Abstract
Bacteriophage (phage) therapy has encountered both enthusiasm and scepticism in the past century. New antimicrobial strategies against lethal pathogens are now a top priority for the World Health Organization, and although compassionate use of phages recently met with significant success, regulated clinical interventions seem unlikely in the near future. The hundredth anniversary of their discovery seems an appropriate time for a revival of phage therapy, particularly as the dilemma of antibiotic resistance grows. Phages are ubiquitous in the environment, on our food and in and on our bodies. Their influence on human health is currently being evaluated, and in this mini‐review, we examine data from recent metagenomic studies that propose a role for phages in the structure of the microbiome and in health and disease. We assess evidence for phages as vehicles for gene transfer in the context of antibiotic resistance and discuss challenges and opportunities along the critical path from phage discovery to a patient‐focused pharmaceutical intervention.
Abbreviations
- ARG
antibiotic resistance genes
- CRISPR
clustered regularly interspaced short palindromic repeats
- FMT
faecal microbiota transplantation
- MTase
methylase
- WHO
World Health Organization
Introduction
In the century since the first published descriptions of bacteriophages (phages) (Twort, 1915; D'Herelle, 1917), many have described the pros and cons associated with their clinical use (O'Flaherty et al., 2009; Loc‐Carillo and Abedon, 2011; Oliveira et al., 2015; Doss et al., 2017; Roach and Debarbieux, 2017). After World War II, interest in phages as therapeutic agents diminished with the advent of antibiotics, and although phage therapy remained popular in Eastern Europe, the West has seemed reluctant to develop phage‐based therapies. It is only since the emergence of antibiotic‐resistant ‘superbugs’ that phage therapy has received renewed interest. Such re‐appraisal is timely, given a recent publication by the World Health Organization (WHO) stressing the need for novel alternative biocontrol agents against ‘priority pathogens’ (Table 1) that pose the greatest threat to human health (Tacconelli and Magrini, 2017).
Table 1.
WHO list of drug‐resistant ‘priority’ pathogens (Tacconelli and Magrini, 2017)
| Critical | High | Medium |
|---|---|---|
| 1. Acinetobacter baumannii – carbapenem‐resistant | 4. Enterococcus faecium – vancomycin‐resistant | 10. Streptococcus pneumoniae – penicillin‐non‐susceptible |
| 2. Pseudomonas aeruginosa – carbapenem‐resistant | 5. Staphylococcus aureus – methicillin‐resistant, vancomycin‐intermediate and resistant | 11. Haemophilus influenzae – ampicillin‐resistant |
| 3. Enterobacteriaceae – carbapenem‐resistant, ESBL‐producing | 6. Helicobacter pylori – clarithromycin‐resistant | 12. Shigella spp., − fluoroquinolone‐resistant |
| – | 7. Campylobacter spp., – fluoroquinolone‐resistant | – |
| – | 8. Salmonellae – fluoroquinolone‐resistant | – |
| – | 9. Neisseria gonorrhoeae – cephalosporin‐resistant, fluoroquinolone‐resistant | – |
A handful of small companies are paving the way to manufacture phage preparations for health, food, veterinary, agriculture and aquaculture applications. While such preparations are permitted in the USA as food additives, from a therapeutic perspective, there are no phage‐specific regulatory guidelines yet in place (Oliveira et al., 2015; Palfrene et al., 2016; Fauconier, 2017), making the application of phage therapy currently extremely challenging. Therefore, more research, awareness, acceptance and agreement on appropriate regulations for phage therapy would serve not only to target bacteria on the WHO priority pathogen list but also to save lives. In a recent Food and Drug Administration‐approved emergency case in the USA, intravenous administration of phages is claimed to have saved the life of a near‐death patient with a multi‐drug‐resistant bacterial infection. Although this case is encouraging, approved application of phage therapy still faces several fundamental challenges (LaFee and Buschman, 2017).
