Abstract
As a prototype cellular sensor, the TRPV1 cation channel undergoes a closed-to-open gating transition in response to various physical and chemical stimuli including noxious heat. Despite recent progress, the molecular mechanism of heat activation of TRPV1 gating remains enigmatic. Toward decrypting the structural basis of TRPV1 heat activation, we performed extensive molecular dynamics simulations (with cumulative simulation time of ∼11 μs) for the wild-type channel and a constitutively active double mutant at different temperatures (30, 60, and 72°C), starting from a high-resolution closed-channel structure of TRPV1 solved by cryo-electron microscopy. In the wild-type simulations, we observed heat-activated conformational changes (e.g., expansion or contraction) in various key domains of TRPV1 (e.g., the S2-S3 and S4-S5 linkers) to prime the channel for gating. These conformational changes involve a number of dynamic hydrogen-bond interactions that were validated with previous mutational studies. Next, our mutant simulations observed channel opening after a series of conformational changes that propagate from the channel periphery to the channel pore via key intermediate domains (including the S2-S3 and S4-S5 linkers). The gating transition is accompanied by a large increase in the protein-water electrostatic interaction energy, which supports the contribution of desolvation of polar/charged residues to the temperature-sensitive TRPV1 gating. Taken together, our molecular dynamics simulations and analyses offered, to our knowledge, new structural, dynamic, and energetic information to guide future mutagenesis and functional studies of the TRPV1 channels and development of TRPV1-targeting drugs.
Introduction
The transient receptor potential (TRP) channels comprise a superfamily of cation channels (1, 2) activated by various physical and chemical stimuli such as heat (3, 4), cold (5, 6, 7), voltage (8), acid (9, 10), force (11, 12, 13), and exogenous ligands (such as capsaicin (4)). The TRP channels make promising drug targets (14, 15) owning to their key roles in various signaling pathways linked to diseases (16, 17). As a prototype TRP channel, TRPV1 forms a homo-tetramer, with each subunit comprised of a six-helix (S1–S6) transmembrane domain (TMD) and an intracellular domain (ICD) (see Fig. 1). Similar to the voltage-gated ion channels (18, 19), the TMD of TRPV1 consists of two structural modules—the S1-S4 module on the channel periphery and the S5-S6 pore module enclosing a central pore (see Fig. 1). Unlike the voltage-gated ion channels, the TRP channels lack a charged S4 helix and their gating features weak voltage dependence (20). The N-terminal portion of ICD forms an ankyrin repeats domain (ARD) (21). The C-terminal domain (CTD) of ICD contains a stretch of highly conserved residues forming a so-called TRP helix (22), which interacts with other proteins and ligands (23, 24). At the TMD-ICD interface is a membrane proximal domain (MPD), which was found to contribute to heat activation in the TRPV subfamily (25). Alternative heat-sensing regions were proposed, including the CTD (26, 27, 28), the outer pore, and the pore domain (29, 30, 31, 32). It remains controversial what specific roles are played by these sites in heat activation (e.g., whether they directly sense heat or indirectly mediate gating events downstream of heat sensing (33)). Additionally, several linker domains contribute to the allosteric couplings underlying the heat/agonist activation of TRPV1, including the MPD linker (residues 400–415), the S2-S3 linker (residues 498–512), and the S4-S5 linker (residues 559–576) (see Fig. 1).
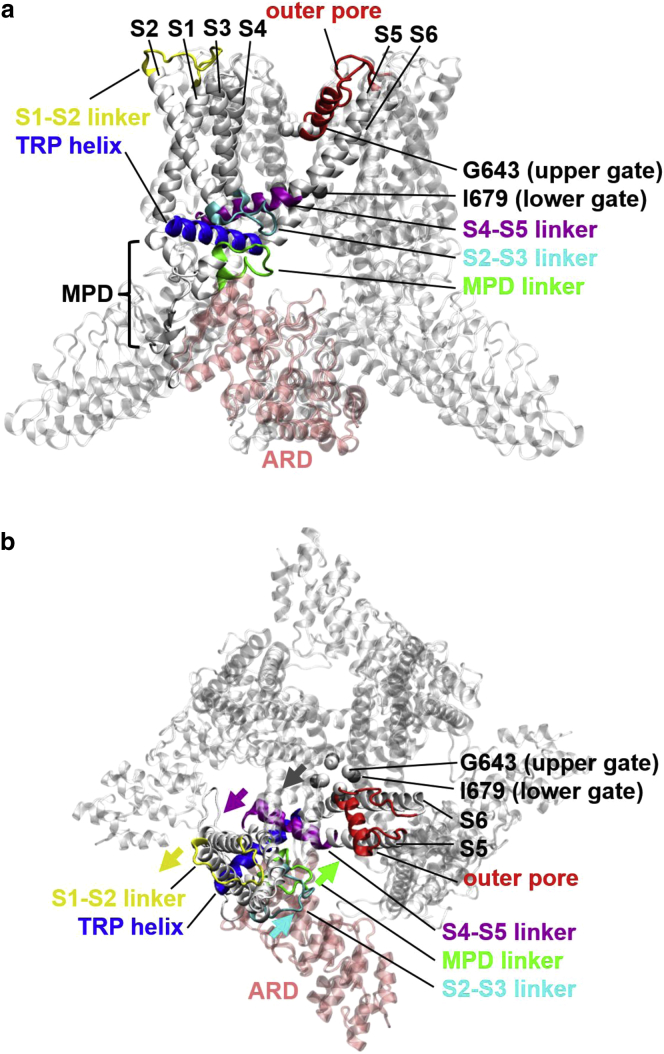
Figure 1.
Structural architecture of TRPV1 tetramer (a) in the side view and (b) in the top view. A representative subunit is colored by domain: the ARD (pink), the MPD linker (green), the S1-S2 linker (yellow), the S2-S3 linker (cyan), the S4-S5 linker (purple), the outer pore (including pore loop and pore helix, red), and the TRP helix (blue). Residues G643 and I679 at the upper and the lower gate are shown as spheres colored in light and dark gray, respectively. In (b), the heat-activated motions of the MPD linker, the S1-S2 linker, the S2-S3 linker, and the S4-S5 linker are marked by arrows colored by domain. To see this figure in color, go online.
High-resolution structures of full-length TRP channels are required to elucidate the TRP-channel activation mechanism. Electron microscopy and x-ray crystallography methods were previously used to solve low-resolution structures of TRP channels (34, 35, 36, 37, 38, 39, 40) and high-resolution structures of their truncated domains (41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55). Recently, the labs of Cheng and Julius solved and later refined the first high-resolution (∼3.3 Å) structures of a minimal functional construct of rat TRPV1 in detergents (56, 57) and in lipid nanodisk (58) by cryo-electron microscopy (cryo-EM), capturing distinct conformations of TRPV1 with a closed or open channel. Two TRPV2 and one TRPV6 structures were later solved at 3–5 Å resolutions (59, 60, 61). These structures revealed a dual-gate channel pore with two constrictions (see Fig. 1)—an upper gate (selectivity filter) near the outer pore (residues G643 and M644) and a lower gate in the lower S6 helix (residue I679). Both gates are closed (open) in the closed (open) structures (56, 57, 58). These detailed structures have paved the way for quantitative structure-driven studies of the TRPV1 gating mechanism. However, because these structures were solved at low temperature under nonphysiological conditions, they did not directly illuminate the heat activation mechanism of TRPV1. Therefore, it remains unclear what is the structural and energetic basis for the large enthalpy change that dictates the high temperature sensitivity of TRPV1 gating (62), although various models were proposed (e.g., solvation/desolvation of nonpolar/polar residues (63, 64), or denaturation of a specific heat-sensing domain (65)).
Molecular dynamics (MD) simulation is the method of choice for investigating protein dynamics and energetics under physiological conditions with atomic details (66). MD has been widely used to simulate various ion channels (67, 68, 69, 70), including truncated domains (12, 71, 72, 73, 74) and homology models (26, 71, 75) of the TRP channels. However, MD simulation is highly expensive, demanding the use of a massively parallelized or special-purpose supercomputer (70) in the past. Thanks to recent developments in computing hardware and software, one can now routinely simulate a large biomolecular system (with ∼105 atoms) at a speed of 1–10 ns/day on a single computer node. However, it remains challenging for MD to access the microsecond-to-millisecond timescales relevant to many biomolecular transitions (including the gating transition in TRPV1). To overcome the timescale limit of MD simulation, coarse-grained modeling (e.g., the elastic network model (76, 77, 78)) has been developed using reduced protein representations (e.g., one bead per amino acid) and simplified force fields (e.g., harmonic potential) (79, 80). The coarse-grained modeling was applied to TRPV1 to explore its key motional modes and gating transition pathway (74), although the energetics of TRPV1 gating transition cannot be accurately analyzed by such modeling.
