Abstract
Background:
The 15q24-25 loci contain genes (CHRNA5 and CHRNA3) encoding nicotinic acetylcholine receptor subunits. We here determined for the first time the association of genetic variants rs16969968 and rs3743074 in CHRNA5 and CHRNA3, respectively, on nicotine dependence and lung cancer risk in a North Indian population by a case-control approach.
Methods:
Venous blood samples were obtained from 324 participants (108 lung cancer patients and 216 healthy individuals). DNA was extracted and PCR amplified with primers flanking the SNPs rs16969968 and rs3743074. Amplicons were subjected to sequencing and logistic regression was used to analyze association between variables.
Results:
The risk variant SNP rs16969968 in both heterozygous and homozygous forms appeared to exert a significant effect on nicotine dependence [GA (OR=2.77) and AA (OR=2.53)]. As expected, smoking was strongly associated with lung cancer (OR= 2.62). Risk allele rs16969968 in CHRNA5 also showed a significant association with increased lung cancer risk in our cohort, alone (OR= 4.99) and with smoking as a co-variable (OR= 4.28). Comparison of our analysis with other populations suggested that individuals with rs16969968 risk allele in the Indian population are more susceptible to lung cancer.
Conclusion:
Overall, the results strongly indicated that, in our cohort North Indian population, the genetic variant rs16969968, but not rs3743074, is significantly associated with both nicotine dependence and increased risk of lung cancer. While the results are significant, there is further need to increase the sample size and improve precision of our risk prediction.
Keywords: Lung cancer, smoking, SNP, rs16969968, CHRNA5, north Indian
Introduction
Lung cancer comprises 21.8% of all cancer related deaths in developed nations. Based on a recent report from the International Agency for Research on Cancer, majority of new lung cancer cases (58%) occurred in the less developed regions. In India, about 6.9% of all cancer cases and 9.3% of all cancer related deaths can be attributed to lung cancer (Ferlay et al., 2012) Recent reports from Indian Council of Medical Research (ICMR) had estimated 114 thousands new lung cancer cases to be reported in 2016 likely to increase to 140 thousands in 2020, which would make it the second most common type of cancer in India. Of all the cancer cases, only 12.5% are detected at an early stage (http://icmr.nic.in/icmrsql/archive/2016/4.pdf), diminishing the chances of disease-free progression considerably. Therefore, there is an urgent need to identify predisposing genetic factors along with other preventive health measures, to help decrease the disease burden in different populations. Unfortunately, despite its incidence there are very few studies on predisposition markers for lung cancer from India.
Tobacco abuse has a tremendous impact on public health, with an estimated 6 million deaths per year, roughly half of its users (http://www.who.int/mediacentre/factsheets/fs339/en/). Unlike western countries, in India tobacco is used in various forms such as cigarette, bidi, hookah, chillum, gutka and pan masala. Bidi smoking, which is extremely common in rural India, carries a higher risk of lung cancer compared to cigarette smoking (Pathak et al., 2004). Smoking, being the single largest factor responsible for lung cancer development, contributes to 80% of all lung cancer cases. However, only 10–20% of smokers develop lung cancer (Young et al., 2009), suggesting that predisposition to onset and progression of the disease may arise due to genetic variation. In 2008, three independent genome wide association studies (GWAS) conducted on Caucasian population concluded that the 15q24–25 variant increases lung cancer risk either directly or indirectly through smoking (Amos et al., 2008; Hung et al., 2008; Thorgeirsson et al., 2008). This 15q24–25 locus spanning 203 kb contains genes encoding the a5, a3, and b4 nicotinic acetylcholine receptor (nAChR) subunits (CHRNA5/A3/B4) (Bierut et al., 2008). Shriashi et al., (2009) observed that CHRNA5 SNP-rs16969968 is significantly associated with lung cancer in Japanese population even though the risk allele is less frequent (<2%). In contrast, while rs16969968 variants was found to play little role in risk of lung cancer and nicotine dependence in Chinese population, four new SNPs in the 15q region were significantly associated with both (Wu et al., 2009). In African-American and European populations too, various SNPs in CHRNA5, CHRNA3 and CHRNB4 gene cluster were found to be associated with lung cancer in the genome-wide scan (Broderick et al., 2009; Amos et al., 2010; Hansen et al., 2010).
The strong biological consistency but variable arrangement of genetic correlation between SNPs of 15q24-25 region with lung cancer and nicotine dependence in different populations encouraged us to determine the yet unknown association in the Indian population. In this paper, we have used the term nicotine dependence and urge to smoke, interchangeably. To the best of our knowledge from published literature, no other study has examined the two SNPs rs16969968 and rs3743074, and analyzed their effect on nicotine dependence and lung cancer risk in an Indian population.
