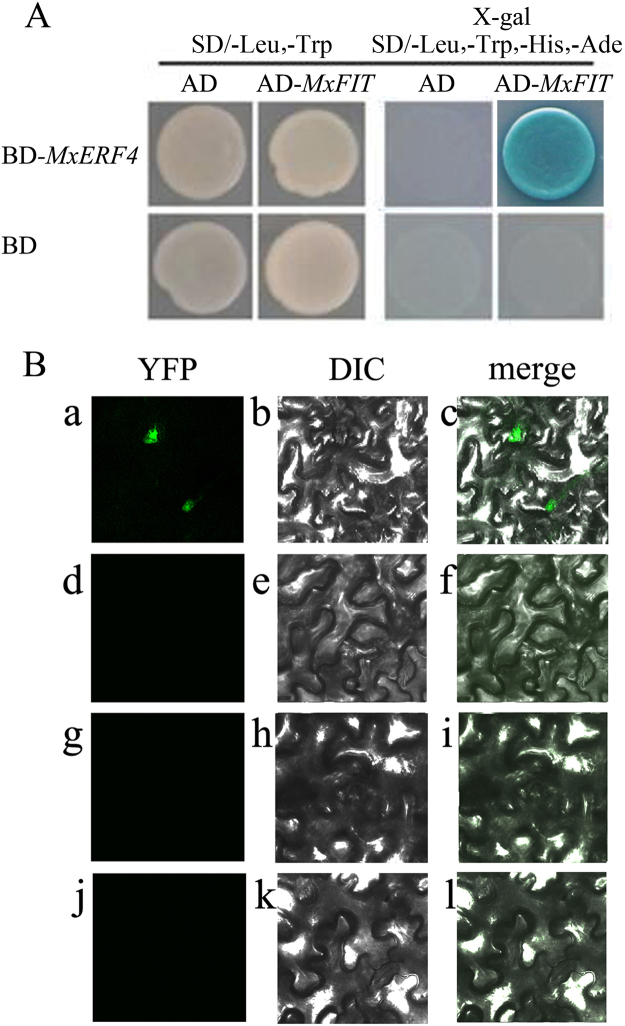
The MxERF4 EAR repression domain inhibits transcriptional activity. (A) The EAR-motif point mutation structure diagram of MxERF4 wEAR-motif (LDLNL) mutated as mEAR-motif (ADANA). (B) Analysis of X-gal assay of MxERF4 in yeast to identify transactivation activity and β-galactosidase activity. Growth status of yeast AH109 harboring MxERF4 full length (GAL4BD-MxERF4), carrying MxERF4 EAR motif of VP16 (GAL4BD-wMxEAR-VP16), MxERF4 EAR motif point mutation of VP16 (GAL4BD-mMxEAR-VP16) and VP16 (GAL4BD-VP16) on minimal medium/-Trp and minimal medium/-Trp,-His, pBD-GAL4 Cam vector (GAL4BD) as control; Transactivation activity by an X-gal assay on minimal medium/-Trp,-His. Values are the means ± SE (n = 9).
MxERF4 participates the FIT interaction network and suppresses MxIRT1 expression
Given that the expression pattern of MxIRT1 was opposite to that of MxERF4, we hypothesized that MxERF4 might serve as a suppressor of MxIRT1 by binding to its promoter. In support of this idea, the results of a yeast one-hybrid (Y1H) assay (Fig. 5A,B) suggested that both MxERF4 and MxFIT bound to the promoter of MxIRT1 (ProIRT1). Then We investigated the regulation of the MxIRT1 promoter by MxERF4 and MxFIT using a β-glucuronidase (GUS) transactivation assay in wild tobacco (Nicotiana benthamiana) leaves involving co-transformation with the Pro35S: MxERF4/Pro MxIRT1:GUS or Pro35S: MxFIT/Pro MxIRT1:GUS constructs. Compared with the pCAMBIA1301 control, when Pro35S: MxFIT was co-transformed with MxIRT1:GUS, MxIRT1 promoter activity increased, while MxERF4 decreased MxIRT1 promoter activity (Fig. 5C). We further hypothesized that since the expression pattern of MxFIT was opposite to that of MxERF4, the interaction between MxFIT and MxERF4 might be responsible for repression of FIT function. To test for an interaction between MxFIT and MxERF4, we used a targeted yeast two-hybrid (Y2H) assay with the full-length MxFIT open reading frame cloned into the activation domain (AD), named MxFIT-pGADT7, and the MxERF4 coding sequence cloned into the pGBKT7 (BD) vector which harbored the binding domain. We observed an interaction between MxFIT and MxERF4 (Fig. 6A), but no interactions were apparent using BD-MxERF4 together with an empty AD vector, or an empty BD vector with AD-MxFIT, which served as negative controls. To test whether an interaction between MxERF4 and MxFIT would take place in plant cells, we performed an in planta bimolecular fluorescence complementation (BiFC) analysis45. Following transient transformation of Nicotiana benthamiana leaves, yellow fluorescent protein (YFP) signal was detected in the nuclei of leaves expressing YFP-MxFIT and YFP-MxERF4 (Fig. 6B), showing that there is interaction between MxERF4 and MxFIT. Further, by using transient transformation we found that the interaction of MxERF4/MxFIT with the MxIRT1 promoter inhibited the activity of GUS, respectively, compared with that in the MxFIT with the MxIRT1 promoter, indicating that MxERF4 acts as an MxFIT interaction partner to serve as a suppressor of MxIRT1 (Fig. 5C).
