Abstract
Background:
Human epidermal growth factor receptor 2 (HER2) is overexpressed in several human malignancies and numerous studies have indicated that it plays important roles in the development and maintenance of the malignant phenotype. Targeting of HER2 molecules with monoclonal antibodies (mAbs) is a promising therapeutic approach. However, anti-HER2 mAbs affect cancer cells differently, depending on the distinct epitopes which are the targets.
Methods:
Reactivity of a panel of 8 mouse anti-HER2 mAbs was investigated by ELISA and Western blotting using different subdomains of the extracellular domain (ECD) of HER2. All subdomains, including I, II, III, IV, I+II, III+IV and full HER2-ECD were constructed and expressed in CHO cells. Cross-reactivity of the mAbs with other members of the human HER family and Cynomolgus HER2 was also studied by ELISA. The mAbs were also tested by immunohistochemistry (IHC) using HER2 positive breast cancer tissues.
Results:
Our results demonstrated that 3 out of 8 mAbs detected conformational epitopes (1T0, 2A8 and 1B5), while 5 mAbs identified linear epitopes (1F2, 1H9, 4C7, 1H6 and 2A9). Three of the mAbs recognized subdomain I, one reacted with subdomain I+II, 2 recognized either subdomain III or IV and 2 recognized subdomain III+IV. However, none of our mAbs recognized the subdomain II alone. The mAbs displayed either inhibitory or stimulatory effects on HER2-overexpressing tumor cells and did not react with other members of the human HER family. The pattern of IHC results implied better reactivity of the mAbs recognizing linear epitopes.
Conclusions:
Our findings suggest that paired subdomains of HER2 are essential for mapping of mAbs recognizing conformational epitopes. Moreover, there seems to be no association between subdomain specificity and antitumor activity of our anti-HER2 mAbs.
Keywords: HER2, extracellular subdomains, monoclonal antibody, epitope mapping, tumor inhibition
Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death in females (Torre et al., 2015). It has been shown that approximately 15-30 percent of patients with breast cancer overexpress human epidermal growth factor receptor2 (HER2) (Burstein, 2005). HER2 is a member of EGFRs superfamily which includes HER1 (EGFR) or ERBB1, HER2 or ERBB2, HER3 or ERBB3 and HER4 or ERBB4. These receptors belong to the large family of receptor tyrosine kinases. HER2 is an orphan receptor and no specific ligand has so far been identified for this receptor (Rubin and Yarden, 2001; Yarden, 2001; Puglisi et al., 2016). Many studies show that HER2 has an integral role in tumor invasiveness and poor prognosis of breast cancer (Rubin and Yarden, 2001; Yarden, 2001; Ménard et al., 2003; Gutierrez and Schiff, 2011; Puglisi et al., 2016). Moreover, it has long been shown that homodimerization and heterodimerization of HER2 with other HERs, especially HER3 result in tumor cells proliferation and survival mediated by downstream signaling through MAPK and AKT pathways (Wallasch et al., 1995; Lemmon and Schlessinger, 2010; Serra et al., 2011). Each member of this family is composed of three domains which include a C-terminal intracellular kinase domain, an intramembrane domain and N-terminal extracellular domain (ECD) (Cho et al., 2003; Maruyama, 2014).
Monoclonal antibodies (mAb) have been shown to play important role in treatment of various cancers by targeting tumor associated antigens (TAAs) (Adams and Weiner, 2005). Since the HER2 receptor is overexpressed in a proportion of patients with breast cancer, it has been considered as a promising target for cancer therapy. Trastuzumab and Pertuzumab, the two FDA approved anti-HER2 therapeutic mAbs, have been shown to induce significant anti-tumor effect when used alone or together as first line therapy (Swain et al., 2015). The anti-HER2 mAbs can also be used in immunohistochemistry (IHC) as an approved method along with flourescence in situ- hybridization (FISH) for diagnosis of HER2 over-expression in patients with breast cancer (Couturier et al., 2000; Owens et al., 2004). Some findings imply that binding of mAbs to certain subdomains of HER2-ECD could have unique anti-tumor effects (Cho et al., 2003; Franklin et al., 2004). The ECD domain of HER2 and other HER family members are composed of four subdomains. Trastuzumab can disrupt ligand-independent HER2/HER3 complexes, whereas Pertuzumab inhibits ligand-dependent HER2/HER3 complexes in HER2 overexpressing tumor cells (Junttila et al., 2009). These different effects can be well explained by Trastuzumab and Pertuzumab binding to subdomain IV and subdomain II, respectively (Junttila et al., 2009).
We previously produced a panel of 8 mAbs which have different effects on proliferation of HER2 overexpressing tumor cells (Kazemi et al., 2011; Tahmasebi et al., 2013). In the present study, we investigated whether the inhibitory or stimulatory activity of these mAbs is associated with their subdomain specificity. For this purpose, different recombinant extracellular subdomains of HER2 including I, II, III, IV, I+II and III+IV along with the full extracellular domain of HER2 (HER2-ECD) were produced in eukaryotic CHO-K1 host cells and employed to localize the subdomains recognized by these mAbs.
