Abstract
Objective
To determine the capacity, effectiveness, efficiency, and reliability of select tissue dissociation methods to isolate mononuclear cells from the CNS of mice with experimental autoimmune encephalomyelitis (EAE).
Methods
As part of an assay qualification, we tested the isolation method Percoll PLUS vs a commercially available enzymatic Neural Tissue Dissociation Kit (Kit), and the enzymes accutase and papain in C57BL/6 mice with active EAE. In a stepwise approach, we applied the following 4 criteria to each dissociation method: (1) mononuclear cell viability post-processing was required to be ≥80% per brain or spinal cord sample, (2) absolute live mononuclear cell numbers was required to be ≥5 × 105 per brain or spinal cord sample of mice with clinical EAE, (3) test-retest reliability had to be verified, and (4) the absolute mononuclear cell numbers in brain and spinal cord had to correlate with the EAE disease course.
Results
Enzymatic dissociations allowed for greatly increased cell yield and specifically allowed for downstream assays from individual brains and spinal cords in C57BL/6 mice with EAE. All enzymatic dissociations provided a more efficient and effective method for isolating mononuclear cells from brains and spinal cord. Only the Kit assay provided a significant correlation between absolute mononuclear cell numbers in the spinal cord and EAE disease severity.
Conclusions
Enzymatic dissociation of CNS tissue of C57BL/6 mice with active EAE with the Kit should be the standard method. The identification of optimized CNS dissociation methods in EAE has the potential to identify cellular events that are pertinent to MS pathogenesis.
Percoll PLUS gradients are commonly used to isolate mononuclear cells from the lipid enriched environment of the CNS.1,2 However, this method does not provide a high cell yield, and requires a large number of mice to perform analytical assays with the isolated cells.3–5 A standard assay should be efficient, effective, reliable, and should allow for further applications with minimal artificial manipulation of isolated cells.
We selected enzymes that are relatively specific for the extracellular matrix of the CNS, including laminins, proteoglycans, tenascins, reelin, agrin, netrins, and slits.6,7 Papain is a cysteine peptidase C1 protease that digests many of the above-mentioned substrates better than pancreatic proteases.8 For enzymatic tissue dissociation, papain has proven more efficient and less destructive than other proteases, including for the isolation of viable, morphologically intact, cortical rat neurons.9 Accutase is a ready-to-use liquid consisting of a proprietary mixture of proteolytic enzymes. It maintains an intact morphology, and equivalent gene expression markers of human embryonic stem cells,10 and human neural stem cells.11
The following criteria were applied to each dissociation method: (1) mononuclear cell viability post-processing was required to be ≥80% per brain or spinal cord sample, (2) absolute live mononuclear cell numbers were required to be ≥5 × 105 per brain or spinal cord sample of mice with clinical experimental autoimmune encephalomyelitis (EAE), (3) test-retest reliability had to be verified, and (4) the absolute mononuclear cell numbers in brain and spinal cord had to correlate with EAE disease activity.
Methods
Peptides
Mouse myelin oligodendrocyte glycoprotein35–55 (MOGp35–55) (MEVGWYRSPFSRVVHLYRNGK) was synthesized by CS Bio (Menlo Park, CA).
Animals
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME), or from a mouse facility at The University of Texas Southwestern Medical Center (UTSW). All mice were maintained in a pathogen-free mouse colony according to the guidelines set forth by the NIH and UTSW. All experiments were approved by the Institutional Animal Care and Use Committee at UTSW.
Induction of EAE
The active EAE induction model was chosen to immunophenotype a broad array of immune competent cells from the CNS. In contrast, adoptive transfer EAE is typically induced by transferring either polarized T-cell receptor transgenic CD4+ T cells, or purified wild-type donor CD4+ T cells into recipient mice. These methods were published by our group previously.12,13 Briefly, 8–12 week old male C57BL/6 mice were anesthetized with tribromoethanol (Avertin; Sigma-Aldrich, St. Louis, MO) 250 mg/kg intraperitoneally (i.p.), and subsequently immunized subcutaneously with MOGp35–55 (100 μg/mouse), emulsified in an equal volume of complete Freund adjuvant14 containing 4 mg/mL H37Ra Mycobacterium tuberculosis (Difco; BD, Franklin Lakes, NJ) in each flank. At the time of immunization and 48 hours later, mice received an i.p. injection of 200 ng pertussis toxin in 200 μL PBS. For all experiments, individual animals were observed daily based on the EAE clinical scoring system as follows: 0 = no clinical disease, 1 = loss of tail tone, 2 = mild paraparesis, 3 = paraplegia, 4 = hindlimb and forelimb paralysis, and 5 = moribund or death.