Materials and Methods
Sample Collection
Of the 324 participants in this case-control study, 108 were patients diagnosed for lung cancer at the Outpatient Department clinic and ward at Department of Pulmonary Medicine and Sleep Disorders, All India Institute of Medical Sciences (AIIMS), New Delhi which attracts patients from various parts of North India. Diagnosis was made by cytological, imaging and histopathological examinations. Control subjects include 216 age-matched individuals with no history of lung cancer or any chronic lung-related disease, visiting AIIMS and Department of Biochemistry, Ram Manohar Lohia (RML) Hospital, New Delhi. Patients diagnosed with any other cancer were excluded from this study. A detailed questionnaire was filled up by each subject providing information on age, sex, smoking status, residential region, and family history of cancer. This study was approved by the institutional ethics committee of Delhi University South Campus, AIIMS and RML Hospital and due diligence was followed to explain the study to all participants before taking their consent for participation.
DNA isolation from blood samples
About 1ml of blood was drawn into tubes containing EDTA from all study subjects. DNA was isolated from the whole blood with the Sigma’s GenElute™ Blood Genomic DNA kit (Sigma Aldrich, USA) as per the manufacturer’s instructions. DNA was quantitated spectrophotometrically (NanoDrop, 2000, Thermo Scientific, US). 260/280 nm ratio was used to confirm quality. Extracted DNA showed a 260/280 nm of > 1.5.
Tag single nucleotide polymorphism (tSNP) selection and genotyping
The Gujarati Indian Houston (GIH) data given in HapMap project was used for SNP selection. The linkage disequilibrium (LD) structure was constructed with the help of Haploview version 4.2, where pairwise LD between all the SNPs within 70-kb stretch of 15q25 region (chr15:76650000 -76720000) was measured as r2 and represented as different color (increasing shades of grey) at the intersection of the diagonals from each SNP (Barrett et al., 2005). White represents no LD (r2 = 0), black signifies maximum LD (r2 =1), and different shades of gray correspond to intermediate LDs (Figure 1). Tag SNP selection was done at r2 >0.8 and MAF > 0.2 and two SNPs rs16969968 and rs3743074 were chosen for the study which were in LD with 19 of 31 SNPs (51%) of the region with r2>0.8.
Figure 1.
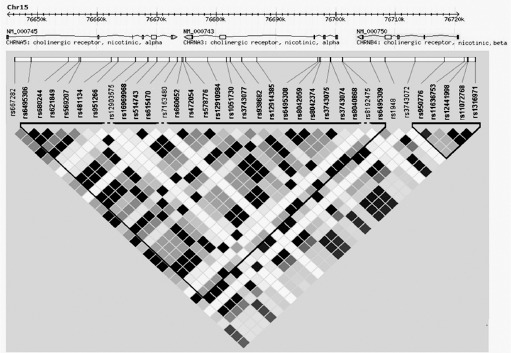
Linkage disequilibrium (LD) structure of 70kb 15q25 region in individuals of GIH ancestry. Boxes are shaded according to r2 values derived in Haploview (v3.2). Triangular border areas denote haplotype blocks assigned according to the method of Gabriel et al
Amplification, purification and sequencing
PCR product was generated using primers for the two selected SNPs rs16969968 in CHRNA5 {forward (5’-3’) GCTGGTATCCGTATGTCACTTAC and reverse (5’-3’) GTCAACAATTCTGGCCCTC} and rs3743074 in CHRNA3 {forward (5’-3’) AAGGAAGGCCAGGTTTTAAGCACAG and reverse (5’-3’) ATCATGGCATGGATGTGGTTCGG}, using Taq DNA polymerase (New England Biolabs, USA). PCR product was purified with the help of QIAquick PCR purification kit (Qiagen, Germany) and sequenced using an automated sequencing system (3130x1 Genetic Analyzer; Applied Biosystems Divisions, Foster City, CA).
Statistical Analysis
For each SNP allele frequencies were tested for departure from Hardy–Weinberg equilibrium (HWE) using Chi-square Hardy-Weinberg equilibrium test calculator (Rodriguez et al., 2009). The Bioinformatics Institute’s Online Sample Size Estimator (OSSE) was used to compute the statistical power and calculate minor allele frequency (MAF) in cases and controls. Logistic regression models were built to estimate contribution of smoking and genetic variants of the two SNPs with lung cancer, and on nicotine dependence. The effects were estimated using odds ratios (OR), chi (χ2) square test and corresponding 95% confidence intervals (CI). To compare the validity of different models, the areas under the Receiver Operating Characteristic (ROC) curve (AUC) were computed (Supplementary figures 1A and 1B). The statistical analysis was done using the computer software R, version 3.3.1.