An understanding of the natural occurrence of phages in humans, particularly their roles in the onset or prevention of illness, could help to ameliorate scepticism and promote greater acceptance of phage therapy in Western society. As the Western lifestyle is associated with serious metabolic health disorders, much attention has focused on the influence of the gut microbiota in health and disease (Sekirov et al., 2010). Microbial balance (as well as presence or absence of certain microbial species) is proposed to be important for maintaining gut health, although whether or not beneficial or detrimental health effects result from population shifts remains conjectural. Scientists are now turning their attention to the more neglected components of the microbiota (including fungi and viruses/phages) and their complex relationships with bacteria in the gut. Specifically, Mirzaei and Maurice (2017) made reference to the ‘ménage ā trois’ that exists between bacteria, phages and the human gut, stressing that understanding of the phage component could help to improve disease outcomes. Here, we assemble the evidence supporting phages as key actors on the microbiome stage, with a focus on metagenomic and transplant studies of the gut microbiota. We explore the evidence for bacteriophages as vehicles for carrying antibiotic resistance and address concerns over phage resistance as limiting factor for clinical intervention. We also discuss phage pharmacology and the practices needed to prepare commercial phage therapy products.
Phages of life
Phages (Figure 1) outnumber all forms of life on our planet (Clockie et al., 2011). They undergo either a lytic cycle (virulent) whereby they infect and destroy a bacterial cell to release progeny phages, or a lysogenic (temperate) cycle whereby they integrate into the bacterial genome until conditions provoke them to undergo lytic replication. Erez et al. (2017) recently demonstrated the existence of a ‘communication’ system in phages that actually controls lysis‐lysogeny decisions. For phage therapy, the preference is to use well‐characterized lytic phages with a broad host range and to avoid lysogenic phages which are less likely to eliminate the target. They also have the capacity to exchange genetic information between bacteria, possibly transferring undesirable traits such as antibiotic resistance or virulence (Figure 2).
Figure 1.
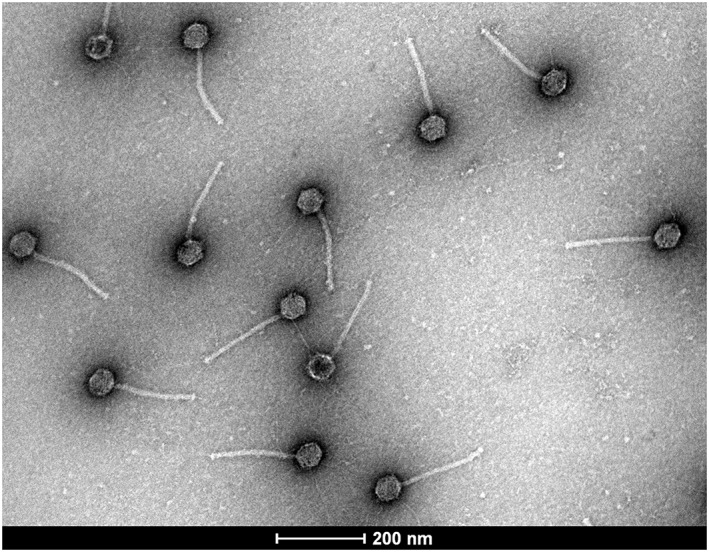
Transmission electron micrograph of bacteriophages isolated from the human gastrointestinal tract. Genotypic and morphological characteristics indicate that these phages belong the family Siphoviridae.
Figure 2.
Some considerations for phage therapy on the path to pharma.