Materials and Methods
Production of anti-HER2 monoclonal antibodies
Eight murine mAbs (1B5, 1F2, 1H6, 1H9, 1T0, 2A8, 2A9, 4C7) were raised against HER2-overexpressing cell line as described elsewhere (Kazemi et al., 2011). The profiles of isotype, affinity and effects of these mAbs on growth and proliferation of HER2-overexpressing tumor cells have already been reported (Kazemi et al., 2011; Tahmasebi et al., 2014)
Construction of recombinant full extracellular domain and subdomains of HER2
The HER2-pCMV-XL4 construct (OriGene Technologies, Rockville, MD, USA) was used as a template for subcloning of extracellular subdomains and HER2-ECD into the pSecTag2A eukaryotic expression vector (OriGene). Amplification of the extracellular subdomains (DI, DII, DIII, and DIV) with 90-bp overlap and DI+DII, DIII+DIV and HER2-ECD was performed by PCR using specific primer sequences (Table 1). Schematic representation of the amplified HER2-ECD domain and its subdomains is shown in Figure 1. The amplification of HER2-ECD and other subdomains was conducted using pfu DNA polymerase. After initial denaturation at 95 °C for 3 min, 35 cycles were run at the following conditions: denaturation at 93 °C for 30s, annealing at 58 °C for 45s, and extension at 72 °C for 1.5 min; and finally, extension at 72 °C for 5 min. The amplicons were visualized by agarose gel electrophoresis containing ethidium bromide, and the bands were then extracted by DNA extraction kit (Vivantis, Cinnagen Co, Tehran, Iran). Extracted PCR products were digested by HindIII and XhoI restriction enzymes (Invitrogen, Carlsbad, CA, USA) and subcloned in pSecTag2A vector digested with the same enzymes. Ligation was performed by T4 DNA ligase (Promega, Madison, WI, USA) according to the instructions of the manufacturer. The constructs were subsequently transformed in the competent JM109 Escherichia coli strain (Novagen, Madison, WI, USA) by thermal shock as previously described (Sadri-Ardalani et al., 2015). The HER2-ECD and subdomains transformed cells were selected by culturing on Luria–Bertani (LB) agar plates containing ampicillin (100 μg/ml) and confirmed by colony PCR and sequencing. The HER2-ECD domain and subdomains constructs were purified by the miniprep kit (Vivantis, Cinnagen, Tehran, Iran) and applied for transfection.
Table 1.
Specific Primers Used for Subcloning of Extracellular Domain and Subdomains of HER2
| Domain/ | Primer | Sequence | Amplicon size (bp) |
|---|---|---|---|
| Subdomain | |||
| DI | DI-Psec-S: | 5′-GGTTTTTCTTAAGCTTGACCCAAGTGTGCACCG-3′ | |
| DI-Psec-AS: | 5′-AAGAAAAAAACTCGAGCGCGGTTGGTGTC-3′ | 512 | |
| DII | DII- Psec -S: | 5′-GGTTTTTCTTAAGCTTGCCCCAGCTCTGCTAC-3′ | |
| DII- Psec -AS: | 5′-AAGAAAAACCCTCGAGCCTTGCTGCACTTCTC-3′ | 548 | |
| DIII | DIII- Psec -S: | 5′-GGTTTTTCTTAAGCTTGGACGTGGGATCCTGC-3′ | |
| DIII- Psec –AS: | 5′-AAGAAAAACCCTCGAGCCACACACTCGTCC-3′ | 611 | |
| DIV | DIV- Psec -S: | 5′-GGTTTTTCTTAAGCTTGTTCGTGCACACGGTGC-3′ | |
| DIV- Psec -AS: | 5′-AAGAAAAAAAACTCGAGCCGTCAGAGGGC-3′ | 545 | |
| Full ECD | Full ECD-Psec-S | 5′-GGTTTTTCTTAAGCTTGACCCAAGTGTGCACCG-3′ | |
| Full ECD-Psec-As | 5′-AAGAAAAAAAACTCGAGCCGTCAGAGGGC-3′ | 2014 |
S, sense;AS, anti-sense;underlined sequences denote restriction enzyme sites
Figure 1.
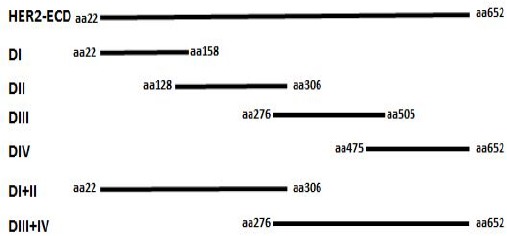
Schematic Representation of the Full Extracellular HER2 (HER2-ECD) Domain and Its Subdomains Employed in This Study
Expression of HER2-ECD domain and subdomains
CHO-K1 cells (National Cell Bank of Iran, Pasteur Institute, Iran) were cultured in RPMI-1640 medium (Gibco, California, USA) containing 10% heat- inactivated fetal bovine serum (Gibco, California, USA) penicillin (100 IU/mL) and streptomycin (100 μg/mL) (Gibco). Cells were transiently transfected using Lipofectamine 3,000 (Invitrogen, California, USA) as recommended by the manufacturer. A sub-confluent monolayer of CHO-K1 cells were washed with culture medium without antibiotics and 150 μL of Opti-MEM (Invitrogen) was added. DNA constructs were diluted in 50 μL of Opti-MEM and 3 μL of P3000 reagent (Invitrogen) was added. Then diluted DNA was added to 1.5 μL Lipofectamine 3,000 pre-diluted in 50 μL of Opti-MEM and kept at room temperature for 5 min. The Lipofectamine-DNA complex was added to the cells and the cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Cell culture supernatants were harvested 48 h later and assessed by ELISA. Positive transfected cells were kept in 0.7 mg/ml Hygromycin (Pasteur Institute, Tehran, Iran) for 2 weeks to select stable transfectants. Stable transfected cells were subcloned three times to select stable high producing clones.
