Abstract
Although the broad and unique differentiation potential of pluripotent stem cells relies on a complex transcriptional network centered around Oct4, Sox2, and Nanog, two well-distinct pluripotent states, called “naive” and “primed”, have been described in vitro and markedly differ in their developmental potential, their expression profiles, their signaling requirements, and their reciprocal conversion. Aiming to determine the key features that segregate and coordinate these two states, data-driven optimization of network models is performed to identify relevant parameter regimes and reduce network complexity to its core structure. Decision dynamics of optimized networks is characterized by signal-dependent multistability and strongly asymmetric transitions among naive, primed, and nonpluripotent states. Further model perturbation and reduction approaches reveal that such a dynamical landscape of pluripotency involves a functional partitioning of the regulatory network. Specifically, two overlapping positive feedback modules, Klf4/Esrrb/Nanog and Oct4/Nanog, stabilize the naive or the primed state, respectively. In turn, their incoherent feedforward and negative feedback coupling mediated by the Erk/Gsk3 module is critical for robust segregation and sequential progression between naive and primed states before irreversible exit from pluripotency.
Introduction
Pluripotent stem cells retain a broad range of differentiation potential in contrast to lineage-restricted cells. This ability is regulated by an extended transcriptional program centered around a core set of pluripotent factors known as Oct4, Sox2, and Nanog. Pluripotency has further been subdivided into two main states characterized by distinct developmental potential and regulatory machineries (1). The naive state of pluripotency characterizes pluripotent stem cells that are able to differentiate into lineages of all germ layers and the germline and to contribute to blastocyst chimeras. Naive cells are typically mouse embryonic stem cells (mESC) derived by in vitro culture from the inner cell mass of the blastocyst. They are long-term propagated in vitro in the presence of LIF and pharmacological inhibitors of diverse signaling pathways (2). In contrast, the primed state of pluripotency corresponds to pluripotent cells that do not contribute to blastocyst chimeras and present some differentiation bias due to their expression of some lineage-specific differentiation factors or to their sensitivity to differentiation signals that is higher than in naive cells. Mouse epiblast stem cells (mEpiSC) derived from the epiblast of the prestreak postimplantation embryo and stabilized in vitro by adding Activin and FGF in the culture medium are a common model of primed pluripotency (3, 4). These two states can also be interconverted in vitro under various procedures (5).
The possibility of culturing and monitoring pluripotent stem cells in different states has allowed comparative studies and has helped to reveal markedly different expression profiles and regulatory mechanisms. Although naive/mESC and primed/mEpiSC cells share significant expression levels of the core Oct4, Sox2, and Nanog pluripotent factors, they also drastically differ in the expression levels of some factors, especially the naive-specific factors such as Klfs, Esrrb, Dax1, and Tbx3 that are strongly downregulated in primed/mEpiSC cells (3, 4). However, the recurrent interactions between all these pluripotent and naive-specific factors and their sensitivity to a multiplicity of signaling cues give rise to a highly complex signaling and transcriptional network (6). How such a densely interconnected network can drive the developmental progression through well-distinct pluripotent states before lineage-specific differentiation is not clear, but nevertheless suggests a functional level of network organization in terms of modules (7) and loop motifs (8).
Diverse mathematical modeling approaches have proven valuable to investigate the key features of the pluripotency regulatory network (9, 10). On the one hand, mechanistic approaches typically consist of translating regulatory network models into dynamical system models whose deterministic and stochastic behaviors can be thoroughly analyzed using tools and concepts from numerical analysis, dynamical system theory and statistical physics. Dynamical models of the Oct4-Nanog-Sox2 transcriptional network have contributed to identify key regulatory and stochastic mechanisms that underlie cell fate decisions in mESCs (11, 12, 13, 14, 15, 16, 17). However, the poor quantitative knowledge of biochemical kinetic constants associated with the numerous regulatory mechanisms restricts the possibility of reliable dynamical modeling of detailed regulatory networks. On the other hand, descriptive approaches begin with systematic analysis of large sets of genomic, expression, and phenotypic data to extract relevant information about the regulatory design (18, 19, 20, 21) or the heterogeneity of the cell population (22, 23, 24) without necessarily requiring mechanistic assumptions and quantitative knowledge regarding regulatory kinetics.
This modeling study couples these descriptive and mechanistic approaches to extract pluripotency network regulatory features that are inferred from experimental data and required for a specific cell-fate decision pattern. In this interplay, the choices of experimental data and of ordinary differential equation formalism are motivated by our restricted focus on the deterministic regulatory mechanisms underlying pluripotency decisions, leaving aside the sources of intracellular stochasticity and population heterogeneity (25). Starting with a pluripotency network model of intermediate complexity and a set of perturbation experiments, we use an evolutionary computation method to explore model parameter sets that optimally match these data. Systematic analysis of optimized parameter statistics and sensitivities allows us to assign relative degrees of significance to diverse regulations and to identify a core network architecture. Further dynamical analysis of optimized models reveals the existence of signal-dependent multistability and asymmetric transitions, not only between pluripotent and nonpluripotent states, but also between naive and primed states of pluripotency. Finally, perturbation and reduction approaches recapitulate the modular, feedforward, and feedback organization of the pluripotency network that underlies these pluripotent cell fate decision properties.
Methods
Pluripotency network structure
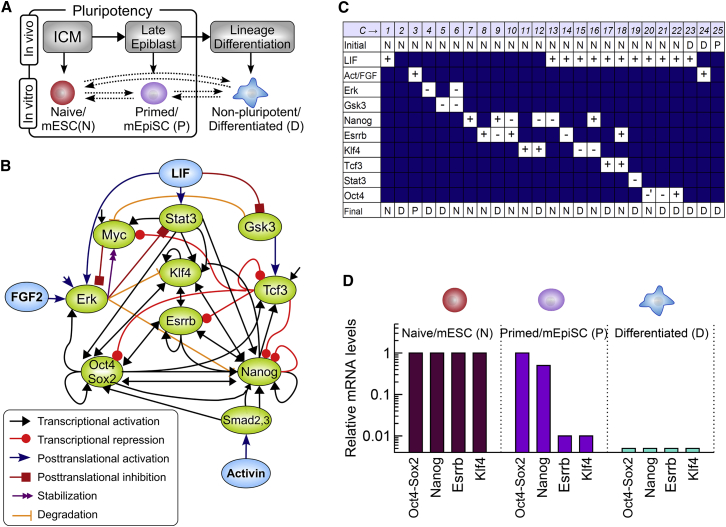
The dichotomy and transitions between the naive and primed states of pluripotency (Fig. 1 A) are studied in a network model (Fig. 1 B) whose architecture is composed of a selected set of proteins and their well-documented posttranslational and transcriptional interactions (Supporting Material; Table S1). Specifically, the network comprises a set of signaling cues (LIF, FGF, and Activin), signaling proteins (Erk, Stat3, Smad2/3, and Gsk3), and pluripotency factors (Oct4-Sox2, Nanog, Esrrb, Klf4, Tcf3, and Myc), most of which have been included in previous pluripotency network models (16, 19, 20, 26) and have been targets of informational perturbation experiments (Fig. 1 C) including reprogramming experiments. This design can be seen as an intermediate level of description (Fig. S1 A) that strikes a balance between the aim to analyze some detailed and nontrivial features of pluripotency network complexity and the need to keep such analysis computationally tractable, given our mechanistic and continuous modeling framework. In striking this balance, it was necessary to leave aside some pluripotency factors often included in larger-scale network representation (such as Dax1, Tbx3, Tcfp2l1, Rex1, Sall4, Mbd3, and Jarid2; see Fig. S1 B) whose signal-mediating or feedback-regulatory roles in pluripotency are presumed either to be comparatively less central or to be more involved in larger-scale epigenetic regulations.
