Abstract
Angola borders and has long-term links with Democratic Republic of Congo (DRC) as well as high levels of Human Immunodeficiency virus (HIV) genetic diversity, indicating a potential role in the initial spread of the HIV-1 pandemic. Herein, we analyze 564 C2V3 and 354 pol publicly available sequences from DRC, Republic of Congo (RC) and Angola to better understand the initial spread of the virus in this region. Phylogeographic analyses were performed with the BEAST software and migration records from the first half of the 20th century in DRC were collected from colonial archives. While our results pinpoint the origin of the pandemic to Kinshasa (DRC) around 1906, our results indicate that the introduction of HIV-1 to Angola could have occurred early between 1910 and 1930. Furthermore, most of the initial HIV-1 migrations out of Kinshasa were directed not only to Lubumbashi (DRC), but also to Angola and Brazzaville. Migration records corroborate these findings, indicating that the early exportation of the virus to Angola might be related to the high number of Angolan immigrants in Kinshasa at that time, originated mostly from the North of Angola.
In summary, our results place Angola at the epicenter of early HIV dissemination, together with DRC and RC.
Keywords: Angola, HIV-1, origin, group M
INTRODUCTION
At the end of 2014, the Human Immunodeficiency Virus (HIV) had caused approximately 78 million infections and 39 million deaths (UNAIDS, 2014). HIV is classified in types 1 and 2, but most infections are caused by HIV-1 group M. This group’s epidemic started in Kinshasa (Democratic Republic of Congo, DRC), and soon spread across the Congo river to Brazzaville located in Republic of Congo (RC), and further to Lubumbashi and Mbuji-Mayi (DRC), around 1937 (Faria et al., 2014; Rambaut et al., 2001; Worobey et al., 2008). The HIV-1 epidemic in Angola displays levels of genetic diversity comparable to DRC, which is consistent with an early viral introduction into this country (Bartolo et al., 2009). Nonetheless, the contribution of Angola to the early dispersal of HIV was never investigated.
METHODS
All sequences for env C2V3 and pol (HXB2 positions 7044–7347 and 2319–3302) sampled in Angola, DRC and RC were downloaded from the LANL (http://www.hiv.lanl.gov/). Duplicates and sequences that did not meet LANL quality control parameters were deleted. City locations were retrieved from the original publications (Afonso et al., 2012a; Bartolo et al., 2005; Bartolo et al., 2009; Djoko et al., 2011; Gao et al., 2001; Guimaraes et al., 2009; Vergne et al., 2000; Vidal et al., 2006). Sequences with unknown year, country or city of sampling were excluded from the analyses. The resulting datasets had 564 C2V3 and 354 pol sequences. The env and pol datasets contained 349 and 190 sequences from DRC, 118 and 50 from RC, 97 and 115 from Angola, respectively. To investigate how sampling affects the results, two env and pol datasets were randomly constructed using Phylogenetic Diversity Analyzer (http://www.cibiv.at/software/pda/), with a similar number of sequences from the most sampled cities and maintaining the temporal span and viral diversity. The resulting env and pol datasets included each time 148 and 254 sequences, respectively.
Sequences were aligned using Muscle (Edgar, 2004), and edited in SeaView (Gouy et al., 2010). The temporal signal of all datasets was evaluated with Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/). To estimate the most recent common ancestor (MRCA), the Bayesian Markov Chain Monte Carlo (MCMC) inference implemented in BEAST v1.7.5. was used (Drummond et al., 2012) with the GTR model, a relaxed uncorrelated Lognormal molecular clock model (Drummond et al., 2006) and a Skygrid coalescent tree prior (Gill et al., 2013). To evaluate the early dispersal and the most important migrations, a discrete phylogeographic model was applied using Bayesian Stochastic Search Variable Selection (BSSVS) analyses and the robust counting approach (Edwards et al., 2011; Lemey et al., 2009; Minin et al., 2008; O'Brien et al., 2009; Talbi et al., 2010). The MCMC chains were run for 100 million generations at least three times with a burn-in of 10%. Convergence was evaluated using Tracer (http://beast.bio.ed.ac.uk/Tracer). The maximum clade credibility (MCC) tree was summarized with TreeAnnotator after removal of the burn-in, and visualized with FigTreev1.4.2 (http://tree.bio.ed.ac.uk).
RESULTS
The MRCA of group M was in 1906 [Bayesian Credible Interval (BCI):1892–1921] using env and 1906 [BCI:1878–1930] using pol. Kinshasa was indicated as the origin of this pandemic for both genomic regions [Posterior Probability =1] (Fig.1A–B).
