Abstract
The stimulator-of-interferon-genes (STING) protein is involved in innate immunity. It has recently been shown that modulation of STING can lead to an aggressive antitumor response. DMXAA is an antitumor agent that had shown great promise in murine models but failed in human clinical trials. The molecular target of DMXAA was subsequently shown to be murine STING (mSTING); however, human STING (hSTING) is insensitive to DMXAA. Molecular dynamics simulations were employed to investigate the differences between hSTING and mSTING that could influence DMXAA binding. An initial set of simulations was performed to investigate a single lid region mutation G230I in hSTING (corresponding residue in mSTING is an Ile), which rendered the protein sensitive to DMXAA. The simulations found that an Ile side chain was enough to form a steric barrier that prevents exit of DMXAA, whereas in WT hSTING, the Gly residue that lacks a side chain formed a porous lid region that allowed DMXAA to exit. A second set of molecular dynamics simulations compared the tendency of STING to be in an open-inactive conformation or a closed-active conformation. The results show that hSTING prefers to be in an open-inactive conformation even with cGAMP, the native ligand, bound. On the other hand, mSTING prefers a closed-active conformation even without a ligand bound. These results highlight the challenges in translating a mouse active STING compound into a human active compound, while also providing avenues to pursue for designing a small-molecule drug targeting human STING.
Introduction
The innate immune system provides the initial line of defense against infectious pathogens. Recent studies have shown that the stimulator-of-interferon-genes (STING) protein plays a central role in this response by mediating type I interferon (INF-α and INF-β) production through both NF-κ-B and IRF3 transcription pathways in response to intracellular dsDNA, intracellular pathogens, and mitochondrial damage (1, 2, 3, 4, 5, 6). STING acts as a direct sensor of cyclic dinucleotides (CDNs) such as cyclic GMP-AMP (cGAMP) (1), and STING variants have evolved to distinguish noncanonical CDNs produced by mammalian cGAS from conventional (3′-5′) CDNs produced primarily by bacteria (7). Additionally, the innate immune system plays a role in both pro- and antitumor immunity (8, 9). It has been demonstrated that intratumoral administration of STING agonists in the tumor microenvironment triggers an antitumor immune T cell response in multiple mouse tumor models (10, 11). As such, STING has recently emerged as an exciting target for both immunological conditions (i.e., STING inhibition) and oncology (i.e., STING activation).
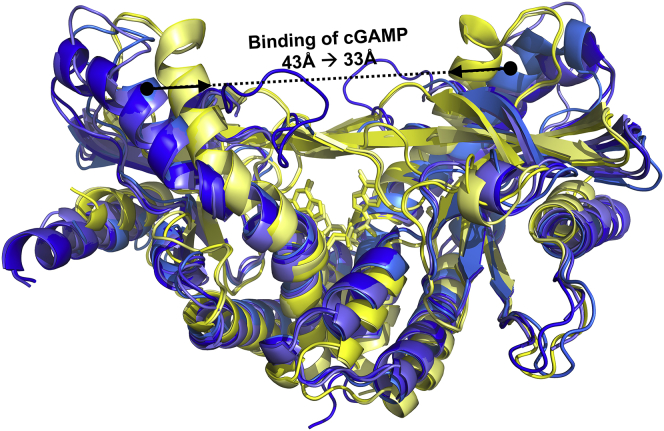
STING is a transmembrane protein consisting of an N-terminal transmembrane region, a C-terminal domain that includes the dimerization domain, and as a carboxy-terminal tail. STING exists as a homodimer in cells. Crystal structures of the C-terminal domain reveal that the symmetrical STING dimer resembles a butterfly with the ligand binding site for cGAMP located deep in the cleft at the dimer interface of the two protomers. Apo hSTING structures show an open conformation (Fig. 1). Upon cGAMP binding, the two protomers shift slightly relative to each other and clamp down onto the ligand binding domain, whereas a β-sheet lid region forms over the binding site, thus forming a closed conformation.
Figure 1.
Comparison of apo- and cGAMP-bound hSTING crystal structures. In hSTING, the configuration of the protomers clamp down, closing the protein upon cGAMP binding (overlay of open apo structures shown in blue, PDB: 4F5W, 4EF5, 4EMU, and 4F9E; closed cGAMP-bound structures are shown in yellow, PDB: 4LOH and 4KSY). In addition to the closing of the hSTING protein, cGAMP binding also results in the formation of a β-sheet lid region over the active site.
The vascular disrupting agent Vadimezan, 5,6-dimethylxanthenone-4-acetic acid (DMXAA), was identified as a potential cancer therapeutic and in combination with paclitaxel and carboplatin was evaluated in phase II clinical trials against non-small-cell lung cancer (12). However, DMXAA failed in human phase III trials (12). It was later discovered that DMXAA is a direct activator of murine STING (mSTING) signaling but not human STING (hSTING) (13, 14, 15). As mSTING and hSTING have high sequence identity (68% amino acid identity and 81% similarity) and identical residues within the DMXAA binding pocket (shared with cGAMP), the reason for this difference in species sensitivity to DMXAA was not immediately evident.
