Abstract
HCV induced hepatitis and hepatocellular carcinoma as its sequel are major health problems world-wide and especially in Egypt. For diagnosis and during treatment of liver diseases, liver functions are monitored through determination of serum levels of liver enzymes and α-fetoprotein although the obtained information is generally not sufficient for either early detection of hepatic insult or effective follow up of therapeutic effects. More sensitive biomarkers may help to achieve these goals. MiRNAs are small non-coding RNAs that have an important role in gene expression and regulation. Many, such as miR-224, miR-215, miR-143 are correlated with tumor appearance and with the degree of fibrosis in lung, breast and colon cancer. This study was performed to estimate the level of these miRNAs in serum of patients with HCV-associated hepatitis and HCC in relation to grade of hepatitis, stage of fibrosis and differentiation of tumor tissue. In addition, correlations between serological and tissue levels were assessed. A total of 80 patients were examined, out of which 50 were included in the study. Blood samples and tissue specimens from malignant tumor and corresponding non-tumor tissue of HCV hepatitis patients were collected. Blood samples from 20 healthy volunteers were also obtained as controls. It was found that miRNAs profiles differed in HCC patients compared to controls and HCV-associated hepatitis cases. Distinction of tumor grade and fibrosis stage of patients as well as between different grades of tumor differentiation proved possible, making miRNAs promising biomarkers for diagnosis and assessment of treatment response of HCC patients.
Keywords: Hepatocellular carcinoma, micro RNA, diagnosis
Introduction
Hepatocellular carcinoma (HCC) is considered one of the most violent diseases in the world (Chang-Hao et al., 2015)-and is a major health problem in Egypt representing 13% of all cancers in Egypt (Zeeneldin et al., 2015). HCC is the second most common cancer in men. (Ran et al., 2016)
The hepatocellular carcinoma’s crucial risk factor that is responsible for its development is cirrhosis (Taned and Kannika, 2015). The long periods of chronic liver illness is what causes cirrhosis and is categorized by a reduction in hepatocyte proliferation, representing the tiredness of the regenerative capacity of the liver (Detlef and Nezam, 2008).
MicroRNAs (miRNAs) are class of non-coding RNAs, about 18–22 nucleotides long. These miRNAs account for only 1% of the human genome, they are highly conserved in nearly all organisms and play a crucial role in the regulation of gene expression (Shrivastava et al., 2015). The main advantage of microRNAs being potential biomarkers is that they can be easily obtained by non-invasive procedures and have potential high accurate biomarkers for tumor diagnostics (Albert et al., 2015). MiRNAs play critical roles in almost all cellular pathways involved in human malignancies, such as: cancer progression, carcinogenesis, cell survival, cell metastasis and invasion, and reaction to therapeutic drugs (Minet al., 2013).
MiR-224 plays an important role in cell proliferation, migration, invasion, and anti-apoptosis in HCC by directly binding to its gene targets, such as CDC42, CDH1, PAK2, BCL-2, and MAPK1 (Zhang et al., 2013). However, miR-215was downregulated and functioned as a potential tumor suppressor in several cancers including, colorectal cancerand associated with the metastasis and progression of renal cell carcinoma. (Jianet al., 2017). In addition, miR-215 was preferentially upregulated in gastric cancer. Changes in miR-143 expression have been frequently implicated in HCC through downregulating the expressions of TLR2, NF-κB, MMP-2 and MMP-9 (Zhu-qinget al., 2014).
The expression of miR-143 is highly downregulated in colorectal cancer, lung, bladder, and gastric cancers (Zhanget al., 2013). the expression of miR-143 is up-regulated in pancreatic stellate cells cancer and esophageal cancer however, few reports about the relationship between miR-143 expression and the diagnosis of HCC ((Zhu-qing et al., 2014).