In a recent study (81), we performed extensive MD simulations (with cumulative simulation time of 3 μs), starting from the high-resolution closed and open structures of TRPV1 solved by cryo-EM (56, 57). In the closed (C)-state simulations at 30°C, we observed a stably closed channel constricted at the lower gate (near residue I679), whereas the upper gate (near residues G643 and M644) is dynamic and undergoes flickery opening/closing. In the open (O)-state simulations at 60°C, we found higher conformational variation consistent with a large entropy increase required for the heat activation, and both the lower and upper gates are dynamic with transient opening/closing. Through ensemble-based structural analyses of the C-state versus the O-state, we revealed pronounced closed-to-open conformational changes involving the MPD linker, the outer pore, and the TRP helix, which are accompanied by breaking/forming of a network of C/O-state specific hydrogen bonds. By comparing the C-state simulations at 30 and 60°C, we observed heat-activated conformational changes in the MPD linker, the outer pore, and the TRP helix that resemble the closed-to-open conformational changes, along with partial formation of the O-state specific hydrogen bonds. However, owing to the shortness of simulation time, we did not directly observe the closed-to-open transition of TRPV1. Following the recent publication of improved TRPV1 structures in nanodisk (with some key domains better resolved, including the S2-S3 linker and the CTD) (58), it is necessary to extend our MD simulations using the new structures and an effective simulation strategy to accelerate the TRPV1 gating transition.
In this study, we have conducted more extensive MD simulations (with cumulative simulation time of ∼11 μs), starting from the new closed structure of WT TRPV1 (58) at different temperatures (30, 60, and 72°C), and then using a constitutively active TRPV1 double mutant. Our goal is twofold: first, elucidate the heat-activated conformational changes in various domains of the WT channel at the residue level of details. Second, directly observe the closed-to-open gating transition in the TRPV1 mutant within limited simulation time (∼200 ns). Encouragingly, the mutant simulations revealed a series of domain motions that lead to the lower-gate opening, which agree well with the heat-activated conformational changes observed in the WT simulations. Our MD simulations also supported the contribution of protein-water electrostatic interaction to the high temperature sensitivity of TRPV1 gating.
Materials and Methods
MD simulation setup
We downloaded preoriented PDB files of the closed structures of TRPV1 (PDB: 5IRZ and 3J5P) from the OPM database (82). We completed PDB: 5IRZ by adding the ARD residues (residues 110–334) of PDB: 3J5P onto PDB: 5IRZ after superimposing along residues 335–384. We introduced F640L/T641S double mutations to PDB: 3J5P using the Mutator module of the VMD program (83). We used the MODLOOP webserver (84) to model the missing S2-S3 linker (residues 503–507) in PDB: 3J5P. We used the Membrane Builder function (85) of the CHARMM-GUI webserver (86) to embed TRPV1 in a bilayer of 1-palmitoyl-2-oleoyl phosphatidylcholine lipids surrounded by a box of water and ions (with a buffer distance of 15 Å). To ensure 0.15 M ionic concentration and zero net charge, 174 K+ and 199 Cl− ions were added. The entire system contains ∼265,000 atoms. Following our previous study (81), to maintain the key intersubunit contacts between each ARD and the MPD of an adjacent subunit, we added intersubunit harmonic restraints between the Cα atoms of residue pairs 376–245 and 375–210 with a spring constant of 500 kJ/mol/nm2. After energy minimization, six steps of equilibration were performed (with gradually reduced harmonic restraints applied to protein, lipids, water, and ions). Finally, we conducted production MD runs in the NPT ensemble. The Nosé-Hoover method (87) was used with temperature T = 30 or 60 or 72°C. The Parrinello-Rahman method (88) was used for pressure coupling. A 10 Å switching distance and a 12 Å cutoff distance were used for nonbonded interactions. The particle-mesh Ewald method (89) was used for electrostatics calculations. The LINCS algorithm (90) was used to constrain the hydrogen-containing bond lengths, which allowed a 2-fs time step for MD simulation. The energy minimization and MD simulation were carried out with the GROMACS program (91, version 5.0.3, using the CHARMM36 force field (92) and TIP3P water model (93).
Root mean square fluctuation analysis of flexibility
To assess the flexibility of TRPV1 at individual residue positions during our MD simulation, we calculated the root mean square fluctuation (RMSF) as follows: first, we saved 1500 snapshots from ten 200-ns MD trajectories (with the first 50 ns of each trajectory discarded) to build the C-state ensemble at a given temperature (named C30, C60, or C72); second, we superimposed the Cα coordinates of residues 400–710 onto the initial structure with a minimal root mean square deviation (RMSD); finally, we calculated the following RMSF at residue position n within the range [400, 710]: , where is the Cα position of residue n in snapshot m, is the average Cα position of residue n, and M is the total number of snapshots in the C-state ensemble. We then calculated the average RMSFn and its SE for four equivalent residue positions n of the TRPV1 tetramer.
Rg analysis of inward/outward domain motions
To probe inward/outward domain motions in TRPV1 at the residue level of detail, we used the measure command of the VMD program (83) to calculate the radius of gyration Rg (n, t, T) based on the Cα coordinates of a set of four equivalent residue positions n at time t and temperature T.
Based on the equilibrium MD simulations of the C-state at T = 30, 60, or 72°C, we averaged Rg (n, t, T) over ten 200-ns MD trajectories with 50 ns ≤ t ≤ 200 ns, and then computed the change of average Rg (n) from T = 30 to 60 or 72°C to quantify the heat-activated expansion/contraction at residue position n.
To monitor the progress of gating transition in the TRPV1 mutant, we averaged Rg (n, t, T) over six 200-ns MD trajectories that exhibited channel opening. Then we calculated the Pearson correlation (PC) between the average Rg (n, t, T) and the average Rg (679, t, T) at the lower gate to assess the temporal correlation between the inward/outward motions at residue position n and the lower gate.
Pairwise distance matrix analysis of interdomain motions
To assess interdomain motions at the residue level of detail, we calculated the pairwise Cα-Cα distance between residues i and j within 20 Å for a MD snapshot at time t and temperature T (denoted D (i, j, t, T)). Based on the equilibrium MD simulations of the C-state at T = 30, 60, or 72°C, we averaged D (i, j, t, T) over ten 200-ns MD trajectories with 50 ns ≤ t ≤ 200 ns, and then computed the change of average D (i, j, t, T) from 30 to 60 or 72°C to quantify the heat-activated opening/closing motion between residues i and j. Here we focused on residue pairs within the same subunit, and averaged the change of average D (i, j, t, T) over four subunits.
Hydrogen bond analysis of dynamic interdomain interactions
We used the following geometric criteria to identify a hydrogen bond (HB) between two polar nonhydrogen atoms (i.e., acceptor and donor): the donor-acceptor distance is <3.5 Å, and the deviation of the donor-hydrogen-acceptor angle from 180° is <60°. The above criteria may also be satisfied by some salt bridges between oppositely charged residues (as identified using the criteria of 4 Å maximal distance between two charged atoms like oxygen or nitrogen). We used the Hbonds plugin of the VMD program (83) to identify and calculate the occupancy of each HB, and summed up the occupancies of all HBs between each pair of residues (see Table 1). For intraprotein HBs, we only considered those HBs that dynamically form/break between key TRPV1 domains with occupancy ≥30%. To count the number of protein-water HBs at residue position n, we summed up occupancies of all HBs between water molecules and four equivalent residues at position n and divided it by 4.
Table 1.