Although DMXAA is not active against wild-type (WT) hSTING, a single point mutation in the lid region, G230I (Fig. 2), has been identified that renders DMXAA partially active against hSTING (16). Interestingly, the corresponding lid region residue in mSTING is an Ile. As reported by Gao et al. (16), IFN-β induction activity of DMXAA in hSTING G230I is threefold less than that of mSTING, whereas WT hSTING has no interferon induction. They additionally report that in isothermal titration calorimetry (ITC) experiments, the KD of DMXAA binding to mSTING to be 0.49 μM, whereas there is no detectable binding in WT hSTING. A KD for DMXAA binding to hSTING G230I is not reported, but the KD hSTINGgroup 2, a construct that contains the G230I mutation as well as additional mouse mutations, is reported to have similar IFN-β induction activity to hSTING G230I, which has a KD of 3.12 μM.
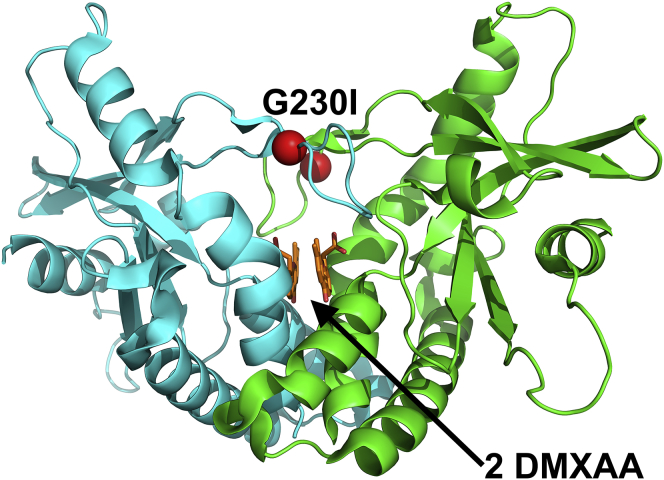
Figure 2.
Crystal structure of hSTING with lid residue mutation G230I (PDB: 4QXP (16)) making it sensitive to DMXAA. The STING dimer is colored in blue and green cartoon representation. The two DMXAAs bind deep in the cleft formed by the two STING protomers, whereas the G230I lid mutation is highlighted by the red spheres.
How G230I confers activity is very perplexing, as it is located in the lid region and not in proximity to the natural ligand binding site shared by cGAMP and DMXAA. It was hypothesized by Gao et al. (16), that upon closing of the lid, the Ile side chain becomes buried in a hydrophobic pocket stabilizing the protein conformation (Fig. 3). As this hydrophobic interaction is not possible in wild-type hSTING but is in wild-type mSTING, it may explain the difference in DMXAA sensitivity.
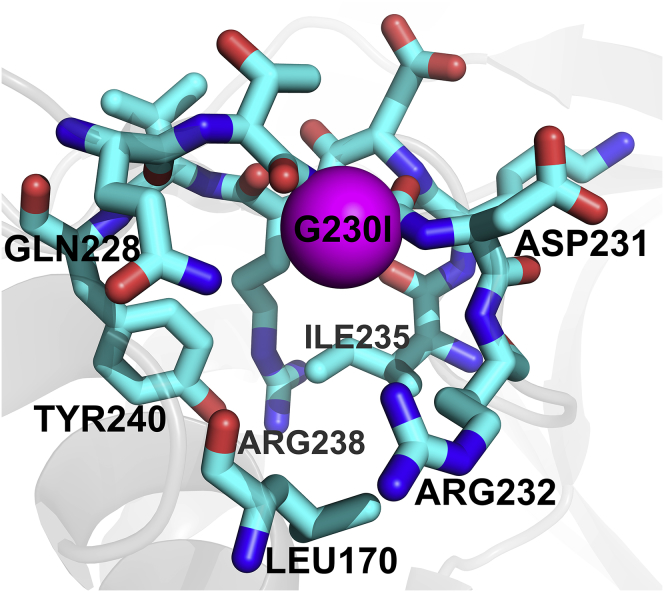
Figure 3.
Schematic showing residues within 5 Å, surrounding the key G230I mutation (shown as a pink sphere), which leads to the differing hSTING sensitivities to DMXAA (PDB: 4QXP). Residue 230 sits in a small hydrophobic pocket formed by Leu170, Ile235, and Tyr240. The remaining residues are charged or polar including Arg238, which sits within the DMXAA binding pocket and makes specific protein-ligand interactions (see Fig. 4).
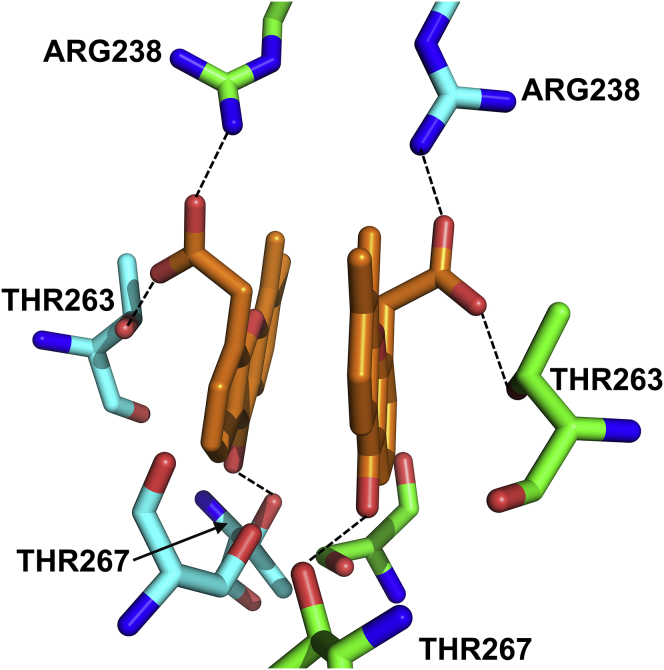
Two DMXAA molecules are found within the ligand binding site of STING (Fig. 4). These two DMXAA molecules form a π-stacking interaction with each other, whereas the carboxylic acid moieties form a charge-charge interaction with Arg238 and a hydrogen bonding interaction with Thr263. The carbonyl group forms hydrogen bonding interactions with Thr267 and Ser162. The G230I mutation that modulates the sensitivity of hSTING to DMXAA is not within 5 Å of the DMXAA binding site, the cutoff typically employed for identifying nonbonded interactions between a protein and ligand.