Results
We have performed this study on 324 individuals, including 108 lung cancer patient samples and 216 controls. Both groups belong to similar age interval, 40-75 years. The main clinical characteristics of patients and controls are summarized in Supplementary Table 1.
Supplementary Table 1.
Patient Characteristics
| Variables | Cases | Controls |
|---|---|---|
| Total No. | 108 | 216 |
| Age Interval (years) | 40-75 | 40-75 |
| Sex | ||
| Male | 96 (88.9%) | 164 (75.9%) |
| Female | 12 (11.1%) | 52 (24.1%) |
| Smoking Status | ||
| Non-Smoker | 34 (31.5%) | 118 (54.6%) |
| Smoker | 74 (68.5%) | 98 (45.4%) |
Minor allele frequency is in agreement with the GIH population
In GIH population, the 70-kb region on 15q25 locus splits into 2 blocks and at r2 >0.8 and MAF > 0.2 was tagged by 5 SNPs (Figure 1). rs11636753 not being in LD with the other SNPs and rs6495308 and rs12441998 having MAF (>0.45) were not taken into analysis in the absence of adequate statistical power (>80%) for our sample size. The two SNPs rs16969968 and rs3743074 chosen for the study were in LD with 19 of 31 SNPs (51%) of the 15q25 region with r2 >0.8. Both the cases and the controls were examined separately for distributions within the parameters of HWE and χ2 test and revealed no evidence of deviation from HWE for either of the chosen SNPs (Table 1). The minor allele frequency of both SNP as present in our population is similar to GIH population and very near to South Asian populations but differ drastically from other HapMap population (Table 2).
Table 1.
Chi-sq (χ2 ) Hardy-Weinberg Equilibrium (HWE) Test for Cases and Controls
| SNP | Genes | HWE χ2 (Cases) | HWE χ2 (Controls) |
|---|---|---|---|
| rs16969968 | CHRNA5 | 1.12 | 0.1 |
| rs3743074 | CHRNA3 | 0.07 | 0.26 |
Table 2.
Minor Allele Frequencies of rs169969968 and rs3743074 in Different HapMap Populations
| Label | Population | rs16969968 | rs3743074 |
|---|---|---|---|
| AFR | African | 0.023 | 0.43 |
| AMR | American | 0.209 | 0.199 |
| EAS | East Asian | 0.027 | 0.447 |
| EUR | European | 0.366 | 0.343 |
| SAS | South Asian | 0.182 | 0.343 |
| GIH | Gujarati Indians in Houston, Texas | 0.204 | 0.325 |
| NI | North Indians, India (present study) | 0.213 | 0.329 |
Genetic models built for association study using logistic regression
The minor allele of each SNP, that is ‘A’ in rs16969968 and ‘G’ in rs3743074 were hypothesized as risk alleles as compared to their wild type counterparts and their association with smoking (Table 3) and lung cancer (Table 4) were analyzed separately in different models. In genotype model individuals with heterozygous risk allele and homozygous risk allele were compared separately to the wild type counterpart. In dominant model the risk allele carrier groups were combined together and compared with wild type individuals. The recessive model involved the comparison of the homozygous risk allele carrying individuals with the major allele carrier group combined together.
Table 3.
Association of rs169969968 and rs3743074 with Smoking
| SNP ID | Variables | GENOTYPE MODEL | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | SE | Z score | p Value | ||
| rs16969968 | Het (GA) vs Common Hz (GG) | 2.53 | 1.52-4.20 | 0.65504 | 3.573 | 0.0004 |
| Rare Hz (AA) vs Common Hz (GG) | 2.77 | 1.11-6.92 | 1.29294 | 2.187 | 0.0287 | |
| rs3743074 | Het (AG) vs Common Hz (AA) | 1.21 | 0.73-2.00 | 0.30998 | 0.736 | 0.4617 |
| Rare Hz (GG) vs Common Hz (AA) | 1.27 | 0.56-2.87 | 0.55947 | 0.57 | 0.5687 | |
| Intercept | 0.68 | 0.44-1.05 | 0.154 | -1.746 | 0.0808 | |
| SNP ID | Variable | DOMINANT MODEL | ||||
| OR | 95% CI | SE | Z score | p Value | ||
| rs16969968 | (GA+AA) vs GG | 2.55 | 1.57-4.13 | 0.62666 | 3.799 | 0.0001 |
| rs3743074 | (AG+GG) vs AA | 1.21 | 0.75-1.95 | 0.29398 | 0.772 | 0.4401 |
| Intercept | 0.69 | 0.44-1.05 | 0.15 | -1.732 | 0.0832 | |
| SNP ID | Variable | RECESSIVE MODEL | ||||
| OR | 95% CI | SE | Z score | p Value | ||
| rs16969968 | AA vs (GA+GG) | 1.7 | 0.73-3.95 | 0.73123 | 1.234 | 0.2172 |
| rs3743074 | GG vs (AG+AA) | 0.79 | 0.38-1.66 | 0.29774 | -0.615 | 0.5386 |
| Intercept | 1.11 | 0.87-1.41 | 0.13644 | 0.858 | 0.3909 | |
Table 4.