Tissue isolation
At the peak of EAE disease, or typically within 12 days of active disease induction, MOGp35–55 immunized mice were perfused through the left ventricle using cold PBS. Brains and spinal cord were dissected and immediately placed in cold PBS for processing.
Percoll PLUS density gradient
The Percoll PLUS (GE Healthcare Bio-Sciences, Pittsburgh, PA) gradient was performed as previously described4; briefly, brains and spinal cords were individually passed through a 70 μm sterile filter to generate a single-cell suspension, and then washed in cold PBS. After washing, the single cell suspension was processed using 2 mL of 37% Percoll PLUS, and a 4 mL/2 mL 30%/70% Percoll PLUS gradient. Cells were then counted using a hemocytometer with Trypan Blue for the discrimination of dead cells. Cells from 3 mice were pooled for subsequent analyses.
Enzymatic digestions
Brains and spinal cords were first finely minced using a sterile scalpel, washed with cold PBS, then processed based on the specific enzymes used. The commercially available Neural Tissue Dissociation Kit (P) (Kit) was used following the manufacturer's protocol (Neural Tissue Dissociation Kit (P); Miltenyi Biotec, San Diego, CA). One milliliter of accutase (Global Cell Solutions, Charlottesville, VA) was added to the tissues at 1 mL per tissue and incubated at room temperature for 15 (accutase 15), 30 (accutase 30), or 60 minutes (accutase 60). Twenty units of papain (Sigma-Aldrich) was added per tissue sample and incubated for 15 (papain 15), 30 (papain 30), or 60 minutes (papain 60) at 37°C. Immediately following the incubation, papain was quenched using 150 μg ovomucoid (Sigma-Aldrich). For the combination of accutase and papain (accutase 15/papain 30), 20 U of papain was first added to the brain isolates and incubated for 30 minutes at 37°C. After 30 minutes, the papain was quenched using 1.5 μg ovomucoid. The brains were then washed with cold PBS, and 1 mL of accutase was added for 15 minutes at room temperature. Following all enzymatic dissociation methods, brains were washed with cold PBS, and then subjected to 1 wash with 37% Percoll PLUS to remove remaining myelin. The myelin-free single cell suspensions were counted using a hemocytometer.
Flow cytometry: Extracellular staining
A total of 1 × 106 cells were stained with 1 μL Ghost Dye Red 780 (Tonbo Biosciences, San Diego, CA) and incubated for 15–30 minutes at room temperature, following manufacturer's recommendations. Cells were then washed using 2% FBS-PBS (FACs buffer) at 1,500 rpm for 5 minutes, followed by incubation with 1 μg FcBlock (anti-CD16/32; Tonbo Biosciences) for 15 minutes at 4°C. Cells were then stained with CD45-Alexa Fluor 700 (BioLegend, San Diego, CA), CD3-Pacific Blue (BD Bioscience, San Jose, CA), CD19-PerCP-Cy5.5, CD11c-APC, CD11b-BV650, CD8-FITC, and CD4-PE-Cy7 (BioLegend) for 35–40 minutes at 4°C. Cells were then gated on time to verify no clog occurred during acquisition (figure e-1A at links.lww.com/NXI/A29). Leukocytes were gated according to morphology side scatter (SSC-A) vs sorward scatter (FSC-A) (figure e-1B, links.lww.com/NXI/A29). Doublets were excluded (FSC-H vs FSC-W and SSC-H vs SSC-W) (figure e-1, C and D, links.lww.com/NXI/A29). Live leukocytes were selected using GhostDye and CD45 staining (figure e-1, E and F, links.lww.com/NXI/A29). Immune cells were gated based on the staining of CD11b, CD11c, CD3, CD4, CD8, and CD19 markers (figure e-1, G–J, links.lww.com/NXI/A29). Starting regions to distinguish granulocytes and microglial cells by side scatter (SSC) vs CD45 (figure e-1K, links.lww.com/NXI/A29).4 Data were acquired using a BD FACS LSR Fortessa (The Moody Foundation Flow Cytometry Facility, Children's Medical Center Research Institute at UT Southwestern). Data were analyzed using FlowJo v10 (FlowJo, Ashland, OR).
Statistical analyses
Groups were compared for normality by using the Kolmogorov-Smirnov test. Normally distributed values were compared using the unpaired two-sided Student t test. Values that were not normally distributed were compared using the non-parametric test Mann-Whitney U test. The criteria for significance (α) was set at *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Each experiment was repeated at least once.