Figure 4.
Key hydrogen bonding interactions (dashed lines) within the ligand binding site of hSTING (blue and green) in complex with DMXAA (orange). The G230I mutation that renders DMXAA active in hSTING is not within the immediate (5 Å) binding site.
Molecular dynamics (MD) has been used in the past to study the dynamics and conformational changes that occur in protein structures. A number of techniques have been used to describe the collective motion of proteins as it relates to their function and folding motions, including MD and principal component analysis (PCA). The application of these methods to various systems is reviewed in Berendsen and Hayward (17). Specifically related to lid region movements, a number of MD studies have evaluated the flexibilities of lid regions. In one such study, Hornak et al. (18) performed unrestrained, all-atom MD simulations of the open form of HIV-1 protease and demonstrated that the flap region of the protein closes down over the binding pocket upon manual placement of the ligand into the site. Hence, the simulations were able to reproduce the known, bound form found in over 200 crystal structures. Furthermore, upon removal of the ligand, the authors were also able to demonstrate the closed and semiopen conformations observed in bound and apo crystal structures, respectively, in addition to a more open state that has not been observed experimentally (19). In another such study in Adenylate kinase (20), MD simulations were able to show the effect of atomic fluctuations that occurred on the pico- to nanosecond timescale within the hinge region and led to larger and much slower lid motions that were needed for a catalytically competent state. In a third MD study, the rotational motor motion of ATP synthase (21) was described in atomic detail, showing that the energy produced by ATP hydrolysis led to concerted motions in the V1-ring and is consumed by the rotation of the central stalk. Lastly, there are precedents in using protein dynamics to engineer novel drugs. MD studies by the Ortoleva group (22, 23) showed that epitope fluctuations in HPV were under dynamic allosteric control. Simulations showed that restricting the fluctuations of the h4 helix was crucial in rigidifying epitopes, which is key to a robust immune response. However, restricting the fluctuations of the h4 helix were found to not impact the structure of the epitope carrying loops. This insight into allosteric control was further exploited into a computationally driven vaccine design strategy. Here, we use MD to probe the mechanism behind the selectivity of DMXAA for mSTING over hSTING. Understanding the steps that occur during the binding event is crucial for the design of compounds that are active against hSTING.
Methods
A series of MD simulations using AMBER 14 (24) were performed to investigate the dynamics of human and mouse STING and the influence of a single residue mutation on the binding of DMXAA. The simulations performed are listed in Table 1 and were initiated from crystallographic coordinates. In Simulation 2, all coordinates from the PDB: 4QXP crystal structure (hSTING I230 with DMXAA (16)) were used, but residue 230 was mutated back into the wild-type Gly residue. In Simulation 6, all coordinates from the PDB: 4LOH crystal structure (hSTING H232 with cGAMP) were used, but H232 was mutated back into the wild-type Arg residue. Schrodinger’s Prime module was used to model the mutant proteins using other hSTING crystal structures with R232 as guidance.
Table 1.
Listing of STING MD Simulations Performed
| Simulation | Description | Crystal Structure (PDB ID) | Mutationa | Ligand | Replicates | Simulation Length (ns) |
|---|---|---|---|---|---|---|
| 1 | hSTING G230I | 4QXP (16) | — | DMXAA | 3 | 150 |
| 2 | WT hSTING | 4QXP (16) | G230 | DMXAA | 3 | 150/450b |
| 3 | WT mSTING | 4LOL (38) | — | DMXAA | 3 | 150 |
| 4 | WT hSTING | 4F5W (35) | — | apo | 3 | 150 |
| 5 | WT mSTING | 4KC0 (37) | — | apo | 3 | 150 |
| 6 | WT hSTING | 4LOH (38) | R232 | cGAMP | 3 | 150 |
The mutation column specifies the mutated residue in reference to the crystal structure construct.
For the WT hSTING PDB: 4QXP with DMXAA simulation, one of the three replicates were simulated out to 450 ns.
Each MD simulation was run for 150 ns and repeated three times, except for one of the simulations of WT hSTING with DMXAA, which was extended out to 450 ns. Simulations were run using AMBER 14 (24, 25, 26) on nVIDIA Titan X GPUs made available at the San Diego Supercomputing Center’s COMET machine. FF14SB (27) was used to simulate the proteins, whereas the ANTECHAMBER module (24) and GAFF with AM1-BCC charges were used to obtain parameters for DMXAA and cGAMP. Simulations used an implicit-solvent generalized Born approach with temperature maintained using Langevin dynamics and a timestep of 1 fs. All simulations systems were minimized with 1000 steps each of restrained minimization in which all hydrogen atoms were restrained with 50, 10, and 2 kcal/mol/A2 restraints, followed by restraints on only the protein CA of 50, 10, 2, and 0.5 kcal/mol/A2. Next the MD systems were allowed to be heated to 300 K, with 5000 steps each of gradually decreasing restrained dynamics with 50, 10, 2, and 0.5 kcal/mol/A2; initially, the restraints were applied to all heavy atoms and subsequently applied only to the CA atoms. After this minimization and restrained dynamics procedure, the entire system was allowed to run without constraints for 5000 frames before the start of the production runs. All production simulations were performed without restraints at 300 K. This simulation method is similar to that used for the previously described simulations with HIV protease (18, 19).