Materials and Methods
Patients
This study was conducted on patients diagnosed as HCC on top of HCV undergoing partial hepatectomy. A total of 80 HCC patients were initially examined, out of which 50 were included in the study who were serologically positive for HCV. Five ml venous blood samples and liver tissue specimens from malignant and corresponding non-tumor tissue were collected. In addition, blood samples from 20 healthy volunteers were obtained as controls. The exclusion criteria include patients above 60 years and patients with HBV or HIV infections. This study was carried out in full accordance with the Helsinki Declaration of 1975, as revised in 1983, and was approved by the Ethics Committee of Theodor Bilharz Research Institute and by National Hepatology and Tropical Medicine Research Institute (NHTNRI). A written informed consent was obtained from each participant, in accordance with the institutional guidelines.
Biochemical Investigations
Laboratory tests including alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea (B. Urea), albumin (ALB) and alpha-fetoprotein (ALP) were performed for all subjects as routine tests.
MiRNAs extraction
Total RNAs including MiR-224, miR-215 and miR-143 were extracted from 200 μl serum and from 100 mg tissue, modified procedure of Abbott mSample preparation system kit (Abbott Molecular, Inc., Des Plaines, IL). The spike in C. elegans miR-39 as a synthetic RNA oligo nucleotide (Qiagen, Hilden, Germany) was added to all samples before extraction.
Quantitative Real-Time Reverse-Transcription Assay (qRT-PCR)
TaqMan miR-224, miR-215, miR-143 and miR-39 ready-made primers and probes (Applied Biosystems, San Diego, CA) were used to perform qRT-PCR assays according to the manufacturer’s instructions. Briefly, 5 μl aliquot of the RNAs extracted from the serum and tissue samples were reverse transcribed using the microRNA reverse transcription kit (Applied Biosystems, San Diego, CA). Five μl of the cDNA was used for the real time PCR amplification step. All reactions were run in duplicate. The ΔΔCT method was used for the relative quantification of miRNAs in all samples (Akin et al., 2012).
Histopathological Study
Paraffin blocks were made from HCC and from non-tumor chronic hepatitis C tissue for each case.4 microns thick sections were stained with hematoxylin and eosin stain (H and E stain) for routine histopathological examination, grading of hepatitis activity and tumor differentiation as well as Masson’s trichrome stain for assessment of fibrosis stage. Hepatitis activity and staging of fibrosis were evaluated using Ishak scoring system (Ishak et al., 1995) based on;
-Amount of inflammation and hepatocyte suffering (hepatitis activity index “HAI”): with a scale of 18 points
-Stage of fibrosis; representing amount of fibrosis or scarring:
F1toF4: Fibrosis
F5 and F6: Cirrhosis.
HCC grading was done according to the WHO classification of tumors of the liver and intrahepatic bile ducts (Bosman, 2010) into:
Grade 1: (well differentiated).
Grade 2: (Moderately differentiated).
Grade 3: (Poorly differentiated).
Statistical analysis
Analysis of the data was carried out with SPSS 18 statistical software (SPSS Inc., Chicago, IL, USA). The frequency distributions for different selected variables were performed using fisher test. The comparisons of quantitative variables were performed between two groups using Mann Whitney U-test and multiple comparisons between more than two groups have been conducted by Kruskal Wallis ANOVA test. The receiver operating characteristic (ROC) curve analysis determined sensitivity, specificity and the accuracy of miRNAs. Associations between miRNAs expressions and tumor grade were evaluated by chi square test. The P value < 0.05 was considered statistically significant.
Results
Patient’s characteristics and biochemical analysis
Total of fifty HCV post-hepatitic HCC patients and twenty controls were enrolled in this study. Data presented in Table 1 shows the analysis performed for the samples. There was significant difference between patients and controls in terms of age with p≤0.0040 while there was no significant difference in term of sex. Biochemical analysis has showed that B. Urea, ALT, AST and AFP were highly significant in post-hepatitic HCC patients than control group (P ≤0.0001).
Table 1.