Purification of rHER2-ECD protein
Stable clone expressing rHER2-ECD was expanded and cultured in RPMI1640 medium (Gibco) supplemented with 10% heat- inactivated fetal bovine serum and antibiotics (Gibco). The supernatants of cultured cells were collected and rHER2-ECD protein was purified by affinity chromatography using Sepharose gel coupled to 3 anti-HER2 mAbs (as described below). Briefly, the supernatant was centrifuged and then filtered. Bound HER2-ECD protein was eluted by Gly-HCL buffer (0.2M, pH 2.5). The eluted recombinant protein was immediately dialyzed against PBS.
Preparation of anti-HER2 affinity chromatography column
Purified mouse mAbs were used to prepare the column. Briefly, 15 milligrams of three mAbs (1H9, 1F2 and 1T0) was dialyzed against coupling buffer (0.2 M NaHCO3, 0.5 M NaCl, pH 8.3) overnight at 4°C. Two mls of NHS-activated Sepharose 4B (Thermo Fisher Scientific, California, USA) was washed three times with 1mM ice-cold HCl and washed with coupling buffer to remove HCL. Coupling was performed by recirculating the antibody solution overnight at 4°C on a rotator. Blocking of remaining uncoupled reactive groups was made by alternate washing the column with 0.5 M ethanolamine, 0.5 M NaCl (pH 8.3), and 0.1M acetate, 0.5 M NaCl (pH 4.0) according to the instructions of the manufacturer.
Production of anti –HER2 polyclonal antibody
New Zealand white rabbit was immunized intramuscularly with 50µg recombinant HER2-ECD and an equal volume of complete Freund’s adjuvant (Sigma, St Louis, MO, USA) and boosted biweekly 5 times with 20µg of recombinant HER2-ECD in incomplete Freund’s adjuvant (Sigma). The rabbit was bled 7 days after the last injection.
Screening of recombinant HER2 subdomains in supernatant of transfected cells
A sandwich ELISA, using affinity-purified rabbit anti-ECD HER2 antibody and HRP-conjugated anti-His tag antibody (SinaBiotech, Tehran, Iran), was developed for screening of recombinant HER2 subdomains in transfected cell culture supernatant. In brief, 10µg of anti-HER2 polyclonal antibody was coated in 96-well flat bottom microtiter plates (Maxisorp, Nunc, Roskilde, Denmark) and incubated for 90 min at 37°C. After washing three times with PBS-Tween20 (0.05%), plates were blocked with PBS-Tween20 (0.05%) containing 3% skim milk (Merck, Darmstadt, Germany) for 90 min at 37°C. Plates were washed again and 50μL of supernatant of the transfected cells was added to the plates. Following incubation at 37°C for 1 h, the plates were washed and incubated with an appropriate dilution of HRP-conjugated anti-His tag antibody. After incubation for 90 min and washing, the reaction was revealed with tetramethylbenzidine (TMB) substrate (Pishtaz Teb, Karaj, Iran). Finally, the reaction was stopped with 30µl of 1N HCL and the absorbance was measured by an ELISA reader (BioTek, Winooski, VT, USA) at 450 nm.
Epitope mapping of anti-HER2 monoclonal antibodies by ELISA
The mouse anti-HER2 mAbs as well as Trastuzumab and Pertuzumab as controls were epitope mapped using sandwich ELISA. The anti-HER2 mAbs were dissolved in PBS at a final concentration of 5 μg/mL. The wells of 96-well flat bottom microtiter plates (Maxisorp, Nunc, Denmark) were coated with anti-HER2 mAbs (50 μL/well) and incubated for 90 min at 37°C. The wells were then washed three times and blocked as described above. After washing, 50μL of supernatant of the transfected cells was added to the plates. Following incubation at 37°C for 90 min, the plates were washed and incubated with an appropriate dilution of HRP-conjugated anti-His tag antibody for 90 min. After washing, the reaction was revealed with TMB substrate (Pishtaz Teb, Karaj, Iran), stopped with 30µl of 1N HCL and the absorbance was measured by an ELISA reader at 450 nm.