HB-Forming Residue Pairs in the WT and Mutant Simulations
| HB-Forming Residue Pair and Occupancy | |||
|---|---|---|---|
| WT at 30°C | WT at 60°C | WT at 72°C | Mutant at 60°C |
| GLU326 ARG367 0.30a | GLU326 ARG367 0.61a,b | GLU326 ARG367 0.97a,b | LYS318 GLU359 0.47a |
| ARG355 ARG367 0.96 | ARG355 LEU365 0.55b | GLU327 ARG367 0.30a | GLU326 ARG367 1.20a |
| ARG355 LYS368 0.58 | ARG355 ARG367 0.94 | ARG355 LEU365 0.80b | GLU327 ARG367 0.96a |
| GLU356 LYS368 0.76a | ARG355 LYS368 0.61 | ARG355 ARG367 1.00 | ARG355 LEU365 1.01 |
| GLU356 THR370 0.35 | GLU356 LYS368 0.74a | ARG355 LYS368 0.59 | ARG355 ARG367 0.83 |
| ILE357 SER366 0.85 | GLU356 THR370 0.60b | GLU356 LYS368 0.79a | ARG355 LYS368 0.43 |
| GLU397 LYS710 0.80a | ILE357 SER366 0.81 | GLU356 THR370 0.73b | GLU356 LYS368 0.64a |
| GLU405 LYS504 0.37a | GLU397 LYS710 0.78a | ILE357 SER366 0.86 | GLU356 THR370 0.46 |
| ARG409 ASP509 2.86a | GLU405 LYS504 0.32a | GLU397 LYS710 0.82a | ILE357 SER366 0.69 |
| GLN423 ARG701 0.54 | ARG409 ASP509 2.66a | GLU405 LYS504 0.34a | GLU397 ARG500 0.52a |
| LYS425 GLU709 1.09a | GLN423 ARG701 0.64 | ARG409 ASP509 2.65a | GLU397 LYS710 0.56a |
| ARG499 ASP707 1.11a | LYS425 GLU709 1.01a | GLN423 ARG701 0.68 | ARG409 ASP509 1.17a |
| TYR511 GLU570 0.30 | ARG499 ASP707 1.29a | LYS425 GLU709 1.11a | ARG409 GLU692 0.49a |
| PHE559 TRP697 0.58 | SER510 GLU570 0.69b | ARG499 ASP707 1.23a | ASN419 LYS698 0.41 |
| GLN560 ARG701 0.51c | TYR511 GLU570 0.50b | ASP509 LYS571 0.32a | GLN423 ARG701 0.55 |
| ARG575 GLU692 2.02a | SER512 GLU570 0.75b | SER510 GLU570 1.30b | LYS425 GLU709 0.59a |
| ASP576 THR685 0.59c | PHE559 TRP697 0.47 | TYR511 GLU570 0.76b | ARG499 ASP707 0.74a |
| TYR584 THR641 0.82 | GLN560 ARG701 0.30c | SER512 GLU570 1.74b | ASP509 LYS571 0.39a |
| TYR584 TYR666 0.37 | ARG575 GLU692 2.09a | PHE559 TRP697 0.44 | ASP509 ARG575 0.33a |
| LEU637 TYR666 0.68 | ASP576 THR685 0.42c | ARG575 GLU692 2.27a | SER510 GLU570 0.97 |
| THR641 THR670 0.33 | TYR584 THR641 0.77 | TYR584 THR641 0.71 | TYR511 GLU570 0.57 |
| TYR584 TYR666 0.32 | TYR584 TYR666 0.34 | SER512 GLU570 1.24 | |
| LEU637 TYR666 0.59 | LEU637 TYR666 0.54 | PHE559 TRP697 0.37 | |
| THR641 TYR666 0.43b | THR641 TYR666 0.51b | ARG575 GLU684 0.34a | |
| THR641 THR670 0.41 | ARG575 GLU692 1.28a | ||
| TYR584 SER641 0.80 | |||
| LEU637 TYR666 0.44 | |||
| SER641 TYR666 0.35 | |||
The O-state-specific HB-forming residue pairs are underlined (see Table 1 of (81)). For each pair of residues, the sum of occupancies of all HBs formed between them is listed.
These residue pairs also form salt bridges.
These HB-forming residue pairs are heat-activated.
These HB-forming residue pairs are heat-deactivated.
Energetic analysis of nonbonded interaction energy
We used the NAMD Energy plugin of the VMD program (83) to calculate nonbonded energy (including van der Waals (vdW) and electrostatic energy) in the TRPV1-water-membrane system. A 10 Å switching distance and a 12 Å cutoff distance were used for the nonbonded interactions. The CHARMM36 force field was used for the nonbonded parameters.
Results and Discussion
Equilibrium MD simulations show temperature-dependent dynamics in TRPV1
To explore the temperature-dependent conformational dynamics of TRPV1 in the C-state, we conducted three sets of ten 200-ns MD simulations based on the new closed-channel structure (PDB: 5IRZ) at 30, 60, and 72°C (named C30, C60, and C72), in the presence of a lipid bilayer and a box of water and ions (see Materials and Methods). For comparison, we also included ten 200-ns MD simulations based on the old closed-channel structure (PDB: 3J5P) at 30 and 60 (named c30 and c60) that were taken from our previous study (81). Ten MD trajectories of each system were combined to form a structural ensemble of the C-state at a specific temperature (excluding the beginning 50 ns when the initial structure undergoes relaxation and equilibration; see Fig. S1). As in our previous study (81), we did not observe channel opening at 60 and 72°C, which could be attributed to limited simulation time. However, these simulations are still useful for elucidating the heat-activated conformational dynamics of TRPV1 in the C-state, which may hint for early structural and dynamic events toward the gating transition (see below).
To assess the conformational stability of TRPV1 in MD simulations, we calculated the RMSD of Cα atoms in the core TRPV1 structure (residues 400–710, which encompass the MPD linker, the TMD, and the TRP helix) for each MD trajectory. Overall, we observed relatively low RMSD in the C-state (average RMSD is 1.9 Å at 30°C, 2.1 Å at 60°C, and 2.4 Å at 72°C; see Fig. S1), which slightly increases with temperature. These RMSD values are comparable to previous MD simulations of the voltage-gated channels (3–4 Å) (94, 95, 96) and the old TRPV1 structure (2.3–2.5 Å) (81), supporting the stability and quality of our simulations.
To further assess the conformational flexibility at individual residue positions, we calculated the RMSF for three C-state ensembles (see Materials and Methods). The RMSF profiles exhibit several pronounced peaks (see Fig. S2 a) in the MPD linker (at residue S404), the S1-S2 linker (at residue N467), the S2-S3 linker (at residue K504), the outer pore (near residues K603 and T650), and the C terminus of the TRP helix (at residue K710). Most peaks become more pronounced as the temperature increases, hinting for heat sensitivity in these flexible regions. To further quantify the temperature-dependence of RMSF, we calculated the fractional change in RMSF from 30 to 60/72°C (denoted ΔRMSF/RMSF, see Fig. S2 b). For control, a temperature increase from 30 to 60–72°C would increase RMSF by 5–7% due to an unspecific thermal effect. Therefore, we deem an RMSF difference to be significant if ΔRMSF/RMSF ≫ 7%. Using the above criterion, we found significant RMSF increases in the MPD linker and pre-S1 helix, the S1-S2 linker, the S2-S3 linker, the outer pore, the lower S6 helix, and the TRP helix (see Fig. S2 b), which were previously found to increase significantly from the C-state to the O-state (81). Previous studies supported the functional significance of the above flexible regions, including the MPD linker (25), the S2-S3 linker (97), the outer pore (9, 29, 30, 31, 32, 98), and the TRP helix (99). Our results further suggest their specific role in dynamically sensing temperature change.
Channel pore analysis indicates a more dynamic upper gate than the lower gate
Following our previous study (81), we analyzed the channel pore dynamics by using the HOLE program (100) to calculate the pore radius for snapshots of each C-state ensemble. We plotted the pore radius at the upper and lower gate (denoted Rug and Rlg, which are defined as the minimal pore radius of the upper and lower half of the channel pore, respectively; see Fig. S3).
The pore radius (recorded as mean ± SD) at the lower gate is 0.72 ± 0.14 Å at 30°C, 0.81 ± 0.22 Å at 60°C, and 0.80 ± 0.17 Å at 72°C, with 0–10% of snapshots being water permeable (i.e., with pore radius > 1.15 Å). This suggests the lower gate is largely closed in the C-state at all three temperatures (see Fig. S3 a), which agrees with our previous study (81). At the upper gate, the pore radius is 0.86 ± 0.25 Å at 30°C, 0.80 ± 0.27 Å at 60°C, and 1.06 ± 0.33 Å at 72°C, with 7–28% of snapshots being water permeable. This finding suggests that the upper gate is more dynamic than the lower gate, particularly at 72°C (with larger SDs in pore radius; see Fig. S3, a and b), which agrees with our previous study (81). Given that the lower gate impedes water permeation more than the upper gate, we infer the lower gate is likely the activation gate of TRPV1 allosterically controlled by various stimuli including heat.
Rg analysis reveals heat-activated expansion/contraction of TRPV1 domains
By comparing the various cryo-EM structures (56, 58) and MD-averaged structures of TRPV1 in the C/O state (81), we have observed extensive conformational changes involving various key domains (e.g., inward motions of the outer pore, the MPD linkers, and the TRP helices; see Fig. S4 and Fig. 1 of (81)), which could drive the channel activation. However, because the open-channel structures of TRPV1 were obtained in complex with agonist/toxin under the cryogenic condition (56, 58), it is uncertain if any of these cryo-EM-observed conformational changes are relevant to the heat activation of TRPV1. Therefore, it is critical to directly observe heat-activated conformational changes in TRPV1 upon temperature increase under physiological conditions. To this end, we have analyzed conformational changes from 30 to 60/72°C as observed by our MD simulations. For simplicity, we focused on those symmetric inward/outward domain motions potentially relevant to gating (e.g., channel pore contraction/expansion). We calculated Rg at every residue position of the core TRPV1 structure (see Materials and Methods). Although insensitive to some motional modes (e.g., perpendicular to the membrane or in the tangential direction parallel to the membrane), the Rg analysis offers a simple and intuitive way to probe those symmetric contraction/expansion motions that could effectively couple the channel periphery to the channel pore at the residue-level of detail. We focused our Rg analysis on the following key domains of TRPV1 (see Fig. 2).