Phages are natural entities found in all ecological niches. In the human body, they are dominant members of the microbiome, which is dispersed across four major habitats: the oral cavity, gastrointestinal tract, vagina and skin (Wahida et al., 2016). Within the gastrointestinal tract, phages have been described as the ‘movers and shakers’ of microbial communities, critically affecting the balance between health and disease (Mills et al., 2013). The mechanisms by which phages modulate the gut microbiome are likely to be multi‐factorial and, although studies to date are limited, researchers are now beginning to acquire clearer insights through metagenomic analysis, made possible by advances in high‐throughput sequencing technology. A recent metagenomic survey revealed that the gut bacteriophage community can be categorized as ‘core’, found in more than 50% of people, ‘common’ (20–25% of the population) and ‘rare’, representing phages rarely shared or unique to a person (Manrique et al., 2016). The majority show temperate behaviour, typically associated with healthy gut status, compared with a higher level of virulent phages found in patients suffering from bowel diseases (Norman et al., 2015). The most prevalent and conserved of gut phages is crAssphage, so‐called based on the cross assembly software used to identify it (Dutilh et al., 2014). Although it has not been propagated in the laboratory as yet, crAsspahge is predicted to infect the genus Bacteriodes and is carried by up to 75% of the global population. Despite this, the human gut ‘phageome’ is thought to be highly individual‐specific (Reyes et al., 2010). According to Manrique et al. (2016), the existence of a ‘healthy’ gut phageome suggests that the overall influence of phages in the human gut is beneficial rather than deleterious. They also propose that our ‘core’ phages could potentially be used for clinical therapeutics and controlled manipulation of the human gut ecosystem.
While metagenomics has helped our understanding of microbial communities in the gut, a significant obstacle for phageome research is the lack of suitable phage reference genomes in public databases, meaning that only a fraction of phage sequences can be identified. In addition, a high percentage of lysogenic phages often go undetected, as a result of having been mislabelled in databases as part of bacterial genomes. Furthermore, compared to double‐stranded DNA phages, single‐stranded DNA and RNA phages are more likely to go un‐sequenced, either due to their relative scarcity or the technical difficulties associated with their extraction and amplification. A further complication is contamination of phage fractions with bacterial DNA which, being far more abundant than viral, poses difficulties during sequence analysis (Roux et al., 2013; Bibby, 2014; Bruder et al., 2016; Hayes et al., 2017).
We postulate that the phageome provides a complementary (yet vital) view of the microbiome, but it is imperative that bioinformatics hurdles are overcome to enable greater insights into phage populations, of which greater than 90% are as yet unexplored. Moreover, the composition of phage populations may be more complex than just acting as a simple reflection of bacterial communities and may indeed contribute to health or disease status (Navarro and Muniesa, 2017). Ly et al. (2016) suggested that viral communities are readily shared within a household, having the capacity to shape not only our own microbiomes but also those of our close contacts. Phages have been proposed to alter recipient physiology following faecal microbiota transplantation (FMT), the transfer of stool from one host into the gastrointestinal tract of another to confer a health benefit (Bojanova and Bordenstein, 2016; Broecker et al., 2016; Ott et al., 2017; Zuo et al., 2017). FMT has been used to successfully treat recurrent Clostridium difficile infection, and there are indications that it has therapeutic potential for other intestinal or metabolic disorders (Gupta et al., 2016). Zuo et al. (2017) proposed that donor selection based on virome characteristics should be considered in FMT practice, and Chehoud et al. (2016) reported that temperate phages are significantly more likely to be transferred during FMT, supporting a model that the temperate phage life cycle has evolved partly to optimize phage transfer between environments. Temperate phages are understood to assist in controlling pathogen invasion, modulating community structure and maintaining gut homeostasis (Reyes et al., 2010). They may also modulate immune function in a beneficial manner, with the potential for broader application in clinical transplantation (Górski et al., 2006, 2016). Yet after a century of study where there has been no evidence of phages causing disease in humans, concerns still remain over their therapeutic use, largely due to reports that they can mediate the transfer of antibiotic resistance genes (ARG) and virulence factors. There is also concern that the host can mount an immune response, particularly to intravenously administered phages.
Do phages transfer antibiotic resistance?