Determination of subdomain specificity of monoclonal antibodies by immunoblotting
Cell lysate was prepared from 106 HER2 subdomains transfected cells using the M-PER buffer (Thermo Scientific Co, USA). Protein concentration was determined by the BCA assay (Bio-Rad Laboratories, Hercules, CA, USA). Equivalent amount of proteins was electrophoresed on 10% SDS–polyacrylamide gel under reduced and non-reduced conditions along with molecular weight marker and then transferred to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). After blocking with 5% skim milk (Merck, Germany) the membranes were incubated with 5µg/mL of affinity-purified mouse mAbs, Trastuzumab and Pertuzumab as controls in 5% skim milk overnight at 4°C, followed by extensive washing with PBS/Tween20 (0.05%). The membranes were then incubated with HRP-conjugated rabbit anti-mouse Ig or anti-human Ig (SinaBiotech, Tehran, Iran) at 37°C for 1.5 hours. After washing, the bands were visualized using ECL prime kit (Amersham Pharmacia Biotech, Chalfont, UK).
Reactivity of anti-HER2 monoclonal antibodies with HER2-positive breast cancer tissues by immunohistochemistry
HER2-positive human breast cancer tissue sections were deparaffinized with xylol and rehydrated with graded ethanol. Sections were retrieved with 10 mM sodium citrate at 95 °C for 25 min. Slides were soaked in Tris-buffered saline (TBS) for 15 min. Intracellular peroxidase was blocked with 1 % hydrogen peroxide in TBS for 15 min and washed with TBS for 5 min. Treated sections were blocked with TBS containing 5 % sheep serum, 1 % BSA, and 0.05 % Tween 20 for 20 min at room temperature. Anti-HER2 mAbs including 1H9, 1F2, 2A8, 4C7, 1B5 and 1T0 were then added to the sections and incubated for 1 h at room temperature. Irrelevant isotype matched antibodies were included as negative controls. Following washing step, slides were incubated with 50 μl of HRP-EnVision Polymer (Dako, Glostrup, Denmark) for 25 min. In the next step, 70 μl (DAB) diaminobenzidine substrate (Roche) was added for 10–15 min and the reaction was stopped by distilled water. Finally, the sections were stained with hematoxylin followed by dehydration procedure and mounting. HER2 expression was verified by HercepTest (Dako, Denmark) which was applied as positive control.
Binding assessment of anti-HER2 mAbs to other ErbB family members and Cynomolgus monkey HER2
Cross-reactivity of the mouse anti-HER2 mAbs with other members of ErbB family (EGFR, HER3 and HER4) as well as Cynomolgus monkey HER2 was determined by sandwich ELISA. Recombinant EGFR, HER3 and HER4 (Speed BioSystems, Rockville, MD, USA) and Cynomolgus HER2 (Sino Biological Inc, Beijing, China) proteins were coated at 1µg/ml in 96-well flat bottom microtiter plates (Maxisorp, Nunc, Denmark) for 90 min at 37°C. After washing, the mouse mAbs and Trastuzumab were added to wells in a final concentration of 5µg/ml dissolved in blocking buffer and incubated for 90 min at 37°C. Finally, HRP-conjugated rabbit anti-mouse Ig or anti-human Ig (SinaBiotech, Tehran, Iran) were added to detect mouse and human mAbs, respectively. All recombinant proteins contain a C-terminal 6X His-Tag, thus the HRP anti-His tag antibody was used to check for presence of coated proteins.
Results
Characterization of the anti-HER2 monoclonal antibodies
Production and characterization of the anti-HER2 mAbs have already been reported (Kazemi et al., 2011; Tahmasebi et al., 2013). Some features of these mAbs including isotype, affinity and their effect on tumor cells growth and proliferation are given in Table 2.
Table 2.
Characteristics of the Anti-HER2 Monoclonal Antibodies
| Clone | Isotype | Affinity (M/L) | Growth effect |
|---|---|---|---|
| 1F2 | IgG2a | 5×10-9 | S |
| 1H6 | IgG1 | 1.2×10-9 | S |
| 1T0 | IgG1 | 4.7×10-8 | I |
| 2A9 | IgG2a | 1.6×10-9 | S |
| 2A8 | IgG1 | 1.1×10-9 | I |
| 1B5 | IgG1 | 7.3×10-8 | S |
| 1H9 | IgG1 | 2.4×10-8 | S |
| 4C7 | IgG1 | 1.9×10-8 | S |
*S, stimulatory;I, inhibitory;The results are taken from (Kazemi et al., 2011) and (Tahmasebi et al., 2014)
Reactivity of anti-HER2 monoclonal antibodies with HER2 extracellular subdomains by ELISA
A sandwich ELISA was designed to check reactivity of the mAbs with HER2 extracellular subdomains including I, II, III, IV, I+II, III+IV as well as full HER2-ECD. Trastuzumab, Pertuzumab and a rabbit polyclonal anti HER2-ECD antibody were included as controls. Since all HER2 subdomains are terminated with a His-tag in their C-terminal, an anti-His mAb was used to validate their coating on ELISA plates. Representative results obtained for two of the mAbs (1F2 and 2A8) are shown in Figure 2. Final results are given in Table 3. Accordingly, 3 mAbs (1F2, 1H6 and 2A9) detect unique epitopes on DI subdomain; whereas1H9 and 4C7 react with DIV and DIII, respectively. The other 3 mAbs recognize epitopes expressed either on DI+II (1T0) or DIII+IV (1B5 and 2A8). We have previously shown that these mAbs do not cross-inhibit each other and recognize distinct epitopes (Kazemi et al., 2011).
