Abstract
Bioavailability of flavonoids is low, especially when occurring as rhamnoglucosides. Thus, the hydrolysis of rutin, hesperidin, naringin and a mixture of narcissin and rutin (from Cyrtosperma johnstonii) by 14 selected probiotics was tested. All strains showed rhamnosidase activity as shown using 4-nitrophenyl α-l-rhamnopyranoside as a substrate. Hesperidin was hydrolysed by 8–27% after 4 and up to 80% after 10 days and narcissin to 14–56% after 4 and 25–97% after 10 days. Rutin was hardly hydrolysed with a conversion rate ranging from 0 to 5% after 10 days. In the presence of narcissin, the hydrolysis of rutin was increased indicating that narcissin acts as an inducer. The rhamnosidase activity as well as the ability to hydrolyse flavonoid rhamnoglucosides was highly strain specific. Naringin was not hydrolysed by rhamnosidase from probiotics, not even by the purified recombinant enzyme, only by fungal rhamnosidase. In conclusion, rhamnosidases from the tested probiotics are substrate specific cleaving hesperidin, narcissin and to a small extent rutin, but not naringin.
Electronic supplementary material
The online version of this article (10.1007/s00449-017-1860-5) contains supplementary material, which is available to authorized users.
Keywords: Rhamnosidase, Hesperidin, Naringin, Rutin, Narcissin, Probiotics
Introduction
As secondary plant metabolites with important contents in human diets [1, 2], flavonoids occur in a wide variety of compounds comprising six subclasses [3, 4]. Flavonols, the most common flavonoids in foods include quercetin, its glycoside rutin (quercetin-3-rutinoside), kaempferol, isorhamnetin and its glycoside narcissin (isorhamnetin-3-rutinoside) [5, 6]. Main sources of these compounds are onions and broccoli, but also red wine and tea. Flavanones with their important representatives naringenin (grapefruits) and hesperetin (oranges), and their glycosides, are also found in other human foods such as tomatoes and aromatic plants [4]. The most familiar flavones in diet are apigenin and luteolin which occur in red pepper [5] and celery [5, 7]; isoflavones are present in soybean-derived products or red clover and show commonalities with estrogens [8] and flavanols occur in apricots, red wine, green tea and chocolate [4].
Interest in flavonoids has been growing because of their biological properties such as anti-inflammatory, antioxidant, antimutagenic, antiproliferative and antiatherogenic effects as shown for quercetin, isorhamnetin, naringenin, hesperetin and its glycosides rutin, narcissin, naringin (naringenin-7-neohesperidoside) and hesperidin (hesperetin-7-rutinoside) [9–12]. Thus, those compounds play a role in amelioration of various diseases or disorders with high prevalence globally including neurodegenerative diseases, osteoporosis, cancer or cardiovascular diseases including atherosclerosis, thrombosis and hypertension.
Most flavonoids are found in glycosylated form, often glycosylated by rutinose (6-O-α-l-rhamnopyranosyl-d-glucose), as rutinosides (e.g. rutin, hesperidin, narcissin) or by neohesperidose (2-O-α-l-rhamnopyranosyl-d-glucose) as neohesperidosides (e.g. naringin). Glycosylation by rutinose or neohesperidose hinders the absorption of the flavonoids in the small intestine [6]. Due to a lack of human intestinal α-l-rhamnosidase, a prior enzymatic hydrolysis increases bioavailability and bioactivity of flavonoid glycosides [13–15]. α-l-Rhamnosidase [EC 3.2.1.40] cleaves terminal α-l-rhamnose from several natural products [16] and is applied for de-bittering of fruit juices [17] or enhancing wine aroma [18]. Beside its biotechnological applications, there is an upcoming interest in α-l-rhamnosidase for enhancing the bioavailability of natural glycosides such as flavonoids rhamnoglucosides by hydrolysing the l-rhamnose [19]. Due to their wide availability, high selectivity, low cost and fast reaction behaviour, the use of enzymes to modify the structure and to improve the physicochemical and biological properties of flavonoids has become an interesting alternative [15]. Enzymes from probiotics have the advantage of safety and possible combined health effects of polyphenols and probiotics [20].
Here, we tested the rhamnosidase activity of 14 selected probiotic strains and their ability to hydrolyse naringin, hesperidin, rutin and the main constituents (rutin and narcissin) of an ethanolic extract of Cyrtosperma johnstonii which exerts antioxidant activity and cytotoxicity towards cancer cells as previously shown [21].