Characteristics, Biochemical and Histopathological Analysis of HCC Patients and Controls
| Variables | HCC Patients | Healthy Volunteers | Pa |
|---|---|---|---|
| NO (%) | NO (%) | ||
| Age (years) | |||
| ≤40 | 7 (14%) | 10 (50%) | 0.0040** |
| >40 | 43 (86%) | 10 (50%) | |
| Gender | |||
| Males | 35 (35%) | 7 (33%) | 0.239 NS |
| Females | 15 (65%) | 13 (66%) | |
| B. Urea (mg/dl) | |||
| ≤15 | 5 (10%) | 18 (90%) | <0.0001*** |
| >15 | 45 (90%) | 2 (10%) | |
| ALB (g/dl) | |||
| ≤3.5 | 48 (96%) | 1 (5%) | <0.0001*** |
| >3.5 | 2 (4%) | 19 (95%) | |
| ALT (IU/l) | |||
| ≤40 | 8 (16%) | 16 (80%) | <0.0001*** |
| >40 | 42 (84%) | 4 (20%) | |
| AST (IU/l) | |||
| ≤30 | 7 (14%) | 19 (95%) | <0.0001*** |
| >30 | 43 (86%) | 1 (5%) | |
| AFP (IU/ml) | |||
| ≤10 | 3 (6%) | 18 (90%) | <0.0001*** |
| >10 | 47 (94%) | 2 (10%) | |
| Tumor grade | |||
| G1 | 10 (20%) | ||
| G2 | 18 (36%) | NA | |
| G3 | 22 (44%) | ||
| Fibrosis score | |||
| (0-4) | 26 (52%) | ||
| (5-6) | 24 (48%) | NA | |
Fisher exact test. Statistical Significance;*, Significant;
, highly significant;
, very highly significant;NA, not applicable
Relative expression of miRNAs in serum and tissue samples
MiR 224, miR-215 and miR-143 expressions were quantified in serum and in tissue extract using qRT PCR and compared with control group serological values as shown in Figure 1. The three miRNAs were higher in the serum of HCC patients compared to controls with P≤ 0.045, P≤ 0.032 and P≤ 0.0421 respectively. In liver tissue samples miR-143(P≤0.014) and miR-224 (P≤0.032) were down regulated in tumor tissue compared to non-tumorous hepatitis C tissues. However mir-225 (P≤0.017) were up regulated in tumor tissue compared to non-tumorous tissues.
Figure 1.
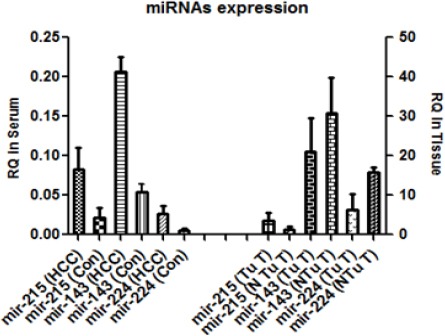
Relative Expression of miR-224, miR-215 and miR-143 in Serum and Liver Tissue of HCC Patients and Sera of Controls. For serum, (HCC) and controls (Con); RQ in the serum are represented as log10 of the results
For serum: (HCC) and controls (Con)
For liver tissues: tumor tissue (Tu T) and non-tumor tissue (N Tu T)
Correlation between miR-224, miR-215 and miR-143 expression levels with fibrosis and tumor grade
MiRNAs expressions were correlated with tumor grade and fibrosis to examine whether the serum miR-224, miR-215 and miR-413 levels were associated with specific stages of the disease as shown in Figure 2. The relative expression of miRNAs was divided into groups according to the tumor grade, stage and the fibrosis. There were significant positive correlation between the expression levels of miR-224 (0.0155*), miR-215(0.0310*) and miR-143 (0.0411*) and tumor grade. Although the expression of miRNAs were higher in patients with cirrhosis (Figures 5-6) compared to patients with fibrosis (Figures 1-4), these correlations were statistically non-significant.
Figure 2.