For the STING simulations containing DMXAA, nonbonded (VDW and electrostatic) interaction energies between selected sets of residues in the STING lid region were calculated using the NAMD Energy plugin in VMD 1.9.3 (28). Energies were calculated every 0.1 ns using a cutoff of 12 Å and a switch of 10 Å. Dynamical network analysis (29, 30) was also performed with additional calculations to determine network communities and critical nodes/edges between communities. Networks were visualized using the NetworkView plugin in VMD (28). Lastly, PCA was performed on the apo hSTING and mSTING simulations to evaluate the primary movement of the STING dimers. PCA was calculated using CPPTRAJ (31) on the CA atoms over the length of the MD trajectories, whereas the STING protomers remained interacting with each other. Once the STING protomers separated, as in the case of the hSTING simulations, those trajectory frames were no longer used for the PCA analysis. The primary principal component was mapped onto the proteins using the nmwiz plugin (32) in VMD (28).
Results and Discussion
STING simulations: DMXAA
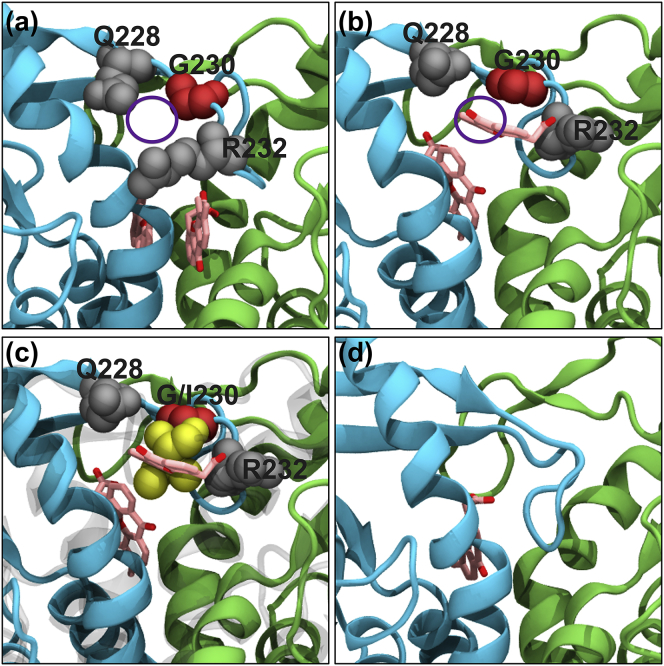
To investigate the effect of the hSTING lid region single G230I mutation in affecting the sensitivity of STING to DMXAA, we carried out several MD simulations with and without the lid region mutation. The 150-ns MD simulations revealed that with the G230I lid mutation in place, the two DMXAA molecules remain in the hSTING-G230I active site and the lid remains in a closed, active conformation (see Movie S1). However, in the simulation with the lid residue converted back to the WT glycine, the lack of a residue side chain results in an opening within the lid (Fig. 5 a). During the MD simulation (see Movie S2), one of the DMXAA molecules is able to exit the binding site exactly where the G230 residue resides (Fig. 5 b). Upon overlay of the G230I simulation, one can easily see that the isoleucine side chain creates a steric barrier that prevents the DMXAA from quickly exiting the binding site. However, the opening provided by the lack of a side chain in the WT hSTING glycine residue allows the DMXAA molecule to slip out of the binding site (Fig. 5 c). Interestingly, when just a single DMXAA molecule is left within the binding site, the STING dimer continues to remain closed and the tip of the lid loop actually comes in and fills the space left by the vacated DMXAA molecule (Fig. 5 d).
Figure 5.
Snapshots from a MD simulation of WT hSTING with DMXAA. (a) At the beginning of the simulation (0 ns), there is a gap in the lid region that results from converting the I230 residue in the PDB: 4QXP crystal structure into WT hSTING with a glycine (highlighted in red). (b) Twenty-eight nanoseconds into the simulation, one of the DMXAA molecules exits the binding site exactly at the opening created by the I230G mutation. (c) An overlay of the I230 residue from a separate MD simulation shows the extra bulk from the isoleucine residue would result in a steric barrier preventing DMXAA from exiting the binding site. (d) At the end of the 150-ns MD simulation, one of the DMXAAs has exited the binding site and the tip of the lid region has come down and filled the space previously occupied by the compound.
In addition to the hSTING simulations, MD simulations of mSTING in complex with DMXAA (PDB: 4LOL) were also performed to determine whether the mouse lid region with a WT I229 residue (equivalent to residue 230 in human) was comparable to the hSTING G230I simulations. And indeed, the mSTING with DMXAA simulation was very similar to those of the hSTING G230I simulations, in which the DMXAA molecules remained in the ligand binding site and the lid regions remained in a closed active-conformation.