Materials and methods
Chemicals, plant extract and bacterial strains
Acetonitrile was purchased from Promochem (Wesel, Germany), recombinant α-l-rhamnosidase from prokaryotic source (190 U/mg) was obtained in purified form from Megazyme (Wicklow, Ireland), β-glycosidase/α-l-rhamnosidase mixture [Cellulase from (A) niger, 0.45 U/mg] from Sigma–Aldrich (St. Louis, MO, USA), flavonoid glycosides, aglycones and all other chemicals from Sigma–Aldrich [or Merck (Darmstadt, Germany)]. Dried powder of C. johnstonii rhizomes (voucher specimen No 010112, gift of Chiang Mai University, Thailand) was extracted with ethanol for 24 h, filtrated and evaporated to obtain a dried crude extract. The probiotic strains used in this study included Lactobacillus (L.) paracasei ssp. paracasei CRL 431 (ATCC 55544), L. paracasei ssp. paracasei (DN114001), L. paracasei ssp. paracasei DSM 20312 (Shirota), L. rhamnosus GG (ATCC 53103), L. reuteri (ATCC 55730), L. acidophilus LA-5 (DSM 13241), Bifidobacterium (B.) animalis ssp. lactis BB12 (DSM 15954), (B) longum ssp. infantis, L. plantarum (ATCC 15697), L. brevis (ATCC 367), L. delbrueckii ssp. bulgaricus (DSM 20081), Lactococcus (Lc.) lactis ssp. lactis (SR 3.54; NCIMB 30117), L. fermentum, Streptococcus (S.) salivarius ssp. thermophilus (ATCC 19258). Man–Rogosa–Sharpe (MRS) liquid medium was prepared as described previously [22].
Hydrolysis of the flavonoid rhamnoglucosides using rhamnosidase
As a positive control, glycosides (10−4 M) were incubated with a recombinant α-l-rhamnosidase (1 U/mL) or an enzyme mixture from A. niger (5 U/mL β-glucosidase with rhamnosidase side activity) in 100 mM sodium phosphate buffer pH = 6.5 at 37 °C for up to 3 h. The negative control was incubated without enzyme; 4-nitrophenyl α-l-rhamnopyranoside (NPRP) was used as reference substance. Samples were drawn at certain time points, diluted with methanol (1:1) to precipitate the enzymes and clarified using centrifugation at 13,000 rpm and 4 °C for 15 min. Supernatants were analysed using high performance liquid chromatography (HPLC). All experiments were performed in independent triplicates.
Analysis of the hydrolysis of flavonol rutinosides by probiotics
Probiotics were cultivated overnight in MRS medium with glucose under anaerobic conditions at 37 °C, diluted with MRS medium without carbohydrates to an OD600 of 0.2 and further incubated for 48 h. Subsequently, the bacterial suspension was diluted to an OD600 of 0.2 in MRS medium without carbohydrate source, supplemented with 10−4 M NPRP, nitrophenylβ-d-glucopyranoside (NPGP), rutin, naringin, hesperidin or with extract of C. johnstonii (main compounds ~ 10−4 M) and incubated at 37 °C for up to 10 days under anaerobic conditions. Samples of NPRP were drawn after 0, 1, 2, 3, 4, 7 days and samples of the rutinosides after 0, 4, 7 and 10 days. The negative control was incubated without bacteria for 10 days. Samples were diluted with methanol (1:1) and clarified using centrifugation at 13,000 rpm and 4 °C for 15 min. Supernatants were analysed using HPLC. All experiments were performed in triplicates.
Analysis of flavonols and their glycosides by HPLC
The extent of hydrolysis was determined using HPLC with ultraviolet detection (HPLC-UV) on an UltiMate3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, USA) connected to a security guard cartridge followed by a Kinetex C-18 column (5 µm C18, 4.6 × 150 mm, Phenomenex, Torrance, CA, USA). The mobile phase consisted of solvent A (water–acetonitrile 95:5, 0.1% trifluoroacetic acid, TFA) and solvent B (acetonitrile, 0.1% TFA). A linear gradient from 0 to 100% B was applied with a flow rate of 0.5 mL/min: from 0 to 2.5 min B was increased to 15%; from 2.5 to 7.5 min B was increased to 20%, until 13.5 min B was increased to 25%; until 14 min B was increased to 30%; until 15 min to 35%; until 17 min to 50%; until 22 min to 100%; after 2 min at 100%, the column was equilibrated with 100% A. The elution profile was recorded using a photodiode array detector PDA-100 set at the respective absorption maxima of the compounds (305 nm for NPRP, NPGP and nitrophenol; 280 nm for naringenin, hesperetin and their glycosides, and 350 nm for quercetin, isorhamnetin and their glycosides). The respective retention times were 11.8 min for NPRP, 7.7 min for NPGP and 12.8 for nitrophenol; 12.9 for rutin, 13.6 for isoquercitrin and 22.1 for quercetin; 14.6 for naringin and 23.8 for naringenin; 15.8 for hesperidin and 24.3 for hesperetin; 15.7 for isorhamnetinrutinosid, 16.6 for isorhamnetin-3-O-glucoside and 24.6 for isorhamnetin. Flavonoids and their glycosides were quantified using Chromeleon software. The results are shown as mean (from a triplicate) ± standard deviation.
Analysis of rhamnosidase by SDS–PAGE
The rhamnosidase expression was detected in the cell lysates of L. acidophilus as an example and the molecular weight was compared to the recombinant rhamnosidase and the enzyme mixture from A. niger using sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE). Bacterial culture samples were taken at different time points and centrifuged at 12,500 rpm and RT for 10 min. The pellets were re-suspended in diluted cell lysis reagent (CelLytic™ B lysis reagent, Sigma Aldrich) and incubated 10 min at RT under mixing at 350 rpm. Samples were centrifuged (12,500 rpm, 10 min, RT) and the supernatants were diluted with Laemmli sample buffer (BIO-RAD). Samples were incubated at 99 °C for 10 min, mixed at 350 rpm and afterward loaded to the gel (12% Mini-PROTEAN® TGX™ precast protein gels, 15-well, 15 µL, BIO-RAD) and run with electrophoreses buffer (10× Tris/Glycine/SDS, BIO-RAD) at 130 V. As protein marker peqGOLD protein marker IV (10–170 kDa, VWR) was used. Proteins were stained with Coomassie brilliant blue R-250.