Figure 1.
The pluripotency model: network architecture and cell fate data. (A) Pluripotency displays specific in vitro and in vivo transition patterns among a naive/mESC pluripotent state, N; a primed/mEpiSC pluripotent state, P; and a nonpluripotent differentiated state, D. (B) The regulatory network model is based on a selected set of signaling and transcription factors interacting with each other through various regulatory mechanisms (arrow legend box). (C) A set of perturbation experiments is used as target experimental dataset for the model optimization procedure. These experiments describe the fate outcome j′ = N,P,D of cells that are initially in a state j = N,P,D and then subjected to specific culture conditions (LIF, Activin, FGF) or to upregulation/downregulation (±) of specific factors. (D) Typical relative mRNA levels of pluripotency factors (Oct4-Sox2, Nanog, Esrrb, and Klf4) are associated with naive/mESC, primed/mEpiSC, and nonpluripotent states, which are determined from experiments (Fig. S4) and used for model optimization. To see this figure in color, go online.
Differential equation model
The dynamics of this pluripotency regulatory network is described by a set of deterministic differential equations (see the Supporting Material):
| (1) |
where the 10 components of are the time-dependent concentrations of the Oct4-Sox2, Klf4, Nanog, Esrrb, Tcf3, Myc, Smad2/3, Erk, Gsk3, and Stat3 proteins. The model parameters consist of signaling parameters associated to the extracellular concentration of LIF, Activin, and FGF2 signaling factors, in 60 regulatory parameters associated to the basal or regulated rates of degradation, transcription, or posttranslational modifications (Tables S1 and S2), and perturbation parameters used for simulating overexpression, knock-down or pharmacological perturbation experiments. Mechanistic assumptions, steady-state approximation, and normalization procedures are made to keep a restricted number of parameters where each regulatory edge is quantified by a single parameter. The lack of quantitative knowledge of many regulatory parameters and their effective description requires us to infer most regulatory parameters from a target set of experimental data.
Data-driven model inference
The target dataset is provided by a collection of in vitro studies describing the fate outcome of naive/mESC cells or primed/mEpiSC cells following the manipulation of proteins that are included in the network model (Fig. 1 C; and see the Supporting Material). In these data, the naive/mESC cells cultured with LIF are considered to a first approximation as a homogeneous phenotype, disregarding the small pool of cells in transition toward the primed state commonly observed in the presence of serum in addition to LIF. Such expression heterogeneity in single-cell data is difficult to address within a mechanistic modeling framework, especially in the absence of knowledge or consensus about the main source of stochasticity. The initial and final cellular states in these experimental studies are therefore restricted to the three classes of naive, primed, and differentiated states, for which we also define typical expression levels estimated from another set of experimental data (Fig. 1 D; Fig. S4; Supporting Material): whereas the naive/mESC state is characterized by the high expression of Oct4, Nanog, Klf4, and Esrrb, the primed/mEPISCs state retains only a high expression of Oct4 and Nanog, which both become downregulated upon exit from pluripotency and lineage-specific differentiation. These datasets define NC = 25 experimental conditions that can be simulated using Eq. 1 with appropriate initial conditions , signaling inputs , and perturbation factors (Table S3). The difference between data and model simulations is quantified by measuring and summing the distances ϕi between the experimental behavioral outputs and the model behavioral outputs for each experimental conditions Ci:
| (2) |
where
| (3) |
The use of least-absolute errors rather than least-square errors aims to provide more robust solutions with respect to the possible outliers in the target dataset. The global error Φ is used as the objective or score function to be minimized in the parameter optimization procedure to generate optimal model parameter sets that best match the target data set. Parameter optimization is performed via a population-based metaheuristic algorithm called “evolution strategy” (see the Supporting Material for details). Evolution strategy is a global and stochastic optimization method that is appropriate to a nonconvex derivative-free optimization problem. In this case, nonconvexity (i.e., multiple local optima) is due to the high nonlinearity of the dynamic network system and the use of a large data sets, and the derivative of the score with respect to parameter variations is difficult to obtain for a score obtained from numerical simulations and for a high-dimensional parameter space.
Sensitivity and similarity measures
The parameter sensitivity measure γi is defined as the relative change of the global error score Φ upon a parameter change Pi → 0 (i.e., disrupted regulation):
| (4) |
The value indicates that the regulation i of strength Pik in the model k can be neglected without significant impact on the model performance in matching experimental data.
The identification of clusters or subclasses within a set of models is performed by computing distance matrices containing the distances, taken pairwise, between specific quantitative features of distinct models (27). In this study, the degree of similarity between optimized models is analyzed by measuring the distances between their condition-specific error scores (ϕ) as well as the parameter and sensitivity values (P and γ) on a logscale. A distance measure between models k and l is defined by
| (5) |
where yi = {ϕi,log10(Pi), log10(γi)}, and N = {NC, NP} depending on the feature considered, whereas the normalization factor Z is the largest distance within the set of optimal models so that Dkl ranges between 0 and 1. The number of clusters or classes is evaluated by measuring the percentage of variance explained.