Figure 5.
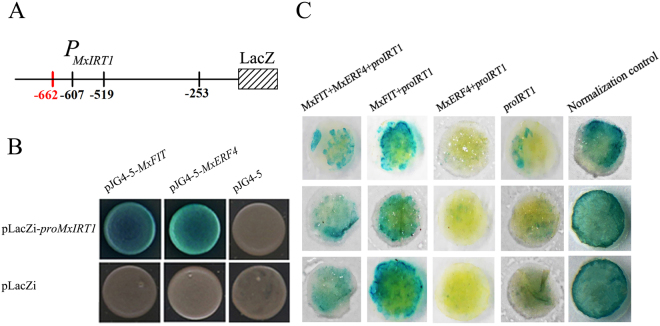
MxERF4 bind the promoter of MxIRT1 and suppress the effect of MxFIT on MxIRT1. (A) The structure of the MxIRT1 promoter (765 bp) showing three GCC-box motifs (−253 bp, −519 bp and −607 bp, in black), one G-box (−662 bp, in red). (B) Yeast one hybrid analysis of MxERF4 and MxFIT with the MxIRT1 promoter. All tests were conducted on minimal medium/-Trp,-Ura. Interactions were determined based on cell growth and were confirmed by an X-gal assay on minimal medium/-Trp,-Ura. (C) The effect of MxFIT and MxERF4 on MxIRT1 promoter. GUS staining of reprentative leaf pieces infiltrated with normalization control (pCAMBIA1301) or only promoter (MxIRT1) or coinfiltrated with the transcriptional factor and the promoter (MxFIT + MxERF4 + proMxIRT1, MxFIT + proMxIRT1, MxERF4 + proMxIRT1).
Figure 6.
There is an interaction between MxERF4 and MxFIT. (A) Yeast two hybrid analysis of the physical interaction between the MxERF4 and MxFIT proteins. The protein interaction was examined using various combinations of prey and bait vectors. All tests were conducted on minimal medium/-Leu,-Trp or on minimal medium/-Leu,-Trp,-His,-Ade. Interactions were determined based on cell growth and were confirmed by an X-gal assay on minimal medium/-Leu,-Trp,-His,-Ade. (B) In planta protein interaction of MxERF4 and MxFIT. bimolecular fluorescence complementation (BiFC) of yellow fluorescent protein (YFP) in transiently transformed tobacco leaf epidermal cells. The left column ([a], [d], [g] and [j]) shows the YFP signal detected by confocal microscopy; the middle column ([b], [e], [h], and [k]) shows differential interference contrast (DIC) microscopy; the right column ([c], [f], [i], and [l]) shows merge images of the fluorescent signal and DIC. YC-MxFIT plus YN-MxERF4 ([a] to [c]); negative control YC plus YN-MxERF4 ([d] to [f]); YC-MxFIT plus negative control YN ([g] to [i]); negative control YC plus negative control YN ([j] to [l]).
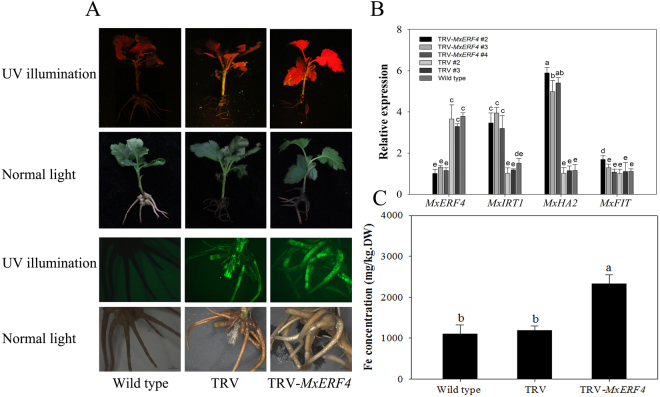
To further test the function of MxERF4 as a regulator, we used virus induced gene silencing (VIGS) to suppress the expression of MxERF4 in M. xiaojinensis plantlets. M. xiaojinensis tissue culture plantlets infiltrated with the virus containing the TRV-GFP-MxERF4 (TRV, for silencing) (Fig. 7), and green fluorescent protein (GFP) fluorescence was observed in leaves, stems and new roots (Fig. 7A), indicating that the virus had spread through the plants. Suppression of MxERF4 expression, using a TRV-GFP-MxERF4 plasmid containing a partial MxERF4 ORF, caused an increase in MxIRT1 and MxHA2 expression; however only minor changes in MxFIT expression were observed (Fig. 7B). And the Fe content in silenced roots (virus containing the TRV-GFP-MxERF4) was higher than wild type and empty vector (Fig. 7B). The results of this study were used to generate a model of the regulatory system involving MxERF4. Exposure of M. xiaojinensis plants to Fe deficiency, induces ethylene production, and while ethylene sensitivity increases in the earlier stages of Fe deficiency it subsequently decreases. MxERF4 act as an MxFIT interacting partner, and the protein complex binds to the MxIRT1 promoter, thereby suppresses MxIRT1 expression which results in a reduction in Fe uptake. In this model, the ethylene response can be slowed down once it has been initiated, and this dampening process may require a transient inhibition of Fe acquisition via MxERF4 action (Fig. 8).