Results
Hydrolysis of flavonoid rhamnoglucosides using purified rhamnosidases
The recombinant rhamnosidase (used as positive control) was highly efficient for the hydrolysis of the flavonoid glycosides and NPRP (Table 1). 50% of rutin were hydrolysed into isoquercitrin after 20 min and 100% after 130 min at an incubation temperature of 37 °C. Naringin was not hydrolysed. Hesperidin showed a lower hydrolysis rate than rutin with 50% of conversion to hesperetin-7-O-glucoside after 90 min, whereas a 100% hydrolysis was not reached. No hydrolysis was observed in the negative controls without enzymes for up to 72 h. The hydrolysis of all tested flavonoid glycosides was higher at 50 °C.
Table 1.
Hydrolysis of rhamnose from NPRP, rutin, naringin and hesperidin by rhamnosidases from different sources: time after which 50 or 100% of the glycosides were cleaved, n = 3
| Hydrolysis | Recombinant rhamnosidase | Rhamnosidase from A. niger | ||||||
|---|---|---|---|---|---|---|---|---|
| 37 °C | 50 °C | 37 °C | 50 °C | |||||
| 50% | 100% | 50% | 100% | 50% | 100% | 50% | 100% | |
| NPRP | < 5 min | ~ 20 min | < 5 min | ~ 20 min | ~ 10 h | ~ 48 h | ~ 6 h | ~ 48 h |
| Rutin | 20 min | 130 min | 15 min | 170 min | ~ 35 h | – | ~ 22 h | – |
| Naringin | – | – | – | – | < 6 h | < 6 h | < 6 h | < 6 h |
| Hesperidin | 90 min | – | 70 min | – | ~ 10 h | – | < 6 h | – |
Rhamnosidase from A. niger is capable of hydrolysing also naringin to naringenin-7-O-glucoside. Using this enzyme, results in the most efficient cleavage of hesperidin followed by naringin and rutin (Table 1).
Rhamnosidase and β-glucosidase activity of probiotics
All probiotic strains exerted a significant rhamnosidase activity as indicated using NPRP as a substrate leading to more than 40% hydrolysis for 8 out of 14 strains after 1 day under anaerobic conditions (Table 2; Fig. 1A). In general, the hydrolysis rate of NPRP ranged between 20% (L. reuteri) and 68% [Lactococcus (Lc.) lactis] after 1 day of incubation and reached a total of > 86% hydrolysis after 4 days with exception of L. reuteri. The highest rhamnosidase activity after 1 day was exerted by Lc. lactis followed by L. fermentum and L. brevis, followed by L. rhamnosus GG, B. longum ssp. infantis and L. paracasei ssp. paracasei DSM 20312. The lowest rhamnosidase activity after 1 day was exerted by L. reuteri, followed by L. delbrueckii ssp. bulgaricus, and S. salivarius ssp. thermophilus.
Table 2.
Hydrolysis of rhamnose (bioconversion) from NPRP, rutin, naringin and hesperidin or hydrolysis of glucose from NPGP at certain time points. The table lists the bioconversion in %
| Incubation | Bioconversion (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| NPRP | NPGP | Rutin | Naringin | Hesperidin | ||||
| 1 day | 4 days | 4 h | 10 days | 10 days | 4 days | 7 days | 10 days | |
| B. animalis ssp. lactis BB12 | 48 ± 0 | 88 ± 7 | 96 | 0 | 0 | 23 ± 3 | 52 ± 4 | 69 ± 9 |
| B. longum ssp. infantis | 50 ± 0 | 95 ± 3 | 100 | 0 | 0 | 17 ± 3 | 52 ± 11 | 54 ± 8 |
| L. acidophilus LA-5 | 33 ± 4 | 87 ± 6 | 100 | 3 ± 0 | 0 | 27 ± 1 | 58 ± 3 | 84 ± 7 |
| L. paracasei ssp. paracasei DN114001 | 43 ± 0 | 88 ± 5 | 100 | 0 | 0 | 8 ± 3 | 15 ± 4 | 25 ± 1 |
| L. paracasei ssp. paracasei DSM 20312 | 49 ± 0 | 92 ± 8 | 100 | 0 | 0 | 20 ± 3 | 36 ± 8 | 56 ± 4 |
| L. paracasei ssp. paracasei CRL 431 | 33 ± 9 | 90 ± 6 | 97 | 0 | 0 | 13 ± 2 | 34 ± 4 | 55 ± 11 |
| L. reuteri | 20 ± 6 | 74 ± 15 | 100 | 0 | 0 | 3 ± 1 | 12 ± 7 | 17 ± 7 |
| L. rhamnosus GG | 52 ± 8 | 97 ± 3 | 28 | 0 | 0 | 6 ± 1 | 8 ± 5 | 18 ± 5 |
| L. plantarum | 32 ± 4 | 98 ± 1 | 100 | 5 ± 0 | 0 | 11 ± 6 | 46 ± 6 | 70 ± 8 |
| L. brevis | 63 ± 5 | 100 ± 0 | 100 | 0 | 0 | 25 ± 3 | 53 ± 4 | 80 ± 5 |
| L. delbrueckii ssp. bulgaricus | 25 ± 9 | 98 ± 0 | 100 | 2 ± 0 | 0 | 21 ± 9 | 34 ± 8 | 48 ± 8 |
| Lc. lactis ssp. lactis | 68 ± 0 | 100 ± 0 | 95 | 0 | 0 | 15 ± 2 | 39 ± 5 | 57 ± 1 |
| L. fermentum | 63 ± 2 | 100 ± 0 | 94 | 0 | 0 | 21 ± 1 | 43 ± 1 | 66 ± 5 |
| S. salivarius ssp. thermophilus | 26 ± 3 | 86 ± 6 | 100 | 4 ± 0 | 0 | 18 ± 5 | 43 ± 6 | 55 ± 5 |
This is defined of the percentages of rhamnose cleaved from the glycosides, n = 3
Fig. 1.