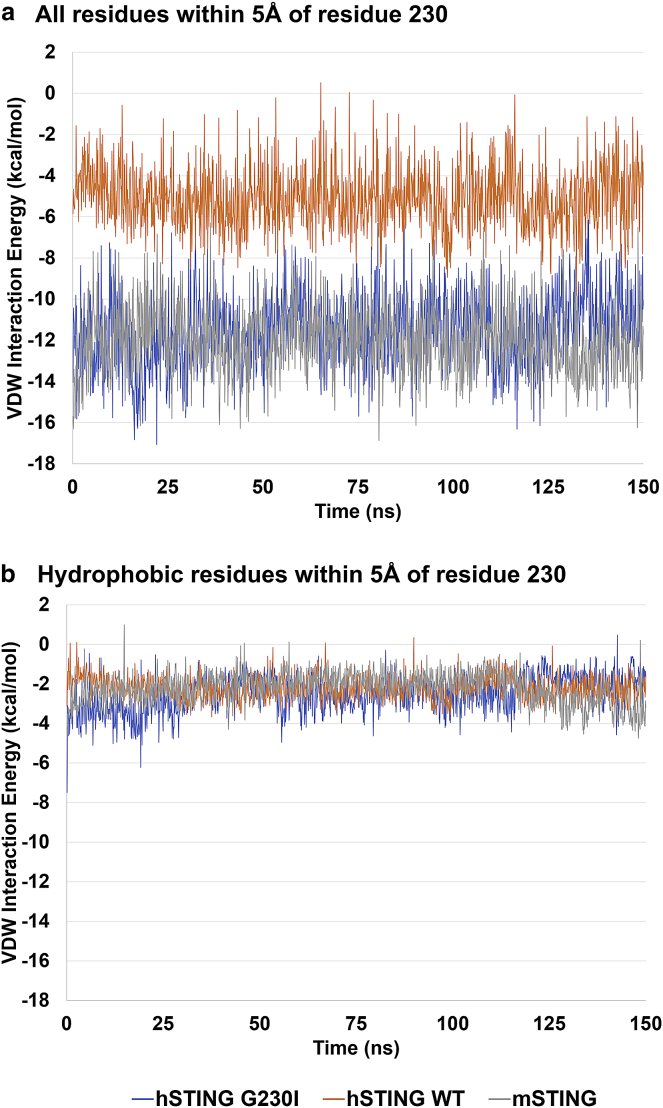
To test the hypothesis by Gao et al. (16) regarding the stabilizing nature of the small hydrophobic pocket near the G230I mutation, VDW interaction energies were calculated for the three STING simulations with DMXAA (Simulations 1–3). Fig. 6 a shows the VDW interaction energies calculated over the course of the MD simulations between residue 230 and all other residues within 5 Å of residue 230. There is a clear difference in VDW interaction energies when comparing the Gly residue in the WT hSTING simulation versus the Ile residue found in the hSTING G230I and mSTING simulations, with the larger hydrophobic side chain of Ile having an 8-kcal/mol stronger interaction than the Gly residue. However, as shown in Fig. 6 b, that difference in VDW interaction energies does not appear to arise from interactions with the hydrophobic residues that form the small hydrophobic pocket near G230I, i.e., residues Leu170, Ile235, and Tyr240, thus suggesting that the hydrophobic pocket is not stabilizing the G230I mutation in hSTING. The total interaction energy between DMXAA and the STING proteins, and DMXAA and the polar residues within the ligand binding site (Arg238, Thr263, Thr267, and Ser162), reveal that DMXAA interacts with the protein primarily through these polar residues in hSTING and mSTING (Fig. S1). A decrease in the interaction between DMXAA and the polar residues leads to the eventual exit of the DMXAA molecule in WT hSTING.
Figure 6.
VDW interaction energy of lid residue I230 (in hSTING G230I), G230 (in WT hSTING), and I229 (in mSTING) with (a) residues within 5 Å and with (b) only hydrophobic residues (Leu170, Ile235, and Tyr240) within 5 Å. The WT hSTING G230 residue has an 8 kcal/mol weaker VDW interaction with its neighboring residues than the Ile found in hSTING G230I and in WT mSTING. However, this difference in VDW interaction energy is not due to the interactions with the neighboring hydrophobic residues as the VDW interaction energies remain similar regardless of whether the residue is a Gly or Ile.