The answer remains controversial. Microbial genomes are in constant flux as a result of horizontal gene transfer events facilitated by mobile genetic elements. Natural horizontal gene transfer processes include conjugation, transformation and transduction, with the last mediated by bacteriophages. A recent review by Touchon et al. (2017) concluded that transduction and lysogenic conversion complement other transfer mechanisms and have the potential to play a key role in spreading adaptive genes between microbial populations. Keen et al. (2017) described two Escherichia coli phage ‘super‐spreaders’ capable of promoting horizontal gene transfer by transformation, with efficient dispersion of ARG. Earlier studies also reported ARG to be enriched in the genomes of antibiotic‐treated phage communities in the lungs of cystic fibrosis patients (Fancello et al., 2011) and the faeces of mice (Modi et al., 2013). However, Enault et al. (2017) disputed these findings on the basis of false positives because the thresholds for in silico detection of ARG were too ‘relaxed’. They suggested that ARG abundances in phage genomes are vastly overestimated, and when they experimentally evaluated in silico‐predicted ARGs in vitro, they failed to confer an antibiotic resistance phenotype. Another study of ARGs in viromes from raw sewage, human faeces, pig faeces, marine and freshwater environments found that their prevalence varied among the groups, with fewest detected in the human‐associated viromes (Lekunberri et al., 2016).
Overcoming phage resistance
Bacteria develop a range of defences to avoid phage predation, including preventing phage attachment, digesting phage nucleic acids and developing abortive infection or clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated systems to survive the attack. However, as phages co‐evolve with their bacterial hosts, they also have the ability to overcome the bacterial defences encountered during an infection. Although phage resistance may not be viewed as problematic as antibiotic resistance for phage therapy, it is still one of the major concerns limiting its application. Several studies describe the use of phage ‘cocktails’ (mixtures of several phages that target the same host) to compensate for the development of phage resistance, although they may not necessarily eliminate it (Chan et al., 2013). Another approach could be to exploit the inherent ability of phages to circumvent bacterial defence mechanisms. Murphy et al. (2013) discussed the potential for phage‐encoded methylases (MTases) in phage therapy. MTases are highly abundant in the prokaryotic world and play important roles in several cellular processes, not least in protecting from invading nucleic acids. They function by transferring a methyl group to a target sequence, protecting the sequence from digestion by a restriction endonuclease. While MTases are often described in the context of restriction‐modification (R‐M) systems, they can exist independently as ‘orphan’ MTases and are predicted to be involved in cell regulation, replication, nucleic acid repair and evolution (Casadesús, 2016). A number of orphan MTases have been found on phage genomes, where it is assumed that they confer a survival benefit and contribute to the emergence of progeny phages with broader host range specificities. Acquisition of an MTase could allow progeny phages to infect additional hosts by overcoming resident R‐M systems. In this regard, the ever‐adapting nature of MTase‐carrying phages to outsmart their bacterial hosts would be highly advantageous for phage therapy applications.
Phages on film
Phages have an impressive ability to degrade biofilms, the communities of microbes that adhere to each other and to surfaces such as plant and animal tissues or medical devices such as catheters. The microbial cells are frequently embedded within extracellular polymers and, as they have heightened resistance to antibiotics, are major causes of persistent infections in clinical settings. Studies have shown that pathogens inside a biofilm can communicate with each other through a process called ‘quorum sensing’, allowing them to detect and join dense populations in their environment and build up the biofilm (Harper et al., 2014). Phages are capable of dispersing biofilms, either through the production of de‐polymerases that degrade the polymers in the biofilm or by ‘quorum quenching’ – enzymatic disruption of the quorum sensing process between bacteria in the biofilm (Pei and Lamas‐Samanamud, 2014). The use of phages as an anti‐biolfilm strategy has tremendous potential for phage therapy, particularly in efforts to eradicate the ever‐challenging multi‐drug resistant bacteria that pose significant threat to human health.
Phages to pharma
Although interest in phage therapy has undergone a resurgence in recent years, fundamental challenges remain for the conversion of phages in laboratory‐based research to patient‐focused pharmaceutical agents. Development of antibiotics by the pharmaceutical industry follows a clear regulatory path, but this path is not as straightforward for phages. Despite many advantages such as their high selectivity, low toxicity, ability to self‐multiply and lack of cross‐resistance with antibiotics, the path to phage‐based therapies for humans is yet to be elucidated. Cooper et al. (2016) detailed the development and approval pathways for new anti‐bacterial drugs, noting the challenges faced for phages and phage‐based products. Limitations exist in terms of development costs and regulations, with sound preclinical data as an essential pre‐requisite. So too is knowledge and experience of phage pharmacology (Abedon, 2014).