Results
Regulatory hierarchies and core circuitries in data-driven network models
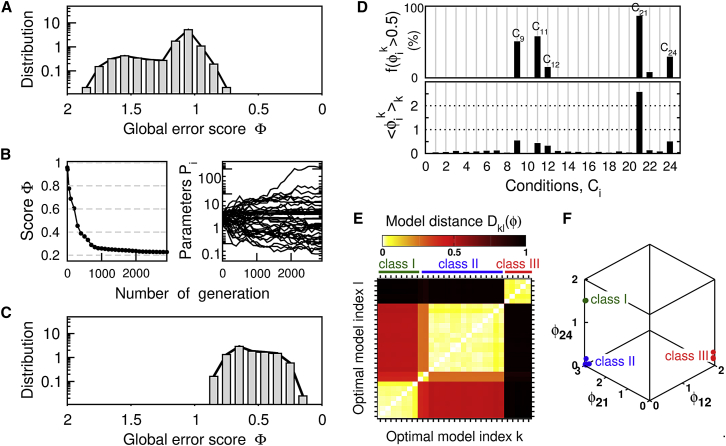
The pluripotency network model exhibits a complex regulatory architecture and its ability to match experimental behaviors is expected to be highly sensitive to the parameter values that set the strength of each regulatory link. As a matter of fact, pluripotency models with random parameters , where ηi,k are uniformly distributed between [−1:1] exhibit a broad distribution of error score values Φ ranging from ∼2 for the worst scores to only ∼0.8 for the best scores (Fig. 2 A). Due to the high dimensionality of the parameter space, a parameter optimization procedure is definitively required to identify the model parameter sets that are the most consistent with the target dataset. Starting from these randomized-parameter models and then optimizing using the evolutionary algorithm significantly decreases the error scores Φ (Fig. 2 B), which now range between ∼0.8 and ∼0.2 for the 103 evolutionary trials (Fig. 2 C). The broad distribution of Φ for parameter-optimized models nevertheless emphasizes the difficulty to reach a global optimum because of the likely existence of many local optima related to the high dimension of the parameter space, the nonlinearity of the network, and a cost function based on heterogeneous datasets.
Figure 2.
Model optimization through computational evolution. (A) The statistical distribution of the global error score Φ is computed over 103 randomized-parameter models. (B) An example of a computational evolutionary trial showing the decreasing score Φ from 0.8 to 0.2 and the parameter set of the fittest model individual of the family of the ith generation. The generation 0 is composed of NI = 20 model individuals k of parameter , where ηi,k values are uniformly distributed between [−1:1]. (C) The statistical distribution of the global error score Φ is computed for the fittest individuals of 103 evolutionary trials. (D) Given here are condition-specific scores ϕik of a set of optimal models satisfying Φ < 0.21: model-averaged value (lower panel) and fraction of models with ϕik > 0.5 (upper panel). (E) Three main classes of optimal models (green, class I; blue, class II; red, class III) are evidenced by the distance matrix of their condition-specific scores (Eq. 5). (F) The three model classes differ in their worst-condition-specific scores. To see this figure in color, go online.
To overcome this difficulty, we investigate and compare the features of a subset of NM = 29 optimized models that have the lowest error scores (Φk < 0.21 that is below ∼10% of the lowest error score). The distribution of their condition-specific scores confirms the existence of several local optima. Most sets of experimental conditions are properly adjusted by all these optimal models 〈ϕik〉k ∼ 0, whereas a few conditions (C9, C11, C12, C21, C24) are suitably adjusted only by a subset of optimal models (Fig. 2 D). The analysis of similarity between the score patterns ϕik of the optimal models reveals in fact the existence of three main classes of models (Fig. 2 E; Table S5), which can be distinguished by their worst-adjusted conditions (Fig. 2 F).
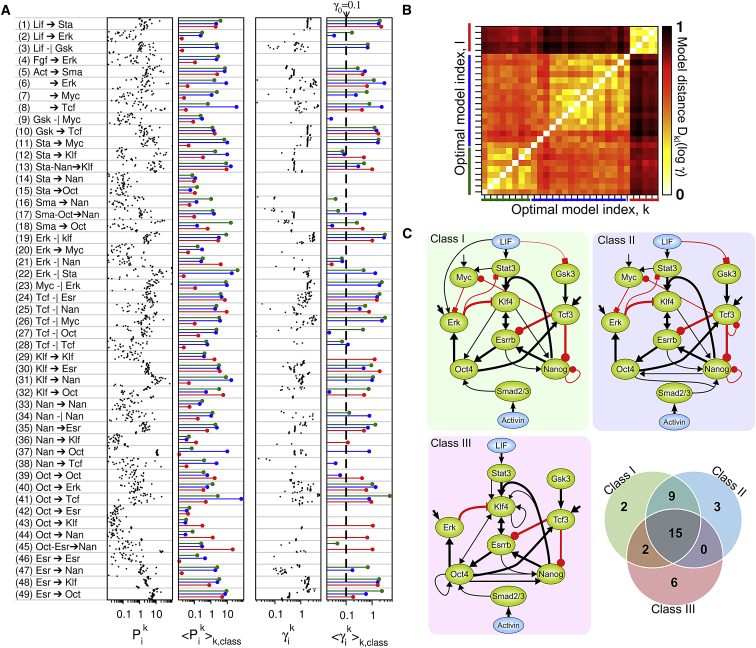
To determine the common and class-specific regulatory properties of this set of optimal models, we perform an extensive statistical and sensitivity analysis of their parameter values Pik (Fig. 3). For each optimal model, distribution and uncertainty of parameters reveals the existence of both highly sensitive and neutral parameters (Fig. 3 A; Fig. S5), which indicates that core networks of reduced complexity can fit the data equally well. Furthermore, a comparative variance and similarity analysis between optimized models (Fig. 3, A and B; Fig. S6 A) shows that the parameter values Pik and sensitivities γik are much more similar between models of a same class than between models of different classes, which indicates that networks of well-distinct topologies are equally consistent with the data. The core network topologies associated to each class are drawn by removing the network edges associated with the most insensitive parameters 〈γik〉k⊂Class < γ0 (Fig. 3 C; Fig. S6 B). These class-specific core circuits share a common set of 15 regulations, but also display some subtle differences. Despite the systematic presence of at least one signaling pathway from LIF or Activin toward pluripotency factors, some models differ in the nature of the main signaling paths, for instance whether the main target of Activin/Smad is Oct4 or Nanog, or in the presence of signaling cross talk such as among the Gsk3/Tcf3, Stat/Myc, and Erk pathways. And although several positive feedback loops are systematically implemented in most models through the reciprocal interactions among Klf4, Esrrb, and Nanog, the self-activation of Klf4 and Oct4 and their reciprocal activation occur only in class-III models. Addressing the significance of both the common and the distinctive topological features in the optimal network models requires us to scrutinize and compare their dynamical properties, beyond their ability to match the target experimental dataset.
Figure 3.
Parameter distribution and core topologies of optimized models. The systematic analysis of the optimized parameters is made for the k = 1,29 optimal models segregated into the three classes identified in Fig. 2E (green, class I; blue, class II; red, class III). (A) The two left panels show parameter values Pik (k points in the ith line) and their class-averaged values 〈Pi〉k⊆class. The two right panels show parameter sensitivities γik (Eq. 4) and their class-averaged values 〈γik〉k⊆class, where the dashed line shows the threshold γ0 = 0.1 between relevant and irrelevant regulations (Fig. S6). (B) The three classes of optimal model differ in their topology, as evidenced by the distance matrix of model parameter sensitivities Dkl(log γi) (Eq. 5). (C) Core pluripotent circuitries for each model class are drawn by including only regulations whose removal results in a significant increase of error score Φ (〈γi〉k⊆class > γ0 in (A)). Model classes share 15 common regulations (thick network edges). To see this figure in color, go online.