To demonstrate reliability, the 95% confidence interval of the mean was calculated to express the precision and repeatability of each dissociation method tested. Then, the coefficient of variation (CV), defined as the ratio of the SD to the mean was computed. The CV shows the extent of variability in relation to the mean of 2 experimental samples. The CV is somewhat arbitrarily set in different experimental settings. Based on our cumulative experience and data in the very heterogeneous EAE model, it was set to ≤25% for the purpose of this study.
All analyses were performed using Prism 7 (GraphPad, La Jolla, CA).
Results
The viability of cells processed from brain and spinal cord tissue of MOGp35–55-immunized mice varies with the type of treatment used for tissue processing
The first requirement in our assay qualification algorithm was a mononuclear cell viability post-processing of ≥80% per brain or spinal cord sample. The identification and exclusion of dead cells is an important aspect in flow cytometric assays. Dead cells possess the potential to bind nonspecifically to many reagents, and they have a tendency to become more autofluorescent. Both of these effects may result in an increase in background fluorescence, and consequently reduce the dynamic range and our ability to detect fluorescent signals that are only weakly positive. In addition, dead cells will eventually release DNA that is notoriously adhesive and may lead to cell aggregation. Finally, some cell subsets may be more prone to death than others in specific experimental settings, and a lower percentage of viable cells may not represent the cellular composition of the original sample.
An extensive search of the scientific literature revealed that there is currently no universally accepted threshold for the percentage of dead mononuclear cells in cellular phenotyping by multiparameter flow cytometry. Therefore, the requirement for a mononuclear cell viability post-processing of ≥80% per brain or spinal cord sample was based on our own extensive experience with Percoll PLUS dissociation of CNS tissue. Using this assay, we have consistently found a cell viability in the 80% range. A further reduction in the percentage of viable cells by other tissue dissociation methods was considered unacceptable.
The cytotoxicity and cell viability of mononuclear cells from immedicately ex vivo brain and spinal cord tissue of mice immunized for EAE was determine by flow cytometry as described in the Method section (figure e-1, A–K, links.lww.com/NXI/A29). For each monoclonal antibody that was used, the laser excitation wavelength, the clone, and the fluorochrome are listed in table e-1 (links.lww.com/NXI/A31). Tissue dissociation of brain tissue with Percoll PLUS, 30 minutes incubation with accutase (accutase 30), or accutase 60 treatments did not reach the pre-defined criterion, and were excluded from further analyses (figure 1A). Spinal cord tissue dissociation with acutase 15, 30, 60, and accutase 15/papain 30 did also not reach the pre-defined criterion, and were excluded from further analyses (figure 1B).
Figure 1. Tissue processing methods and cellular viability.
The first requirement of our assay qualification algorithm was a mononuclear cell viability post-processing of ≥80% per brain or spinal cord sample. (A) Brains and (B) spinal cords from individual mice with experimental autoimmune encephalomyelitis scores >2 were dissociated with Percoll PLUS, Neural Tissue Dissociation Kit, or incubated with accutase or papain for 15, 30, or 60 minutes. Cellular viability was determined with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit. The mean of a minimum of 4 separate tissue dissociations are shown.
Some enzymatic dissociation methods provide sufficient cell numbers for flow cytometric analyses of mononuclear cells from brains tissue, but not spinal cords of mice with sub-clinical EAE
Induction of EAE by active immunization may not lead to clinical disease despite the detectable presence of inflammatory mononuclear cell infiltrates in the brain and spinal cord. It is of great potential interest to determine the absolute threshold of mononuclear cells in these compartments that confers disease susceptibility or disease resistance, and to further identify specific mononuclear cell subsets that are detrimental or protective in CNS autoimmunity. These observations could potentially apply to patients with a so called radiologically isolated syndrome, who meet radiologic criteria for MS, but have not yet experienced a clinical neurologic event.15 Approximately 30% of patients with radiologically isolated syndrome will eventually develop clinically MS, and it would be meaningful to identify boichemical and biological risk factors of disease progression in the EAE model.
To determine whether there are sufficient numbers of live cells for flow cytometric analyses of mice with sub-clinical EAE, live cells from brains and spinal cords of mice immunized with MOGp35–55 and an EAE score of 0 were quantified. Cell enumeration and phenotyping was conducted by multiparameter flow cytometry.