Phage pharmacology is differentiated into two distinct components – pharmacokinetics (how the body affects the phage) and pharmacodynamics (how the phage affects the body, including tissues and microbial flora). Factors such as the half‐life of the phages (a pharmacokinetic property), the virulence of the phage (a pharmacodynamics property) and other components of the host system influence the success of phage treatment. According to Curtright and Abedon (2011), the effect of a standard chemical drug is difficult to predict due to the complexity of the organisms being treated (such as humans), and with anti‐microbial agents (such as phages), this complexity is further increased due to the multi‐faceted nature of the pathogens being targeted. The success of phage therapy ultimately depends on optimal dose, timing, formulation and administration, with pharmacokinetics and pharmacodynamics characterized for each phage or phage cocktail. To date, only a few clinical trials have evaluated the potential of phage or phage‐based therapies in humans (Rhoads et al., 2009; Wright et al., 2009; Sarker and Brüssow, 2016; Jun et al., 2017). There is significant interest in the outcomes of ‘Phagoburn’ (www.phagoburn.eu), a European Union‐funded research and development trial assessing the safety, effectiveness and pharmacodynamics of two therapeutic phage cocktails for burn wounds infected by Escherichia coli and Pseudomonas aeruginosa.
Lytic phages are considered better candidates for therapeutic application than their lysogenic counterparts, because they kill their hosts with high specificity and limited collateral damage to the natural microbiome. Despite being the method of choice, the use of lytic phages poses a health concern because rapid bacterial lysis can result in the release of endotoxin and inflammatory mediators (albeit that bactericidal antibiotics can have similar effects). However, there is no evidence of immunological complications or side effects based on trials conducted to date, implying that phages are safe and well tolerated by humans. Immunological response is reported to vary depend on route of administration, and although phages can elicit specific anti‐phage antibody responses, these are not considered to be problematic for phage therapy practice (Sulakvelidze et al., 2001; Łusiak‐Szelachowska et al., 2014; Cisek et al., 2017; Roach and Debarbieux, 2017).
Phages intended for human use face significant manufacturing challenges. Abedon (2017a, 2017b) listed several phage companies that provide phage‐based products commercially, with a majority focusing on phage therapy applications. Bacteria and phage bank systems (consisting of master cell banks and working cell banks) must first be established and characterized (physiologically and genetically) according to various regulatory principles, and manufacturing processes must adhere to the guidelines of Good Manufacturing Practice (Palfrene et al., 2016). A key challenge is the limited stability of phages in solution, dropping in titre during processing and storage, which is unacceptable if they are to become regulated pharmaceutical agents. Stability of phages is influenced by environmental factors such as temperature, acidity and salinity and, in the perspective of phage therapy, they should be stored at either refrigeration or room temperature. Malik et al. (2017) have discussed the potential for encapsulation to facilitate slow and controlled release of phages, ensuring that phage concentrations remain at a therapeutically effective level over time.
The pharmacology of phage delivery depends to a large extent on the targeted site of action and the route of administration (Bodier‐Montagutelli et al., 2016; Cisek et al., 2017; Malik et al., 2017). The main concern with oral delivery is phage inactivation due to the acidic and proteolytic environment of the stomach (Zelasko et al., 2017). Brown et al. (2016) formulated a cream for topical treatment of Propionibacterium acnes and found that activity was maintained even after prolonged storage. They also suggested that the formulation had a dual effect, acting as a moisturizer while allowing close contact of the phage to P. acnes‐inhabited areas of the skin. The key challenges for delivery of inhalable phages for respiratory tract infection have been outlined by Bodier‐Montagutelli et al. (2016). They pointed out that the sensitivity of phages to external factors is highly variable between and within morphological families, which should be a consideration for cocktails containing different morphotypes. One must also consider the situation that an ideal phage in vitro may not function as well in vivo, so it is essential that preclinical experiments are conducted to the highest standards and reported in a manner that facilitates translation to clinical utility (Abedon, 2017a,b). In essence, phage pharmacology is still a subject of basic research, and progression must be rapidly accelerated in efforts to develop phages as health‐promoting, commercially viable biopharmaceutical agents.