Multistability and transitions among naive, primed, and nonpluripotent states
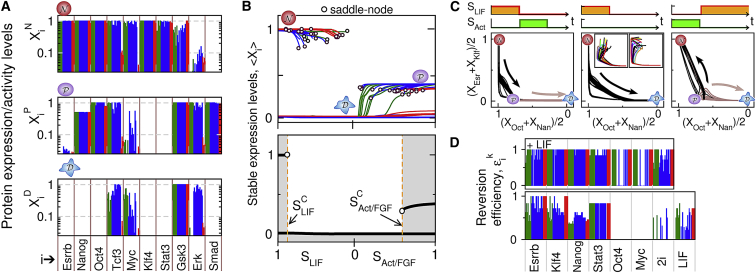
The model adjustment procedure is based on a restricted set of experiments describing the fate outcome of pluripotent cells subjected to specific and discrete perturbations. However, the differential equation framework not only enables parameter fitting of experimental behaviors, but can also provide a broader picture of network dynamics by computing the whole set of stable expression states (solutions of ) and transitional trajectories (Fig. 4). The protein concentration phenotypes of the naive, primed, and differentiated states in their classical signaling contexts are very similar across the optimal models, especially for Oct4, Nanog, Klf, and Esrrb in agreement with the mRNA targets (Fig. 4 A). The manner in which these distinct phenotypic states depend upon signals SLIF and SAct/FGF reveals a very specific pattern of signal-dependent multistability common to most optimal models (Fig. 4 B). A gradual decrease in signaling factor levels has little impact on the concentrations of the stable naive and primed pluripotent states. Instead, such decrease leads to a sudden destabilization of these states through a saddle node bifurcation occurring for some critical signal levels SCLIF and SCAF, toward an always stable nonpluripotent state. Therefore, all optimized models display two distinct bistability domains between each pluripotent state and the nonpluripotent differentiated state (except class-I models that display an ultrasensitive switch from to states) with differences only in the signaling threshold values.
Figure 4.
Multistability domains and asymmetric transitions between naive and primed states. Color code represents the three classes of optimal models (green, class I; blue, II; red, class III). (A) Normalized expression/activity levels Xj∞,j of the i protein species are associated with the stable states j = N,P,D (top, naive state with SLIF = 1; middle, primed state with SAct/FGF = 1; bottom, differentiated state with SLIF = SAct/FGF = 0). (B) Distinct multistable subdomains between these three states are depicted by their average expression levels of pluripotency factors 〈Xik〉i = Oct, Nan, Esr, Klf as a function of SLIF (and SAct/FGF = 0) and SAct/FGF (and SLIF = 0). (Lower panel) An example of multistable property with two bistability domains (shading) and two saddle-node bifurcation points (circles) is illustrated for one optimal model. (C) Dynamical trajectories of optimal models in a 2D section of the concentration space are shown for distinct transition scenarios between naive and primed pluripotent states. (Middle panel, insets) A set of trajectories from a heterogeneous naive state seems attracted to a slow manifold through the primed state. (D) Primed-to-naive reversion efficiencies ϵik = 1/θik are given for the strategy i (overexpression of a given pluripotency factor or 2i, with or without LIF) and optimal model k, where θi is the critical level of upregulation or downregulation αi or of SLIF that is required for successful reversion. To see this figure in color, go online.
Such multistable properties raise the question of the transition dynamics between these diverse states (Fig. 4 C). In particular, the bifurcation diagram alone does not indicate whether naive and primed states can be easily converted upon reciprocal change of their stabilizing signaling cues (SLIF versus SAct/FGF), or instead would be trapped in the strongly stable differentiated steady state. Simulations first show that the naive pluripotent state systematically transits toward a primed state upon the signaling change from SLIF = 1 to SAct/FGF = 1, whereas further removal of Activin/FGF (SAct/FGF → 0) leads, as expected, to a transition toward the differentiated state (Fig. 4 C, left panel). Interestingly, the dynamical trajectory from the destabilized naive state (SLIF → 0) seems to be canalized along a similar path in the concentration space through a primed-like state in a manner that is very robust to heterogeneous initial conditions (Fig. 4 C, insets of middle panel). In sharp contrast with the likely naive-to-primed transition, the opposite primed-to-naive transition is not systematically induced by a signaling change from SAct/FGF = 1 to SLIF = 1 as the outcome is found to be model-dependent (Fig. 4 C, right panel). This result suggests that the primed state of steady-state concentration sits close to the state-space boundary separating the basin of attraction of the naive state and differentiated state in the presence of LIF. Accordingly, the primed-to-naive reversion is sensitive to specific model features, and, more importantly, can be promoted through perturbations that extend the attraction basin of the naive state, such as the overexpression of any naive-specific pluripotent factors. For all optimal models, measuring the reversion efficiency E shows that the overexpression of Esrrb, Klf4, or Stat3 are the most efficient strategies to drive primed-to-naive transition as compared with the limited efficiency of overexpressing Nanog or the null-efficiency of overexpressing Myc or Oct4 (Fig. 4 D). Furthermore, a strong synergetic effect is observed by combining LIF with another input, even with those that are weakly efficient alone such as Oct4 or Myc overexpression, or Erk/Gsk3 inhibition (2i).
Despite their possible structural differences, optimized pluripotency network models share a similar qualitative dynamical landscape characterized with signal-dependent multistability and asymmetric transition between naive and primed states with only minor quantitative discrepancies in signaling thresholds and transition timescales.
Network design principles for segregation of pluripotent states
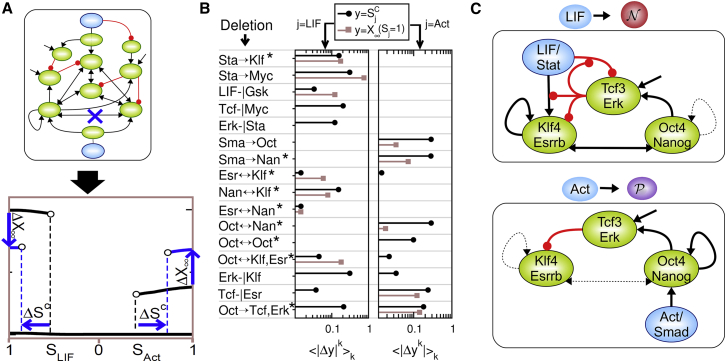
A challenging issue is now to determine the relationship between, on the one hand, the maintenance and transition properties of naive and primed pluripotent states, and, on the other hand, the core regulatory circuitries that have been identified. To address this issue, we evaluate the effect of network perturbations on the attractor landscape of naive and primed pluripotency. The changes of expression state and signaling thresholds associated with naive and primed identity are measured upon the deletion of single or several network edges and averaged over all optimal models (Fig. 5, A and B). The results show that most edge/motif deletions are prone to influence only the naive/mESC state or only the primed/mEpiSC state, suggesting a functional segregation between diverse regulatory motifs. The signaling control of the naive state depends on several LIF-dependent signaling pathways distributed among the Stat3-dependent activation of Klf4, the inhibition of the Gsk3 pathway, and the reciprocal interaction between Stat and Erk, notably through Myc. Its intrinsic regulation is also distributed between various regulatory motifs such as the reciprocal activation between Esrrb and Klf4 and their respective interactions with Nanog and Oct4. In contrast, the primed state is exclusively dependent on regulations centered on Oct4 and Nanog, as likely targets of Activin/Smad signaling, and through their reciprocal interaction or through Oct4 self-activation. Finally, only the inhibitory actions of Erk and Tcf3 toward Klf4 and Esrrb, respectively, and their activation by Oct4, significantly influence the stability and the expression phenotypes of both naive and primed states. The naive/mESC and primed/mEpiSC states are thus proposed to rely on the segregated operations of two regulatory modules featured with their own stabilizing positive-feedback and signaling inputs, and subtly coupled through reciprocal positive interactions and negative interactions mediated by an inhibitory Tcf3/Erk module (Fig. 5 C).