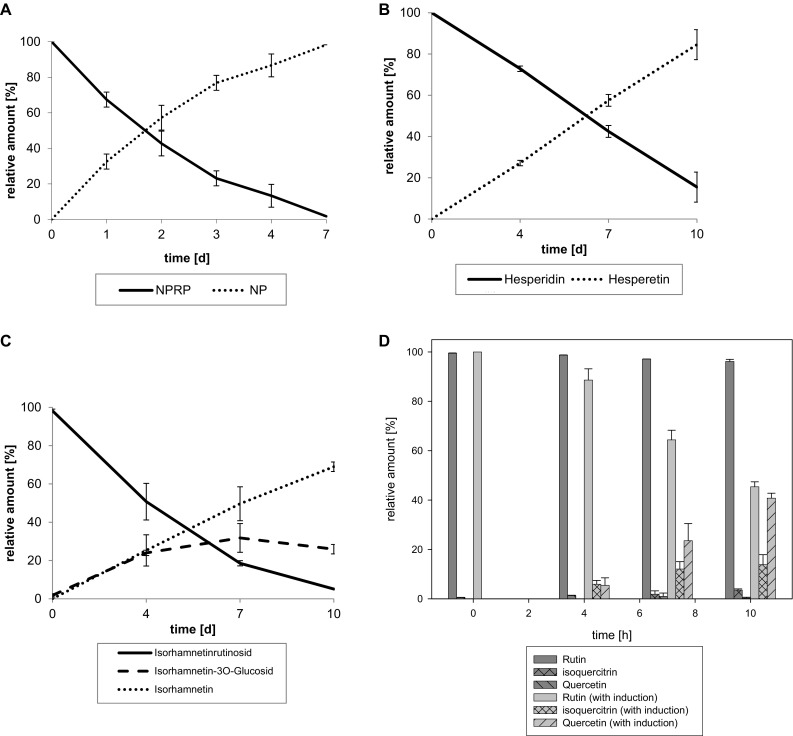
Rhamnosidase activity of L. acidophilus (as example). Deglycosylation of A NPRP, B hesperidin, C narcissin and D rutin without and with induction by narcissin in an extract of C. johnstonii
The β-glucosidase activity is higher than the rhamnosidase activity. In fact, NPGP is converted into the aglycone to an extent of 94–100% already after 4 h (except L. rhamnosus GG) and totally converted after 1 day by all 14 included strains (Table 2; supplements).
Hydrolysis of rutin, hesperidin and naringin by probiotics
The rate of hydrolysis of rhamnose from flavonoid rhamnoglucosides was highly dependent on the aglycon (Table 2). After cleavage of the rhamnose, the glucose is cleaved rapidly.
Rutin was hardly hydrolysed by probiotic rhamnosidases with a conversion (to isoquercitrin, following by hydrolysis to quercetin) ranging from 0 to 2% after 4 days, 0–3% after 7 days and 0–5% after 10 days by four strains, namely L. plantarum, L. delbrueckii ssp. bulgaricus, S. salivarius ssp. thermophilus and L. acidophilus.
The conversion rate of hesperidin was significantly higher ranging from 8% (L. paracasei DN114001) to 27% (L. acidophilus) after 4 days, 9% (L. rhamnosus GG) up to 54% (L. brevis) after 7 days and 25 up to 80% (L. paracasei DN114001) after 10 days (L. brevis) (Table 2). Hesperidin was hydrolyzed most efficiently by L. acidophilus, followed by L. brevis and B. animalis ssp. lactis. The hydrolysis of hesperidin was least efficient by L. reuteri and L. rhamnosus GG. Since the hydrolysis of the glucose after the hydrolysis of the rhamnose is much faster, only the aglycone hesperetin but not the hesperetin-7-O-glucoside can be found (Fig. 1B, supplements).
Naringin was not hydrolysed by any of the used probiotic bacteria.