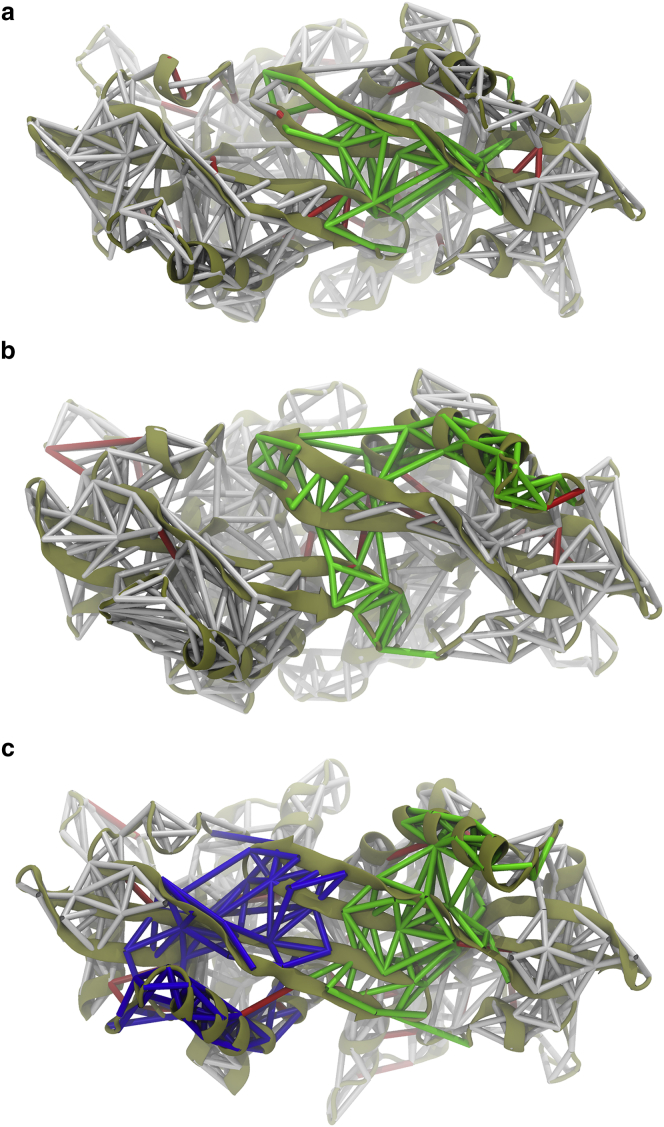
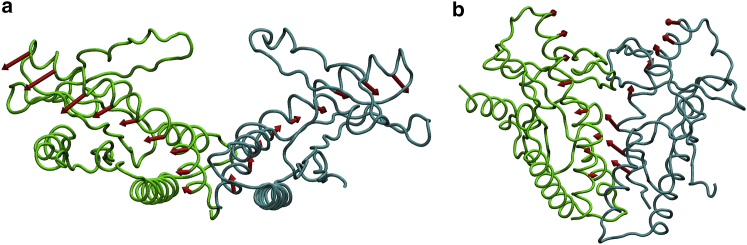
A dynamical network analysis was also performed on Simulations 1–3 (Fig. 7). Selected communities that connect the two protomer lid regions are highlighted in green and blue. For the hSTING with DMXAA simulations, either G230I (Fig. 7 a) or WT (Fig. 7 b), there are a few edges that connect the two lid regions, but there are dramatically fewer compared with the robust network of edges connecting the mSTING lid region (Fig. 7 c). This difference in lid region can be explained by the difference in affinity for DMXAA between mouse and human. Although DMXAA is active in mouse, it is only partially active in human even with the G230I mutation (16). This also suggests that there are additional species differences beyond the simple G230I lid mutation that makes mSTING more susceptible to ligand binding.
Figure 7.
Dynamical network representations for (a) hSTING G230I, (b) WT hSTING, and (c) WT mSTING, all with DMXAA, calculated from Simulations 1–3 and projected on to the protein. Network community edges are shown as sticks with selected lid communities highlighted in green and blue. Critical node edges connecting communities are shown in red. For mSTING with DMXAA, a robust network connects the lids of the two STING monomers, whereas for hSTING, either WT or G230I, there is a much weaker network connecting the two monomer lid regions.
The simulations of WT and G230I hSTING and mSTING with DMXAA reveal a single Gly-to-Ile mutation creates a porous lid region that creates a convenient exit tunnel for the DMXAA molecules. The presence of the larger side chain in the Ile residue of G230I hSTING and mSTING creates a steric block, effectively preventing the escape of DMXAA along that exit vector. The VDW interaction energies calculated between residue 230 and its surrounding residues suggest that the Gly in WT hSTING has fewer stabilizing interactions than the Ile residue in the G230I mutant and in mSTING. However, this decrease in interaction energy is not due to the stabilizing effects of the more hydrophobic Ile residue with the small hydrophobic pocket in which it resides. Rather, as shown from the network analysis, additional differences outside of the G230I mutation exist between hSTING and mSTING that allows mSTING to form a more cohesive closed conformation with a strong cross-protomer interactions stabilizing the lid region. These MD simulations suggest a possible avenue for rationally optimizing the design of DMXAA or other small molecules to gain human activity; as DMXAA is a small flat molecule, bulking up the structure could prevent it from exiting out of the binding site through the exit vector provided from the G230 residue in WT hSTING. Additionally, the compounds would need to increase the cross-protomer interactions of the lid region.