Figure 1. Phylogeographic analysis of the early spread of HIV-1.
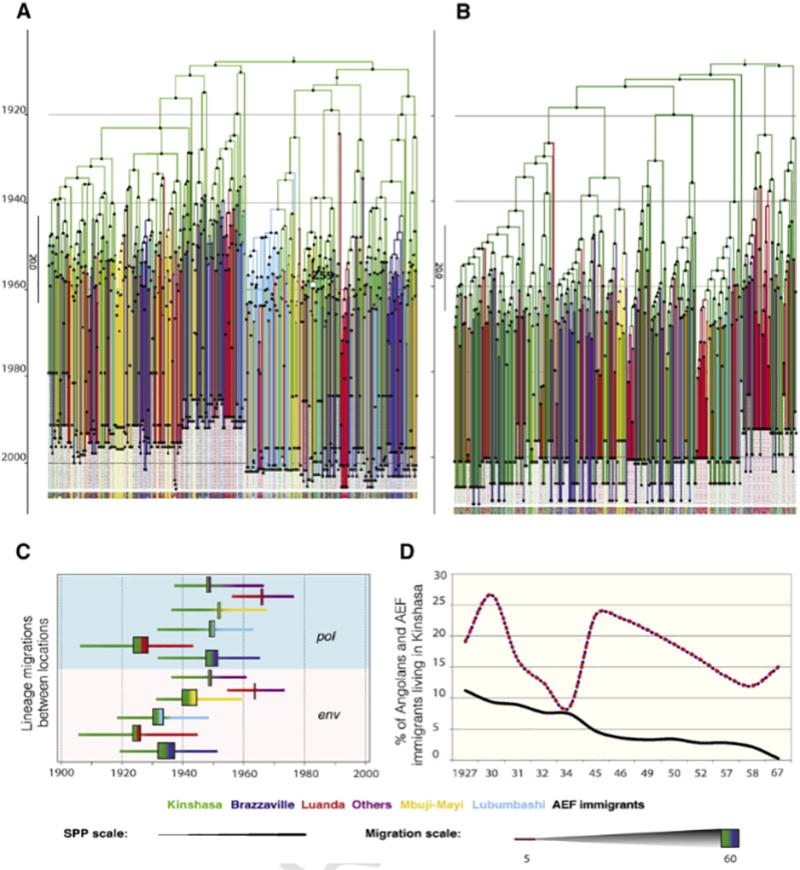
The MCC trees of the C2V3 (A) and pol (B) datasets are shown. The most likely location of the parental node is represented with colors that are explained in the lower panel. The oldest sequence available sampled in DRC in 1959 is represented with a square (ZR59). The C2V3 and pol datasets included the following number of sequences per location (C2V3/pol): Angola: Cabinda (12/19), Cuanza Norte (1/1), Luanda (78/82), Lunda Norte (2/2), Malanje (2/8), Zaire province (2/2); Democratic Republic of Congo: Bwamanda (33/1), Kimpese (0/1), Kinshasa (97/142), Kisangani (23/8), Likasi (24/0), Lubumbashi (76/20), Mbuji-Mayi (96/18); Congo: Brazzaville (97/50), Pointe Noire (21/0). Regions of Angola with low number of sequences such as Cabinda, Cuanza Norte, Malanje, Lunda Norte, and Zaire were grouped into ‘Others’. The posterior probability (PP) > 0.7 is pictured as a circle in the nodes, the color and the size reflects the scale of this probability. (C) The earliest dates of significant lineage migrations in DRC, RC and Angola in env C2V3 and pol are represented by colors as indicated. The rectangle represents the mean of the earliest introduction from the exporter (left) to the importer (right) according to the direction of the migration, whereas the horizontal size of the rectangle is proportional to the number of migrations in each transition. Lines represent the Bayesian Credible intervals. (D) The percentage of Angolans (mainly from the North region, red and violet line) and people from Afrique Équatoriale Française (AEF, black line) living in Leopoldville/Kinshasa according to different censuses of the population (Affaires Indigènes et Main d’Oeuvre (AIMO), 1927; Comissariat de Police de Léopoldville, 1931; Congo Belge, 1930, 1932, 1934, 1949, 1957; Institut National de la Statistique, 1969; Service d'Administration de la Population Noire, 1945, 1946, 1950, 1952; Spitaels, 1959).