Cleavage of rhamnoglucosides from C. johnstonii extract by probiotics
The extract of C. johnstonii contains rutin and narcissin as main components. Narcissin is converted significantly faster as rutin or hesperidin. In fact, narcissin is converted by 14% (L. reuteri) to 56% (L. acidophilus) after 4 days, by 15% (L. reuteri) to 87% (L. fermentum) after 7 days and to more than 80% by ten strains after 10 days (Table 3). The glucoside (isorhamnetin-3-glucoside) and aglycone (isorhamnetin) can be detected in parallel. As expected, the relative amount of aglycone to glucoside increases with the duration of incubation (Fig. 1C, supplements).
Table 3.
Hydrolysis of rhamnose (bioconversion) from narcissin and rutin in C. johnstonii extract at certain time points; percentages of cleaved glycosides. n = 3
| Bioconversion (%) | ||||||
|---|---|---|---|---|---|---|
| Narcissin | Rutin | |||||
| 4 days | 7 days | 10 days | 4 days | 7 days | 10 days | |
| B. animalis ssp. lactis | 36 ± 6 | 72 ± 1 | 93 ± 10 | 3 ± 2 | 28 ± 1 | 57 ± 1 |
| B. longum ssp. infantis | 47 ± 1 | 85 ± 1 | 84 ± 13 | 15 ± 1 | 45 ± 2 | 73 ± 3 |
| L. acidophilus | 56 ± 1 | 77 ± 4 | 86 ± 6 | 11 ± 4 | 36 ± 4 | 55 ± 2 |
| L. paracasei DN114001 | 22 ± 2 | 67 ± 2 | 95 ± 1 | 3 ± 1 | 26 ± 2 | 66 ± 1 |
| L. paracasei DSM 20312 | 33 ± 7 | 54 ± 15 | 89 ± 1 | 8 ± 1 | 21 ± 3 | 50 ± 3 |
| L. paracasei CRL431 | 22 ± 1 | 38 ± 3 | 52 ± 4 | 2 ± 2 | 6 ± 2 | 18 ± 5 |
| L. reuteri | 14 ± 9 | 15 ± 7 | 25 ± 14 | 0 | 0 | 5 ± 2 |
| L. rhamnosus GG | 22 ± 8 | 32 ± 4 | 72 ± 3 | 3 ± 2 | 13 ± 5 | 38 ± 11 |
| L. plantarum | 56 ± 9 | 87 ± 5 | 94 ± 3 | 6 ± 0 | 16 ± 1 | 26 ± 2 |
| L. brevis | 40 ± 5 | 79 ± 7 | 88 ± 8 | 10 ± 2 | 38 ± 11 | 64 ± 11 |
| L. delbrueckii ssp. bulgaricus | 38 ± 4 | 47 ± 3 | 81 ± 15 | 4 ± 1 | 20 ± 6 | 32 ± 6 |
| Lc. lactis ssp. lactis | 31 ± 8 | 69 ± 7 | 98 ± 5 | 6 ± 1 | 45 ± 13 | 62 ± 14 |
| L. fermentum | 42 ± 3 | 87 ± 7 | 97 ± 1 | 12 ± 0 | 51 ± 13 | 78 ± 9 |
| S. salivarius ssp. thermophilus | 42 ± 7 | 60 ± 9 | 71 ± 10 | 7 ± 0 | 21 ± 4 | 33 ± 0 |
Interestingly, in the presence of narcissin, the rhamnosidase activity is induced, so that the conversion of rutin is enabled or enhanced (Table 3; Fig. 1D, supplements). In fact, rutin is converted by up to 15% after 4 days (B. infantis), up to 51% after 7 days (L. fermentum) and up to 78% after 10 days (L. fermentum). Rhamnosidase activity was induced most efficiently in L. fermentum, B. longum ssp. infantis and Lc. lactis. However, rhamnosidase was hardly induced in L. reuteri. The hydrolysis products isoquercitrin and quercetin can be found in parallel with increasing amounts of quercetin with longer incubation duration.
Expression of rhamnosidase in probiotics
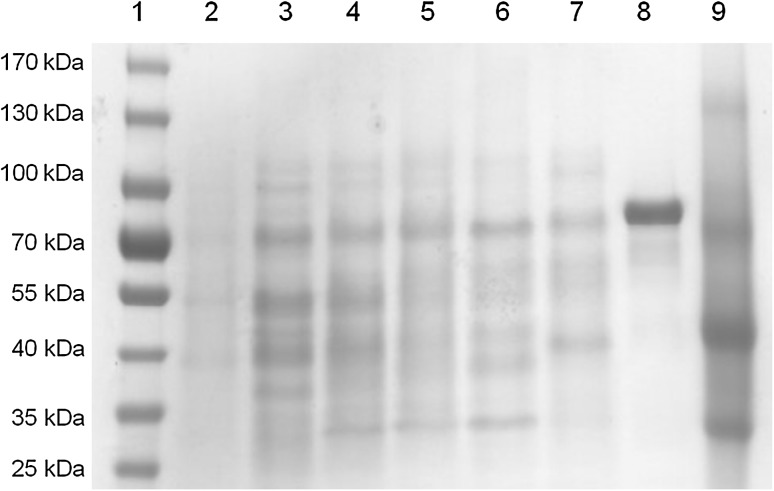
The expression of rhamnosidase is induced by the substrates as shown with NPRP in L. acidophilus (Fig. 2). It could be shown that the amount of rhamnosidase increases with the incubation time. The probiotic rhamnosidase was found to have a molecular weight of 80–90 kDa which is similar to the recombinant rhamnosidase with 90 kDa. The rhamnosidase in the enzyme mixture from A. niger had 80 kDa.