Concluding remarks
Metagenomics is a powerful tool for deciphering the diversity of microbes in the human body. We now predict that the human microbiota, in particular the gut microbiota, plays a major role in health and well‐being. We are in a period of discovery about the role of phages in the overall interplay between host and microbiome and, as we learn more from metagenomic and pharmacological studies, we will be able to identify candidate targets of diagnostic, pharmaceutical and therapeutic significance. Although phage therapy against pathogenic infections of humans has a century‐long history in Eastern Europe and former Soviet countries, approved use in Western society still requires several clinical, manufacturing and regulatory hurdles to be overcome. Ongoing dialogue between scientists, health care professionals, pharmaceutical companies, regulatory authorities and policy‐makers is imperative to implement phage therapy in clinical practice. In light of the WHO's urgency for novel agents to target the world's most life‐threatening superbugs, the question is not whether we should pursue phage therapy as a solution, but whether we can risk not pursuing it.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
This publication emanated from research supported by Science Foundation Ireland (SFI) under grant numbers 12/RC/2273 and 14/SP APC/B032. The authors wish to thank Andrey Shkoporov (University College Cork), Tiina O'Neill and Dmitri Scholtz (University College Dublin) for electron micrographs of intestinal phages.
Forde, A. , and Hill, C. (2018) Phages of life – the path to pharma. British Journal of Pharmacology, 175: 412–418. doi: 10.1111/bph.14106.
References
- Abedon S (2014). Bacteriophages as drugs: the pharmacology of phage therapy In: Borysowski J, Międzybrodzki R, Górski A. (eds). Phage Therapy: Current Research and Applications. Caister Academic Press: U.K. [Google Scholar]
- Abedon S (2017a) Phage companies – companies.phage.org.
- Abedon S (2017b). Information phage therapy research should report. Pharmaceuticals 10 pii: E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K (2014). Improved bacteriophage genome data is necessary for integrating viral and bacterial ecology. Microb Ecol 67: 242–244. [DOI] [PubMed] [Google Scholar]
- Bodier‐Montagutelli E, Morello E, L'Hostis G, Guillon A, Dalloneau E, Respaud R et al (2016). Inhaled phage therapy: a promising and challenging approach to treat bacterial respiratory infections. Expert Opin Drug Deliv 10: 1–14. [DOI] [PubMed] [Google Scholar]
- Bojanova DP, Bordenstein SR (2016). Faecal transplants: what is being transferred? PLoS Biol 14: e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broecker F, Russo G, Klumpp J, Moelling K (2016). Stable core virome despite variable microbiome after faecal transfer. Gut Microbes 9: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TL, Petrovski S, Dyson ZA, Seviour R, Tucci J (2016). The formulation of bacteriophage in a semi solid preparation for control of Propionibacterium acnes growth. PLoS One 11: e0151184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder K, Malki K, Cooper A, Sible E, Shapiro JW, Watkins SC et al (2016). Freshwater metaviromics and bacteriophages: a current assessment of the state of the art in relation to bioinformatic challenges. Evol Bioinform Online 12: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J (2016). Bacterial DNA methylation and methylomes. Adv Exp Med Biol 945: 35–61. [DOI] [PubMed] [Google Scholar]
- Chan BK, Abedon ST, Loc‐Carrillo C (2013). Phage cocktails and the future of phage therapy. Future Microbiol 8: 769–783. [DOI] [PubMed] [Google Scholar]