STING simulations: apo and cGAMP
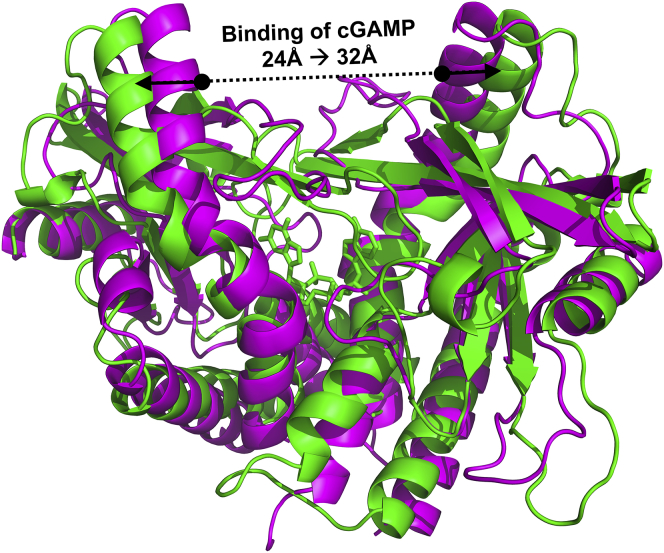
Publicly available crystal structures of hSTING and mSTING reveal that all of the apo hSTING structures (PDB: 4EF5 (33), 4EMU (34), 4F5W (35), and 4F9E (36)), and one with c-di-GMP bound (PDB: 4EMT (34)), are in an open conformation (Fig. 1), whereas the single apo mSTING structure (PDB: 4KC0 (37)) is in a closed conformation (Fig. 8). This finding is very intriguing and although it may be an artifact of what is available in the public domain, it also suggests a preference for a specific conformation state that varies between human and mouse. A second series of MD simulations were performed to examine the differences in dynamics between WT hSTING and mSTING, in particular to observe the preference of being in an open or closed state. One-hundred-fifty-nanosecond simulations of apo WT hSTING starting from the open-inactive conformation of the PDB: 4F5W crystal structure revealed that without a ligand, the STING dimer did not close and furthermore, within the simulation, individual STING proteins within the dimer separate from one another. The separation of the two hSTING protomers is likely an artifact of the simulation system because it does not include the N-terminal transmembrane domain, which would likely anchor the hSTING dimer in place. Next, a 150-ns simulation of apo WT mSTING was performed starting from a pseudo-closed conformation (with the dimer closed down on the active site, but with the lid region not fully formed, PDB: 4KC0 apo mSTING crystal structure). Unlike in the hSTING simulations, the mSTING dimer became fully closed and did not separate. Comparing it with the apo WT hSTING simulation, this suggests that a lower energy barrier may be required for mSTING to assume and maintain a closed conformation than hSTING.
Figure 8.
Comparison of apo- and cGAMP-bound mSTING crystal structures. In mSTING, the apo structure (shown in pink, PDB: 4KC0), unlike in hSTING (Fig. 1), is even more closed than the cGAMP-bound structure (shown in green, PDB: 4LOJ), albeit the β-sheet lid region only forms upon binding of cGAMP.
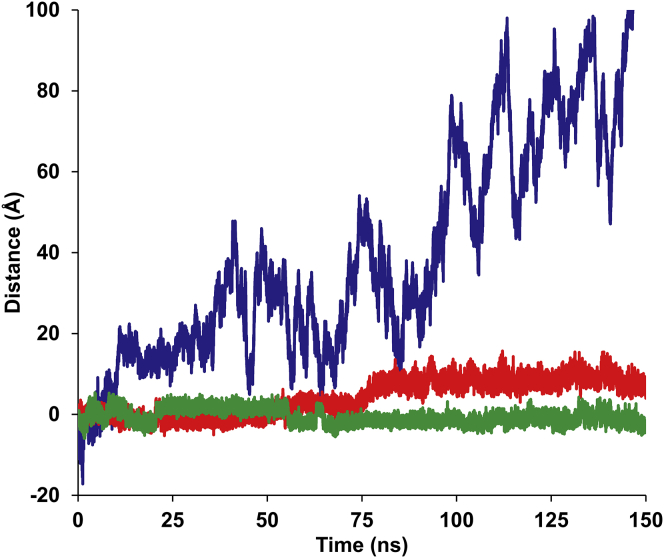
A plot of distance between residue Tyr182 (a residue located at the top of the central long helix, see Fig. 1) on one protomer to the corresponding residue on the other protomer (normalized to their crystallographic distances) for both the apo hSTING and apo mSTING (Fig. 9) shows that in apo hSTING, the two protomers move away from each other throughout the entire simulation until they separate. Indeed, a third simulation of hSTING bound to its native cyclic di-nucleotide substrate, cGAMP (PDB: 4LOL (38)) shows that even with a native substrate bound, the distance between the two protomers still increases. This is in contrast to what is seen in the apo mSTING simulation in which the two protomers remain close to their initial distance, becoming only slightly closer together throughout the simulation. A PCA of the movement of the apo hSTING and mSTING simulations shows the difference in the preferred directionality of movement between the two species of STING protomers. The PCA results shown in Fig. 10 highlight the movement of the long helix (residues 156–184) as a representation of the movement of the protomers relative to each other. In all three apo hSTING simulations, the primary principal components reveal a movement away from each other, with the top of the helices moving and separating at a faster rate than the bottoms of the helices (closest to the N-terminus). The three mSTING simulations showed primary principal component movement in the exact opposite direction for the two long helices, in that the two long helices and in particular, the top of the helices, moved closer.
Figure 9.
MD simulations of apo hSTING, apo mSTING, and hSTING with cGAMP were performed. The preference of the STING dimer to be open or closed was determined based on the measurement between Cα atoms of Tyr182 on each. The plot shows that apo hSTING (blue) has a strong propensity to open and the dimer separates during the simulation, whereas apo mSTING (green) prefers to stay closed. hSTING with a native substrate ligand, cGAMP (red), also trended toward a more open conformation. See Fig. S2 for plots of each of the three replicates for each simulation setup.
Figure 10.
PCA of the movement of CA atoms calculated from the MD simulations of (a) apo hSTING and (b) apo mSTING. The principal components of the selected long helix only are shown for clarity. The PCA analysis shows that the primary variance of the two protomers of hSTING is away from each other, becoming more open, whereas for mSTING, the two protomers move toward each other, becoming more closed.