Fig. 2.
SDS-PAGE: (1) marker; (2) lysate of L. acidophilus without induction by NPRP; lysate of L. acidophilus with induction using NPRP after incubation for (3) 10 days, (4) 7 days, (5) 4 days, (6) one day, (7) without incubation; (8) recombinant rhamnosidase (20 mg/mL); (9) enzyme mixture from A. niger (cellulase; 75 mg/mL)
Discussion
Due to their occurrence as glycosides, the bioavailability and bioactivity of polyphenols are limited, especially for rhamnoglucosides which cannot be cleaved by human intestinal enzymes. A few studies reported the release of rhamnose from flavonoid rhamnoglucoside by gut bacteria [23]. However, information on the rhamnosidase activity of probiotics is still limited. Previous studies have shown the occurrence and the properties of rhamnosidases from L. plantarum, L. acidophilus [24, 25] and Pediococcus acidilactici [26]. Another study has shown the hydrolysis of hesperidin by 6 out of 33 bifidobacteria strains within 48 h [6]. The results from our study provide further evidence of rhamnosidase activity in probiotics and of its substrate specificity. Rhamnosidase activity was found in all strains as shown with NPRP as substrate although with some differences of the activity among the strains. However, naringin could not be cleaved by any strain; rutin was cleaved by four strains to an extent below 5%, whereas the hydrolysis of narcissin and hesperidin was more rapid and to a higher extent. The preferred cleavage of hesperidin in contrast to rutin is consistent with the results from Amaretti et al. [6]. The preference for cleaving hesperidin (1–6 bonds) and no activity on naringin (1–2 bonds) conforms to previous studies with Pediococcus acidilactici or L. plantarum [25, 26]. In contrast to these and our results, Bang et al. [27] isolated a bifidobacterium (B. dentium) from human faeces samples with the ability to hydrolyse also rhamnose from the neohesperidose in naringin. Interestingly, this strain also was more effective in hydrolysing (1–6) bonds than (1–2) linkages of rhamnoglycosides.
In case of hesperidin no glucoside was detected, only the aglycone was found so the cleavage may be at least partly caused by rutinosidase. So far only fungal rutinosidase is known, so the contribution of bacterial rutinosidase has to be confirmed in further studies.
When comparing the results of Amaretti et al. [6] and our study, the conclusion can be drawn that lactobacilli are more effective in the cleavage of rhamnose from rutin and hesperidin than bifidobacteria, at least those used by Amaretti et al. [6]. A high rhamnosidase activity correlates well with the ability of cleaving hesperidin as shown for L. brevis, Lc lactis ssp. lactis and L. fermentum. A low rhamnosidase activity mostly correlates with a low hydrolysis rate of hesperidin as seen for L. reuteri. Interestingly, L. rhamnosus GG has a high rhamnosidase activity but low hydrolysis rate of hesperidin, indicating that the substrate specificity of the rhamnosidase enzyme from different probiotics differs. In strains with a high rhamnosidase activity as determined using NPRP, the probability for an induction of the rhamnose expression by narcissin and a following hydrolysis of rutin is increased. This was shown for Lc. lactis, L. fermentum or B. longum ssp. infantis. For L. reuteri, a low rhamnosidase activity (as shown with NPRP) correlates with a very low induction of the rhamnosidase using narcissin and a low following cleavage of rutin. Again, for L. rhamnosus GG no high induction of the rhamnosidase activity using narcissin was found even if the rhamnosidase activity using NPRP was high.
Here, the recombinant α-l-rhamnosidase was active towards hesperidin consistent to [28] and rutin, both of them show α-1,6 linkages to the β-d-glucosides. However, the enzyme was not active towards naringin, in which the L-rhamnose is α-1,2 linked to the β-d-glucoside. Interestingly, naringin is cleaved by rhamnosidase from A. niger. This goes conform with previous studies showing rhamnosidases from fungi to hydrolyse naringin [29]. The molecular weight of the rhamnosidase from A. niger and the probiotics or the recombinant one are different, potentially indicating a distinct glycosylation pattern which may lead to different substrate specificities.
Hydrolysis of the rhamnose from the main compounds (rutin, narcissin) of a C. johnstonii extract may increase bioactivity and bioavailability. The hydrolysis of narcissin by probiotics is efficiently and interestingly induces the activity of probiotic rhamnosidase so that rutin is also cleaved by all strains. Narcissin may be used as inducers for other flavonoid rhamnoglucosides as well. An induction of rhamnosidase activity by glycosides containing l-rhamnosyl moieties was suggested previously and shown for rutin by gut bacteria [25, 30]. In contrast to these previous studies, one easily hydrolysed compound of a plant extract helps to induce the cleavage of another compound in this extract. In such a way, one compound in the extract increases the bioavailability of another compound in this extract having a synergistic effect.