- Chehoud C, Dryga A, Hwang Y, Nagy‐Szakal D, Hollister EB, Luna RA et al (2016). Transfer of viral communities between human individuals during faecal microbiota transplantation. MBio 7: e00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek AA, Dąbrowska I, Gregorczyk KP, Wyżewski Z (2017). Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages. Curr Microbiol 74: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clockie MRJ, Millard AD, Letarov AV, Heaphy S (2011). Phages in nature. Bacteriophage 1: 31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CJ, Khan Mirzaei M, Nilsson AS (2016). Adapting drug approval pathways for bacteriophage‐based therapeutics. Front Microbiol 7: 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtright AJ, Abedon S (2011). Phage therapy: emergent property pharmacology. J Bioanal Biomed S6: 002. [Google Scholar]
- D'Herelle F (1917). Sur un microbe invisible antagoniste des Bacillus dysenterique. CR Acad Sci Paris 165: 373–375. [Google Scholar]
- Doss J, Culbertston K, Hahn D, Camacho J, Bareki N (2017). A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Virus 9: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L et al (2014). A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5: 4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J 11: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez Z, Steinberger‐Levy I, Shamir M, Doron S, Stokar‐Avihail A, Peleg Y et al (2017). Communication between viruses guides lyis‐lysogeny decisions. Nature 541: 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancello L, Desnues C, Raoult D, Rolain JM (2011). Bacteriophages and diffusion of genes encoding antimicrobial resistance in cystic fibrosis sputum microbiota. J Antimicrob Chemoter 66: 2448–2454. [DOI] [PubMed] [Google Scholar]
- Fauconier A (2017). Regulating phage therapy. EMBO Rep 18: 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski A, Kniotek M, Perkowska‐Ptasińska A, Mróz A, Przerwa A, Gorczyca W et al (2006). Bacteriophages and transplantation tolerance. Transplant Proc 38: 331–333. [DOI] [PubMed] [Google Scholar]
- Górski A, Międzybrodzki R, Weber‐Dąbrowska B, Fortuna W, Letkiewicz S, Rogóż P et al (2016). Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front Microbiol 7: 1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Allen‐Vercoe E, Petrof EO (2016). Faecal microbiota transplantation: in perspective. Ther Adv Gastroenterol 9: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper DR, Parracho HMRT, Walker J, Sharp R, Hughes G, Werthén M et al (2014). Bacteriophages and biofilms. Antibiotics 3: 270–284. [Google Scholar]
- Hayes S, Mahony J, Nauta A, van Sinderen D (2017). Metagenomic approaches to assess bacteriophages in various environmental niches. Virus 9 pii: E127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun SY, Jang IJ, Yoon S, Jang K, Yu KS, Cho JY et al (2017). Pharmacokinetics and tolerance of the phage endolysin‐based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother 61 pii: e02629‐16): e02629–e02616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen EC, Bliskovsky VV, Malagon F, Baker JD, Prince JS, Klaus JS et al (2017). Novel ‘superspreader’ bacteriophages promote horizontal gene transfer by transformation. MBio 8: e02115–e02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFee S, Buschman H (2017). Novel therapy saves patient with multi‐drug resistant bacterial infection. Available at: https://health.ucsd.edu/news/releases/Pages/2017‐04‐25‐novel‐phage‐therapy‐saves‐patient‐with‐multidrug‐resistant‐bacterial‐infection.aspx (accessed 25/04/2017).