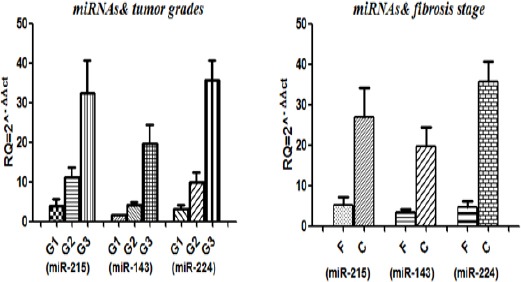
Serum miRNAs Expressions in Relation to Tumor Grade and Liver Fibrosis Score. RQ, relative expression; G1, grade 1; G2, grade 2; G3, grade 3; F, fibrosis; C, cirrhosis
Figure 3.
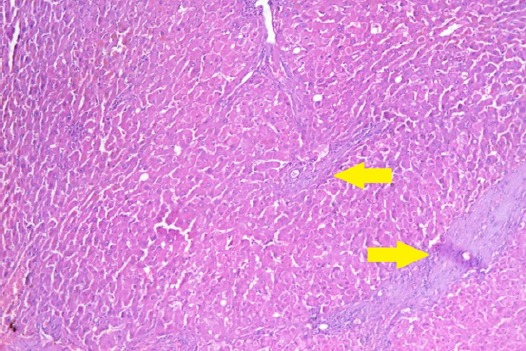
Section in Liver Tissue in a Patient with Mild Degree of Post-Hepatitic Fibrosis (Stage 2) Showing Focal Expantion of Portal Tract by Fibrous Tissue (Arrows). (Hematoxylin and eosin stain, X200).
Figure 4.
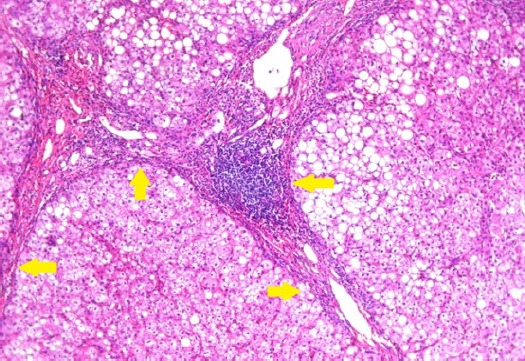
Section in Liver Tissue in a Patient with Marked Degree of Post-Hepatitic Fibrosis (stage 5) Showing Focal Encircling of Regenerating Hepatic Nodules by Fibrous Tissue (Arrows). (Hematoxylin and eosin stain, X200).
Figure 5.
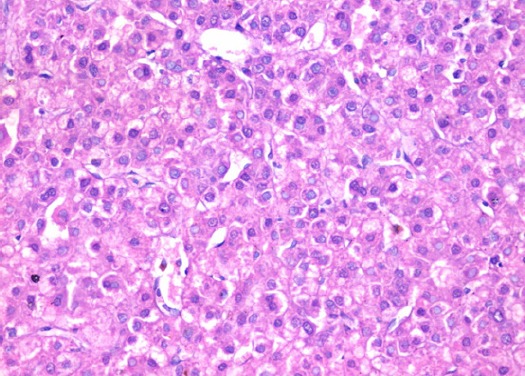
Section in Liver Tissue in a Patient with Low Grade Hepatocellular Carcinoma, Showing Thick Cords, Increased Nucleocytoplasmic Ratio and Focally Prominent Nucleoli. (Hematoxylin and eosin stain, X400).
Figure 6.
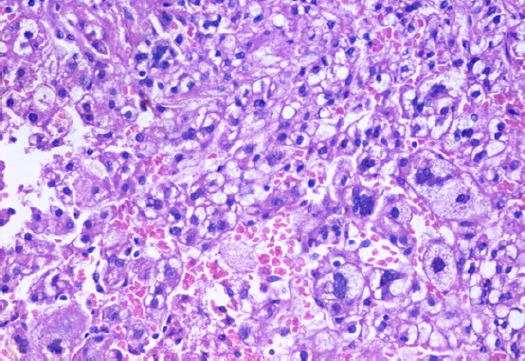
Section in Liver Tissue in A Patient with High Grade Hepatocellular Carcinoma, Showing Marked Variability in Cellular and Nuclear Sizes, Nuclear Hyperchromasia And Prominent Nucleoli. (Hematoxylin And Eosin Stain, X400).