Considering the transit time of food in the gastro-intestinal tract, the velocity of hydrolysis by gut bacteria in the proximal as well as distal colon is relevant for hesperidin and narcissin. A pre-incubation of a flavonol glycoside containing supplement with bacteria may increase the yield of hydrolysis and thus bioavailability tremendously.
Conclusion
14 probiotics hydrolysed hesperidin and narcissin, four probiotics hydrolysed a small amount of rutin, but no strain was able to cleave naringin. The efficiency of hydrolysis and substrate specificity was strain dependent. Recombinant α-l-rhamnosidase cleaved all rhamnoglucosides except naringin which was only cleaved by rhamnosidase from A. niger. One of the main compounds from C. johnstonii extract, narcissin, was not only hydrolysed by probiotic rhamnosidase, but induced rhamnosidase activity for cleavage of rutin. These findings provide further evidence of rhamnosidase activity in probiotics, making them to attractive sources for combined products with plant extracts with increased bioavailability of the active compounds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Open access funding provided by University of Vienna. Thanks to CMU for providing C. johnstonii powder. Parts of this work were supported by short term grant from the ASEAN-European Academic University Network (ASEA-UNINET) and by a grant of the City of Vienna HJST (Hochschuljubiläumsstiftung).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00449-017-1860-5) contains supplementary material, which is available to authorized users.
References
- 1.Middleton E., Jr . Flaonoids in the living system. Berlin: Springer; 1998. Effect of plant flavonoids an immune and inflammatory cell function; pp. 175–182. [DOI] [PubMed] [Google Scholar]
- 2.DuPont MS, Day AJ, Bennett RN, Mellon FA, Kroon PA. Absorption of kaempferol from endive, a source of kaempferol-3-glucuronide, in humans. Eur J Clin Nutr. 2004;58:947–954. doi: 10.1038/sj.ejcn.1601916. [DOI] [PubMed] [Google Scholar]
- 3.Harborne JB, Williams CA. Advances in flavonoid research since 1992. Phytochem. 2000;55:481–504. doi: 10.1016/S0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 4.Manach C, Scalbert A, Morand C, Rémésy C, Jiménez L. Polyphenols: food sources and bioavailability. Am J Clin Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 5.Hertog MGL, Hollman PCH, Venema DP. Optimization of a quantitative HPLC determination of potentially anticarcinogenic flavonoids in vegetables and fruits. J Agric Food Chem. 1992;40:1591–1598. doi: 10.1021/jf00021a023. [DOI] [Google Scholar]
- 6.Amaretti A, Raimondi S, Leonardi A, Quartieri A, Rossi M. Hydrolysis of the rutinose-conjugates flavonoids rutin and hesperidin by the gut microbiota and bifidobacteria. Nutrients. 2015;7:2788–2800. doi: 10.3390/nu7042788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlund I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr Res. 2004;24:851–874. doi: 10.1016/j.nutres.2004.07.005. [DOI] [Google Scholar]
- 8.Manach C, Williamson G, Morand C, Scalbert A, Rémésy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am J Clin Nutr. 2005;81:230S-242S. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
- 9.Mueller M, Hobiger S, Jungbauer A. Anti-inflammatory activity of extracts from fruits, herbs and spices. Food Chem. 2010;122:987–996. doi: 10.1016/j.foodchem.2010.03.041. [DOI] [Google Scholar]
- 10.Russo M, Spagnuolo C, Tedesco I, Bilotto S, Russo GL. The flavonoid quercetin in disease prevention and therapy: facts and fancies. Biochem Pharmacol. 2012;83:6–15. doi: 10.1016/j.bcp.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Parhiz H, Roohbakhsh A, Soltani F, Rezaee R, Iranshahi M. Antioxidant and anti-inflammatory properties of the citrus flavonoids hesperidin and hesperetin: an updated review of their molecular mechanisms and experimental models. Phytother Res. 2015;29:323–331. doi: 10.1002/ptr.5256. [DOI] [PubMed] [Google Scholar]
- 12.Patel K, Singh GK, Patel DK (2012) A review on pharmacological and analytical aspects of naringenin. Chin J Integr Med 1–12 [DOI] [PubMed]
- 13.Nielsen ILF, Chee WSS, Poulsen L, Offord-Cavin E, Rasmussen SE, Frederiksen H, Enslen M, Barron D, Horcajada MN, Williamson G. Bioavailability is improved by enzymatic modification of the citrus flavonoid hesperidin in humans: a randomized, double-blind, crossover trial. J Nutr. 2006;136:404–408. doi: 10.1093/jn/136.2.404. [DOI] [PubMed] [Google Scholar]