Figure 7.
Transient expression of MxERF4 in Malus xiaojinensis. The M. xiaojinensis seedlings were infiltrated with agrobacterium containing a TRV control (pTRV1 + pTRV2-GFP), or TRV carrying a MxERF4 fragment (pTRV1 + pTRV2-GFP-MxERF4). (A) Green fluorescent protein (GFP) imaging of TRV-GFP-MxERF4 infiltrated M. xiaojinensis seedlings by UV and fluorescence microscopy. (B) Quantitative real-time reverse transcription-PCR (qRT-PCR) analysis of MxIRT1, MxHA2 and MxFIT expression in silenced roots. Values are means ± SE of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test). (C) Analysis of Fe concentration in silenced roots. Bars represent means ± SE of three replicates. Different letters represent statistically different means at P < 0.05 (one-way ANOVA analysis with Duncan post-hoc test).
Figure 8.
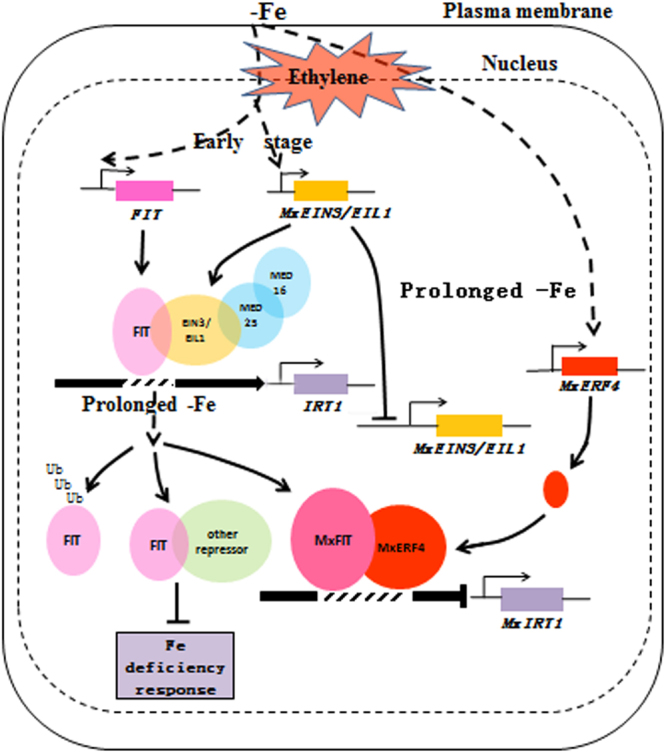
Model of MxERF4 action in suppressing Fe uptake. The early stages of Fe deficiency leads to the induced expression of MxEIN3/EIL1. After prolonged Fe starvation, the expression of MxEIN3/EIL1 is inhibited and the expression of MxERF4 is up-regulated. We propose that MxERF4 can act as a transcriptional repressor through its EAR motif. Ethylene causes an enhanced Fe deficiency adaptive response at the early stages of the response by up-regulating EIN3/EIL1 and the MED complex, but the ethylene response slows down once it has been initiated via the action of MxERF4. MxERF4 eventually interact with MxFIT and suppresses MxIRT1 gene expression.
Discussion
It has long been known that Fe deficiency can induce a wide range of adaptive responses, but the existence of a dampening mechanism associated with these responses has been unclear, particularly in woody plants38. We show here that MxERF4 suppresses Fe uptake by binding to the MxIRT1 promoter and, importantly, that a key factor in modulating the ethylene response is a counterbalanced Fe uptake response.
In different dicotyledonous plants, experiments indicated a physiological connection between iron deficiency signaling and ethylene, in that ethylene production increases with Fe deficiency treatment7,46–48. Applying ethylene precursors to plants promotes Fe deficiency responses, such as the activation of IRT1 and FRO2 gene expression10,48,49. Our data suggest that exogenous application of the ethylene inhibitor, AVG, suppresses the Fe deficiency response, since it prevented Fe deficiency-induced ethylene accumulation and concomitantly blocked the up-regulation of Fe(III) reductase activity and the root proton extrusion, while addition of ethephon (ACP) reversed these physiological responses (Fig. 2). However, once the ethylene signal has been initiated, the mechanism by which the ethylene response can counteract prolonged Fe treatment is unknown.
FIT can interact with transcription factors, such as basic helix-loop-helix (bHLH) proteins50,51, the EIN3 and EIL123and the mediator subunit MED1625, which play a central role that governs Fe deficiency responses. And all these interactions were shown to promote FIT activity23,25,50,51. Conversely, ZAT12 from Arabidopsis thaliana, was reported to suppress Fe acquisition21. In our study, the identification of MxERF4 as a suppressor of MxIRT1 expression provides insights into counterbalanced effects at the molecular level. Most of the ERF proteins characterized to date have been shown to act as transcriptional activators28–33,52–54; however, some ERF proteins with EAR motif can function as transcriptional repressors, although little is known about their physiological roles. From our previously published transcriptional profile data44, we identified an EAR motif ERF (MxERF4) as a regulator capable of modulating ethylene response to counterbalance the Fe response. Using VIGS, in combination with an activity repression analysis, we speculated that MxERF4 directly binds to the MxIRT1 promoter and that the EAR motif of MxERF4 was required for this suppression. Upon Fe deficiency, MxERF4 expression levels was inhibited during the first 6 days, but was induced at day 9. With prolonged Fe deficiency treatment, MxERF4 could be served as a negative regulator. In the later Fe deficiency stage, in order to prevent damage from any over accumulation of other metals, downregulation of the metal uptake machinery is required, which are transported by IRT155. It may be that the suppression ability of MxERF4 binding to MxIRT1 promoter.