Enzymatic tissue dissociation of brain tissue from experimental animals with subclinical EAE with papain 15 of brain tissue did not reach the pre-defined criterion for mice with clinical EAE, namely an absolute live mononuclear cell numbers of ≥5 × 105 per brain or spinal cord sample (figure e-2A, links.lww.com/NXI/A30). In the spinal cord, none of the dissociation methods reached the pre-defined criterion for mice with clinical EAE except papain 60 (figure e-2B, links.lww.com/NXI/A30).
Enzymatic dissociation methods allow downstream processing of mononuclear cells from mice with clinical EAE
The second requirement of our assay qualification algorithm was an absolute live mononuclear cell numbers of ≥5 × 105 per brain or spinal cord sample in mice with clinical EAE. In animals with clinical EAE, the numbers if mononuclear cells in the brain and spinal cord were determined. All enzymatic dissociation methods of brain tissue yielded high enough total live cell numbers to meet our second criterion, and accutase 15/papain 30 showed significantly higher numbers of live cells than accutase 15, papain 15, papain 30, and papain 60. After enumeration of total live cells, Kit treatments yielded a higher median cell number than accutase 15/papain 30 (figure 2A; table e-2, links.lww.com/NXI/A32).
Figure 2. Mononuclear cell numbers after enzymatic dissociation methods in mice with clinical experimental autoimmune encephalomyelitis.
(A) All enzymatic dissociation methods yielded high enough total live cell numbers to meet our second criterion, and accutase 15/papain 30 showed significantly higher numbers of live cells than accutase 15, papain 15, papain 30 and papain 60. After enumeration of total live cells, Kit treatments yielded a higher median cell number than accutase 15/papain 30. (B) Treatment of spinal cord tissue of mice with clinical experimental autoimmune encephalomyelitis with different dissociation methods demonstrated that the Kit and papain 60 yielded sufficient cell numbers for further flow cytometiic analyses as pre-defined by the second criterion. (C) When mononuclear cells isolated from the brain were gaited on CD45+ cells, Kit enzymatic dissociation yielded a higher median cell number than other methods, including accutase 15/papain 30. (D) In the spinal cord, numbers of CD45+ cells were similar between Kit and papain 60. Groups were compared for normality by using the Kolmogorov-Smirnov test. Values were compared using the non-parametric test Mann-Whitney U test. *p < 0.05, **p < 0.01, ***p < 0.001.
Treatment of spinal cord tissue of mice with clinical EAE with different dissociation methods demonstrated that the Kit and papain 60 yielded sufficient cell numbers for further flow cytometric analyses as pre-defined by the second criterion (figure 2B; table e-3, links.lww.com/NXI/A33).
When mononuclear cells isolated from the brain were gated on CD45+ cells, Kit enzymatic dissociation yielded a higher median live cell number than other methods, including accutase 15/papain 30 (figure 2C; table e-2, links.lww.com/NXI/A32). In the spinal cord, number of CD45+ cells were similar between Kit and papain 60 (figure 2D; table e-3, links.lww.com/NXI/A33).
Some enzymatic dissociation methods provide reliable test results when processing of mononuclear cells from brain and spinal cord
The third pre-defined criterion of our assay qualification algorithm was test reliability. As stated in the Method section, the 95% confidence interval of the mean was calculated to express the precision and repeatability of each dissociation method tested. Then, the CV, defined as the ratio of the SD to the mean was computed. The CV shows the extent of variability in relation to the mean of 2 experimental samples. In this study, the CV was set to ≤25, given the heterogeneity of EAE disease activity.
Following the enzymatic dissociation of brains from mice with clinical EAE, Kit, accutase 15, and papain 60 met the criteria (figure 3A; table e-3, links.lww.com/NXI/A33). Kit and papain 60 were sufficiently reliable when used to dissociate spinal cords (figure 3B; table e-3, links.lww.com/NXI/A33).
Figure 3. Test reliability of enzymatic dissociation methods.