- 14.Amaro IM, Rocha J, Vila-Real H, Eduardo-Figueira M, Mota-Filipe H, Sepodes B, Ribeiro MH. Anti-inflammatory activity of naringin and the biosynthesised naringenin by naringinase immobilized in microstructured materials in a model of DSS-induced colitis in mice. Food Res Int. 2009;42:1010–1017. doi: 10.1016/j.foodres.2009.04.016. [DOI] [Google Scholar]
- 15.de Araújo ME, Moreira Franco YE, Alberto TG, Sobreiro MA, Conrado MA, Priolli DG, Frankland Sawaya AC, Ruiz AL, de Carvalho JE, de Oliveira Carvalho P. Enzymatic de-glycosylation of rutin improves its antioxidant and antiproliferative activities. Food Chem. 2013;141:266–273. doi: 10.1016/j.foodchem.2013.02.127. [DOI] [PubMed] [Google Scholar]
- 16.Kaur A, Singh S, Singh RS, Schwarz WH, Puri M. Hydrolysis of citrus peel naringin by recombinant α-l-rhamnosidase from Clostridium stercorarium. J Chem Technol Biotechnol. 2010;85:1419–1422. doi: 10.1002/jctb.2433. [DOI] [Google Scholar]
- 17.Soria F, Ellenrieder G. Thermal inactivation and product inhibition of Aspergillus terreus CECT 2663 α-l-rhamnosidase and their role on hydrolysis of naringin solutions. Biosci Biotechnol Biochem. 2002;66:1442–1449. doi: 10.1271/bbb.66.1442. [DOI] [PubMed] [Google Scholar]
- 18.Spagna G, Barbagallo RN, Martino A, Pifferi PG. A simple method for purifying glycosidases: α-l-rhamnopyranosidase from Aspergillus niger to increase the aroma of Moscato wine. Enz Microbial Technol. 2000;27:522–530. doi: 10.1016/S0141-0229(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 19.Yadav V, Yadav PK, Yadav S, Yadav KDS. α-l-Rhamnosidase: a review. Process Biochem. 2010;45:1226–1235. doi: 10.1016/j.procbio.2010.05.025. [DOI] [Google Scholar]
- 20.Gupta C, Prakash D. Phytonutrients as therapeutic agents. J Complement Integr Med. 2014;11:151–169. doi: 10.1515/jcim-2013-0021. [DOI] [PubMed] [Google Scholar]
- 21.Okonogi S, Khonkarn R, Mankhetkorn S, Unger FM, Viernstein H. Antioxidant activity and cytotoxicity of Cyrtosperma johnstonii extracts on drug sensitive and resistant leukemia and small cell lung carcinoma cells. Pharm Biol. 2013;51:329–338. doi: 10.3109/13880209.2012.729064. [DOI] [PubMed] [Google Scholar]
- 22.Mueller M, Reiner J, Fleischhacker L, Viernstein H, Loeppert R, Praznik W. Prebiotic effect of fructans depending on polymerization degree and structure. J Funct Foods. 2016;24:264–275. doi: 10.1016/j.jff.2016.04.010. [DOI] [Google Scholar]
- 23.Yu K, Jang I, Kang K, Sung C, Kim D. Metabolism of saikosaponin c and naringin by human intestinal bacteria. Arch Pharm Res. 1997;20:420–424. doi: 10.1007/BF02973933. [DOI] [PubMed] [Google Scholar]
- 24.Beekwilder J, Marcozzi D, Vecchi S, de Vos R, Janssen P, Francke C, van Hylckama Vlieg J, Hall R. Characterization of Rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl Environ Microbiol. 2009;75:3447–3454. doi: 10.1128/AEM.02675-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avila M, Jaquet M, Moine D, Requena T, Peláez C, Arigoni F, Jankovic I. Physiological and biochemical characterization of the two alpha-l-rhamnosidases of Lactobacillus plantarum NCC245. Microbiol. 2009;155:2739–2749. doi: 10.1099/mic.0.027789-0. [DOI] [PubMed] [Google Scholar]
- 26.Michlmayr H, Brandes W, Eder R, Schümann C, del Hierro A, Kulbe K. Characterization of two distinct glycosyl hydrolase family 78 alpha-l-rhamnosidases from Pediococcus acidilactici. Appl Environ Microbiol. 2011;77:6524–6530. doi: 10.1128/AEM.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bang S, Hyun Y, Shim J, Hong S, Kim D. Metabolism of rutin and poncirin by human intestinal microbiota and cloning of their metabolizing α-l-rhamnosidase from Bifidobacterium dentium. J Microbiol Biotechnol. 2015;25:18–25. doi: 10.4014/jmb.1404.04060. [DOI] [PubMed] [Google Scholar]
- 28.Zhang R, Zhang BL, Xie T, Li GC, Tuo Y, Xiang YT. Biotransformation of rutin to isoquercitrin using recombinant α-l-rhamnosidase from Bifidobacterium breve. Biotechnol Lett. 2015;37:1257–1264. doi: 10.1007/s10529-015-1792-6. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Lu L, Xiao M. Cell surface engineering of α-l-rhamnosidase for naringin hydrolysis. Bioresour Technol. 2012;123:144–149. doi: 10.1016/j.biortech.2012.05.083. [DOI] [PubMed] [Google Scholar]
- 30.Shin NR, Moon JS, Shin SY, Li L, Lee YB, Kim TJ, Han NS. Isolation and characterization of human intestinal Enterococcus avium EFEL009 converting rutin to quercetin. Lett Appl Microbiol. 2015;62:68–74. doi: 10.1111/lam.12512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.