Figure 5.
Identification of a modular network through systematic perturbation approach. (A) The approach consists in quantifying the effect of diverse network motif perturbations on the signaling thresholds and expression levels of the naive and primed states. (B) Model-average alterations 〈|ΔYk|〉k of expression levels ( and signaling thresholds (Y = SCj) of the naive state (left, j = LIF) and the primed state (right, j = Act/FGF) are computed upon the deletion of a single or multiple (∗) regulatory links (a ↔ b: a → b ∪ b → a; a → b,c : a → b ∪ a → c). (C) Functional modular circuits are inferred from (B) regulating the naive and primed states. (Black arrows) Activatory regulation; (red circles) inhibitory regulation. (Solid lines) Effective regulation; (dashed line) ineffective regulation. To see this figure in color, go online.
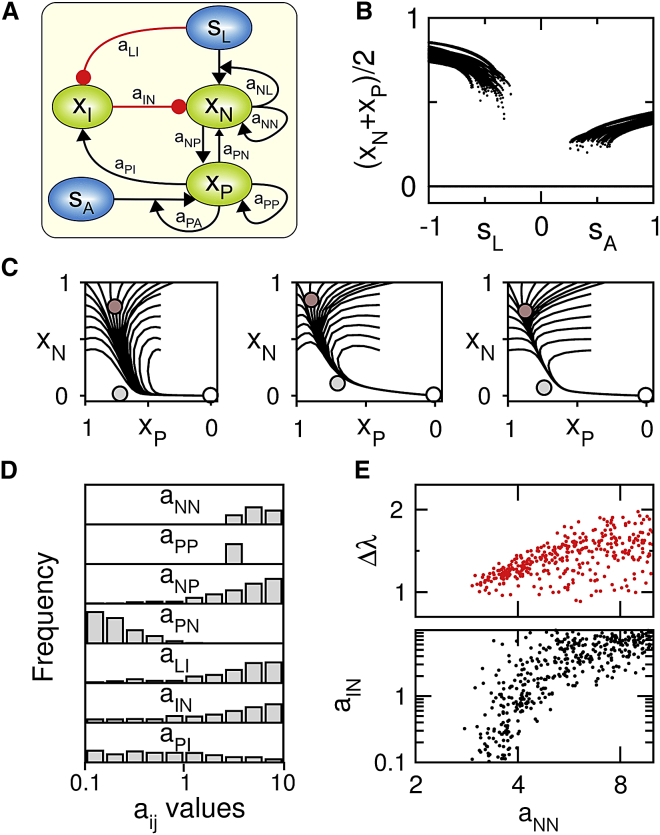
To assess the significance of the modular architecture depicted in Fig. 5 C, a computational strategy consists in designing a coarse-grained model of such architecture (Fig. 6 A) where the activities of the naive module N (Klf/Esrrb), the primed module P (Nanog/Oct4/Sox2), and the inhibitory module I (Tcf3/Erk) are described by normalized variables xN,I,P, whereas their interactions are described by effective transcriptional activations and repressions (Supporting Material):
| (6) |
The differential equation system of Eq. 6 contains few parameters which quantify the strength of the various regulatory edges and loop motifs. Systematic exploration of the parameter space identifies a subset of model parameters for which the modular model exhibits a signal-dependent multistability among three states (high xN and xp; high xN and low xp; low xN and xp) that is very similar to that observed in the optimal set of detailed models (compare Fig. 6 B with Fig. 4 B; Fig. S7). It turns out that these suitably parameterized modular models also display similar transition dynamics from a heterogeneous naive state toward a differentiated state canalized through the primed state (compare Fig. 6 C with middle panel insets of Fig. 4 C). The distribution of their model parameters is relatively broad, but always peaks around high values (except aPN) (Fig. 6 D). These high parameter values support the premise that this modular circuit design is both necessary and sufficient to achieve the signal-dependent multistability observed in detailed and optimized pluripotent models, which is confirmed by the significant alterations of the dynamical landscape when any parameter is set to zero (Fig. S7). The broadness of the parameter distribution reflects in part the existence of correlation between parameters, especially between aNN and aIN, whose concomitant high values also coincide with a higher signal-dependent stability of the naive-like state (Fig. 6 E; Fig. S7). The coexistence of strong naive-specific positive feedback loop (aNN high) counterweighted by tightly regulated inhibitory pathways of strength aIN thus promotes the possibility to switch the naive state from being very stable to very unstable upon suitable signal changes (Fig. S7 C).
Figure 6.
A low-dimensional modular model reproduces the pluripotent dynamics of detailed models. (A) Minimal circuit corresponding to Eq. 6 consists of regulatory modules xi = N,I,P, extrinsic signals si = L,A, and regulatory parameters aii′. (Black arrows) Activatory regulation (red circles) inhibitory regulation. (B) A signal-dependent multistability among three discrete states similar to Fig. 4B is found for 381 parameter sets from a sample of 106 random parameter sets for which log10(aii′) values are uniformly distributed between [−1:1] except aNL = 1 and aPA = 1. (See Fig. S7A for the quantitative multistability criteria used for parameter set selection.) (C) Three examples of canalized trajectories where heterogeneous initial conditions around the naive-like state (dark shaded circle) are quickly attracted to a slow 1D manifold that passes near the primed-like state (light shaded circle) toward the differentiated-like state (blank circle). (D and E) The 381-parameter set satisfying the multistability criteria in (B) shows specific distribution profiles for each parameter with peaks for high values (D), and significant positive correlations between the parameters aNN and aIN (lower panel of E) and between the parameters aNN and Δλ (upper panel of E). Δλ is a measure of stability of the naive state (Fig. S7, B and C). To see this figure in color, go online.