Association of rs169969968 and rs3743074 with Lung Cancer; A, with Crude Odds Ratio ; B, with Odds Ratio Adjusted for Smoking
| A | ||||||||||||
| SNP ID | Variable | GENOTYPE MODEL | ||||||||||
| OR | 95% CI | SE | Z score | p Value | ||||||||
| rs16969968 | Het (GA) vs Common Hz (GG) | 1.66 | 0.98-2.81 | 0.4462 | 1.872 | 0.0612 | ||||||
| Rare Hz (AA) vs Common Hz (GG) | 4.99 | 1.97-12.64 | 2.36624 | 3.39 | 0.0007 | |||||||
| rs3743074 | Het (AG) vs Common Hz (AA) | 0.92 | 0.54-1.56 | 0.24781 | -0.313 | 0.7543 | ||||||
| Rare Hz (GG) vs Common Hz (AA) | 0.72 | 0.28-1.88 | 0.35331 | -0.661 | 0.5086 | |||||||
| Intercept | 0.38 | 0.24-0.61 | 0.09148 | -4.019 | <0.0001 | |||||||
| SNP ID | Variable | DOMINANT MODEL | ||||||||||
| OR | 95% CI | SE | Z score | p Value | ||||||||
| rs16969968 | Het + Rare Hz (GA+AA) vs GG | 2 | 1.21-3.28 | 0.50547 | 2.728 | 0.0064 | ||||||
| rs3743074 | Het + Rare Hz (AG+GG) vs AA | 0.79 | 0.48-1.30 | 0.19944 | -0.95 | 0.3421 | ||||||
| Intercept | 0.4 | 0.26-0.37 | 0.09441 | -3.879 | <0.0001 | |||||||
| SNP ID | Variable | RECESSIVE MODEL | ||||||||||
| OR | 95% CI | SE | Z score | p Value | ||||||||
| rs16969968 | Rare Hz (AA) vs (GA+GG) | 4.09 | 1.75-9.60 | 1.77136 | 3.256 | 0.0011 | ||||||
| rs3743074 | Rare Hz (GG) vs (AG+AA) | 0.61 | 0.25-1.46 | 0.27148 | -1.117 | 0.2640 | ||||||
| Intercept | 0.46 | 0.36-0.60 | 0.06088 | -5.862 | <0.0001 | |||||||
| B | ||||||||||||
| SNP ID | Variable | GENOTYPE MODEL | ||||||||||
| OR | 95% CI | SE | Z score | p Value | ||||||||
| Smoking | 2.4 | 1.45-3.98 | 0.61933 | 3.404 | 0.0007 | |||||||
| rs16969968 | Het (GA) vs Common Hz (GG) | 1.38 | 0.80-2.39 | 0.38566 | 1.167 | 0.2432 | ||||||
| Rare Hz (AA) vs Common Hz (GG) | 4.29 | 1.65-11.09 | 2.07795 | 3.003 | 0.0027 | |||||||
| rs3743074 | Het (AG) vs Common Hz (AA) | 0.88 | 0.51-1.51 | 0.2424 | -0.462 | 0.6441 | ||||||
| Rare Hz (GG) vs Common Hz (AA) | 0.68 | 0.26-1.80 | 0.33705 | -0.78 | 0.4354 | |||||||
| Intercept | 0.26 | 0.15-0.44 | 0.071 | -4.924 | <0.0001 | |||||||
| SNP ID | Variable | DOMINANT MODEL | ||||||||||
| OR | 95% CI | SE | Z score | p Value | ||||||||
| Smoking | 2.37 | 1.44-3.91 | 0.60464 | 3.396 | 0.0007 | |||||||
| rs16969968 | Het + Rare Hz (GA+AA) vs GG | 1.68 | 1.01-2.81 | 0.44027 | 1.984 | 0.0473 | ||||||
| rs3743074 | Het + Rare Hz (AG+GG) vs AA | 0.75 | 0.45-1.24 | 0.19414 | -1.115 | 0.2649 | ||||||
| Intercept | 0.27 | 0.16-0.46 | 0.0737 | -4.811 | <0.0001 | |||||||
| SNP ID | Variable | RECESSIVE MODEL | ||||||||||
| OR | 95% CI | SE | Z score | p Value | ||||||||
| Smoking | 2.54 | 1.55-4.16 | 0.64166 | 3.68 | 0.0002 | |||||||
| rs16969968 | Rare Hz (AA) vs (GA+GG) | 3.89 | 1.63-9.27 | 1.72306 | 3.065 | 0.0022 | ||||||
| rs3743074 | Rare Hz (GG) vs (AG+AA) | 0.63 | 0.26-1.53 | 0.286 | -1.023 | 0.3063 | ||||||
| Intercept | 0.27 | 0.18-0.41 | 0.0559 | -6.332 | <0.0001 | |||||||
Significant association of single minor allele at SNP rs16969968 with smoking