Arabidopsis, FIT is not able to activate IRT1 promoter on its own but only in combination with another bHLH of the Ib subgroup50,51. Compared with model plant Arabidopsis, woody plants have high level of genetic variation; the use of highly heterozygous woody plant ‘Malus xiaojinensis’ has allowed us to demonstrate that FIT can activate IRT1 promoter from our results. Also our experiment showed the interaction of MxIRO2/MxFIT with the MxIRT1 promoter enhanced the activity of GUS (Fig. S2). This regulation will be the target of future studies.
Several plant signaling molecules are known to modulate the Fe deficiency response, either by inducing it, such as nitric oxide10,56–58 and auxin38,58,59, or by suppressing it, as is the case with cytokinin60. Ethylene and auxin have been shown which can regulate iron-deficiency responses from the pharmacological experiments and use of some plant mutants6,7,38,58,59,61. Application of ethylene precursor or auxin mimicked iron deficiency in inducing the iron adaptive responses6–8,61. Furthermore, our research showed that IAA plays a key role to mediate the iron deficiency responses in M. xiaojinensis38. Blum et al. showed the balance between the ethylene and auxin leads to an almost continuous IRT1 presence under iron deficiency condition62. Dominant ethylene presence induces IRT1 expression in the root with iron deficient treatment62. Thus, we proposed that both ethylene and auxin play crucial characters in iron deficiency responses and the cross-talks among ethylene and auxin need to be further explored.
Previous studies have suggested a molecular link between Fe deficiency responses and hormone signaling regulation, and we report here a suppression mechanism that dampens the plant responses. This model provides a basic molecular framework for future investigation of the regulatory systems that allow environmental adaption.
Materials and Methods
Plant material and growth conditions
Malus xiaojinensis seedlings were cultivated until stem lignification in Murashige and Skoog (MS) medium with 0.3 mg/L indolebutyric acid (IBA) + 0.3 mg/L 6-Benzyl Aminopurine (6-BA), and were then transferred to rooting medium (1/2 MS + 0.5 mg/L IBA) for a month, until white roots were visible. The seedlings were removed from the culture medium and washed with distilled water three times to remove the agar. After pretreatment, the seedlings were cultivated in a standard nutrition solution58, with the pH adjusted to 6.0 using 1 M NaOH. The nutrition solution was replaced every week. Plants were grown for 60 d in a growth room at 25 ± 2 °C day/17 ± 2 °C night with a 16 h photoperiod at a light intensity of 250 µmol·m−2·s−1. After a variable precultivation period, some plants were transferred to nutrient solutions representing Fe-deficient (0 µM Fe), Fe-deficient with added ethylene inhibitor (0 µM Fe + 10 µM AVG), Fe-sufficient (40 µM Fe) and Fe-sufficient with added ethephon (40 µM Fe + 100 mg/L ACP) conditions. For RNA extraction, roots were harvested after 0, 2, 6 and 9 d of the above treatments.
Isolation of total RNA and quantitative real-time reverse transcription-PCR
Total RNA was extracted from the roots using a modified CTAB method63 and treated with DNase I to remove DNA contamination (TaKaRa Biotechnology Co., Ltd., Dalian, China). To generate cDNA, the RNA samples were reverse-transcribed using an oligo-dT primer and reverse transcriptase (TaKaRa Biotechnology Co., Ltd., Dalian, China), according to the manufacturer’s instructions. The relative expression levels of genes were detected using an Applied Biosystems 7500 real-time PCR system. The housekeeping gene β-actin which is degenerate primer (MDP0000896590, MDP0000428264 and MDP0000170174) was used as the control. The primers were designed using the Primer Premier 5 software (Premier Biosoft, USA)64. The efficacy of the primers was confirmed by qRT-PCR. Amplifications using these primers yielded 100–200 bp products, which were subjected to melting curve analyses. The relative expression was calculated according to the 2−ΔΔCT method65. Primers are listed in Supplemental Table S1. Each reaction was performed in triplicate.
Chlorophyll content analysis
Leaf samples (0.2 g) were collected and transferred to 80% acetone for 24 h. Chlorophyll content was detected by measuring the absorbance of the solution at A645 and A663 using a spectrophotometer66.
Fe content determination
Samples were collected and dried at 105 °C for 20 min, followed by 70 °C for 6 days, and separately ground to a powder using grinding rods. Subsequently, 0.1 g of the powder was transferred to 10 mL 1 M HCl and the samples were shaken at 180 rpm for 5 h. The Fe content was measured by atomic absorption spectrometry after filtration (Z-5000, Hitachi, Tokyo, Japan)67,68.
Measurements of net H+ fluxes with non-invasive microtest technique (NMT)
Net H+ fluxes were measured using a BIO-IM Series NMT system (Younger USA) at the Xuyue Beijing NMT Research Service Center, China. H+ ion-selective microelectrodes were placed 2 mm from the root cap and the samples were placed in the testing solution69 at pH 6.0. Only electrodes with a Nernstian slope >50 mV/decade were used in this study. H+ fluxes were calculated using the JCal V3.3 (a free MS Excel spreadsheet, youngerusa.com or xuyue.net). Ion flux was calculated by Fick’s law of diffusion, which demonstrated in the NMT Experiments instruction in xuyue.net.