The ability of the reduced model to capture the dynamical landscape confirms the notion that the subset of detailed model of slightly distinct parameterization and topologies share a common high-level network organization. The critical feature of this organization is the manner in which two positive-feedback modules are differentially controlled by signaling inputs, and are reciprocally coupled through both mutual activation and negative feedback interaction.
Discussion
This modeling study dissects the complexity of the pluripotency network to extract the core regulatory properties that underlie its naive and primed pluripotent cell decisions. Data-driven network inference approaches have proven valuable to hierarchize, clusterize, and reevaluate the functional role of many pluripotent factors (19, 20, 21, 23). This study further seeks to evaluate how the large-scale feedback and feedforward organization of the pluripotency network shapes its dynamical landscape including multistable and transitional properties. This issue requires the use of a dynamical modeling framework that is confronted, for large-scale networks, to the difficult challenge of the determination of relevant parameter regimes (28, 29). Parameter optimization and sensitivity methods were first devoted to identifying a subset of model parameters and network topologies consistent with experimental data. Bifurcation analysis and reduction methods then established some dynamical and network mechanisms underlying pluripotent decisions that are shared within this subset of models despite and beyond their structural differences. This two-step approach has proved to be very effective to identify a modular regulatory pattern within the densely interconnected pluripotency networks, and to assign a dynamic and functional role to this pattern in underlying robust and controllable transition from a naive to a primed state of pluripotency before lineage differentiation.
Feedforward and feedback topology of the pluripotency network
Computational studies have focused on diverse topological aspects of pluripotency network ranging from core circuitries, functional clusters, regulatory hubs, feedback motifs, sensitive reprogramming nodes, and concurrent topologies (13, 14, 17, 19, 20, 21, 26, 30). In this study, the emphasis is made on the functional interplay and hierarchy among a plethora of feedback loops, either positive or negative, and feedforward loops, either coherent or incoherent. In the context of cell-fate decisions, the common interplay of multiple positive feedback loops has been proposed to contribute either to robust switches using dual time mechanisms (31) or to binary fate decisions based on tristability (32, 33). In the case of the pluripotency regulatory network, the many reciprocal activatory regulations between several transcriptional factors give rise to a particularly large number of intricate positive feedback loops. Some of these loops are shown to be functionally segregated between redundant naive-stabilizing loops, consisting in the many reciprocal activatory interactions among Esrrb, Klf4, and Nanog, and the primed-stabilizing loops, involving the self- and reciprocal interactions of Oct4/Sox2 and Nanog. Such a segregation was not apparent in the network architecture, and is shown to rely in fact on how these distinct modules are differentially coupled with activatory (Stat3 and Smad2/3) and inhibitory (Erk and Gsk3) signaling pathways. First, some positive feedback loops operate in synergy with signaling pathways consistently with the acknowledged interaction between Smad2/3 and Oct4 (34) or between Stat3 and Nanog (35), which allows a sharper signal-dependent maintenance and exit of naive or primed pluripotent states toward an attracting nonpluripotent state. Second and even more important, the Erk and Gsk3/Tcf3 pathways play a pivotal role by inhibiting only the naive-specific factors Klf4 and Esrrb, respectively (36, 37), while being regulated negatively by LIF signals and positively by Oct4. This set of regulations contributes to several feedforward and feedback motifs that counterweight the strong stabilizing role of the many naive-specific positive feedback loops. In particular, coherent feedforward loops in LIF signaling pathways contribute to restricting the naive state stability to a LIF signaling context. In addition, the incoherent feedforward and negative feedback loops mediated by Oct4-dependent upregulations of the Erk/Tcf3 pathways promote naive-state destabilization toward a well-distinct primed state upon LIF removal.
Although these network design principles explain some key multistable and transitional properties in pluripotency, their implementation by a specific set of factors and regulations can nevertheless differ between models depending on prior network design assumptions. This is the case for the Oct4-dependent negative feedback and incoherent feedforward loops whose presence and role have been acknowledged to account for culture-dependent expression profile and variability in mESCs (12, 13, 14, 17, 21, 38). Yet, these models propose diverse realizations of these loops where the main inhibitory regulation either consists in the Oct4-dependent repression of Nanog (12, 16), the Oct4-dependent repression of Tcfp2l1 (20), the Nanog-dependent repression of Sall4 (21), the Tcf3-dependent repression of Oct4 (19), or the Erk-dependent inhibition of Nanog (17), to compare with the Tcf3-dependent repression of Esrrb and Erk-dependent degradation of Klf4 in this study. These discrepancies between models support the notion that the functional organization of complex regulatory networks can be reliably inferred without necessarily identifying with certainty the full set of regulatory mechanisms or factors involved.
Distinct and ambivalent roles for Oct4 and Nanog
Although Nanog and Oct4 are expressed at significant levels in both mESC and mEpiSC states (3), these two master pluripotent factors perform markedly distinct regulatory functions. Nanog is part of a large number of mutual activatory interactions with many other pluripotent factors, whereas Oct4 exerts both activatory and inhibitory influences, which are restricted to a subset of pluripotency factors and are tuned in a context-dependent manner. Besides their participation to a sophisticated feedback and feedforward network organization required for specific pluripotency decision making, these distinct regulatory roles make sense to a large number of experimental observations among which some are apparently paradoxical. The contribution of Nanog distributed into multiple positive feedback loops suggests a role in fine-tuning the level of pluripotency stability, while being dispensable for both mESC maintenance (39) and mEpiSC maintenance (40). In sharp contrast, the role of Oct4 drastically changes throughout pluripotency, from an ambivalent dose-dependent role in mESC self-renewal to a strongly supporting role in mEpiSC maintenance. The importance of an inhibitory role of Oct4 toward naive-specific factors in the model coincides with the two key assumptions that its overexpression promotes mESC differentiation while its moderate downregulation stabilizes the mESC state (41, 42), and that Oct4 is likely to upregulate Tcf3 (43) and Erk (44, 45). In contrast, the pluripotency-sustaining role of Oct4 becomes the most effective, and apparent mainly toward supporting the primed/mEpiSC state, consistent with the observation that Oct4 mediates Activin/Smad signaling as both a target (46) and a coactivator (34) of Smad2/3 transcriptional activity. Supplementing the model with regulations of the targets of Oct4 and their redistribution between mESCs and mEpiSCs (47, 48) should refine the mechanisms contributing to the dose-dependent and stage-dependent roles of Oct4.