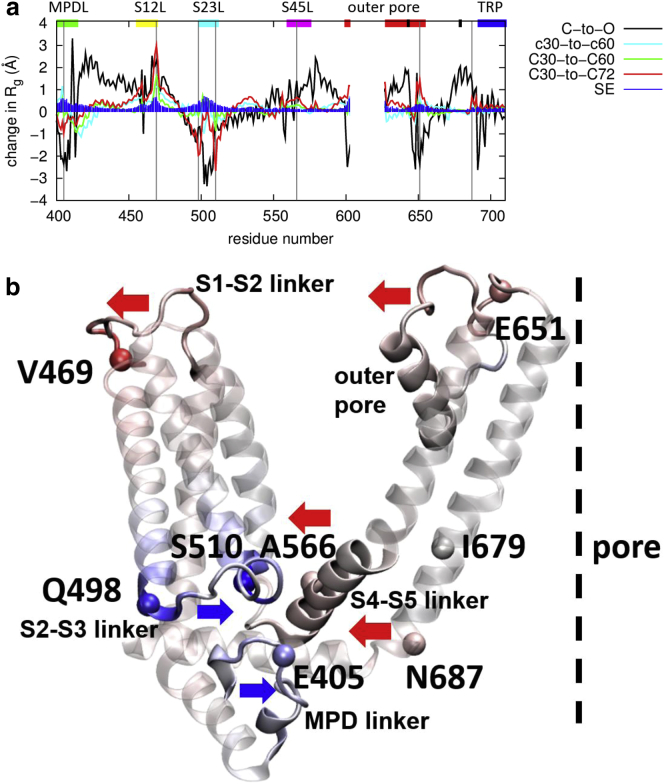
Figure 2.
The Rg analysis of heat-activated conformational changes in TRPV1. (a) Shown here is the temperature-dependent change in position-specific Rg (cyan, c30-to-c60; green, C30-to-C60; red, C30-to-C72; blue, SEs of Rg change due to averaging over MD trajectories), in comparison with the Rg change from the closed structure to the open structure of TRPV1 (C-to-O, colored in black). Marker positions (E405, V469, Q498, S510, A566, E651, and N687) for key domains are marked by vertical lines. The residue positions corresponding to the MPD linker, the S1-S2 linker (S12L), the S2-S3 linker (S23L), the S4-S5 linker (S45L), the outer pore, the TRP helix, and the upper/lower gate are marked by horizontal bars colored in green, yellow, cyan, purple, red, blue, and black, respectively. (b) Shown here is a TRPV1 subunit colored by the C30-to-C72 Rg change (with positive/negative change corresponding to red/blue). Outward/inward motions are indicated by arrows colored in red/blue. Marker positions for key domains are shown as spheres and labeled. To see this figure in color, go online.
In the MPD linker, we observed a heat-activated inward motion in the C-terminal region at 60°C, while the entire MPD linker moves inward at 72°C (with a negative peak at E405; see Fig. 2, a and b). The contraction of MPD linker was also apparent between the closed and open structures of TRPV1 (see Fig. 2 a; Fig. S4), suggesting its involvement in TRPV1 gating as noted in our previous study (81).
In the S1-S2 linker, we observed a larger heat-activated expansion at 72°C than at 60°C (with a sharp peak at V469; see Fig. 2, a and b). The expansion of S1-S2 linker was also visible between the closed and open structures of TRPV1 (see Fig. 2 a; Fig. S4), whereas the S1-S4 module changes little (56).
In the S2-S3 linker, we observed a more pronounced heat-activated contraction at 72°C than at 60°C near the N-/C-termini (with two negative peaks at Q498 and S510; see Fig. 2, a and b). The inward motion of the S2-S3 linker (fully resolved in the new TRPV1 structures (58)) was also observed between the closed and open structures of TRPV1 (see Fig. 2 a; Fig. S4).
In the S4-S5 linker, we observed a larger heat-activated expansion at 72°C than at 60°C (with a broad peak at A566; see Fig. 2, a and b), which is diminished toward the C-terminus. Such expansion was also detected between the closed and open structures of TRPV1 (see Fig. 2 a; Fig. S4), supporting its putative role in TRPV1 gating (56, 58).
In the outer pore, we observed a larger heat-activated expansion at 72°C than at 60°C (with a sharp peak at E651; see Fig. 2, a and b). However, such expansion does not propagate to the nearby upper-gate residue G643, which remains closed. This expansion is in contrast to the observed contraction of the outer pore between the closed and open structures of TRPV1 (see Fig. 2 a; Fig. S4), which may be attributed to toxin binding in the open structures (56, 58) but not in our MD simulations.
In the S6 helix, we found no heat-activated change in Rg near the lower-gate residue I679, which is in contrast to the observed S6 expansion between the closed and open structures of TRPV1 (see Fig. 2 a; Fig. S4). Nevertheless, we did observe a significant peak at N687 near the junction between S6 and the TRP helix (see Fig. 2, a and b), suggesting heat-activated dilation in the pore just below the lower gate.
In the TRP helix, we observed little heat-activated change in Rg (see Fig. 2 a). This is in contrast to the observed rotation and inward motion of TRP helix between the closed and open structures near the N-terminus (see Fig. 2 a; Fig. S4). Therefore, our finding does not support a functional role of TRP helix in pulling S6 outward upon heat activation.
In sum, our MD simulations have observed significant heat-activated expansion/contraction in a number of key domains that exceed the equilibrium fluctuations in Rg (see Fig. 2 a). These domain motions could allosterically couple the channel periphery to the channel pore (see Figs. 1 b and 2 b) via intermediate domains (e.g., S2-S3 linker and S4-S5 linker). Reassuringly, these heat-activated motions qualitatively resemble some of the observed conformational changes between the closed and open structures (see Fig. 2 a), supporting their relevance to channel gating. Notably, the largest heat-activated expansion and contraction was found in the S1-S2 linker and the S2-S3 linker, respectively. Therefore, we postulate that these heat-activated domain motions, especially in the S1-S2 linker and the S2-S3 linker, represent early structural events that prime the channel for subsequent gate opening. There are also some discrepancies between the MD-observed heat-activated changes and the cryo-EM-observed closed-to-open changes in some regions (e.g., the pre-S1, S1, S5, outer pore, S6, and TRP helix; see Fig. 2 a), which could be attributed to slow heat-activated changes not observed by our MD simulations, or differences between heat- and agonist-activated changes in TRPV1.
Analysis of the pairwise distance matrix uncovers heat-activated interdomain motions
To complement the Rg analysis of inward/outward domain motions, we further probed the relative motions (opening or closing) between the various TRPV1 domains upon heat activation. To this end, we calculated from MD simulations the temperature-dependent changes in the pairwise distance matrix (see Materials and Methods) from 30 to 60/72°C. We focused our discussion on the following key domains of TRPV1 (see Fig. 3).
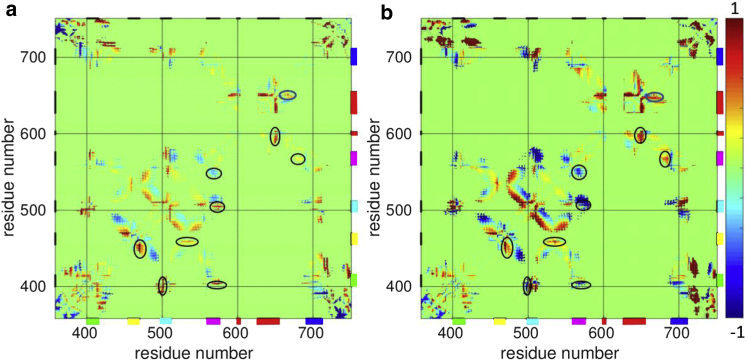
Figure 3.
The heat-activated changes in pairwise distance matrix of TRPV1. (a) C30-to-C60; (b) C30-to-C72. Distance change clamped at ±1 Å is colored using the blue-green-red colormap. Selected regions of the distance matrix discussed in the main text are circled. The residue positions corresponding to the MPD linker, the S1-S2 linker, the S2-S3 linker, the S4-S5 linker, the outer pore, and the TRP helix are marked by horizontal/vertical bars colored in green, yellow, cyan, purple, red, and blue, respectively. Only pairwise distance changes that exceed the SEs (due to averaging over MD trajectories) are colored in blue/red. To see this figure in color, go online.
The MPD linker moves away from and closer to the N-terminus of S2-S3 linker at 60 and 72°C, respectively (see Fig. 3, a and b). Similarly, the N-terminal region of the MPD linker moves away from and closer to the S4-S5 linker at 60 and 72°C, respectively (see Fig. 3, a and b). These findings are consistent with the observation of partial and full heat-activated contraction of the MPD linker at 60 and 72°C, respectively (see Fig. 2 a).
At 60 and 72°C, the C-terminus and N-terminus of S1-S2 linker move away from S1 and S3/S4, respectively (see Fig. 3, a and b), which is consistent with its heat-activated outward expansion (see Fig. 2 a).
The S2-S3 linker moves away from and closer to the S4-S5 linker at 60 and 72°C, respectively (see Fig. 3, a and b). This is consistent with the observation of larger heat-activated contraction of the S2-S3 linker at 72°C than at 60°C (see Fig. 2 a).
The S4-S5 linker moves away from the lower S6 helix and closer to the S4 helix, which is more pronounced at 72°C than at 60°C (see Fig. 3, a and b). This is consistent with its heat-activated expansion revealed by the Rg analysis (see Fig. 2 a).