Exportations of HIV-1 from DRC to Angola were observed soon after, in 1912 [BCI:1905–1944] using C2V3 and 1919 [BCI:1906–1943] using pol; yet the PP for such observations was low (<0.5). These virus migrations preceded others, including to Brazzaville and Lubumbashi, inferred with similarly low PP values (1926 [BCI:1919–1952] using C2V3 and 1946 [BCI:1930–1963] using pol; 1929 [BCI:1920–1960] using C2V3 and 1948 [BCI:1932–1963] using pol; respectively). The earliest exportations of the virus out of Kinshasa to Angola and Brazzaville were almost simultaneous when high PP values were considered. The introduction to Angola was estimated around 1946 [BCI:1932–1960, PP: 0.98] using C2V3 and 1944 [BCI:1927–1959, PP: 0.97] using pol, whereas introduction to Brazzaville-RC was around 1941 [BCI:1928–1955, PP: 0.90] and 1950 [BCI:1934–1964, PP: 0.90], respectively (Fig.1C).
Angola and Brazzaville-RC were not only the earliest but also the most substantial targets for HIV-1 migrations out of Kinshasa (BF>1000; Angola: 29 and 52 transitions using C2V3 and pol, respectively, [BCI: 20–37 and 47–59]; Brazzaville-RC: 60 and 42 transitions in C2V3 and pol, respectively, [BCI: 52–70 and 36–47]). Within DRC, Lubumbashi and Mbuji-Mayi were the cities with the highest number of migrations from Kinshasa (BF>1000; Lubumbashi: 38 and 19 transitions using C2V3 and pol, respectively, [BCI: 27–47 and 15–21]; Mbuji-Mayi: 52 and 10 transitions using C2V3 and pol, respectively, [BCI: 40–61 and 7–14]). Significant migrations to Lubumbashi happened around 1933 and 1948, whereas to Mbuji-Mayi around 1943 and 1953, using C2V3 and pol [PP>0.9].
Limited migrations were found between Luanda and other cities of Angola (mainly Cabinda), but this is not surprising given the scarcity of sequences from other regions than Luanda (BF>1000, 12 [BCI: 6–16] and 8 [BCI: 5–11] in env and pol, respectively). However, the ongoing historical population movement between different areas of the country would suggest that our sampling most likely reflects a broader geographic region of the country. A significant migration link was also found between Kinshasa and other regions of Angola around 1948 using C2V3 and around 1955 using pol. These regions were mainly Cabinda and Lunda Norte. When the two smaller env and pol random datasets were analyzed, the results were mostly consistent with the main analyses. However, the number of migrations out of Kinshasa to Angola and to Brazzaville-RC was comparable and slightly different from the main analyses, indicating a potential effect of the sampling in the quantification of migrations.
Analysis of archival data about immigration records in that region in the early 20th century indicated that Angolan immigrants constituted an important percentage of the population in Kinshasa (Fig.1D). The number of Angolans living in Kinshasa reached 26% of the city population in 1930 and 23% in the 1940s (Fig.1D). Specifically, these Angolan immigrants, in the 1930s–40s, originated mostly from the area of Maquela do Zombo, which lies in the Zaire province of Angola (Congo Belge, 1930–1957) Comparatively, this population of immigrants was much higher than the number immigrants originated from Afrique Équatoriale Française (Fig.1D).
DISCUSSION
Herein we analyze for the first time the role of Angola in the early dissemination of the HIV-1 group M epidemic. Our results indicate that the earliest estimated exportations of the virus out of Kinshasa occurred quasi concurrently to Angola and Brazzaville in the early 20th century, around the same time as to other locations in DRC. The exportation of HIV-1 to Angola accounted for a major part of HIV-1 migrations out of Kinshasa, as much as to Brazzaville or other cities of DRC. The large proportion of Angolan immigrants living in Kinshasa at that time could explain these findings, at least partly.
Together, our results indicate an important role of Angola in the early dispersal of the epidemic. These findings are consistent with the similar levels of genetic diversity found in Kinshasa and Luanda (Bartolo et al., 2009), and with their profound historical connectivity. Intense native population movements across the borders of colonial Angola and DRC are related with shared ethnicities; for example, the former Kongo Kingdom that had its capital in the North of Angola (M'Banza Kongo in the Zaire province) became divided by colonial borders. The export of HIV to Angola coincided with a time frame where a large proportion of Kinshasa’s population consisted of Angolan immigrants, likely related to labor migration movements as a result of the late 19th-early 20th century expansion of infrastructures, agriculture and mining industries in both Angola and DRC (Henriques, 2004; Vansina, 1966).