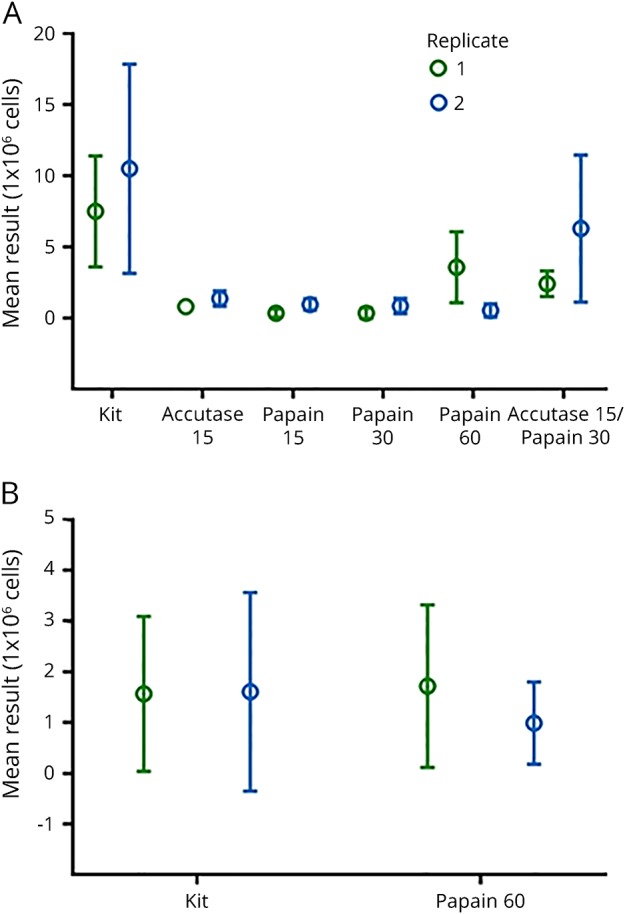
The third pre-defined criterion of our assay qualification algorithm was test reliability. As stated in the method section, the 95% confidence interval of the mean was calculated to express the precision and repeatability of each dissociation method tested. Then, the coefficient of variation (CV), defined as the ratio of the SD to the mean was computed. The CV shows the extent of variability in relation to the mean of 2 experimental samples. In this study, the CV was set to ≤25%, given the heterogeneity of experimental autoimmune encephalomyelitis (EAE) disease activity. (A) Following the enzymatic dissociation of brains from mice with clinical EAE, Kit, accutase 15, and papain 60 met the criteria. (B) Kit and papain 60 were sufficiently reliable when used to dissociate spinal cords.
Enzymatic dissociation allow leukocyte subset isolation from brain tissue
The number of mononuclear cell subsets isolated from brains of mice with EAE was compared across all enzymatic dissociation methods. Regarding the absolute numbers of CD45+ mononoculear cells, there were some significant differences between tissue dissociastion methods (table e-4, links.lww.com/NXI/A34). Accutase 15/papain 30 and Kit yielded the highest CD3+ T cells numbers, and the difference between accutase 15/papain 30 and papain 15 was significant (figure 4A; table e-4, links.lww.com/NXI/A34). There were no differences between any of the experimental groups with regard to CD4+ T cell numbers (figure 4B), or CD8+ T-cell numbers (figure 4C). Enzymatic tissue dissociation with Kit offered the highest amounts of CD19+ B cells, and the increase in cell yield compared to accutase 15, papain 15, and papain 30 was significant (figure 4D; table e-5, links.lww.com/NXI/A35). Brain dissociation with Kit yielded a higher number of CD11b+ myeloid cells as compared to papain 15 and papain 30, and accutase 15/papain 30 treatment provided significantly higher cell numbers than papain 15 and papain 30 (figure 4E; table e-5, links.lww.com/NXI/A35). Accutase 15 tissue dissociation resulted in signficantly higher CD11b+ cell numbers than papain 15 and papain 30, and papain 60 in signficantly higher CD11b+ cell numbers than papain 30 (figure 4E; table e-5, links.lww.com/NXI/A35).
Figure 4. Leukocyte subset isolation from brain tissue by enzymatic dissociation.
(A) Accutase 15/papain 30 and Kit yielded the highest CD3+ T cells numbers, and the difference between accutase 15/papain 30 and papain 15 was significant. (B) There were no differences between any of the experimental groups regarding to CD4+ T cell numbers, or (C) CD8+ T cell numbers. (D) Enzymatic tissue dissociation with Kit offered the highest amounts of CD19+ B cells, and the increase in cell yield compared to accutase 15, papain 15, and papain 30 was significant. (E) Brain dissociation with Kit yielded a higher number of CD11b+ myeloid cells as compared to papain 15 and papain 30, and accutase 15/papain 30 treatment provided significantly higher cell numbers than papain 30. Groups were compared for normality by using the Kolmogorov-Smirnov test. Values were compared using the non-parametric test Mann-Whitney U test. *p < 0.05, **p < 0.01, ***p < 0.001.