The N-terminus and C-terminus of the outer pore move apart at 60 and 72°C (see Fig. 3, a and b), which is consistent with its heat-activated expansion (see Fig. 2 a). Additionally, the upper-gate region (near G643) moves away from the upper S6 helix at 60 and 72°C (see Fig. 3, a and b).
In sum, we observed heat-activated interdomain motions (see Fig. 3), which are consistent with the heat-activated domain expansions/contractions (see Fig. 2). Together, these motions suggest the following heat activation pathway in TRPV1: upon temperature increase, the MPD linker and S2-S3 linker move inward to interact with the S4-S5 linker, causing the latter to move outward and away from the pore-forming S6 helix, which subsequently expands for gating. Additional expansions in the S1-S2 linker and the outer pore may further contribute to gating. This putative pathway overlaps with a previously proposed vanilloid-activated pathway involving the S2-S3 linker (97) and the S4-S5 linker (56), which is consistent with the notion that vanilloid and heat activation of TRPV1 can cross-fertilize each other.
Dynamic hydrogen bonds form and break to enable heat-activated conformational changes
In our previous study (81), we identified hydrogen bonds (HB) specifically formed in the C/O-state ensembles as critical to the gating transition of TRPV1. However, it is uncertain if these O-state-specific HBs are relevant to the heat-activated O-state. In this study, to probe dynamic interactions that enable the heat-activated conformational changes in TRPV1, we have analyzed HBs (with occupancy >0.3; see Table 1) specifically formed in three C-state ensembles at 30, 60, and 72°C. Some of those HB-forming residue pairs are oppositely charged and can form salt bridges that could further enhance these key interactions (see Table 1). Following our previous study (81), we focused on those intrasubunit HBs that couple between key domains of TRPV1 (including the ARD, the MPD, the S2-S3 linker, the S4-S5 linker, the S5 helix, the outer pore, the S6 helix, and the TRP helix; see Fig. 1).
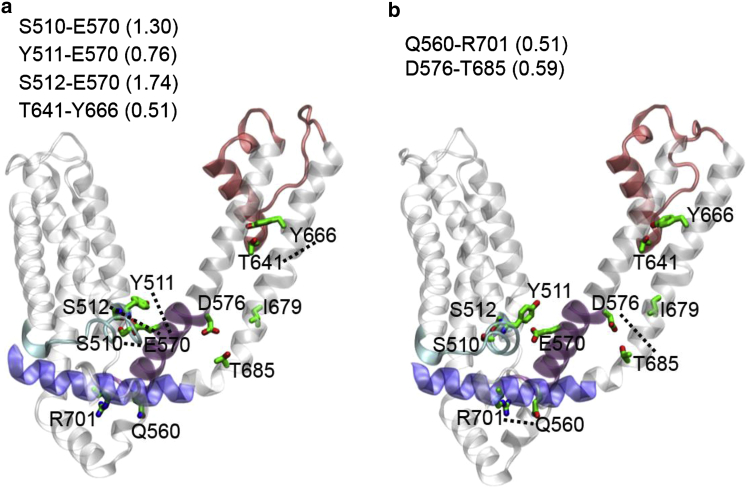
We found heat-activated HBs (i.e., with higher occupancy at 60/72°C than at 30°C; see Fig. 4; Table 1) between the following residue pairs: E326-R367, R355-L365, E356-T370, S510-E570, Y511-E570, S512-E570, and T641-Y666. Three of them couple the S2-S3 linker (S510, Y511, and S512) to the S4-S5 linker (E570), which is consistent with the observed heat-activated motions between these linkers (see Figs. 2 and 3). The above three HBs also had higher occupancy in the c60 and O-state ensembles than in the c30 ensemble in our previous study (81).
Figure 4.
Temperature-dependent HB-forming residues in a representative snapshot of a TRPV1 subunit at (a) 72°C and (b) 30°C. The HB-forming residues are labeled, connected by dotted lines, and colored by atom type (carbon, green; nitrogen, blue; oxygen, red). Key domains are colored as follows: S2-S3 linker (cyan), S4-S5 linker (purple), outer pore (red), and TRP helix (blue). The S2-S3 linker interacts more closely with the S4-S5 linker in (a) than in (b). Key HB-forming residue pairs are shown along with their occupancies (see also Table 1). To see this figure in color, go online.
We found two heat-deactivated HBs (i.e., with higher occupancy at 30°C than at 60/72°C; see Fig. 4; Table 1) between the following residue pairs: Q560-R701 and D576-T685. These HBs serve to anchor the N-/C-termini of S4-S5 linker (Q560 and D576) on the TRP helix (R701) and the lower S6 helix (T685) in the C state, so their weakening could allow the heat-activated outward motion of the S4-S5 linker relative to the TRP helix and the S6 helix (see Figs. 2 and 3).
For validation, many of the above HB-forming residues were shown to be functionally important according to previous mutational studies. The Q560H/R mutations caused gain of function to TRPV1 and weak toxicity when expressed in Saccharomyces cerevisiae (101), which is consistent with its predicted role in stabilizing the C-state. Mutations in E570 and D576 compromised heat activation by greatly reducing the temperature coefficients Q10 (102). The T641S mutant displayed large constitutive channel activation and caused toxicity when expressed in S. cerevisiae (101). The Y666A mutation resulted in a nonfunctional channel (71). The R701A mutation resulted in strongly reduced PIP2-dependent activation (26), suggesting that this residue may bind with PIP2 to destabilize the R701-Q560 interaction. The mutations Y511A and S512Y resulted in wild-type heat response (97), which is consistent with our finding that the observed HBs between Y511/S512 and E570 involve their backbone amide nitrogen (see Fig. 4). Other mutations that change the charge of Y511, S512, or E570 will be desirable to test the functional importance of the above interactions.
By coupling between functionally critical residues, the above dynamic HBs break and form to facilitate heat-activated motions of the S2-S3 linker and the S4-S5 linker, thereby mediating the heat activation pathway in TRPV1. Additionally, we also found some high-occupancy HBs that are temperature-insensitive (including R409-D509, K425-E709, R499-D707, and R575-E692; see Table 1), and they may serve to stabilize an allosteric network for transmitting activation signals. Besides the above intrasubunit HBs, other intraprotein, protein-water, or protein-membrane interactions may also be involved in the heat activation of TRPV1 gating (see below).
MD simulation of fast gating transition in a TRPV1 double mutant
To test the prediction that the above-observed heat-activated conformational changes (see Fig. 2) actually occur during TRPV1 gating, we must directly observe the gating transition of TRPV1 within limited simulation time (e.g., hundreds of nanoseconds). To this end, we conducted MD simulations of a F640L/T641S double mutant of TRPV1 (see Materials and Methods). The F640L mutant of TRPV1 is known to be constitutively active and hypersensitive to thermal stimulus while preserving an intact gating machinery (101). Similarly, the T641S mutant also exhibited constitutive channel activation (101). These mutations likely shift the thermodynamic equilibrium from the C-state toward the O-state without altering the gating transition mechanism (101). Indeed, in another ion channel (i.e., ligand-gated acetylcholine receptor), gain-of-function mutations were successfully introduced to directly investigate the slow unliganded gating transition, which was found to occur by the same reaction mechanism as the fast diliganded gating of the WT channel (103). Therefore, we expect the double mutant of TRPV1 to undergo a similar gating transition more readily and rapidly than the WT TRPV1, making it feasible for MD simulation to directly probe this gating conformational transition. Here we assume the above two mutations favor channel opening with a similar mechanism and their effects are additive. Indeed, we tried MD simulation for the single mutant F640L and did not observe channel opening, suggesting that a single mutation is not enough to drive channel opening within the time frame of 200-ns MD simulation.
We conducted 27 replicates of 200-ns MD simulations of the double mutant at 60°C. Encouragingly, we observed channel opening at the lower gate in six mutant simulations, with the pore radius increasing to ∼2 Å at the lower gate (see Fig. S3 c), which is comparable to the O-state MD simulations (81). We then plotted a cumulative density map of K+ ions within the channel pore as sampled by these mutant simulations (see Fig. 5 a). Reassuringly, the entire channel pore is accessible by K+ ions without any gap, which is consistent with a conducting channel. Additionally, we observed water and ion permeation events in our mutant simulation (see Movies S1 and S2). In a representative snapshot of the open-channel mutant, we observed asymmetric outward motions of I679 and the S4-S5 linkers accompanied by inward motions of the MPD linkers and the S2-S3 linkers relative to the initial closed structure (see Fig. 5 b). These gating motions agree well with the heat-activated expansions/contractions of these domains observed in the WT MD simulations (see Fig. 2). This agreement strongly supports the relevance of the heat-activated domain motions to the gating transition. As revealed by the analysis of interdomain HBs formed in the mutant simulations, most of the O-state-specific HBs identified previously (81) were partially formed in the mutant (see Table 1), suggesting the mutant indeed undergoes a gating transition toward the O-state ensemble of the WT channel.