Measurement of Fe3+ reductase activity and rhizosphere pH
The solution pH of different treatments (0 µM Fe, 0 µM Fe + 10 µM AVG, 40 µM Fe and 40 µM Fe + 100 mg/L ACP) was original adjusted to 6.3, whereafter the pH of the solutions was measured when the plants were incubated in the solution for 1.5 month. Root Fe3+ reductase activity was determined as described by Schikora and Schmidt59 and Li et al.70, with some modifications. The plant roots were soaked in a saturated CaSO4 solution for 5 minutes, washed with distilled water, and the plants were then transferred to the nutrient solution described above, with the addition of Fe-EDTA (0.1 mM) and 2,2-bipyridyl (0.4 mM), then incubated in the dark for 2 h. The environmental conditions during the measurements were the same as those for normal growth. The reducing capacity of the solution was determined by measuring the concentration of the Fe2+-dipyidyl complex formed at A520 using a spectrophotometer (Unico UV-2012). Each experiment was repeated at least three times. For some treatments, the location of the ferric reductase activity along the roots was visualized on agar (0.7%, w/v) plates with a ferric reduction assay solution consisting of Fe-EDTA (0.5 mM) and FerroZine (0.5 mM).
Yeast one hybrid (Y1H) and two hybrid (Y2H) assays
The CDS of MxERF4 and MxFIT were ligated into the pJG4-5 vector, MxIRT1 promoter was cloned into the pLacZi vector which contains LacZ reporter gene71. And the Y1H assay was conducted as described in the Yeast Protocols Handbook (Clontech). Transformants were grown on a minimal medium/-Ura,-Trp containing X-gal (5-bromo-4–chloro-3–indolyl-β–D–galactopyranoside) to observe the color development of yeast colonies. Vector construction primers are listed in Supplemental Table S1.
For the Y2H assay, full-length cDNAs of MxERF4 and MxFIT were cloned into the pGBKT7 and pGADT7 vectors from Clontech. All constructs were transformed into yeast (S. cerevisiae) strain AH109 using the lithium acetate method, and yeast cells were grown on a minimal medium/-Leu,-Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were plated onto a minimal medium/-Leu,-Trp,-His,-adenine containing 20 mg/mL X-gal to test for possible interactions. Primers used for vector construction are listed in Supplemental Table S1.
For the yeast transcription activation assay, full-length cDNAs of MxERF4 and the MxERF4 EAR sequence were separately cloned into a strong transcriptional activation vector pBD-GAL4-VP1628. Constructs were transformed into yeast (S. cerevisiae) strain AH109 using the lithium acetate method, as above, and yeast cells were grown on a minimal medium/-Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were plated onto a minimal medium/-Trp,-His containing 20 mg/mL X-gal and ortho-nitrophenyl-β-D-galactopyranoside (ONPG) as substrates. Primers used for vector construction are listed in Supplemental Table S1. Experiments were repeated three times.
BiFC Assays
To generate the BiFC constructs, the full-length cDNA sequences of MxERF4 and MxFIT were cloned into the pSPYNE-35S and pSPYCE-35S vectors72. The primers used for the BiFC assays are listed in Supplemental Table S1. Coexpression was observed in tobacco (Nicotiana tabacum) leaves as described by Schütze et al.45. The fluorescence of the fusion proteins was detected 3 days after Agrobacterium infiltration. Fluorescence images were acquired using a Nikon D-ECLIPSE C1 spectral confocal laser-scanning system. YFP and brightfield images were generated by excitation at 488 and 543 nm, respectively. A Nikon ECLIPSE TE2000-E Inverted fluorescence microscope was used for the fluorescence analysis.
Transient transformation analysis in tobacco leaf
The full-length coding regions (CDS) of MxERF4 and MxFIT were cloned into the KpnI and SalI sites of the pCAMBIA2300 vector to form the pCAMBIA2300- MxERF4 and pCAMBIA2300-MxFIT constructs. The pCAMBIA1301 plasmid was used as normalization control which contains the GUS reporter driven by the 35S promoter, the promoter of MxIRT1 was fused into pCAMBIA1301 and replaced the 35S promoter. Then the four groups (pCAMBIA2300-MxFIT/MxERF4 + pCAMBIA1301-proMxIRT1; pCAMBIA2300-MxFIT + pCAMBIA1301-proMxIRT1; pCAMBIA2300-MxERF4 + pCAMBIA1301-proMxIRT1; pCAMBIA1301-proMxIRT1) were respectively infiltrated tobacco (Nicotiana tabacum) leaves, tobacco plants were grown for 2 d after infiltration, and then collected the transformed leaves soaked in GUS staining solution at 37 °C. Total removal of chlorophyll using 70% alcohol decolorization, and observed the expression of GUS.