Figure 2.
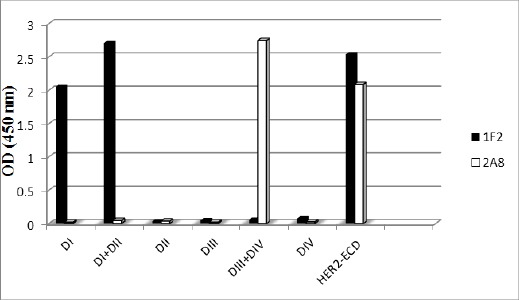
Representative Profile of Reactivity of Anti-HER2 mAbs with Extracellular Subdomains of HER2 by ELISA
Table 3.
Binding Profiles of mAbs to Recombinant Subdomains of HER2-ECD by ELISA
| mAbs | DI | DI+DII | DII | DIII | DIII+DIV | DIV | HER2-ECD |
|---|---|---|---|---|---|---|---|
| 1F2 | ++ | ++ | - | - | - | - | ++ |
| 1T0 | - | ++ | - | - | - | - | ++ |
| 2A8 | - | - | - | - | ++ | - | ++ |
| 2A9 | ++ | ++ | - | - | - | - | ++ |
| 1H6 | ++ | ++ | - | - | - | - | ++ |
| 1H9 | - | - | - | - | ++ | ++ | ++ |
| 1B5 | - | - | - | - | ++ | - | ++ |
| 4C7 | - | - | - | + | ++ | - | ++ |
| Pertuzumab | - | - | - | - | - | - | ++ |
| Trastuzumab | - | - | - | - | ++ | - | ++ |
| Anti-HER2 polyclonal Ab | ++ | ++ | - | + | ++ | ++ | ++ |
| Anti –His Ab | + | ++ | ++ | + | ++ | ++ | ++ |
Trastuzumab, Pertuzumab and anti-HER2 polyclonal antibodies are used as positive controls. Full HER2-ECD is also employed as positive control in the coating layer. Anti-His mAb was used to verify the coated recombinant proteins. The results represent OD values of <0.2 (-), 1.0-2.0 (+) and >2.0 (++)
Determination of specificity of monoclonal antibodies by immunoblotting
Immunoblotting results indicate that 3 mAbs including 1T0, 1B5 and 2A8 detect conformational epitopes, whereas the other 5 remaining mAbs detect linear epitopes at both non-reduced and reduced conditions (Figure 3). The pattern of reactivity of all mAbs by immunoblotting was similar to the results obtained by ELISA (Table 3), with the exception of 4C7 mAb which did not react with DIII subdomain by Western blot technique.
Figure 3.
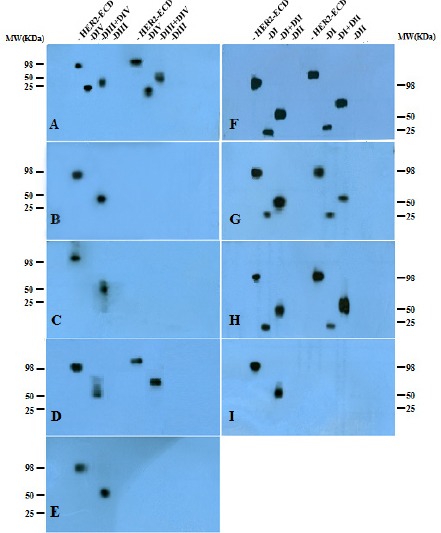
Immunoblotting Reactivity of Anti-HER2 Monoclonal Antibodies with Recombinant Subdomains of Extracellular HER2; Crude lysates of CHO cells transfected with different HER2 extracellular subdomains were boiled for 5 min and then electrophoresed on 10% gel under reduced and non-reduced conditions along with molecular weight markers. For antibodies that recognized DI or DI+II in ELISA, immunoblotting was carried out with HER2-ECD, DI, DI+II and DII subdomains whereas mAbs which reacted with DIII or DIV by ELISA were checked using HER2-ECD, DIV, DIII+DIV and DIII subdomains under both reduced (lanes 1-4) and non-reduced (lanes 5-8) conditions. (A) 1H9, (B) 2A8, (C) 1B5, (D) 4C7, (E) Trastuzumab, (F) 1F2, (G) 2A9, (H) 1H6 and (I) 1T0 mAbs. Isolated and paired subdomains displayed ~ 25 KD and ~ 50 KD band size, respectively, whereas full HER2-ECD displayed ~ 95 KD molecular weight.
Reactivity of anti-HER2 monoclonal antibodies with HER2-positive breast cancer tissues by immunohistochemistry (IHC)
Six mAbs together with the commercial Hercep antibody as a positive control and an irrelevant isotype matched mAb as a negative control were tested. Breast tissue sections obtained from 2 patients with 3+ HER2 staining pattern were employed. Two of the mAbs were not tested due to shortage of tissue sections. The results show significant reactivity of 1H9 and 1F2 mAbs similar to the Hercep antibody (Figure 4).