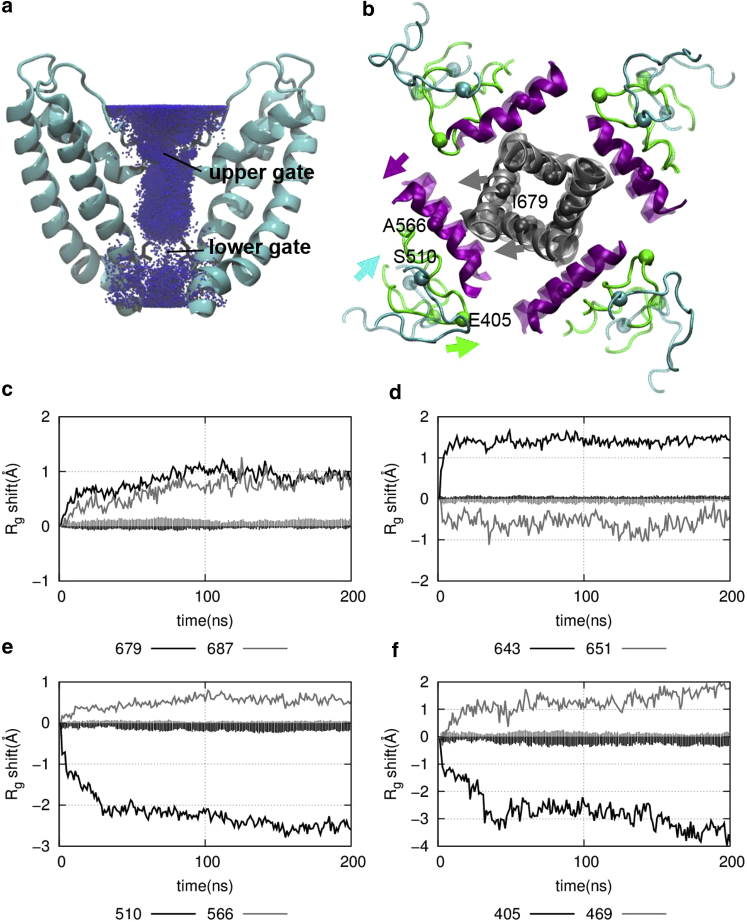
Figure 5.
The results of TRPV1 mutant simulations. (a) Shown here is a cumulative density map of K+ ions (blue dots) based on six mutant MD trajectories that exhibited channel opening. (b) Shown here is an open-channel conformation (opaque) superimposed on the closed structure of TRPV1 (transparent), where key domains are colored as follows: MPD linkers (green), S2-S3 linkers (cyan), S4-S5 linkers (purple), and S6 helices (gray). Marker residues of these domains (E405, S510, and A566) and the lower-gate residue I679 are shown as spheres and labeled. Motions of these domains are indicated with arrows colored by domain. (c–f) Shown here is the time-dependent shift of Rg during the mutant MD simulations at eight marker residue positions (E405 of the MPD linker, V469 of the S1-S2 linker, S510 of the S2-S3 linker, A566 of the S4-S5 linker, I679 and N687 of the S6 helix, G643 and E651 of the outer pore). The vertical impulses indicate the SEs of Rg shift due to averaging over MD trajectories. To see this figure in color, go online.
To decrypt a sequence of domain motions leading to channel opening, we temporally correlate the expansion/contraction motions of various domains as observed during the mutant simulations. To this end, we plotted Rg(t) (averaged over the six mutant trajectories at the same time t) at selected residue positions in the MPD linker, S1-S2 linker, S2-S3 linker, S4-S5 linker, outer pore, lower S6 helix, and lower gate (see Fig. 5, c–f).
The lower-gate residues I679, along with nearby residues N687, undergo a gradual opening from 0 to 100 ns, suggesting that the heat-activated expansion at N687 (see Fig. 2) is likely coupled to the lower-gate opening in the mutant simulations. The upper-gate residues G643 undergo a rapid opening accompanied by an equally rapid contraction at residues E651 of the outer pore. This agrees with the outer-pore contraction observed between the closed and open structures of TRPV1 (see Fig. S4), but differs from the heat-activated expansion of the outer pore observed in the WT simulations (see Fig. 2). Notably, the upper-gate Rg at G643 increases more than the lower-gate Rg at I679 (see Fig. 5, c and d), suggesting a more open and accessible upper gate than lower gate (see Fig. 5 a).
Outside the channel pore, residues A566 of the S4-S5 linkers undergo a gradual opening from 0 to 100 ns in accord with the lower-gate opening, supporting a strong coupling between the S4-S5 linker and the lower S6 helix. In contrast, residues S510 of the S2-S3 linkers undergo fast contraction from 0 to 30 ns, which is consistent with our proposal that an early inward motion of the S2-S3 linker subsequently causes the S4-S5 linker to move outward.
At the TMD periphery, residues E405 of the MPD linkers undergo a fast contraction from 0 to 30 ns, which is similar to the S2-S3 linkers. Similarly, residues V469 of the S1-S2 linkers undergo a fast expansion from 0 to 30 ns.
To quantitatively compare the correlations between the above domain motions and the lower-gate opening, we calculated the PC values between Rg (n, t) and Rg (679, t) (n = 405, 510, 469, 566, and 687; see Materials and Methods). As expected, residues E405 (PC = −0.57) and S510 (PC = −0.72) correlate negatively with the lower-gate opening, whereas residues V469 (PC = 0.59), A566 (PC = 0.81), and N687 (PC = 0.78) correlate positively with the lower-gate opening. By ordering the PC values from low to high (assuming higher PC implies smaller temporal distance to the I679 opening event), we can infer the following signaling pathway: E405 and V469 → S510 → A566 and N687 → I679. This is consistent with our proposed heat-activation pathway from the MPD linkers to the lower gate via the S2-S3 and S4-S5 linkers.
To further probe the energetic basis of the TRPV1 gating transition featuring high temperature sensitivity (62), we calculated gating-associated changes in nonbonded energy in the protein-membrane-water system based on the six mutant simulations that observed channel opening (see Materials and Methods; Table S1). We focused on those nonbonded energy terms that increase upon the gating transition, which may contribute to the large enthalpy increase (∼100 kcal/mol) in TRPV1 gating (62). Notably, we observed a large increase in the protein-water electrostatic interaction energy (5978 ± 509 kcal/mol), which hints for significant desolvation of polar/charged residues during the gating transition. We found a similar large increase in the protein-water electrostatic interaction energy from 30 to 60/72°C in the WT simulations (see Table S1), supporting its role in heat-activated TRPV1 gating. The net contribution of such a large electrostatic energy to thermodynamics should be vastly reduced due to solvent screening (roughly by a factor of ε = 80 to ∼75 kcal/mol). To identify those residues that undergo large desolvation, we counted protein-water HBs, which decrease by 111 ± 24 after the gating transition. Most of these lost HBs involve polar/charged residues of key domains, including the MPD and the S4-S5 linker (see Table S2). We observed a similar loss of protein-water HBs involving these domains from 30 to 60/72°C in the WT simulations (see Table S2). Additionally, we found an increase in the intraprotein vdW energy (157 ± 64 kcal/mol), which is mainly contributed by the ICD (150 ± 38 kcal/mol) in agreement with our previous finding of 35 kcal/mol increase in the vdW energy of an isolated ICD (74). A positive change in the intraprotein vdW energy was also observed from 30 to 60/72°C in the WT simulations (see Table S1). In contrast to the above two energy terms, all the other nonbonded energy terms (e.g., protein-water vdW energy, protein-membrane electrostatic/vdW energy, and intraprotein electrostatic energy; see Table S1) decrease after the gating transition, so they do not make positive contributions to the observed enthalpy change. Furthermore, we analyzed temperature-dependent changes in protein-membrane and protein-water nonbonded interactions involving several key TRPV1 domains (see Table S3). We did not observe robust/significant temperature-dependent changes (for both C30-to-C60 and C30-to-C72) in protein-membrane interactions (except for the electrostatic energy involving the S2-S3 linker; see Table S3). However, we did observe significant C30-to-C60 and C30-to-C72 increase in protein-water electrostatic energy involving the MPD, S2-S3 linker, S4-S5 linker, and TRP helix (see Table S3). This finding suggests that the heat activation of TRPV1 depends more on the protein-water interactions than the protein-membrane interactions.
In sum, our mutant simulations observed gating transitions involving two sets of temporally distinct domain motions: 1) fast (∼30 ns) inward motions of the MPD linkers and the S2-S3 linkers, and outward motion of the S1-S2 linkers; and 2) slow (∼100 ns) outward motion of the S4-S5 linkers and the lower S6 helices. Meanwhile, the upper gate opens rapidly along with the outer-pore contraction, which may be facilitated by the gain-of-function mutations in the outer pore. Additionally, our energetic analysis revealed a large increase in the protein-water electrostatic interaction energy, along with an increase in the intraprotein vdW energy, which may contribute to the large enthalpy change responsible for the high temperature sensitivity of TRPV1 gating.