Silencing of MxERF4 in M. xiaojinensis by VIGS
Silencing of MxERF4 expression by virus induced gene silencing (VIGS) was performed as described by Dai et al.73, with some modifications. A 300-bp fragment at the 3′ UTR of MxERF4 was amplified using a forward primer with an EcoRI restriction site and a reverse primer with a XbaI restriction site. The pTRV1 and pTRV2-GFP vector from Professor Gao and 3′ UTR of MxERF4 cloned into pTRV2-GFP vector (pTRV2-GFP-MxERF4). The vectors of pTRV1, pTRV2-GFP and pTRV2-GFP-MxERF4 were transformed individually into Agrobacterium tumefaciens strain GV3101, and the transformed A. tumefaciens lines were cultured for 24 h in Luria–Bertani medium55 supplemented with 50 mg/ml kanamycin and 50 mg/ml rifampicin. The cultures were harvested, and suspended in infiltration buffer (10 mM MgCl2, 1.5 mM acetosyringone, 10 M MES, pH 5.6) to a final OD600 of approximately 1.0. Mixtures of cultures containing an equal ratio (v/v) of pTRV1 and pTRV2-GFP (control), pTRV1 and pTRV2-GFP-MxERF4 were placed at room temperature in the dark for 3–4 h before vacuum infiltration. Tissue culture seedlings were collected and vacuum infiltration was performed by immersing them in the bacterial suspension solution and infiltrating under a vacuum at 0.7 MPa. After release of the vacuum, seedlings were washed in deionized water and kept in deionized water for 3 days at 8 °C, followed by cultivation in a standard nutrition solution67. The nutrition solution was replaced every week. Plants were grown in a growth room at 25 ± 2 °C day/17 ± 2 °C night with a 16 h photoperiod at a light intensity of 250 µmol·m−2·s−1. After 12 d, fluorescence in the transformed plants was observed using a fluorescence microscope (ZEISS), and the roots were used for RNA isolation.
Statistical analysis
The Statistical Product and Service Solutions (SPSS) software (IBM Co, Armonk, USA) was used for statistical analysis. All experimental data were tested using one-way analysis of variance (ANOVA) and Duncan’s multiple-range test.
Electronic supplementary material
Acknowledgements
Financial support was provided by the National Key Research and Development Program of China (2016YFD0201103), the National Natural Science Foundation of China (No. 31572097), The National Key Technology R&D Program (2013BAD02B01), the earmarked fund for China Agriculture Research System (CARS-27), the 111 Project (B17043), Beijing Municipal Education Commission (CEFF-PXM2017_014207_000043) and Key Labs of Nutrition and Physiology for Horticultural Crops. We thank PlantScribe (www.plantscribe.com) for editing this manuscript.
Author Contributions
Y.W. and Z.H. designed the research and edited the manuscript. W.L., T.W. and Q.L. conducted the experiments. X.Z., X.X. and T.L. contributed reagents and analytical tools. W.L. and T.W. wrote the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Wei Liu and Ting Wu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19518-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhenhai Han, Email: rschan@cau.edu.cn.
Yi Wang, Email: wangyi@cau.edu.cn.
References
- 1.Marschner H, Römhelda V, Kissela M. Different strategies in higher plants in mobilization and uptake of iron. Journal of Plant Nutrition. 1986;9:695–713. doi: 10.1080/01904168609363475. [DOI] [Google Scholar]
- 2.Briat JF, et al. Cellular and molecular aspects of iron metabolism in plants. Biology of the Cell. 1995;84:69–81. doi: 10.1016/0248-4900(96)81320-7. [DOI] [Google Scholar]
- 3.Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology. 2012;63:131–152. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- 4.Landsberg EC. Organic acid synthesis and release of hydrogen ions in response to Fe deficiency stress of mono-and dicotyledonous plant species. Journal of Plant Nutrition. 1981;3:579–591. doi: 10.1080/01904168109362862. [DOI] [Google Scholar]
- 5.Romheld V, Marschner H. Mobilization of iron in the rhizosphere of different plant species. Advances in Plant Nutrition. 1986;2:155–204. [Google Scholar]
- 6.Romera FJ, Alcántara E. Iron-deficiency stress responses in cucumber (Cucumis sativus L.) roots (A possible role for ethylene?) Plant Physiology. 1994;105:1133–1138. doi: 10.1104/pp.105.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romera FJ, Alcántara E, DelaGuardia MD. Ethylene production by Fe-deficient roots and its involvement in the regulation of Fe-deficiency stress responses by Strategy I plants. Annals of Botany. 1999;83:51–55. doi: 10.1006/anbo.1998.0793. [DOI] [Google Scholar]
- 8.Schmidt W, Schikora A, Pich A, Bartels M. Hormones induce an Fe-deficiency-like root epidermal cell pattern in the Fe-inefficient tomato mutant fer. Protoplasma. 2000;213:67–73. doi: 10.1007/BF01280506. [DOI] [Google Scholar]
- 9.Schmidt W, Schikora A. Different path ways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiology. 2001;125:2078–2084. doi: 10.1104/pp.125.4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García MJ, Lucena C, Romera FJ, Alcántara E, Pérez-Vicente R. Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. Journal of Experimental Botany. 2010;61:3885–3899. doi: 10.1093/jxb/erq203. [DOI] [PubMed] [Google Scholar]