MiR-224, miR-215 and miR-143 as diagnostic biomarkers in serum validation test
Table 2 shows the Receiver Operator Characteristic (ROC) analysis which used to determine the optimum cut-off value, sensitivity and specificity of the qRT-PCR of miR-224, miR-215 and miR-143 calculated in the serum.
Table 2.
The Validation of miR-215, miR-143 and miR-224) as a Diagnostic Biomarker in the Serum for HCC Patients
| miRNA | Cut off value | Specificity | Sensitivity | AUC |
|---|---|---|---|---|
| miR-215 | 8.137 | 78.66% | 81.14% | 0.754 |
| miR-143 | 3.177 | 80.31% | 75.44% | 0.855 |
| miR-224 | 5.785 | 75.16% | 82.22% | 0.823 |
AUC, area under curve;the results according to the ROC curve analysis.
Discussion
Hepatocellular carcinoma (HCC) is an aggressive malignant tumor of the liver, and it is one of the most common disorders worldwide, and known to be the second cancer related universal (Julius et al., 2016). Biomarkers are beneficial in the early detection of cancer at an early stage which is important since early detection has a direct effect on clinical outcomes and prognosis. Serum alpha-fetoprotein (AFP) is a useful tumor marker for the detection and monitoring of HCC, but unfortunately the false negative rate with AFP level alone may be as high as 40% for patients with early stage HCC (Nobuhiro et al., 2015).
MicroRNAs are small, single stranded non-coding RNA molecules nucleotides, 19-23 in lengths that may possibly serve as novel non-invasive molecular biomarkers for cancer diagnosis (Nina and Damjan, 2013). Several studies have examined their role in the regulation of different biological processes such as: protein synthesis, energy production, apoptosis, and differentiation (Anders et al., 2013). This study focused on three microRNAs; miR-215, miR-143 and miR-224.
This study aims to estimate the level of these miRNAs in serum of HCV induced hepatitis and HCC in relation to grades of hepatitis, stages of fibrosis and differentiation of tumor tissue and to correlate these serological levels with their counterparts extracted from liver tissue. Patient’s clinical data was analyzed among 50 patients and there were strong positive correlations between blood urea, ALT, AST and AFP levels in post-hepatitic HCC patients compared to healthy controls.
Firstly, the expression of the three miRNAs (miR-215, miR-143 and miR-224) was evaluated in the tumor and non-tumor liver tissues of post-hepatitic HCC patients. All the three miRNAs were decreased in tumor tissue as opposed to non-tumorous HCV chronic hepatitis tissues. It was reported that the expression of miR-143 is up-regulated in pancreatic stellate cells cancer and esophageal cancer and there are few reports about the relationship between miR-143 expression and the diagnosis of HCC. In addition miR-215 had a significantly lower expression in nephroblastomas regardless of the subtype compared with mature kidney measured by quantitative real-time-PCR (Zhu-qing et al., 2014). Anin vitro study showed that miR-215 might contribute to kidney cancer metastasis through different biological processes (Yang et al., 2017).
Secondly miR-215, miR-143 and miR-224 expressions were evaluated in the serum of HCV associated HCC patients and in contrast to control serum samples from healthy volunteers. We found that the three miRNAs were up regulated in the serum of HCC patients compared to control samples.
The expression of the 3 miRNAs in the serum was evaluated in relation to tumor grade (G1, G2 or G3). It was found that the expression of the three miRNAs was significantly higher in the serum of G3 patients than G2 and G1 patients. This revealed that the expression of miRNAs in the serum may be used to predict the tumor grade.