This MD data lends support to the crystallography data in suggesting that hSTING has a natural bias toward an open conformation, whereas mSTING prefers the closed conformation. This difference creates a higher conformational energy barrier for small molecule modulators of hSTING to overcome than mSTING. Additional evidence from prior hSTING mutational studies (38), coupled with MD simulations by Che et al. (39), have shown that mutations within the ligand binding site (S162A) can confer hSTING with DMXAA sensitivity. They additionally show that a strong cross-protomer correlation between the small molecule(s) and two protein protomers is needed to achieve hSTING potency, whereas disruptions to this cross-protomer correlation, as in the case of WT hSTING and DMXAA, render the small molecules ineffective (39). The difference in inherent preference of conformational states between human and mouse, in addition to the need for a strong cross-protomer correlation, may contribute to challenges in designing a human active drug for hSTING activation.
Conclusions
The MD simulations reveal potential dynamic structural differences between human and mouse STING that compound difficulties in designing a small-molecule human active drug. The single residue difference within the lid region of hSTING versus mSTING (Gly versus Ile) makes the human STING have a porous lid. Thus, any potential small molecule drug would likely need to be bulky enough to allow it to remain in the binding site. Additionally, given the inherent bias of hSTING to be in an open-inactive conformation, when a strong cross-protomer interaction is likely needed (39), a small molecule drug would likely need to increase its interactions with the protein to help stabilize it in a closed state. Therefore, a compound must bind with enough favorable interactions to help stabilize the closed-active conformation as well as contain enough bulk to discourage the compound from easily slipping out of the binding site through the lid region.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Editor: Emad Tajkhorshid.
Footnotes
Two figures and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31147-5.
Supporting Material
References
- 1.Burdette D.L., Monroe K.M., Vance R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corrales L., Glickman L.H., Gajewski T.F. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdette D.L., Vance R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 5.Zhong B., Yang Y., Shu H.B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Sun W., Li Y., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diner E.J., Burdette D.L., Vance R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakoff-Nahoum S., Medzhitov R. Toll-like receptors and cancer. Nat. Rev. Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 9.Rakoff-Nahoum S., Medzhitov R. Role of toll-like receptors in tissue repair and tumorigenesis. Biochemistry (Mosc) 2008;73:555–561. doi: 10.1134/s0006297908050088. [DOI] [PubMed] [Google Scholar]
- 10.Woo S.R., Fuertes M.B., Gajewski T.F. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng L., Liang H., Weichselbaum R.R. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara P.N., Jr., Douillard J.Y., Scagliotti G. Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent Vadimezan (ASA404) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2011;29:2965–2971. doi: 10.1200/JCO.2011.35.0660. [DOI] [PubMed] [Google Scholar]
- 13.Prantner D., Perkins D.J., Vogel S.N. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J. Biol. Chem. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conlon J., Burdette D.L., Fitzgerald K.A. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S., Li L., Mitchison T.J. Anticancer flavonoids are mouse-selective STING agonists. ACS Chem. Biol. 2013;8:1396–1401. doi: 10.1021/cb400264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao P., Zillinger T., Patel D.J. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell Reports. 2014;8:1668–1676. doi: 10.1016/j.celrep.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berendsen H.J., Hayward S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 18.Hornak V., Okur A., Simmerling C. HIV-1 protease flaps spontaneously close to the correct structure in simulations following manual placement of an inhibitor into the open state. J. Am. Chem. Soc. 2006;128:2812–2813. doi: 10.1021/ja058211x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornak V., Okur A., Simmerling C. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henzler-Wildman K.A., Lei M., Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 21.Singharoy A., Chipot C., Schulten K. Chemomechanical coupling in hexameric protein-protein interfaces harnesses energy within V-type ATPases. J. Am. Chem. Soc. 2017;139:293–310. doi: 10.1021/jacs.6b10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi H., Lewis K., Ortoleva P.J. Epitope engineering and molecular metrics of immunogenicity: a computational approach to VLP-based vaccine design. Vaccine. 2013;31:4841–4847. doi: 10.1016/j.vaccine.2013.07.075. [DOI] [PubMed] [Google Scholar]
- 23.Singharoy A., Polavarapu A., Ortoleva P. Epitope fluctuations in the human papillomavirus are under dynamic allosteric control: a computational evaluation of a new vaccine design strategy. J. Am. Chem. Soc. 2013;135:18458–18468. doi: 10.1021/ja407489r. [DOI] [PubMed] [Google Scholar]
- 24.Case D.A., Babin V., Cheatham I. University of California; San Francisco, CA: 2014. AMBER 14. [Google Scholar]
- 25.Götz A.W., Williamson M.J., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Grand S., Goetz A.W., Walker R.C. SPFP: speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 27.Maier J.A., Martinez C., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 29.Black Pyrkosz A., Eargle J., Luthey-Schulten Z. Exit strategies for charged tRNA from GluRS. J. Mol. Biol. 2010;397:1350–1371. doi: 10.1016/j.jmb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sethi A., Eargle J., Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. USA. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 32.Bakan A., Meireles L.M., Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang S., Song X., Liu Z.J. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu C., Yi G., Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang G., Zhu D., Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 36.Yin Q., Tian Y., Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin K.H., Tu Z.L., Chou S.H. Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING. Acta Crystallogr. D Biol. Crystallogr. 2013;69:352–366. doi: 10.1107/S0907444912047269. [DOI] [PubMed] [Google Scholar]
- 38.Gao P., Ascano M., Patel D.J. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Che X., Du X.X., Gao Y.Q. Single mutations reshape the structural correlation network of the DMXAA-human STING complex. J. Phys. Chem. B. 2017;121:2073–2082. doi: 10.1021/acs.jpcb.6b12472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.