Both CHRNA5 and CHRNA3 are associated with nicotine dependence (Saccone et al., 2007), therefore we assessed the relationship between SNP rs16969968 in CHRNA5 gene and rs3743074 in CHRNA3 gene with smoking in Indian population (Table 3). Our analysis shows that rs3743074 has no significant association with smoking (Z score < 2). However, both GA and AA genotype at rs16969968 were found to be significantly associated with smoking in our genotype model. The dominant model suggests a strong association of rs16969968 SNP with nicotine dependence compared to others. Also, the smoking outcome for different alleles indicate that while a single copy of allele (GA) is enough to create an urge to smoke, the presence of an additional allele (AA) does not increase the nicotine dependence in our dataset.
Significant association of smoking with lung cancer risk
Cigarette smoking being the strongest determinant of lung cancer is a key indicator of both high-risk populations and high-risk individuals (Haiman et al., 2006). Association of smoking with lung cancer in our population was significantly associated with increased risk for lung cancer (OR= 2.62, 95% CI=1.61-4.26, Z-score= 3.882, p value = 0.000104) irrespective of other factors.
Significant associations of AA genotype at rs16969968 with lung cancer
To explore the association of rs16969968 and rs3743074 with lung cancer, the genotype distribution between lung cancer cases and controls were compared and although no significant association (Z-score < 2) was established between rs3743074 and lung cancer risk in Indian population, rs16969968 in CHRNA5 was found to be strongly associated with elevated risk for lung cancer as indicated by various models. The ORs (Table 4A) and adjusted ORs (with smoking included as a variable in the model) (Table 4B) of genotype model showed that AA genotype at rs16969968 conferred significantly increased ORs for lung cancer risk (OR= 4.99, 95% CI=1.97-12.63, Z-score= 3.39), while a single copy of rs16969968 risk allele (present in GA) conferred no significant effect on lung cancer risk. Adjusted ORs, too, indicated that AA genotype at rs16969968 in CHRNA5 conferred elevated risk for lung cancer (OR= 4.28, 95% CI=1.66-11.08, Z-score= 3.003). Similarly, rs16969968 in CHRNA5 showed increased crude and adjusted ORs in recessive model, suggesting its strong association with increased lung cancer risk in Indian population. Conversely, both the crude OR (OR= 1.996, 95% CI=1.21-3.28, Z-score= 2.728) and adjusted OR (OR= 1.68, 95% CI=1.00-2.81, Z-score= 1.984) as identified by dominant model suggests that presence of single copy of minor allele of rs16969968 was insufficient for enhancing the lung cancer risk; implying a recessive effect of risk allele on lung cancer. For rs16969968, among nonsmokers, 14 of 34 (41.18%) lung cancer patients and 37 of 118 (31.36%) control were heterozygous (GA) and/or homozygous minor allele (AA). For rs3743074, 17/34 (50%) patients and 62/118 (52.54%) controls were heterozygous (AG) and/or homozygous (GG) for the minor allele in the nonsmoking group. While there is also an increase in association of minor allele with lung cancer without the effect of smoking in rs16969968, a larger sample size is required to statistically state a direct association.