Kit and papain 60 are efficient in isolating mononuclear cell subsets from spinal cord of mice with EAE
Next, the cellular yield of mononuclear cell subsets after enzymatic tissue dissociation of spinal cord tissue from mice with EAE with Kit and papain 60 was assessed. There was no significant difference between the 2 experimental groups regarding CD3+ T cells (figure 5A; table e-4, links.lww.com/NXI/A34), CD4+ T cells (figure 5B; table e-4, links.lww.com/NXI/A34), CD8+ T cells (figure 5C; table e-4, links.lww.com/NXI/A34), CD19+ B cells (figure 5D; table e-4, links.lww.com/NXI/A34), and CD11b+ myeloid cells (figure 5E; table e-4, links.lww.com/NXI/A34).
Figure 5. Mononuclear cell subset isolation from spinal cord of mice with experimental autoimmune encephalomeylitis.
There was no significant difference between the 2 experimental groups regarding (A) CD3+ T cells, (B) CD4+ T cells, (C) CD8+ T cells, (D) CD19+ B cells, and (E) CD11b+ myeloid cells. Finally, the fourth requirement of our assay qualification algorithm, namely a correlation between absolute mononuclear cell numbers in brain and in spinal cord and experimental autoimmune encephalomyelitis (EAE) disease course was tested. (F) There was a significant correlation between live mononuclear cell numbers isolated from spinal cord tissue with Kit and clinical EAE disease severity. Analyses of mononuclear cell subsets in the brain or spinal cord did not demonstrate any significant correlations with EAE disease activity, regardless of the tissue dissociation method that was utilized. Each experiment was repeated at least once. Representative results are shown. Groups were compared for normality by using the Kolmogorov-Smirnov test. Values were compared using the non-parametric test Mann-Whitney U test.
There is a correlation between EAE disease severity and total number of live mononuclear cells in the spinal cord
The fourth requirement of our assay qualification algorithm was a correlation between absolute mononuclear cell numbers in brain and spinal cord and EAE disease course. The EAE scores of each mouse were plotted against the total number of mononuclear cell subsets that were isolated from brain and spinal cord tissue with all dissociation methods that were tested. There was only one significant correlation between the total live mononuclear cell numbers isolated from spinal cord tissue with Kit and clinical EAE disease severity (figure 5F). In contrast, analyses of mononuclear cell subsets did not demonstrate any significant correlations.
Dissociation methods differ significantly regarding the cellular compositions of the tissue extract
To determine whether the different tissue dissociation methods would yield a similar cellular composition of mononuclear cells from brain tissue, the percent of CD11b+, CD11c+, CD4+, CD8+, and CD19+ cells gated from live CD45+ cells (figure 6A), and subsequently normalized to 100% (figure 6B). The same mononuclear cell subsets from spinal cord were then gated from the live CD45+ population (figure 6C), and normalized to 100% (figure 6D).
Figure 6. Cellular composition after tissue dissociation.
(A) To determine whether the different tissue dissociation methods would yield a similar cellular composition of mononuclear cells from brain tissue, the percent of CD11b+, CD11c+, CD4+, CD8+, and CD19+ cells gated from live CD45+ cells, and (B) subsequently normalized to 100%. (C) The same mononuclear cell subsets from spinal cord were then gated from the live CD45+ population, and (D) normalized to 100%. The mean of a minimum of 4 separate tissue dissociations are shown.
Numerous significant differences in absolute live CD45+ cells, CD3+ T cells, CD19+ B cells, and CD11b+ myeloid cells were observed with different tissue dissociation methods (table e-5, links.lww.com/NXI/A35).
The commercially available kit is the most expensive dissociation method
We also intended to determine the cost of each dissociation method per brain or per spinal cord. While the commercially available Kit outperformed the other methods regarding many of the pre-defined criteria, it is also by far the most expensive way to dissociate brain and spinal cord tissue (table e-6, links.lww.com/NXI/A36). Specifically, the Kit assay per experimental animal was up to 11-fold higher than some of the other dissociation methods.
Discussion
In this study, we aimed to analyze and identify dissociation methods that are efficient, effective, reliable, inert, non-toxic, readily available, and affordable for the isolation of mononuclear cells from individual brains and spinal cords of mice with EAE. Specifically, we applied a 4-step algorithm to determine potentially feasible methods of tissue dissociation that include effects on mononuclear cell viability post-processing of brain or spinal cord sample, the absolute live mononuclear cell numbers of per brain or spinal cord sample in C57BL/6 mice with clinical EAE, test-retest reliability, and a correlation between absolute mononuclear cell numbers in brain and spinal cord and EAE disease course. We also assessed the cost of each method.