Figure 4.
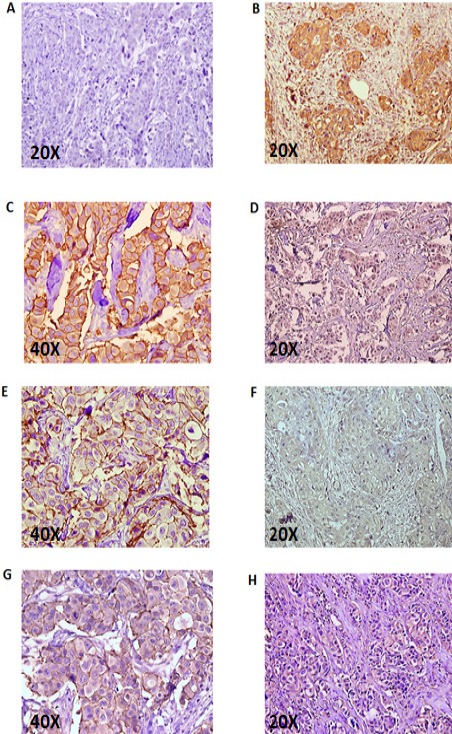
Immunohistochemistry Profile of Anti-HER2 mAbs with Human HER2+ Breast Cancer Tissues; A) negative control; B) Hercep-test ; C) 1H9; D) 1T0; E) 1F2; F) 4C7; G) 2A8 and H) 1B5. The sections were taken from breast tissue of two breast cancer patients with 3+ HER2 staining profile.
Cross-reactivity of anti-HER2 monoclonal antibodies with human HER family members and Cynomolgus monkey HER2
The ELISA results showed that all mAbs react with HER2 of Cynomolgus monkey, similar to human HER2. However none of the mAbs recognized other members of the human HER family, including HER1, HER3 and HER4. The results are summarized in Table 4.
Table 4.
Cross- Reactivity of Anti-HER2 mAbs with Human HER Family Members and Cynomolgus Monkey HER2
| mAbs | HER1 | HER2 | HER3 | HER4 | HER2 (Cynomolgus) |
|---|---|---|---|---|---|
| 1F2 | - | + | - | - | + |
| 1T0 | - | + | - | - | + |
| 2A9 | - | + | - | - | + |
| 1H6 | - | + | - | - | + |
| 1H9 | - | + | - | - | + |
| 2A8 | - | + | - | - | + |
| 4C7 | - | + | - | - | + |
| 1B5 | - | + | - | - | + |
| Trastuzumab | - | + | - | - | + |
| Anti-His Ab | + | + | + | + | + |
The results represent OD values of <0.2 (-) and >2.0 (+)
Discussion
Some reports suggest that the binding specificity of anti-HER2 mAbs may affect their biological function and mechanism of action (Schmitz and Ferguson, 2009; Zhou et al., 2011; Meng et al., 2016). Trastuzumab and Pertuzumab, two humanized mAbs bind to DIV and DII subdomains of human HER2-ECD, respectively (Cho et al., 2003; Franklin et al., 2004). Cho et al showed that Trastuzumab binds to DIV and subsequently induces anti-tumor activity in HER2 overexpressing breast cancer cells via diverse mechanisms including, decreasing proliferative signaling, increasing HER2 endocytosis, inhibition of HER2 shedding, steric hindrance and also intruption of HER2 homo and heterodimerization (Cho et al., 2003; Valabrega et al., 2007). On the other hand, binding of Pertuzumab to DII subdomain, which is responsible for HER2 dimerization, specifically acts by inhibiting of receptor dimerization (Franklin et al., 2004). Although both therapeutic mAbs inhibit receptor dimerization, Pertuzumab inhibits ligand-dependent HER2/HER3 complexes, whereas Trastuzumab disrupts ligand-independent heterodimerization (Junttila et al., 2009) and shows its anti-tumor effect only when HER2 is overexpressed (3+ level by IHC staining) (Hudis, 2007; Valabrega et al., 2007). Interestingly, the combined use of these two antibodies increases anti-tumor activity with significant tumor regression in metastatic breast cancer (Swain et al., 2015). Similarly, combination of two of our own anti-HER2 mAbs (1T0 and 2A8) together or with Trastuzumab induces a synergistic inhibition on tumor cell growth in vitro (Tahmasebi et al., 2013). The chimeric form of 1T0 was also found to display the same biological features (Amiri et al., 2013). Given that each of our anti-HER2 mAbs recognized distinct non-overlapping epitope on HER2 (Kazemi et al., 2011) and also considering that these mAbs induce either inhibitory or stimulatory proliferative signals in tumor cells (Tahmasebi et al., 2013), mapping the epitopes recognized by these mAbs is of considerable importance in understanding the characteristics and underlying mechanisms of action of these antibodies.
To localize the epitopes recognized by each mAb we constructed the coding sequences of all four subdomains of HER2-ECD as well as the paired domains of DI+DII and DIII+DIV to cover all linear and conformational epitopes within this region. The recombinant proteins were expressed in a mammalian system (CHO cells) to preserve the native configuration and the post translational modifications of HER2. Since all proteins were terminated by a His-tag, screening of the secreted recombinant proteins in culture supernatant of transfected cells was performed by an anti-His mAb, which confirmed successful production of all proteins (Table 3).