Discussion
Nicotine acetylcholine receptors (nAChRs) are ligand-gated cation channels that form membrane bound pentameric structures assembled from a family of subunits that include CHRNA1-A10 and CHRNB1-B4 and gets activated by the endogenous neurotransmitter acetylcholine and exogenous agonists, such as nicotine or tobacco-specific nitrosamines NNN or NNK (Lindstrom, 1996; Lindstrom, 1997; Millar, 2003). Apart from their expression in brain and a classical role in neuronal signaling particularly in addiction pathways (Leslie et al., 2013), nAChRs are expressed in numerous other cell type including the bronchial epithelium (Proskocilet al., 2004; Grando et al., 2007; Millar and Gotti, 2009) where they can modulate proliferation, survival, angiogenesis, migration and invasion pathway via intracellular calcium influx (Improgo et al., 2011). Signaling pathways activated by nAChR explain plausible associations between SNPs in 15q24 region, smoking behaviors, and lung cancer risk. 15q24-25 region has been widely studied for association with smoking and lung cancer in various populations (Amos et al., 2008; Hung et al., 2008;Thorgeirsson et al., 2008; Shiraishi et al., 2009; Wu et al., 2009; Hansen et al., 2010 ; Islamet al., 2013). However till date, no such study has been conducted in an Indian population, which often behaves differently from other South Asian population (Majumder, 2010).
In this cohort of North Indian population, we observed that SNP rs16969968 in chromosome 15q24-25 region posed as a risk allele for nicotine dependence and lung cancer. Individuals with either one or both copies of rs16969968 risk allele were likely to be more addicted to smoking. Both the genotype model and the dominant model were a significantly better fit than its recessive model for nicotine dependence. Since, smoking is the largest confounding factor associated with lung cancer, we therefore, studied the effect of smoking on lung cancer in Indian population. The calculated ORs indicated that smokers are, ceteris paribus, 2.6 times more likely to have lung cancer than nonsmokers in this North Indian population.
Many population based studies suggest an association of 15q24-25 region SNPs with lung cancer (Table 5). Population based differences in the smoking related risk of lung cancer have also been determined (Schwartz and Swanson, 1997; Stellman, 2003). Haiman and colleagues have reported that African American and Native Hawaiians have greater risk for lung cancer than Caucasian, Asian and other United States based ethnic groups (Haiman et al., 2006).
Table 5.
Association of key SNPs in CHRNA3 and CHRNA5 with Lung Cancer in Different Populations
| Gene | SNP | Risk allele | MAF | Population | OR (p value) | Relation with lung cancer | Reference |
|---|---|---|---|---|---|---|---|
| CHRNA3 | rs1051730 | A>G | 0.36 | USA and UK | 1.8 (4.6 X 10-12) | association | Amos et al., (2008) |
| CHRNA3 | rs1051730 | A>G | 0.36 | European | 1.7 (1.1X10-7) | association | Thorgeirsson et al., (2008) |
| CHRNA5 | rs16969968 | G>A | 0.37 | European | 1.3 (1X10-20) | association | Hung et al., (2008) |
| CHRNA5 | rs16969968 | G>A | 0.04 | Chinese | 0.84 (0.347) | no association | Wu et al., (2009) |
| CHRNA5 | rs16969968 | G>A | 0.04 | African American | 1.27 (0.145) | association | Hansen et al., (2010) |
| CHRNA5 | rs17486278 | T>G | 0.29 | 1.28 (0.008) | association | ||
| CHRNA5 | rs16969968 | G>A | 0.001 | Japanese | 2.2 (0.00015) | association | Shiraishi et al., (2009) |
| CHRNA3 | rs1051730 | A>G | 0.014 | 2.4 (0.000088) | association | ||
| CHRNA5 | rs16969968 | G>A | 0.21 | Bangladeshi | 1.28 (not scored) | no association | Islam et al., (2013) |
| CHRNA5 | rs16969968 | G>A | 0.21 | Indian | 4.29 (0.0027) | association | Our study |
| CHRNA3 | rs3743074 | A>G | 0.33 | 0.68 (0.4354) | no association |
From our study population, we can conclude that CHRNA5 rs16969968 polymorphism is strongly associated with an increased risk of lung cancer. The recessive model emerged as a highly significant representative of the association of rs16969968 polymorphism with lung cancer risk. This was also reiterated in the genotype model where a single copy of risk allele does not show significant association to lung cancer, while two copies of risk allele confer approximately 4.5 times higher risk of lung cancer to the individual. A comparison of our analysis with others suggests that individuals with rs16969968 risk allele in Indian population are more susceptible to lung cancer than other populations. This may also be attributed to unique and different smoking habits of Indians, which includes bidi smoking. While rs16969968 has been shown to have direct correlation to lung cancer, this varies across populations. Our data-set has lesser number of nonsmoking cancer cases, and hence it will not be statistically appropriate to either establish or rule out a direct correlation of this SNP with lung cancer. We hope to address this in further studies. The effect of other environmental factors such as exposure to urban pollution, passive smoke, occupational factors, exposure to smoke from different cooking fuels, etc are not accounted for in the interpretation of these studies.