Developmental and induced pluripotent transitions
The robust developmental progression from a naive to a primed state before differentiation has been corroborated by in vitro observations that the forward naive-to-primed transition is much more likely than the reverse primed-to-naive transition (5). In particular, the naive/mESC cells subjected to LIF removal are prone to transiently acquire a phenotype resembling primed/mEpiSC cells before differentiation, as evidenced by the transient upregulation of Fgf5 and the maintenance or reactivation of Oct4 and Nanog expression (40, 49). Without intending to match these data, optimized models are nevertheless found to exhibit a transitional trajectory through a primed-like state, which is highly robust to heterogeneities and where the primed state stability can be finely tuned by extrinsic signals. This dynamical behavior reflects the existence of a low-dimensional attracting slow manifold, which is prone to occur in a network with multiple strong and antagonistic feedback regulations and which has been proposed to contribute to the developmental canalization through successive states against extrinsic and intrinsic sources of noise (50, 51). Methods for reconstructing global potential landscapes would be valuable for a more comprehensive analysis of transition paths and rates (26, 52).
In sharp contrast, the reverse primed-to-naive transition is performed in vitro by a fastidious process that requires multiple passaging of mEpiSC in mESC signaling culture conditions (53) and whose efficiency can be increased by using 2i (54) or upregulating specific pluripotent factors such as Klf4 (55), Esrrb (56), or Stat3 (57, 58). Model simulations confirm that LIF stimulation in primed/mEpiSCs cells is often not sufficient to restore a naive/ESC state and needs to be supplemented with 2i or overexpression of naive-specific factors to optimally succeed. However, the reversion efficiency seems much lower in experiments than in simulations, which may be explained by the acknowledged role of epigenetic locks and barriers opposing the reversion toward the naive/mESC state (59). The control of pluripotency factor expression by various epigenetic mechanisms operating at the level of gene transcription (60) or of mRNA stability (61) has been proposed to contribute to the stringent temporal control of sequential and irreversible pluripotency decisions. In this respect, it is conceivable that the modular organization of the pluripotency transcriptional network would also facilitate dynamic interplay with epigenetic mechanisms to drive both flexible and robust fate decisions during development, notably by avoiding spurious states (30).
Author Contributions
B.P., C.K., and B P. designed the research and wrote the manuscript. B.P. performed the research, developed analytic tools, and analyzed the data.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS) grant PEPS-PTI-2012-2013 (to B.P.), the Agence National de Recherche (ANR) grants CRB-ANIM-ANR-11-INBS-0003 and ECLAIRE-ANR-14-CE16-0002-01 (to B.P.), and the Institut National de Recherche Agricole (INRA) grant credit incitatif PHASE-2012-2013 (to C.K. and B.P.).
Editor: Reka Albert.
Footnotes
Supporting Materials and Methods, eight figures, and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31160-8.
Supporting Citations
References (62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95) appear in the Supporting Material.
Supporting Material
References
- 1.Weinberger L., Ayyash M., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 2.Ying Q.-L., Wray J., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tesar P.J., Chenoweth J.G., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 4.Brons I.G., Smithers L.E., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 5.Festuccia N., Osorno R., Chambers I. The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr. Opin. Genet. Dev. 2013;23:504–511. doi: 10.1016/j.gde.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J., Chu J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartwell L.H., Hopfield J.J., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 8.Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 9.Macarthur B.D., Ma’ayan A., Lemischka I.R. Systems biology of stem cell fate and cellular reprogramming. Nat. Rev. Mol. Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herberg M., Roeder I. Computational modelling of embryonic stem-cell fate control. Development. 2015;142:2250–2260. doi: 10.1242/dev.116343. [DOI] [PubMed] [Google Scholar]
- 11.Chickarmane V., Troein C., Peterson C. Transcriptional dynamics of the embryonic stem cell switch. PLoS Comput. Biol. 2006;2:e123. doi: 10.1371/journal.pcbi.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalmar T., Lim C., Martinez Arias A. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glauche I., Herberg M., Roeder I. Nanog variability and pluripotency regulation of embryonic stem cells—insights from a mathematical model analysis. PLoS One. 2010;5:e11238. doi: 10.1371/journal.pone.0011238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chickarmane V., Olariu V., Peterson C. Probing the role of stochasticity in a model of the embryonic stem cell: heterogeneous gene expression and reprogramming efficiency. BMC Syst. Biol. 2012;6:98. doi: 10.1186/1752-0509-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herberg M., Kalkan T., Roeder I. A model-based analysis of culture-dependent phenotypes of mESCs. PLoS One. 2014;9:e92496. doi: 10.1371/journal.pone.0092496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakatos D., Travis E.D., Czirok A. Autocrine FGF feedback can establish distinct states of Nanog expression in pluripotent stem cells: a computational analysis. BMC Syst. Biol. 2014;8:112. doi: 10.1186/s12918-014-0112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herberg M., Glauche I., Roeder I. Dissecting mechanisms of mouse embryonic stem cells heterogeneity through a model-based analysis of transcription factor dynamics. J. R. Soc. Interface. 2016;13:20160167. doi: 10.1098/rsif.2016.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacArthur B.D., Sevilla A., Lemischka I.R. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat. Cell Biol. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H., Ang Y.S., Ma’ayan A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput. Biol. 2014;10:e1003777. doi: 10.1371/journal.pcbi.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn S.J., Martello G., Smith A.G. Defining an essential transcription factor program for naïve pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papatsenko D., Darr H., Lemischka I.R. Single-cell analyses of ESCs reveal alternative pluripotent cell states and molecular mechanisms that control self-renewal. Stem Cell Reports. 2015;5:207–220. doi: 10.1016/j.stemcr.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y., Lim C.L., Wernisch L. Cell signalling regulates dynamics of Nanog distribution in embryonic stem cell populations. J. R. Soc. Interface. 2013;10:20120525. doi: 10.1098/rsif.2012.0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott J., Hayashi K., Martinez-Arias A. Dissecting ensemble networks in ES cell populations reveals micro-heterogeneity underlying pluripotency. Mol. Biosyst. 2012;8:744–752. doi: 10.1039/c1mb05398a. [DOI] [PubMed] [Google Scholar]
- 24.Sasai M., Kawabata Y., Terada T.P. Time scales in epigenetic dynamics and phenotypic heterogeneity of embryonic stem cells. PLoS Comput. Biol. 2013;9:e1003380. doi: 10.1371/journal.pcbi.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez Arias A., Brickman J.M. Gene expression heterogeneities in embryonic stem cell populations: origin and function. Curr. Opin. Cell Biol. 2011;23:650–656. doi: 10.1016/j.ceb.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Li C., Wang J. Quantifying cell fate decisions for differentiation and reprogramming of a human stem cell network: landscape and biological paths. PLoS Comput. Biol. 2013;9:e1003165. doi: 10.1371/journal.pcbi.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henkel R., Hoehndorf R., Waltemath D. Notions of similarity for systems biology models. Brief. Bioinform. 2016;3:1–12. doi: 10.1093/bib/bbw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutenkunst R.N., Waterfall J.J., Sethna J.P. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzkudun M., Marcon L., Sharpe J. Data-driven modelling of a gene regulatory network for cell fate decisions in the growing limb bud. Mol. Syst. Biol. 2015;11:815. doi: 10.15252/msb.20145882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang A.H., Li H., Mehta P. Epigenetic landscapes explain partially reprogrammed cells and identify key reprogramming genes. PLoS Comput. Biol. 2014;10:e1003734. doi: 10.1371/journal.pcbi.