Mapping of the mAbs was performed by both ELISA and immunoblotting techniques. Both assays gave essentially the same results with one exception. The 4C7 mAb failed to react with isolated DIII subdomain by immunoblotting, but not ELISA. The ELISA results showed reactivity of 4C7 mAb with both paired DIII+IV and to a lesser extent isolated DIII subdomain. Lack of reactivity with isolated DIII subdomain could be due to lower sensitivity of the immunoblotting technique. Immunoblotting was performed on reduced and non-reduced proteins. The pattern of reactivity indicates that 5 mAbs (1F2, 1H6, 1H9, 2A9 and 4C7) recognized linear epitopes and the remaining 3 mAbs bind to conformational epitopes (Figure 3). Interestingly, all linear epitopes recognizing mAbs reacted both with isolated and paired subdomains, whereas mAbs recognizing conformational epitopes reacted only with paired subdomains, with the exception of 4C7 which reacted only with DIII isolated subdomain by ELISA. This mAb reacted with paired DIII+DIV subdomain and full HER2-ECD by immunoblotting at both reduced and non-reduced conditions.
Three of our mAbs recognized epitopes on DI (1F2, 1H6 and 2A9) because they could recognize isolated DI and DI+DII but not isolated DII, one mAb reacted with either DIII (4C7) or DIV (1H9) and the remaining three mAbs reacted with paired DI+DII (1T0) or DIII+DIV (2A8 and 1B5) subdomains (Figure 2 and Table 3). The fine subdomain specificity of the latter three mAbs which recognize conformational epitopes could not be elucidated with the present approach and needs further investigations. None of the mAbs reacted with the isolated DII subdomain. The polyclonal antibody and pertuzumab also failed to bind to DII. This clearly shows the influence of other subdomains on the integrity of this subdomain. Indeed DI and DIII subdomains were found to be necessary for the maintenance of DII in its native folding for binding to Pertuzumab (Franklin et al., 2004). Similar to our work, Hu and coworker (Hu et al., 2008) characterized a new anti-HER2 mAb (A21) by using several eukaryotic recombinant single and paired subdomains of HER2 and showed that this mAb binds to DI+II but not to isolated subdomain DI or DII (Hu et al., 2008).
Similarly, Trastuzumab did not bind to isolated subdomain DIV, it seems that the conformational epitope recognizing by Trastuzumab is created only when subdomain DIII and DIV are expressed together. Controversial results have also been reported showing that Trastuzumab could bind to isolated DIV subdomain (Ceran et al., 2012; Ko et al., 2015). However, this subdomain was either as a transmembrane protein (Ceran et al., 2012) or possibly as Fc-fusion (Ko et al., 2015) protein which may alter its three dimensional structure and provides more native conformation as compared to isolated protein.
These findings indicate importance of the use of paired subdomains for mapping studies, particularly for mAbs recognizing conformational epitopes and also importance of employment of different methodological approaches (such as ELISA and immunoblotting) to extend and clarify the results. The IHC pattern of reactivity of our mAbs could also be interpreted in support of this notion. The best reactive mAbs (1H9 and 1F2) recognize linear epitopes on DIV and DI subdomains, respectively (Figure 3).
Since human HER2 is 40-45% homologous to other members of the HER family (HER1, HER3 and HER4) (Franklin et al., 2004), we checked reactivity of our mAbs with these proteins. No cross-reactivity was observed with any of the HER family members (Table 4). However, all mAbs strongly reacted with Cynomolgus monkey HER2 which is highly homologous (~99%) to the human counterpart. Due to this significant homology with human HER2, this animal has been considered as the most relevant and appropriate animal model to assess pharmacokinetics, toxicity and safety of therapeutic anti-HER2 mAbs (Adams et al., 2006).
In the next step we sought to find any correlation between subdomains specificity and biological function of our panel of mAbs. We have previously demonstrated that two of our mAbs (1T0 and 2A8) induce inhibitory effect on tumor cell proliferation, whereas the remaining mAbs were stimulatory (1H9, 1F2, 1B5, 1H6, 4C7 and 2A9) (Tahmasebi et al., 2013). Both inhibitory mAbs recognize conformational epitopes on either paired DI+DII (1T0) or DIII+DIV (2A8) subdomains. The stimulatory mAbs, however, bind to either linear or conformational epitopes on different HER2 subdomains. Considering the limited number of mAbs, our results are not conclusive and do not seem to support the association between subdomain specificity and tumor inhibitory or stimulatory activity of the mAbs.
In conclusion we localized the extracellular subdomains recognized by a panel of 8 anti- human HER2 mAbs using eukaryotic recombinant HER2-ECD subdomains. The mAbs reacted with DI (n=3), DI+DII (n=1), DIII (n=1) and DIII+DIV (n=3) subdomains. None of the mAbs cross-reacted with other members of the human HER family. Although, all DI-specific mAbs failed to inhibit tumor cell proliferation, no clear correlation could be assigned between subdomain specificity and anti-tumor activity of the anti-HER2 mAbs.