Overall our study shows that the nAchR subunit, rs16969968 polymorphism is strongly associated with nicotine dependence and an increased risk of lung cancer in our North Indian population. It was marked that the missense SNP rs16969968 having a change from aspartic acid to asparagines at codon 398 in CHRNA5 can cause 2-fold increase in risk of developing nicotine dependence once exposed to tobacco smoking (Saccone, et al., 2007) and thus can cause an increase in smoking dependent lung cancer risk. nAChRs are expressed in adenocarcinoma and small cell carcinoma tissue samples and CHRNA7 was found to promote cell proliferation in poorly differentiated NSCLC cells by mediating the pro-proliferative activity of nicotine (Medjber et al., 2015). We have also observed that nicotine increases the expression of nAChR (Supplementary Figure 2) and can activate nAChR mediated pathway involving upregulation of genes contributing to progression of lung cancer (Mousa and Mousa, 2006; Dasgupta et al., 2006; Nishioka et al., 2010; Dasgupta et al., 2011; Lin et al., 2012). Nicotine is detrimental to existing therapeutic regimens and can inhibit chemotherapy induced cell death (Zhang et al. 2009; Wang et al. 2013; Li et al. 2015). nAChRs play a significant role in pathogenesis of lung cancer and further insight into its significance in lung cancer susceptibility will help in development of nAChR antagonist as potential drug for lung cancer therapy.
While most parameters of the analyzed genetic models have statistical significance, our findings will be greatly strengthened with an increase in the sample numbers. This will also ensure the scope of our analysis for preclinical prognosis in the future.
Abbreviations
NSCLC, non small cell lung cancer; GWAS, genome wide association studies; CHRNA5, nicotinic acetylcholine receptor subunit alpha 5; SNP, single nucleotide polymorphism; GIH, Gujarati Indian Houston; HWE, Hardy–Weinberg equilibrium; OR, odds ratio
Author Contributions
NP designed and performed experiments, data analysis and manuscript writing. SP did data analysis and manuscript writing. LKS, RG and AM helped in sample collection. TS designed experiments, data analysis and manuscript writing.
Funding
This work was funded by research grant from University of Delhi R&D grant and Department of Biotechnology to TS. We also thank UGC for SAP (BRS III), DST for FIST (level II) program and DU- DST PURSE (Phase II) for supporting our work.
Acknowledgments
NP is a recipient of UGC-SAP Meritorious Award from University of Delhi. We thank Ms. Shabana Ansari and Mr Krishna Gopal for technical help. Sequencing was performed at the Central Instrumentation Facility at University of Delhi South Campus.
References
- Amos CI, Gorlov IP, Dong Q, et al. Nicotinic acetylcholine receptor region on chromosome 15q25and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102:1199–205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–41. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta P, Rastogi S, Pillai S, et al. Nicotine induces cell proliferation by beta-arrestin-mediatedactivation of Src and Rb-Raf-1 pathways. J Clin Invest. 2006;116:2208–17. doi: 10.1172/JCI28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta P, Rizwani W, Pillai S, et al. ARRB1-mediated regulation of E2F target genes innicotine-induced growth of lung tumors. J Natl Cancer Inst. 2011;103:317–33. doi: 10.1093/jnci/djq541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Ervik M, et al. Globocan 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; 2013; 2014. [[accessed on January 21, 2014]]. http://globocan.iarc.fr . [Google Scholar]
- Grando SA, Kawashima K, Kirkpatrick CJ, et al. Recent progress in understanding the non-neuronal cholinergic system in humans. Life Sci. 2007;80:2181–5. doi: 10.1016/j.lfs.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Grucza RA, Wang JC, Stitzel JA, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–9. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Stram DO, Wilkens LR, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354:333–42. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- Hansen HM, Xiao Y, Rice T, et al. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African-Americans. Hum Mol Genet. 2010;19:3652–61. doi: 10.1093/hmg/ddq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Improgo MR, Tapper AR, Gardner PD. Nicotinic acetylcholine receptor-mediated mechanisms in lung cancer. Biochem Pharmacol. 2011;82:1015–21. doi: 10.1016/j.bcp.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Islam MS, Ahmed MU, Sayeed MS, et al. Lung cancer risk in relation to nicotinic acetylcholine receptor, CYP2A6 and CYP1A1 genotypes in the Bangladeshi population. Clin Chim Acta. 2013;416:11–9. doi: 10.1016/j.cca.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Leslie FM, Mojica CY, Reynaga DD. Nicotinic receptors in addiction pathways. Mol Pharmacol. 2013;83:753–8. doi: 10.1124/mol.112.083659. [DOI] [PubMed] [Google Scholar]