- 11.Stein RJ, Waters BM. Use of natural variation reveals core genes in the transcriptome of iron-deficient Arabidopsis thaliana roots. Journal of Experimental Botany. 2012;63:1039–1055. doi: 10.1093/jxb/err343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Wang B, Tian Q, Wang T, Zhang WH. Medicago truncatula ecotypes A17 and R108 differed in the irresponse to iron deficiency. Journal of Plant Physiology. 2014;171:639–647. doi: 10.1016/j.jplph.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Lauter ANM, et al. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. Bmc Genomics. 2014;15:1–25. doi: 10.1186/1471-2164-15-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Rourke JA, et al. Microarray analysis of iron deficiency chlorosis innear-isogenic soybean lines. Bmc Genomics. 2007;8:476. doi: 10.1186/1471-2164-8-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García MJ, et al. Hypoxia and bicarbonate could limit the expression of iron acquisition genes in Strategy I plants by affecting ethylene synthesis and signaling in different ways. Physiologia Plantarum. 2014;150:95–106. doi: 10.1111/ppl.12076. [DOI] [PubMed] [Google Scholar]
- 16.Bauer P, et al. Grosse I Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiology. 2004;136:4169–4183. doi: 10.1104/pp.104.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colangelo EP, Guerinot ML. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. Febs Letters. 2004;577:528–534. doi: 10.1016/j.febslet.2004.10.062. [DOI] [PubMed] [Google Scholar]
- 19.Yuan YX, Zhang J, Wang DW, Ling HQ. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Research. 2005;15:613–621. doi: 10.1038/sj.cr.7290331. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov R, Brumbarova T, Bauer P. Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Molecular Plant. 2012;5:27–42. doi: 10.1093/mp/ssr065. [DOI] [PubMed] [Google Scholar]
- 21.Le CT, et al. Zinc Finger of Arabidopsis Thaliana12 (ZAT12) interacts with Fer-Like Iron Deficiency-Induced Transcription Factor (FIT) linking iron. Plant Physiology. 2016;170:540–557. doi: 10.1104/pp.15.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García MJ, et al. A new model involving ethylene, nitricoxide and Fe to explain the regulation of Fe-acquisition genes in Strategy I plants. Plant Physiology & Biochemistry. 2011;49:537–544. doi: 10.1016/j.plaphy.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Lingam S, et al. Interaction between the bHLH transcription factor FIT and Ethylene Insensitive3/Ethylene Insensitive3-Like1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell. 2011;23:1815–1829. doi: 10.1105/tpc.111.084715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, et al. The Arabidopsis mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. The Plant Journal. 2014;77:838–851. doi: 10.1111/tpj.12440. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytologist. 2014;203:770–783. doi: 10.1111/nph.12860. [DOI] [PubMed] [Google Scholar]
- 26.Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohta M, Ohme-Takagi M, Shinshi H. Three ethylene-responsive transcriptional factors in tobacco with distinct transactivation functions. Plant Journal. 2000;22:29–38. doi: 10.1046/j.1365-313x.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- 29.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Plant Biology. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:391–406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by Ethylene-Insensitive3 and Ethylene-Response-Factor1. Genes & Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menke FLH, Champion A, Kijne JW, Memelink J. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. Embo Journal. 1999;18:4455–4463. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu K, Tian L, Hollingworth J, Brown D, Miki B. Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiology. 2002;128:30–37. doi: 10.1104/pp.010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biological Chemestry. 1998;379:633–46. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 35.Büttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proceedings of the National Academy of Sciences. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuma Y, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 37.Han ZH, Shen T, Korcak RF, Baligar VC. Iron absorption by iron-efficient and -inefficient species of apples. Journal of Plant Nutrition. 1998;21:181–190. doi: 10.1080/01904169809365392. [DOI] [Google Scholar]
- 38.Wu T, et al. Induction of root Fe(III) reductase activity and proton extrusion by iron deficiency is mediated by auxin-based systemic signaling in Malus xiaojinensis. Journal of Experimental Botany. 2012;63:859–870. doi: 10.1093/jxb/err314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zha Q, Wang Y, Zhang XZ, Han ZH. Both immanently high active iron contents and increased root ferrous uptake in response to low iron stress contribute to the iron deficiency tolerance in Malus xiaojinensis. Plant Sciece. 2014;214:47–56. doi: 10.1016/j.plantsci.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Chaney RL, Brown JC, Tiffin LO. Obligatory reduction of ferric chelates in iron uptake by soybeans. Plant Physiology. 1972;50:208–213. doi: 10.1104/pp.50.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buckhout TJ, Bell PF, Luster DG, Chaney RL. Iron-stress induced redox activity in tomato (Lycopersicum esculentum Mill.) is localized on the plasma membrane. Plant Physiology. 1989;90:151–156. doi: 10.1104/pp.90.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grusak MA, Pezeshgi S. Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiology. 1996;110:329–334. doi: 10.1104/pp.110.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brüggemann W, Moog PR, Nakagawa H, Janiesch P, Kuiper PJC. Plasma membrane-bound NADH: Fe3+-EDTA reductase and iron deficiency in tomato (Lycopersicon esculentum). Is there a Turbo reductase? Physiologia Plantarum. 1990;79:339–346. doi: 10.1111/j.1399-3054.1990.tb06751.x. [DOI] [Google Scholar]
- 44.Wang S, et al. Transcriptomic analysis demonstrates the early responses of local ethylene and redox signaling to low iron stress in Malus xiaojinensis. Tree Genetics & Genomes. 2014;10:573–584. doi: 10.1007/s11295-014-0705-5. [DOI] [Google Scholar]