Acknowledgements
This study was partially supported by grants from the Food and Drug Organization of the Ministry of Health and Medical Education of Iran, Tehran University of Medical Sciences (grant number 33245) and Avicenna Research Institute.
References
- Adams CW, Allison DE, Flagella K, et al. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol Immunother. 2006;55:717–27. doi: 10.1007/s00262-005-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol. 2005;23:1147–57. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- Amiri MM, Jeddi-Tehrani M, Kazemi T, et al. Construction and characterization of a new chimeric antibody against HER2. Immunotherapy. 2013;5:703–15. doi: 10.2217/imt.13.67. [DOI] [PubMed] [Google Scholar]
- Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353:1652–4. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- Ceran C, Cokol M, Cingoz S, et al. Novel anti-HER2 monoclonal antibodies: synergy and antagonism with tumor necrosis factor-α. BMC Cancer. 2012;12:450. doi: 10.1186/1471-2407-12-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-S, Mason K, Ramyar KX, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–60. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- Couturier J, Vincent-Salomon A, Nicolas A, et al. Strong correlation between results of fluorescent in situ hybridization and immunohistochemistry for the assessment of the ERBB2 (HER-2/neu) gene status in breast carcinoma. Mod Pathol. 2000;13:1238–43. doi: 10.1038/modpathol.3880228. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Carey KD, Vajdos FF, et al. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–28. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- Gutierrez C, Schiff R. HER2: biology, detection, and clinical implications. Arch Pathol Labo Med. 2011;135:55–62. doi: 10.1043/2010-0454-RAR.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Zhu Z, Li L, et al. Epitope mapping and structural analysis of an anti-ErbB2 antibody A21: Molecular basis for tumor inhibitory mechanism. Proteins. 2008;70:938–49. doi: 10.1002/prot.21551. [DOI] [PubMed] [Google Scholar]
- Hudis CA. Trastuzumab-mechanism of action and use in clinical practice. N Engl J Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- Junttila TT, Akita RW, Parsons K, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–40. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Kazemi T, Tahmasebi F, Bayat AA, et al. Characterization of novel murine monoclonal antibodies directed against the extracellular domain of human HER2 tyrosine kinase receptor. Hybridoma. 2011;30:347–53. doi: 10.1089/hyb.2011.0023. [DOI] [PubMed] [Google Scholar]
- Ko B-K, Lee S-Y, Lee Y-H, et al. Combination of novel HER2-targeting antibody 1E11 with trastuzumab shows synergistic antitumor activity in HER2-positive gastric cancer. Mol Oncol. 2015;9:398–408. doi: 10.1016/j.molonc.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama IN. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells. 2014;3:304–30. doi: 10.3390/cells3020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard S, Pupa SM, Campiglio M, et al. Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003;22:6570–8. doi: 10.1038/sj.onc.1206779. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zheng L, Yang Y, et al. A monoclonal antibody targeting ErbB2 domain III inhibits ErbB2 signaling and suppresses the growth of ErbB2-overexpressing breast tumors. Oncogenesis. 2016;5:e211. doi: 10.1038/oncsis.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin. Breast Cancer. 2004;5:63–9. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- Puglisi F, Fontanella C, Amoroso V, et al. Current challenges in HER2-positive breast cancer. Crit Rev Oncol Hematol. 2016;98:211–21. doi: 10.1016/j.critrevonc.2015.10.016. [DOI] [PubMed] [Google Scholar]
- Rubin I, Yarden Y. The basic biology of HER2. Ann Oncol. 2001;12:3–8. doi: 10.1093/annonc/12.suppl_1.s3. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardalani F, Shabani M, Amiri MM, et al. Antibody response to HER2 extracellular domain and subdomains in mouse following DNA immunization. Tumor Biol. 2015;37:1–11. doi: 10.1007/s13277-015-3897-x. [DOI] [PubMed] [Google Scholar]
- Schmitz KR, Ferguson KM. Interaction of antibodies with ErbB receptor extracellular regions. Exp Cell Res. 2009;315:659–70. doi: 10.1016/j.yexcr.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Scaltriti M, Prudkin L, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–57. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Baselga J, Kim S-B, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724–34. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi F, Kazemi T, Amiri MM, et al. In vitro assessment of the effects of anti-HER2 monoclonal antibodies on proliferation of HER2-overexpressing breast cancer cells. Immunotherapy. 2014;6:43–9. doi: 10.2217/imt.13.156. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Valabrega G, Montemurro F, Aglietta M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Ann Oncol. 2007;18:977–84. doi: 10.1093/annonc/mdl475. [DOI] [PubMed] [Google Scholar]
- Wallasch C, Weiss F, Niederfellner G, et al. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995;14:4267. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61:1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zha Z, Liu Y, et al. Structural Insights into the down-regulation of overexpressed p185her2/neu protein of transformed cells by the antibody chA21. J Biol Chem. 2011;286:31676–83. doi: 10.1074/jbc.M111.235184. [DOI] [PMC free article] [PubMed] [Google Scholar]