Although Angola contributed to the early ignition of the epidemic, its role seems to have been limited for the spread of the worldwide predominant subtypes B and C (data no shown). While for subtype B this is expected, for subtype C seems to have been mostly imported to Angola from the 1960s onwards (Afonso et al., 2012b). More comprehensive sampling of HIV-1 patients in the North of Angola would be important to better understand the early epidemic of HIV-1 and the spread of different subtypes.
In conclusion, herein we show for the first time that Angola played an important role in the early dissemination of HIV-1 group M. Including its sequences in future analyses is therefore crucial to better understand the origin of HIV or its spread through population movements during colonial times.
Acknowledgments
Role of funding source
This study was supported by European Funds through grant ‘Bio-Molecular and Epidemiological Surveillance of HIV Transmitted Drug Resistance, Hepatitis Co-Infections and Ongoing Transmission Patterns in Europe -BEST HOPE- (project funded through HIVERA: Harmonizing Integrating Vitalizing European Research on HIV/Aids, grant 249697); by L’Oréal Portugal Medals of Honor for Women in Science 2012 (financed through L’Oréal Portugal, Comissão Nacional da Unesco and Fundação para a Ciência e Tecnologia (FCT-http://www.fct.pt)); by FCT for funds to GHTM– UID/Multi/04413/2013; by the Fonds voor Wetenschappelijk Onderzoek – Flanders (FWO) grant G.0692.14, by a National Institutes of Health (NIH) grant AI087520, by FCT (grants PTDC/SAU-EPI/122400/2010,VIH/SAU/0029/2011 and PTDC/AFR/100646/2008) and by NEH- Prof. William Schneider - An International Collaboration on the Political, Social, and Cultural History of the Emergence of HIV/AIDS. The computational resources and services used in this work were provided by the Hercules Foundation and the Flemish Government – department EWI-FWO Krediet aan Navorsers (Theys, KAN2012 1.5.249.12.). I.B. is supported by a post-doc fellowship (SFRH/BPD/76225/2011) from FCT. K.T. is supported by a postdoctoral grant from FWO.
Footnotes
Authors’ Contributions
APP: data analysis, data interpretation, figures, writing; JV: data analysis, literature review, writing; JS: data collection, writing; KT: writing; IB: data collection, writing; TL: writing; NT: data collection, writing; AMV: data interpretation, writing; AA: study design, data analysis, data interpretation, figures, writing.
References
- Affaires Indigènes et Main d’Oeuvre (AIMO) Enquête sur la Main d’œuvre, District Urbain de Léopoldville. Léopoldville, Belgian Congo: AIMO, Province du Congo-Kasai. Afrika Archief (FO-BZBHO, Brussels), Series GG, Folder GG 16186; 1927. [Google Scholar]
- Afonso JM, Bello G, Guimaraes ML, Sojka M, Morgado MG. HIV-1 genetic diversity and transmitted drug resistance mutations among patients from the North, Central and South regions of Angola. PloS one. 2012a;7:e42996. doi: 10.1371/journal.pone.0042996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso JM, Morgado MG, Bello G. Evidence of multiple introductions of HIV-1 subtype C in Angola. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012b;12:1458–1465. doi: 10.1016/j.meegid.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Bartolo I, Epalanga M, Bartolomeu J, Fonseca M, Mendes A, Gama A, Taveira N. High genetic diversity of human immunodeficiency virus type 1 in Angola. AIDS research and human retroviruses. 2005;21:306–310. doi: 10.1089/aid.2005.21.306. [DOI] [PubMed] [Google Scholar]
- Bartolo I, Rocha C, Bartolomeu J, Gama A, Marcelino R, Fonseca M, Mendes A, Epalanga M, Silva PC, Taveira N. Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: new insights into the origins of the AIDS pandemic. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2009;9:672–682. doi: 10.1016/j.meegid.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Comissariat de Police de Léopoldville. Récensement de la Cité Indigène de Léopoldville. Est. Afrika Archief (FO-BZBHO, Brussels), Inventory A39, Box RA/CB GG 239; 1931. [Google Scholar]
- Congo Belge. Census of Léopoldville, discriminating regions of origin of inhabitants. Afrika Archief: Federale Overheidsdients – Buitenlandse Zaken Buitenlandse Handel en Ontwikkelingssamenwerking (FO-BZBHO), Series GG Folder GG 5408, Brussels; 1930, 1934, 1949, 1957. [Google Scholar]