- 45.Schütze K, Harter K, Chaban C. Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods in Molecular Biology. 2009;479:189–202. doi: 10.1007/978-1-59745-289-2_12. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Li C. Is ethylene involved in regulation of root ferric reductase activity of dicotyledonous species under iron deficiency? Plant & Soil. 2004;261:147–153. doi: 10.1023/B:PLSO.0000035536.79001.60. [DOI] [Google Scholar]
- 47.Zuchi S, Cesco S, Varanini Z, Pinton R, Astolfi S. Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta. 2009;230:85–94. doi: 10.1007/s00425-009-0919-1. [DOI] [PubMed] [Google Scholar]
- 48.Lucena C, et al. Ethylene could influence ferric reductase, iron transporter and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. Journal of Experimental Botany. 2006;57:4145–4154. doi: 10.1093/jxb/erl189. [DOI] [PubMed] [Google Scholar]
- 49.Waters BM, et al. Ethylene involvement in the regulation of the H+-ATPase CsHA1 gene and of the new isolated ferricreductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiology & Biochemistry. 2007;45:293–301. doi: 10.1016/j.plaphy.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Yuan Y, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research. 2008;18:385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- 51.Wang N, et al. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Molecular Plant. 2013;6:503–513. doi: 10.1093/mp/sss089. [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. Embo Journal. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van dFL, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- 54.Oñate-Sánchez L, Singh KB. Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiology. 2002;128:1313–1322. doi: 10.1104/pp.010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Molecular Biology. 1999;40:37–44. doi: 10.1023/A:1026438615520. [DOI] [PubMed] [Google Scholar]
- 56.Graziano M, Beligni MV, Lamattina L. Nitric oxide improves internal iron availability in plants. Plant Physiology. 2002;130:1852–1859. doi: 10.1104/pp.009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graziano M, Lamattina L. Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant Journal. 2007;52:949–960. doi: 10.1111/j.1365-313X.2007.03283.x. [DOI] [PubMed] [Google Scholar]
- 58.Chen WW, et al. Nitric oxide acts down stream of auxin to trigger root ferric-chelate reductase activity inresponse to iron deficiency in Arabidopsis thaliana. Plant Physiology. 2010;154:810–819. doi: 10.1104/pp.110.161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schikora A, Schmidt W. Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiology. 2001;125:1679–1687. doi: 10.1104/pp.125.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Séguéla M, Briat JF, Vert G, Curie C. Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant Journal. 2008;55:289–300. doi: 10.1111/j.1365-313X.2008.03502.x. [DOI] [PubMed] [Google Scholar]
- 61.Schikora A, Schmidt W. Formation of transfer cells and H+-ATPase expression in tomato roots under P and Fe deficiency. Planta. 2002;215:304–311. doi: 10.1007/s00425-002-0738-0. [DOI] [PubMed] [Google Scholar]
- 62.Blum A, Brumbarova T, Bauer P, Ivanov R. Hormone influence on the spatial regulation of IRT1 expression in iron-deficient Arabidopsis thaliana roots. Plant Signaling & Behavior. 2014;9:e28787. doi: 10.4161/psb.28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang XX, Tian WM, Li YX. Development of an efficient protocol of RNA isolation from recalcitrant tree tissues. Molecular Biotechnology. 2008;38:57–64. doi: 10.1007/s12033-007-0073-6. [DOI] [PubMed] [Google Scholar]
- 64.Zhang M, et al. Hsp70 and HSF-1 expression is altered in the tissues of pigs transported for various periods of times. Journal of Veterinary Science. 2012;13:253–259. doi: 10.4142/jvs.2012.13.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi J, et al. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in chinese cabbage (Brassica rapa L. ssp. pekinensis) Plant Molecular Biology Reporter. 2010;28:597–604. doi: 10.1007/s11105-010-0185-1. [DOI] [Google Scholar]
- 66.Aono M, Kubo A, Saji H, Tanaka K, Kondo N. Enhanced tolerance to photo-oxidative stress of transgenic Nicotiana tabacum with high chloroplastic glutathione reductase activity. Plant & Cell Physiology. 1993;34:129–135. [Google Scholar]
- 67.Han ZH, Shen T, Korcak RF, Baligar VC. Screening for iron efficient species in the genus Malus. Journal of Plant Nutrition. 1994;17:579–592. doi: 10.1080/01904169409364751. [DOI] [Google Scholar]
- 68.Takkar PN, Kaur NP. HCl method for Fe2+ estimation to resolve iron chlorosis in plants. Journal of Plant Nutrition. 1984;7:81–90. doi: 10.1080/01904168409363176. [DOI] [Google Scholar]
- 69.Sun J, et al. NaCl-Induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiology. 2009;149:1141–1153. doi: 10.1104/pp.108.129494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li C, Zhu X, Zhang F. Role of shoot in regulation of iron deficiency responses in cucumber and bean plants. Journal of Plant Nutrition. 2000;23:1809–1818. doi: 10.1080/01904160009382144. [DOI] [Google Scholar]
- 71.Lü PT, et al. RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. The Plant Journal. 2014;78:578–590. doi: 10.1111/tpj.12494. [DOI] [PubMed] [Google Scholar]
- 72.Han Y, et al. Sucrose Nonfermenting1-Related Protein Kinase2.6, an Ortholog of Open Stomata1, is a negative regulator of strawberry fruit development and ripening. Plant Physiology. 2015;167:915–930. doi: 10.1104/pp.114.251314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai F, et al. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiology. 2012;160:2064–2082. doi: 10.1104/pp.112.207720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.