Conclusions
Based on the improved cryo-EM structure of TRPV1 in the C-state (58), we have conducted extensive MD simulations (total ∼11 μs) at 30, 60, and 72°C. Besides confirming the findings of a previous MD study of the older TRPV1 structures in the C/O states (56, 57) (e.g., temperature-sensitive domain flexibility, distinct upper/lower-gate dynamics, etc.; see (81)), the present MD simulations focused on elucidating the heat-activated conformational changes in terms of expansion/contraction of key domains (see Fig. 2) and interdomain motions (see Fig. 3) at the residue-level of detail. Although these WT simulations did not observe channel opening, they did reveal extensive heat-activated domain motions that couple the channel periphery to the channel pore via intermediate domains (e.g., the MPD linker, the S2-S3 linker, and the S4-S5 linker; see Fig. 2), which may prime the channel for subsequent pore opening. These heat-activated domain motions only partially resemble the observed conformational changes from the closed structures to the agonist/toxin-bound open structures (with marked differences in the S2-S3 linker, the outer pore, and the TRP helix; see Fig. 2 a), thus supporting the importance of using MD simulations to probe the heat activation of TRPV1 under physiological conditions. The predicted heat-activated domain motions are accompanied by dynamic HBs forming/breaking between key residues on the S2-S3 linker and the S4-S5 linker (see Fig. 4), which were validated against past mutational studies and offered promising targets for future experiments.
To directly observe the TRPV1 gating transition by MD simulation, we exploited a gain-of-function F640L/T641S double mutant expected to open faster than the WT channel (101). Indeed, our mutant simulations observed asymmetric channel opening involving fast and slow domain motions that resemble the heat-activated domain motions observed in the WT simulations (see Fig. 2), hinting for a heat-evoked pathway propagating from the MPD linkers and the S2-S3 linkers to the S4-S5 linkers and then the lower gate. Notably, the gating transition results in a large increase in the protein-water electrostatic interaction energy, which may contribute to a large enthalpy increase associated with TRPV1 gating (62), and support the proposal that desolvation of polar/charged residues accounts for the temperature-sensitive TRPV1 gating. We have identified a small set of polar/charged residues critically involved in such desolvation (see Table S2), which will be tested in future mutational and functional studies. The gating transition also led to an increase in the intraprotein vdW energy as found previously (74). Future studies will test if both of these energy contributions are important to the gating thermodynamics of TRPV1.
Whereas our previous study (81) emphasized the importance of the MPD linkers and the TRP helices in the C-to-O gating transition, this study highlighted the key roles of the MPD linkers, the S2-S3 linkers, and the S4-S5 linkers in heat activation, with additional contributions from the S1-S2 linkers and the outer pore. Future longer simulations of the gating transition in the WT channel, although highly challenging, will be needed to ultimately validate the structural, dynamic, and energetic changes observed in the mutant simulations.
Author Contributions
W.Z. designed research. W.Z. and H.W. performed research. W.Z. and H.W. analyzed data. W.Z. wrote the manuscript.
Acknowledgments
Computational support was provided by the Center for Computational Research at the University at Buffalo and the Biowulf system at the National Institutes of Health.
We appreciate funding support from the American Heart Association (14GRNT18980033 and 17GRNT33690009).
Editor: Bert de Groot.
Footnotes
Four figures, three tables, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31161-X.
Supporting Material
The water molecule is colored by atom type: red for oxygen and white for hydrogen.
The K+ ion is shown as a blue sphere.
References
- 1.Clapham D.E. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 2.Voets T., Talavera K., Nilius B. Sensing with TRP channels. Nat. Chem. Biol. 2005;1:85–92. doi: 10.1038/nchembio0705-85. [DOI] [PubMed] [Google Scholar]
- 3.Caterina M.J., Rosen T.A., Julius D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature. 1999;398:436–441. doi: 10.1038/18906. [DOI] [PubMed] [Google Scholar]
- 4.Caterina M.J., Schumacher M.A., Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 5.Story G.M., Peier A.M., Patapoutian A. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 2003;112:819–829. doi: 10.1016/s0092-8674(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 6.Nilius B., Appendino G., Owsianik G. The transient receptor potential channel TRPA1: from gene to pathophysiology. Pflugers Arch. 2012;464:425–458. doi: 10.1007/s00424-012-1158-z. [DOI] [PubMed] [Google Scholar]
- 7.Karashima Y., Talavera K., Voets T. TRPA1 acts as a cold sensor in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2009;106:1273–1278. doi: 10.1073/pnas.0808487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voets T., Droogmans G., Nilius B. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature. 2004;430:748–754. doi: 10.1038/nature02732. [DOI] [PubMed] [Google Scholar]
- 9.Jordt S.E., Tominaga M., Julius D. Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc. Natl. Acad. Sci. USA. 2000;97:8134–8139. doi: 10.1073/pnas.100129497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tominaga M., Caterina M.J., Julius D. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 1998;21:531–543. doi: 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 11.Howard J., Bechstedt S. Hypothesis: a helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors. Curr. Biol. 2004;14:R224–R226. doi: 10.1016/j.cub.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 12.Sotomayor M., Corey D.P., Schulten K. In search of the hair-cell gating spring elastic properties of ankyrin and cadherin repeats. Structure. 2005;13:669–682. doi: 10.1016/j.str.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Yin J., Kuebler W.M. Mechanotransduction by TRP channels: general concepts and specific role in the vasculature. Cell Biochem. Biophys. 2010;56:1–18. doi: 10.1007/s12013-009-9067-2. [DOI] [PubMed] [Google Scholar]
- 14.Gunthorpe M.J., Szallasi A. Peripheral TRPV1 receptors as targets for drug development: new molecules and mechanisms. Curr. Pharm. Des. 2008;14:32–41. doi: 10.2174/138161208783330754. [DOI] [PubMed] [Google Scholar]
- 15.Nilius B. Transient receptor potential TRP channels as therapeutic drug targets: next round! Curr. Top. Med. Chem. 2013;13:244–246. doi: 10.2174/1568026611313030002. [DOI] [PubMed] [Google Scholar]
- 16.Nilius B., Voets T., Peters J. TRP channels in disease. Sci. STKE. 2005;2005:re8. doi: 10.1126/stke.2952005re8. [DOI] [PubMed] [Google Scholar]
- 17.Nilius B. TRP channels in disease. Biochim. Biophys. Acta. 2007;1772:805–812. doi: 10.1016/j.bbadis.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Catterall W.A. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catterall W.A. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J. Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baez D., Raddatz N., Latorre R. Gating of thermally activated channels. Curr. Top. Membr. 2014;74:51–87. doi: 10.1016/B978-0-12-800181-3.00003-8. [DOI] [PubMed] [Google Scholar]
- 21.Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Mol. Biosyst. 2008;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montell C. Physiology, phylogeny, and functions of the TRP superfamily of cation channels. Sci. STKE. 2001;2001:re1. doi: 10.1126/stke.2001.90.re1. [DOI] [PubMed] [Google Scholar]
- 23.Numazaki M., Tominaga T., Tominaga M. Structural determinant of TRPV1 desensitization interacts with calmodulin. Proc. Natl. Acad. Sci. USA. 2003;100:8002–8006. doi: 10.1073/pnas.1337252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott E.D., Julius D. A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science. 2003;300:1284–1288. doi: 10.1126/science.1083646. [DOI] [PubMed] [Google Scholar]
- 25.Yao J., Liu B., Qin F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. USA. 2011;108:11109–11114. doi: 10.1073/pnas.1105196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brauchi S., Orta G., Latorre R. Dissection of the components for PIP2 activation and thermosensation in TRP channels. Proc. Natl. Acad. Sci. USA. 2007;104:10246–10251. doi: 10.1073/pnas.0703420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brauchi S., Orta G., Latorre R. A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J. Neurosci. 2006;26:4835–4840. doi: 10.1523/JNEUROSCI.5080-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlachová V., Teisinger J., Vyklický L. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J. Neurosci. 2003;23:1340–1350. doi: 10.1523/JNEUROSCI.23-04-01340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Y., Yang F., Zheng J. Selective disruption of high sensitivity heat activation but not capsaicin activation of TRPV1 channels by pore turret mutations. J. Gen. Physiol. 2012;139:273–283. doi: 10.1085/jgp.201110724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F., Cui Y., Zheng J. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc. Natl. Acad. Sci. USA. 2010;107:7083–7088. doi: 10.1073/pnas.1000357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S.E., Patapoutian A., Grandl J. Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS One. 2013;8:e59593. doi: 10.1371/journal.pone.0059593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandl J., Kim S.E., Patapoutian A. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 2010;13:708–714. doi: 10.1038/nn.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latorre R., Brauchi S., Vargas G. ThermoTRP channels as modular proteins with allosteric gating. Cell Calcium. 2007;42:427–438. doi: 10.1016/j.ceca.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Cvetkov T.L., Huynh K.W., Moiseenkova-Bell V.Y. Molecular architecture and subunit organization of TRPA1 ion channel revealed by electron microscopy. J. Biol. Chem. 2011;286:38168–38176. doi: 10.1074/jbc.M111.288993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huynh K.W., Cohen M.R., Moiseenkova-Bell V.Y. Structural insight into the assembly of TRPV channels. Structure. 2014;22:260–268. doi: 10.1016/j.str.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruyama Y., Ogura T., Sato C. Three-dimensional reconstruction using transmission electron microscopy reveals a swollen, bell-shaped structure of transient receptor potential melastatin type 2 cation channel. J. Biol. Chem. 2007;282:36961–36970. doi: 10.1074/jbc.M705694200. [DOI] [PubMed] [Google Scholar]