- Congo Belge. Récensement de Léopoldville-Est. Afrika Archief: Federale Overheidsdients – Buitenlandse Zaken Buitenlandse Handel en Ontwikkelingssamenwerking (FO-BZBHO), Inventory A39, Box RA/MED 46., Brussels; 1932. [Google Scholar]
- Djoko CF, Rimoin AW, Vidal N, Tamoufe U, Wolfe ND, Butel C, LeBreton M, Tshala FM, Kayembe PK, Muyembe JJ, Edidi-Basepeo S, Pike BL, Fair JN, Mbacham WF, Saylors KE, Mpoudi-Ngole E, Delaporte E, Grillo M, Peeters M. High HIV type 1 group M pol diversity and low rate of antiretroviral resistance mutations among the uniformed services in Kinshasa, Democratic Republic of the Congo. AIDS research and human retroviruses. 2011;27:323–329. doi: 10.1089/aid.2010.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CJ, Suchard MA, Lemey P, Welch JJ, Barnes I, Fulton TL, Barnett R, O'Connell TC, Coxon P, Monaghan N, Valdiosera CE, Lorenzen ED, Willerslev E, Baryshnikov GF, Rambaut A, Thomas MG, Bradley DG, Shapiro B. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol. 2011;21:1251–1258. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Vidal N, Li Y, Trask SA, Chen Y, Kostrikis LG, Ho DD, Kim J, Oh MD, Choe K, Salminen M, Robertson DL, Shaw GM, Hahn BH, Peeters M. Evidence of two distinct subsubtypes within the HIV-1 subtype A radiation. AIDS research and human retroviruses. 2001;17:675–688. doi: 10.1089/088922201750236951. [DOI] [PubMed] [Google Scholar]
- Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 2013;30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular biology and evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guimaraes ML, Vicente AC, Otsuki K, da Silva RF, Francisco M, da Silva FG, Serrano D, Morgado MG, Bello G. Close phylogenetic relationship between Angolan and Romanian HIV-1 subtype F1 isolates. Retrovirology. 2009;6:39. doi: 10.1186/1742-4690-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques IC. Os pilares da diferença : relações Portugal-África : séculos XV–XX 2004 [Google Scholar]
- Institut National de la Statistique. Étude Socio-Démographique de Kinshasa 1967. Kinshasa, DRC: Ministère d’État Chargé du Plan, de la Recherche Scientifique, de l’Amenagement du Territoire et de la Coordination de la Planification. Office National de la Recherche et du Dévéloppement; 1969. [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS computational biology. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Molecular biology and evolution. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JD, Minin VN, Suchard MA. Learning to count: robust estimates for labeled distances between molecular sequences. Molecular biology and evolution. 2009;26:801–814. doi: 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. Human immunodeficiency virus. Phylogeny and the origin of HIV-1. Nature. 2001;410:1047–1048. doi: 10.1038/35074179. [DOI] [PubMed] [Google Scholar]
- Service d'Administration de la Population Noire. Récensement par origine des habitants des 7 quartiers de Léo-Est. Afrika: Archief (FO-BZBHO, Brussels), Inventory A39, Box RA/AIMO 120 annexe; 1945, 1946, 1950, 1952. [Google Scholar]
- Spitaels G. Letter to the Centre de Recherches et d’Informations Socio-Politiques (Brussels) Archives of Afrika Museum (Tervuren, Belgium) 1959 [Google Scholar]
- Talbi C, Lemey P, Suchard MA, Abdelatif E, Elharrak M, Nourlil J, Faouzi A, Echevarria JE, Vazquez Moron S, Rambaut A, Campiz N, Tatem AJ, Holmes EC, Bourhy H. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS pathogens. 2010;6:e1001166. doi: 10.1371/journal.ppat.1001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS. World AIDS Day 2014 Report -Fact sheet, Ending the AIDS epidemic. Joint United Nations Programme on HIV/AIDS; Geneva, Switzerland: 2014. [Google Scholar]
- Vansina J. Kingdoms of the savanna. University of Wisconsin Press; Madison: 1966. [Google Scholar]
- Vergne L, Peeters M, Mpoudi-Ngole E, Bourgeois A, Liegeois F, Toure-Kane C, Mboup S, Mulanga-Kabeya C, Saman E, Jourdan J, Reynes J, Delaporte E. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. Journal of clinical microbiology. 2000;38:3919–3925. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal N, Mulanga C, Bazepeo SE, Mwamba JK, Tshimpaka J, Kashi M, Mama N, Valea D, Delaporte E, Lepira F, Peeters M. HIV type 1 pol gene diversity and antiretroviral drug resistance mutations in the Democratic Republic of Congo (DRC) AIDS research and human retroviruses. 2006;22:202–206. doi: 10.1089/aid.2006.22.202. [DOI] [PubMed] [Google Scholar]
- Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]