- Li H, Wang S, Takayama K, et al. Nicotine induces resistance to erlotinib via cross-talk between α1 nAChR and EGFR in the non-small cell lung cancer xenograft model. Lung Cancer. 2015;88:1–8. doi: 10.1016/j.lungcan.2015.01.017. [DOI] [PubMed] [Google Scholar]
- Lin W, Hirata N, Sekino Y, et al. Role of α7-nicotinic acetylcholine receptor in normal and cancer stem cells. Curr Drug Targets. 2012;13:656–65. doi: 10.2174/1389450111209050656. [DOI] [PubMed] [Google Scholar]
- Lindstrom J. Neuronal nicotinic acetylcholine receptors. Ion Channels. 1996;4:377–450. doi: 10.1007/978-1-4899-1775-1_10. [DOI] [PubMed] [Google Scholar]
- Lindstrom J. Nicotinic acetylcholine receptors in health and disease. Mol Neurobiol. 1997;15:193–222. doi: 10.1007/BF02740634. [DOI] [PubMed] [Google Scholar]
- Majumder PP. The human genetic history of South Asia. Curr Biol. 2010;20:184–7. doi: 10.1016/j.cub.2009.11.053. [DOI] [PubMed] [Google Scholar]
- Medjber K, Freidja ML, Grelet S, et al. Role of nicotinic acetylcholine receptors in cell proliferation and tumour invasion in broncho-pulmonary carcinomas. Lung Cancer. 2015;87:258–64. doi: 10.1016/j.lungcan.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Millar NS, Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology. 2009;56:237–46. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Millar NS. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem Soc Tran. 2003;31:869–74. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- Mousa S, Mousa SA. Cellular and molecular mechanisms of nicotine's pro-angiogenesis activity and its potential impact on cancer. J Cell Biochem. 2006;97:1370–8. doi: 10.1002/jcb.20741. [DOI] [PubMed] [Google Scholar]
- Nishioka T, Guo J, Yamamoto D, et al. Nicotine, through upregulating pro-survival signaling, cooperates with NNK to promote transformation. J Cell Biochem. 2010;109:152–61. doi: 10.1002/jcb.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak AK, Bhutani M, Mohan A, et al. Non small cell lung cancer (NSCLC): current status and future prospects. Indian J Chest Dis Allied Sci. 2004;46:191–203. [PubMed] [Google Scholar]
- Press release report to the nation on the status of cancer in India (as of December 2014) based on the consolidated cancer registry reports by Indian council of medical research through its national cancer registry programme. http://icmr.nic.in/icmrsql/archive/2016/4.pdf .
- Proskocil BJ, Sekhon HS, Jia Y, et al. Acetylcholine is an autocrine or paracrine hormone synthesized and secreted by airway bronchial epithelial cells. Endocrinolog. 2004;145:2498–506. doi: 10.1210/en.2003-1728. [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–14. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AG, Swanson GM. Lung carcinoma in African Americans and whites. A population-based study in metropolitan Detroit, Michigan. Cancer. 1997;79:45–52. doi: 10.1002/(sici)1097-0142(19970101)79:1<45::aid-cncr7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Shiraishi K, Kohno T, Kunitoh H, et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- Stellman SD, Chen Y, Muscat JE, et al. Lung cancer risk in white and black Americans. Ann Epidemiol. 2003;13:294–302. doi: 10.1016/s1047-2797(02)00420-9. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Takayama K, Tanaka K, et al. Nicotine induces resistance to epidermal growth factor receptor tyrosine kinase inhibitor by α1 nicotinic acetylcholine receptor-mediated activation in PC9 cells. J Thorac Oncol. 2013;8:719–25. doi: 10.1097/JTO.0b013e31828b51d4. [DOI] [PubMed] [Google Scholar]
- World Health Organisation (WHO) Tobacco Fact sheet. http://www.who.int/mediacentre/factsheets/fs339/en/
- Wu C, Hu Z, Yu D, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–72. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- Young RP, Hopkins RJ, Hay BA, et al. Lung cancer susceptibility model based on age, family history and genetic variants. PLoS One. 2009;4:e5302. doi: 10.1371/journal.pone.0005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kamdar O, Le W, et al. Nicotine induces resistance to chemotherapy by modulating mitochondrial signaling in lung cancer. Am J Respir Cell Mol Biol. 2009;40:135–46. doi: 10.1165/rcmb.2007-0277OC. [DOI] [PMC free article] [PubMed] [Google Scholar]