Overall, we found that some of the enzymatic dissociation methods provide a more reliable, efficient, and effective method for isolating mononuclear cells from individual EAE brains and spinal cords then Percoll PLUS. Only Kit dissociation met all of our pre-specified criteria, and may provide advantages in identifying biochemical and biological factors that mediate EAE disease susceptibility or resistance.
With improved performance comes an increased price, and the Kit assay per test was up to 11-fold higher than some of the other dissociation methods. In a situation where an EAE score is not a consideration, alternatives to the Kit assay may be feasible, especially if experiments are carried out on a large scale. Otherwise, the authors believe that the best possible technique should be used for CNS tissue dissociation in the active EAE model in C57BL/6 mice.
The authors believe that animal welfare should strongly be considered in every instance an experiment is designed and executed. By broad expert consensus, the guiding principles underlying the humane use of experimental animals in scientific research are termed the “3 Rs”16: (1) to Replace the use of animals with alternative techniques; (2) to Reduce the number of animals used through experimental design to a minimum; (3) to Refine the way experiments are carried out to reduce the suffering of animals as much as possible. Our data suggest ways to reduce the number of animals needed to obtain meaningful biological data in experiments that assess mechanisms underlying CNS autoimmunity and inflammation.
Perhaps not surprisingly, we observed significant differences in the cellular compositions between some of the dissociation methods. It appears intuitive that proteolytic degradation of brain and spinal cord by different methods would favor the yield and viability of mononuclear cell subsets. Ultimately, this observation substantially limits the ability to correlate cellular subsets with clinical EAE phenotypes, or effects on any therapeutic interventions in this animal model, as there is currently no assay that precisely reflects the number and composition of CNS cells in situ just prior to dissociation. Immunohistochemistry/immunofluorescence assays are always semi-quantitative, and as a detection assay are also potentially prone to systemic errors.17 For instance, each detection antibody would have to be validated to ascertain reproducibility.
The authors believe that in the absence of a truly standardized method, investigators should only compare results of CNS dissociation experiments that use the same method. Alternatively, there should be an expert consensus to reach agreement on a unified experimental approach. It appears critical to not alternate between dissociation methods during longitudinal experiments, or when comparing samples in a cross-sectional research design.
Glossary
- CV
coefficient of variation
- EAE
experimental autoimmune encephalomyelitis
- MOGp35–55
myelin oligodendrocyte glycoprotein35–55
- UTSW
University of Texas Southwestern Medical Center
Author contributions
Rehana Z. Hussain participated in study concept and design, acquisition of data, data analysis and interpretation, and critical revision of the manuscript for important intellectual content. William A. Miller-Little participated in acquisition of data, data analysis and interpretation, and critical revision of the manuscript for important intellectual content. Richard Doelger participated in acquisition of data, data analysis and interpretation, and critical revision of the manuscript for important intellectual content. Gary Cutter participated in study concept and design, data analysis and interpretation, and critical revision of the manuscript for important intellectual content. Nicolas Loof participated in study concept and design, acquisition of data, data analysis and interpretation, and critical revision of the manuscript for important intellectual content. Petra D. Cravens participated in study concept and design, acquisition of data, data analysis and interpretation, and critical revision of the manuscript for important intellectual content. Olaf Stüve participated in study concept and design, acquisition of data, data analysis and interpretation, critical revision of the manuscript for important intellectual content, and study supervision.
Study funding
This work was in part supported by a Merit Review grant (federal award document number [FAIN] I01BX001674) from the US Department of Veterans Affairs, Biomedical Laboratory Research and Development (O.S.).
Disclosure
R.Z. Hussain, W.A. Miller-Litter, and R. Doelger report no disclosures. G. Cutter served on a scientific advisory board of AMO Pharmaceuticals, Apotek, Horizon Pharmaceuticals, Modigenetech/Prolor, Merck, Merck/Pfizer, Opko Biologics, Neurim, Sanofi-Aventis, Reata Pharmaceuticals, Receptos/Celgene, Teva, NHLBI, and NICHD; is a statistical reviewer for the Journal of the American Society of Nephrology; consults for Atara Biotherapeutics, Argenix, Bioeg GmBH, Consortium of MS Centers, Genzyme, Genentech, Innate Therapeutics, Klein-Buendel Inc., Medium, MedDay, Novartis, Opexa Therapeutics, Roche, Savara Inc., Somahlution, Teva, Transparency Life Sciences, and Therapeutics; is president of Pythagoras; received research support from Myasthenia Gravis Foundation of America, NIH/NIAID, DOD, NIH/NHLBI, Consortium of MS Centers, NIH/Children's Hospital (Boston), Alabama Department of Commerce, NIH/NICHHD, NIH, National Eye Institute, NIH/National Institute of Neurological Disorders and Stroke, and Patient-Centered Outcomes Research Institute; and is president of Consortium of MS Centers (only expenses covered). N. Loof and P.D. Cravens report no disclosures. O. Stuve served on the scientific advisory board of Pfizer and TG Therapeutics; serves on the editorial board of Therapeutic Advances in Neurological Disorders and Multiple Sclerosis; and received research support from Teva Pharmaceuticals, Opexa Therapeutics, and US Department of Veterans Affairs. Go to Neurology.org/NN for full disclosure forms.