- Lekunberri I, Subirats J, Borrego CM, Balcázar JL (2016). Exploring the contribution of bacteriophages to antibiotic resistance. Environ Pollut 220: 981–984. [DOI] [PubMed] [Google Scholar]
- Loc‐Carillo C, Abedon ST (2011). Pros and cons of phage therapy. Bacteriophage 1: 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łusiak‐Szelachowska M, Żaczek M, Weber‐Dąbrowska B, Międzybrodzki R, Kłak M, Fortuna W (2014). Phage neutralization by sera of patients receiving phage therapy. Viral Immunol 27: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly M, Jones MB, Abeles SR, Santiago‐Rodriguez TM, Gao J, Chan IC et al (2016). Transmission of viruses via our microbiomes. Microbiome 4: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik DJ, Sokolov IJ, Vinner GV, Mancuso F, Cinquerrui S, Vladisavljevic GT et al (2017). Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv Colloid Interface Sci 249: 100–133. https://doi.org/10.1016/j.cis.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ (2016). Healthy human gut phageome. Proc Natl Acad Sci U S A 113: 10400–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP (2013). Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 4: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei MK, Maurice CF (2017). Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15: 397–408. https://doi.org/10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- Modi SR, Lee HH, Spina CS, Collins JJ (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499: 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D (2013). Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol 79: 7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Muniesa M (2017). Phages in the human body. Front Microbiol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC et al (2015). Disease‐specific alterations in the enteric virome in inflammatory bowel disease. Cell 160: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Flaherty S, Ross RP, Coffey A (2009). Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol Rev 33: 801–819. [DOI] [PubMed] [Google Scholar]
- Oliveira H, Silankorva S, Merabishvili M, Kluskens LD, Azeredo J (2015). Unexploited opportunities for phage therapy. Front Pharmacol 6: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Waetzig GH, Rehman A, Moltzau‐Anderson J, Bharti R, Grasis JA et al (2017). Efficacy of sterile faecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152: 799–811. [DOI] [PubMed] [Google Scholar]
- Palfrene E, Willebrand E, Cavaleiro Sanches A, Sebris Z, Cavaleri M (2016). (2016). Bacteriophage therapy: a regulatory perspective. J Antimicrob Chemother 71: 2071–2074. [DOI] [PubMed] [Google Scholar]
- Pei R, Lamas‐Samanamud GR (2014). Inhibition of biofilm formation by T7 bacteriophages producing quorum‐quenching enzymes. Appl Environ Microbiol 80: 5340–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F et al (2010). Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466: 334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A (2009). Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care 18: 240–243. [DOI] [PubMed] [Google Scholar]
- Roach DR, Debarbieux L (2017). Phage therapy: awakening a sleeping giant. Emerg Top Life Sci 1: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Krupovic M, Debroas D, Forterre P, Enault F (2013). Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol 3: 130160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker SA, Brüssow H (2016). From bench to bed and back again: phage therapy of childhood Escherichia coli diarrhoea. Ann N Y Acad Sci 1372: 42–52. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB (2010). Gut microbiota in health and disease. Physiol Rev 90: 859–904. [DOI] [PubMed] [Google Scholar]
- Sulakvelidze A, Alavidze Z, Morris JG (2001). Bacteriophage therapy. Antimicrob Agents Chemother 45: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E, Magrini N (2017). Global priority list of antibiotic‐resistant bacteria to guide research, discovery, and development of new antibiotics. Available at: http://www.who.int/entity/medicines/publications/WHO‐PPL‐Short_Summary_25Feb‐ET_NM_WHO.pdf?ua=1 (accessed 27/02/2017).
- Touchon M, Moura de Sousa JA, Rocha EP (2017). Embracing the enemy: the diversification of microbial gene repertoires by phage‐mediated horizontal gene transfer. Curr Opin Microbiol 38: 66–73. [DOI] [PubMed] [Google Scholar]
- Twort FW (1915). An investigation on the nature of ultra‐microcopic viruses. Lancet 186: 1241–1243. [Google Scholar]
- Wahida A, Ritter K, Horz HP (2016). The Janus‐Face of bacteriophages across human body habitats. PLoS Pathog 12: e1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Hawkins CH, Änggård EE, Harper DR (2009). A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic‐resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol 34: 349–357. [DOI] [PubMed] [Google Scholar]
- Zelasko S, Gorski A, Dabrowska K (2017). Delivering phage therapy per os: benefits and barriers. Expert Rev Anti Infect Ther 15: 167–179. [DOI] [PubMed] [Google Scholar]
- Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W et al (2017). Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut pii: gutjnl‐2017‐313952 : gutjnl‐2017‐313952. [DOI] [PMC free article] [PubMed] [Google Scholar]