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandman O., Ferrell J.E., Jr., Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S., Guo Y.P., Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 33.De Mot L., Gonze D., Dupont G. Cell fate specification based on tristability in the inner cell mass of mouse blastocysts. Biophys. J. 2016;110:710–722. doi: 10.1016/j.bpj.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L.T., Yamaguchi S., Tada T. Nanog co-regulated by Nodal/Smad2 and Oct4 is required for pluripotency in developing mouse epiblast. Dev. Biol. 2014;392:182–192. doi: 10.1016/j.ydbio.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Stuart H.T., van Oosten A.L., Silva J.C. NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr. Biol. 2014;24:340–346. doi: 10.1016/j.cub.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim M.O., Kim S.-H., Dong Z. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat. Struct. Mol. Biol. 2012;19:283–290. doi: 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- 37.Martello G., Sugimoto T., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muñoz Descalzo S., Rué P., Martinez Arias A. A competitive protein interaction network buffers Oct4-mediated differentiation to promote pluripotency in embryonic stem cells. Mol. Syst. Biol. 2013;9:694. doi: 10.1038/msb.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers I., Colby D., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 40.Osorno R., Tsakiridis A., Wilson V. The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development. 2012;139:2288–2298. doi: 10.1242/dev.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karwacki-Neisius V., Göke J., Chambers I. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12:531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radzisheuskaya A., Chia Gle. B., Silva J.C.R. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat. Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi F., Pereira L., Merrill B.J. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan H., Corbi N., Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 45.Li L., Sun L., Jin Y. Stk40 links the pluripotency factor Oct4 to the Erk/MAPK pathway and controls extraembryonic endoderm differentiation. Proc. Natl. Acad. Sci. USA. 2010;107:1402–1407. doi: 10.1073/pnas.0905657107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K.L., Lim S.K., Lim B. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011;7:e1002130. doi: 10.1371/journal.pgen.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun C., Nakatake Y., Yokota T. Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol. Cell. Biol. 2009;29:4574–4583. doi: 10.1128/MCB.01863-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buecker C., Srinivasan R., Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trott J., Martinez Arias A. Single cell lineage analysis of mouse embryonic stem cells at the exit from pluripotency. Biol. Open. 2013;2:1049–1056. doi: 10.1242/bio.20135934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manu S., Surkova S., Reinitz J. Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors. PLoS Comput. Biol. 2009;5:e1000303. doi: 10.1371/journal.pcbi.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeuty B., Kaneko K. Requirements for efficient cell-type proportioning: regulatory timescales, stochasticity and lateral inhibition. Phys. Biol. 2016;13:026007. doi: 10.1088/1478-3975/13/2/026007. [DOI] [PubMed] [Google Scholar]
- 52.Wang J., Zhang K., Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:8257–8262. doi: 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao S., Tang F., Surani M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva J., Nichols J., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo G., Yang J., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Festuccia N., Osorno R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J., van Oosten A.L., Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Oosten A.L., Costa Y., Silva J.C.R. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat. Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rais Y., Zviran A., Hanna J.H. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 60.Hemberger M., Dean W., Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 61.Geula S., Moshitch-Moshkovitz S., Hanna J.H. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 62.Boeuf H., Hauss C., Kedinger C. Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J. Cell Biol. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bourillot P.-Y., Aksoy I., Savatier P. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- 64.Carey B.W., Finley L.W., Thompson C.B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cartwright P., McLean C., Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 66.Chew J.L., Loh Y.H., Ng H.H. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cole M.F., Johnstone S.E., Young R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Do D.V., Ueda J., Fu X.Y. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013;27:1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Filipczyk A., Marr C., Schroeder T. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat. Cell Biol. 2015;17:1235–1246. doi: 10.1038/ncb3237. [DOI] [PubMed] [Google Scholar]
- 70.Greber B., Wu G., Schöler H.R. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 71.Hall J., Guo G., Smith A. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Hann S.R., Eisenman R.N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol. Cell. Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hishida T., Nozaki Y., Okuda A. Indefinite self-renewal of ESCs through Myc/Max transcriptional complex-independent mechanisms. Cell Stem Cell. 2011;9:37–49. doi: 10.1016/j.stem.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 74.Huang G., Yan H., Ying Q.-L. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem Cells. 2014;32:1149–1160. doi: 10.1002/stem.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Illich D.J., Zhang M., Schöler H.R. Distinct signaling requirements for the establishment of ESC pluripotency in late-stage EpiSCs. Cell Reports. 2016;15:1–14. doi: 10.1016/j.celrep.2016.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang J., Chan Y.-S., Ng H.-H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 77.Loh Y.H., Wu Q., Ng H.H. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 78.Mayshar Y., Rom E., Benvenisty N. Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem Cells. 2008;26:767–774. doi: 10.1634/stemcells.2007-1037. [DOI] [PubMed] [Google Scholar]
- 79.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 80.Paling N.R., Wheadon H., Welham M.J. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- 81.Pereira L., Yi F., Merrill B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Relógio A., Westermark P.O., Herzel H. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput. Biol. 2011;7:e1002309. doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reynolds N., Latos P., Hendrich B. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodda D.J., Chew J.-L., Robson P. Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 85.Sears R., Nuckolls F., Nevins J.R. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suresh B., Lee J., Ramakrishna S. The importance of ubiquitination and deubiquitination in cellular reprogramming. Stem Cells Int. 2016;2016:6705927. doi: 10.1155/2016/6705927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turco M.Y., Furia L., Lanfrancone L. Cellular heterogeneity during embryonic stem cell differentiation to epiblast stem cells is revealed by the ShcD/RaLP adaptor protein. Stem Cells. 2012;30:2423–2436. doi: 10.1002/stem.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vallier L., Mendjan S., Pedersen R.A. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van den Berg D.L.C., Zhang W., Poot R.A. Estrogen-related receptor β interacts with Oct4 to positively regulate Nanog gene expression. Mol. Cell. Biol. 2008;28:5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watanabe S., Umehara H., Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 91.Weidgang C.E., Seufferlein T., Mueller M. Pluripotency factors on their lineage move. Stem Cells Int. 2016;2016:6838253. doi: 10.1155/2016/6838253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wray J., Kalkan T., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang X., Zhang J., Pei D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J. Biol. Chem. 2008;283:35825–35833. doi: 10.1074/jbc.M803481200. [DOI] [PubMed] [Google Scholar]
- 94.Zhang P., Andrianakos R., Lu W. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 2010;285:9180–9189. doi: 10.1074/jbc.M109.077958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang K., Li L., Jing N. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development. 2010;137:2095–2105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.