References
- 1.Neeley SP, Conley FK. Extraction and immunocytochemical characterization of viable mononuclear inflammatory cells from brains of mice with chronic Toxoplasma gondii infection. J Neuroimmunol 1987;15:159–172. [DOI] [PubMed] [Google Scholar]
- 2.Dick AD, Pell M, Brew BJ, Foulcher E, Sedgwick JD. Direct ex vivo flow cytometric analysis of human microglial cell CD4 expression: examination of central nervous system biopsy specimens from HIV-seropositive patients and patients with other neurological disease. AIDS 1997;11:1699–1708. [DOI] [PubMed] [Google Scholar]
- 3.Cravens PD, Hussain RZ, Zacharias TE, et al. Lymph node-derived donor encephalitogenic CD4+ T cells in C57BL/6 mice adoptive transfer experimental autoimmune encephalomyelitis highly express GM-CSF and T-bet. J Neuroinflammation 2011;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cravens PD, Kieseier BC, Hussain R, et al. The neonatal CNS is not conducive for encephalitogenic Th1 T cells and B cells during experimental autoimmune encephalomyelitis. J Neuroinflammation 2013;10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cravens PD, Hussain RZ, Miller-Little WA, et al. IL-12/IL-23p40 is highly expressed in secondary lymphoid organs and the CNS during all stages of EAE, but its deletion does not affect disease perpetuation. PLoS One 2016;11:e0165248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruoslahti E. Brain extracellular matrix. Glycobiology 1996;6:489–492. [DOI] [PubMed] [Google Scholar]
- 7.Barros CS, Franco SJ, Muller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol 2011;3:a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furst DE, Emery P. Rheumatoid arthritis pathophysiology: update on emerging cytokine and cytokine-associated cell targets. Rheumatology (Oxford) 2014;53:1560–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huettner JE, Baughman RW. Primary culture of identified neurons from the visual cortex of postnatal rats. J Neurosci 1986;6:3044–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bajpai R, Lesperance J, Kim M, Terskikh AV. Efficient propagation of single cells accutase-dissociated human embryonic stem cells. Mol Reprod Dev 2008;75:818–827. [DOI] [PubMed] [Google Scholar]
- 11.Hall PE, Lathia JD, Caldwell MA, Ffrench-Constant C. Laminin enhances the growth of human neural stem cells in defined culture media. BMC Neurosci 2008;9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arellano B, Hussain R, Miller-Little WA, et al. A single amino acid substitution prevents recognition of a dominant human aquaporin-4 determinant in the context of HLA-DRB1*03:01 by a murine TCR. PLoS One 2016;11:e0152720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain RZ, Miller-Little WA, Lambracht-Washington D, et al. Laquinimod has no effects on brain volume or cellular CNS composition in the F1 3xTg-AD/C3H mouse model of Alzheimer's disease. J Neuroimmunol 2017;309:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippi M, Dousset V, McFarland HF, Miller DH, Grossman RI. Role of magnetic resonance imaging in the diagnosis and monitoring of multiple sclerosis: consensus report of the White Matter Study Group. J Magn Reson Imaging 2002;15:499–504. [DOI] [PubMed] [Google Scholar]
- 15.Kantarci OH, Lebrun C, Siva A, et al. Primary progressive multiple sclerosis evolving from radiologically isolated syndrome. Ann Neurol 2016;79:288–294. [DOI] [PubMed] [Google Scholar]
- 16.Flecknell P. Replacement, reduction and refinement. ALTEX 2002;19:73–78. [PubMed] [Google Scholar]
- 17.Taylor CR, Levenson RM. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology 2006;49:411–424. [DOI] [PubMed] [Google Scholar]