- 37.Mio K., Ogura T., Sato C. The non-selective cation-permeable channel TRPC3 is a tetrahedron with a cap on the large cytoplasmic end. Biochem. Biophys. Res. Commun. 2005;333:768–777. doi: 10.1016/j.bbrc.2005.05.181. [DOI] [PubMed] [Google Scholar]
- 38.Mio K., Ogura T., Sato C. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. J. Mol. Biol. 2007;367:373–383. doi: 10.1016/j.jmb.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 39.Moiseenkova-Bell V.Y., Stanciu L.A., Wensel T.G. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shigematsu H., Sokabe T., Nagayama K. A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. J. Biol. Chem. 2010;285:11210–11218. doi: 10.1074/jbc.M109.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inada H., Procko E., Gaudet R. Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry. 2012;51:6195–6206. doi: 10.1021/bi300279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin X., Touhey J., Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J. Biol. Chem. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]
- 43.Lishko P.V., Procko E., Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 44.McCleverty C.J., Koesema E., Kreusch A. Crystal structure of the human TRPV2 channel ankyrin repeat domain. Protein Sci. 2006;15:2201–2206. doi: 10.1110/ps.062357206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phelps C.B., Huang R.J., Gaudet R. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 2008;47:2476–2484. doi: 10.1021/bi702109w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi D.J., Ye S., Wang K. Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function. Protein Cell. 2013;4:942–950. doi: 10.1007/s13238-013-3091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landouré G., Zdebik A.A., Sumner C.J. Mutations in TRPV4 cause Charcot-Marie-Tooth disease type 2C. Nat. Genet. 2010;42:170–174. doi: 10.1038/ng.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ihara M., Hamamoto S., Yamashita A. Molecular bases of multimodal regulation of a fungal transient receptor potential (TRP) channel. J. Biol. Chem. 2013;288:15303–15317. doi: 10.1074/jbc.M112.434795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi H., Matsushita M., Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol. Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- 50.Fujiwara Y., Minor D.L., Jr. X-ray crystal structure of a TRPM assembly domain reveals an antiparallel four-stranded coiled-coil. J. Mol. Biol. 2008;383:854–870. doi: 10.1016/j.jmb.2008.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y., Ulbrich M.H., Yang J. Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc. Natl. Acad. Sci. USA. 2009;106:11558–11563. doi: 10.1073/pnas.0903684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molland K.L., Paul L.N., Yernool D.A. Crystal structure and characterization of coiled-coil domain of the transient receptor potential channel PKD2L1. Biochim. Biophys. Acta. 2012;1824:413–421. doi: 10.1016/j.bbapap.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Yu Y., Ulbrich M.H., Yang J. Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex. Nat. Commun. 2012;3:1252. doi: 10.1038/ncomms2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petri E.T., Celic A., Hodsdon M.E. Structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity. Proc. Natl. Acad. Sci. USA. 2010;107:9176–9181. doi: 10.1073/pnas.0912295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schumann F., Hoffmeister H., Kalbitzer H.R. Ca2+-dependent conformational changes in a C-terminal cytosolic domain of polycystin-2. J. Biol. Chem. 2009;284:24372–24383. doi: 10.1074/jbc.M109.025635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao E., Liao M., Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao M., Cao E., Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao Y., Cao E., Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huynh K.W., Cohen M.R., Moiseenkova-Bell V.Y. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zubcevic L., Herzik M.A., Jr., Lee S.Y. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saotome K., Singh A.K., Sobolevsky A.I. Crystal structure of the epithelial calcium channel TRPV6. Nature. 2016;534:506–511. doi: 10.1038/nature17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao J., Liu B., Qin F. Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys. J. 2010;99:1743–1753. doi: 10.1016/j.bpj.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clapham D.E., Miller C. A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. USA. 2011;108:19492–19497. doi: 10.1073/pnas.1117485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chowdhury S., Jarecki B.W., Chanda B. A molecular framework for temperature-dependent gating of ion channels. Cell. 2014;158:1148–1158. doi: 10.1016/j.cell.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin F. Demystifying thermal channels: driving a channel both forwards and backwards with a single gear? Biophys. J. 2013;104:2118–2120. doi: 10.1016/j.bpj.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 67.Nury H., Poitevin F., Baaden M. One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc. Natl. Acad. Sci. USA. 2010;107:6275–6280. doi: 10.1073/pnas.1001832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nury H., van Renterghem C., Corringer P.J. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 69.Zhu F., Hummer G. Pore opening and closing of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. USA. 2010;107:19814–19819. doi: 10.1073/pnas.1009313107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen M.O., Jogini V., Shaw D.E. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 71.Susankova K., Ettrich R., Vlachova V. Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1) J. Neurosci. 2007;27:7578–7585. doi: 10.1523/JNEUROSCI.1956-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darré L., Furini S., Domene C. Permeation and dynamics of an open-activated TRPV1 channel. J. Mol. Biol. 2015;427:537–549. doi: 10.1016/j.jmb.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 73.Hanson S.M., Newstead S., Sansom M.S. Capsaicin interaction with TRPV1 channels in a lipid bilayer: molecular dynamics simulation. Biophys. J. 2015;108:1425–1434. doi: 10.1016/j.bpj.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng W., Qin F. A combined coarse-grained and all-atom simulation of TRPV1 channel gating and heat activation. J. Gen. Physiol. 2015;145:443–456. doi: 10.1085/jgp.201411335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedretti A., Labozzetta A., Vistoli G. Exploring the activation mechanism of TRPM8 channel by targeted MD simulations. Biochem. Biophys. Res. Commun. 2011;414:14–19. doi: 10.1016/j.bbrc.2011.08.134. [DOI] [PubMed] [Google Scholar]
- 76.Atilgan A.R., Durell S.R., Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tama F., Sanejouand Y.H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 78.Zheng W., Doniach S. A comparative study of motor-protein motions by using a simple elastic-network model. Proc. Natl. Acad. Sci. USA. 2003;100:13253–13258. doi: 10.1073/pnas.2235686100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tozzini V. Coarse-grained models for proteins. Curr. Opin. Struct. Biol. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 80.Tozzini V. Minimalist models for proteins: a comparative analysis. Q. Rev. Biophys. 2010;43:333–371. doi: 10.1017/S0033583510000132. [DOI] [PubMed] [Google Scholar]
- 81.Wen H., Qin F., Zheng W. Toward elucidating the heat activation mechanism of the TRPV1 channel gating by molecular dynamics simulation. Proteins. 2016;84:1938–1949. doi: 10.1002/prot.25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 83.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 84.Fiser A., Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 85.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 87.Martyna G.J., Hughes A., Tuckerman M.E. Molecular dynamics algorithms for path integrals at constant pressure. J. Chem. Phys. 1999;110:3275–3290. [Google Scholar]
- 88.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 89.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 90.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 91.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 94.Shrivastava I.H., Sansom M.S. Simulations of ion permeation through a potassium channel: molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 2000;78:557–570. doi: 10.1016/S0006-3495(00)76616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kulleperuma K., Smith S.M., Pomès R. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 2013;141:445–465. doi: 10.1085/jgp.201210856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jogini V., Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys. J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jordt S.E., Julius D. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell. 2002;108:421–430. doi: 10.1016/s0092-8674(02)00637-2. [DOI] [PubMed] [Google Scholar]
- 98.Ryu S., Liu B., Qin F. Uncoupling proton activation of vanilloid receptor TRPV1. J. Neurosci. 2007;27:12797–12807. doi: 10.1523/JNEUROSCI.2324-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.García-Sanz N., Valente P., Ferrer-Montiel A. A role of the transient receptor potential domain of vanilloid receptor I in channel gating. J. Neurosci. 2007;27:11641–11650. doi: 10.1523/JNEUROSCI.2457-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smart O.S., Neduvelil J.G., Sansom M.S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. 376. [DOI] [PubMed] [Google Scholar]
- 101.Myers B.R., Bohlen C.J., Julius D. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 2008;58:362–373. doi: 10.1016/j.neuron.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boukalova S., Marsakova L., Vlachova V. Conserved residues within the putative S4-S5 region serve distinct functions among thermosensitive vanilloid transient receptor potential (TRPV) channels. J. Biol. Chem. 2010;285:41455–41462. doi: 10.1074/jbc.M110.145466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Purohit P., Auerbach A. Unliganded gating of acetylcholine receptor channels. Proc. Natl. Acad. Sci. USA. 2009;106:115–120. doi: 10.1073/pnas.0809272106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The water molecule is colored by atom type: red for oxygen and white for hydrogen.
The K+ ion is shown as a blue sphere.