In addition the fibrosis stage of the patients was correlated to the expression level of the 3 miRNAs in the serum. It was found that the expression level of the miRNAs was significantly higher in cirrhosis (stages F5 and F6) compared to fibrosis (stages F1-F4).
According to our findings, all the studied miRNAs; miR-215, miR-143 and miR-224 were significantly upregulated in serum of HCV associated HCC cases and could differentiate HCC positive from HCC negative ones with cut off value 8.137, 3.177 and 5.785 respectively.
In conclusion, we have investigated the expression of miR-215, miR-143 and miR-224in HCV associated HCC patients, and found that all the three miRNAs were up-regulated in the patient’s serum and could promote malignant progression of liver cancer, suggesting that miR-215, miR-143 and miR-224 may function as diagnostic and prognostic markers as well aspossible therapeutic targets for treatment of HCC.
References
- Akin Y, Hacer I, Ebru A, Sevda M. Real-time PCR for gene expression analysis in'polymerase chain reaction'Eds Hernandez P and Gomez A. InTech Journals. 2012;(Chapter 12):229–54. [Google Scholar]
- Albert R, Yujuan D, Ho T, Jun Y. Detection of miRNA as Non-Invasive Biomarkers of Colorectal Cancer. Int J Mol Sci. 2015;16:2810–23. doi: 10.3390/ijms16022810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders R, Leonard J. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol Syst Biol. 2013;9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih-Hao C, Jing Q, David O, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–41. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detlef S, Nezam H. Liver cirrhosis. Lancet. 2009;371:838–51. [Google Scholar]
- Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- Jian Y, Ping Z, Jin L, Wei X. MicroRNA-215 acts as a tumor suppressor in breast cancer by targeting AKT serine/threonine kinase 1. Oncol Lett. 2017;14:1097–1104. doi: 10.3892/ol.2017.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius B, David V, Emad A, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Jiujie C, Keping X. Signaling of MiRNAs-FOXM1 in cancer and potential targeted therapy. Curr Drug Targets. 2013;14:1192–1202. doi: 10.2174/13894501113149990192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nina H, Damjan G. MicroRNAs and long non-coding RNAs: prospects in diagnostics and therapy of cancer. Radiol Oncol. 2013;47:311–8. doi: 10.2478/raon-2013-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuhiro T, Yu S, Itaru E, et al. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10573–83. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Z, Wai-Kay S, Ching-Lung L, Man-Fung Y. Epidemiology of Hepatocellular Carcinoma in the Asia-Pacific Region. Gut Liver. 2016;10:332–9. doi: 10.5009/gnl15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubham S, Robert S, Ranjit R, Ratna B. MicroRNAs: role in hepatitis C virus pathogenesis. Genes Dis. 2015;2:35–45. doi: 10.1016/j.gendis.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taned C, Kannika P. Risk factors for the development of hepatocellular carcinoma in Thailand. J Clin Transl Hepatol. 2015;3:182–8. doi: 10.14218/JCTH.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Yang J, Tianmin X, Shunqing Z, Manhua C. MicroRNA-215 targets NOB1 and inhibits growth and invasion of epithelial ovarian cancer. Am J Transl Res. 2017;9:466–77. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zeeneldin A, Salem S, Darwish A, et al. Untreated hepatocellular carcinoma in Egypt: outcome and prognostic factors. J Hepatocell Carcinoma. 2015;2:3–9. doi: 10.2147/JHC.S73828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Takahashi S, Tasaka A, et al. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:565–75. doi: 10.1111/j.1440-1746.2012.07271.x. [DOI] [PubMed] [Google Scholar]
- Zhenyue H, Jun Y, Xiaolong L, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial–mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15:51. doi: 10.1186/s12943-016-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu-qing Z, Hua M, Nan W, et al. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn Pathol. 2014;9:135. doi: 10.1186/1746-1596-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]


