Abstract
Identifying genes required by pathogens during infection is critical for antimicrobial development. Here, we used a Monte Carlo simulation-based method to analyze high-throughput transposon sequencing data to determine the role of infection site and co-infecting microbes on the in vivo ‘essential’ genome of Staphylococcus aureus. We discovered that co-infection of murine surgical wounds with Pseudomonas aeruginosa results in conversion of ~25% of the in vivo S. aureus mono-culture essential genes to non-essential. Furthermore, 182 S. aureus genes are uniquely essential during co-infection. These “Community Dependent Essential” (CoDE) genes illustrate the importance of studying pathogen gene essentiality in polymicrobial communities.
Keywords: Staphylococcus, Pseudomonas, wound infection, essential genome, CoDE genes, Aggregatibacter
Introduction
To cause infection, bacteria must possess the genetic requirements to colonize and proliferate within a host. These genes can be classified into two primary categories, those that are essential during all growth conditions (in vitro and in vivo) and those that are solely essential in the infection site (in vivo). Identification of essential genes is important for developing new therapeutics against human pathogens1. While most antimicrobials target factors essential under all growth conditions, there is increasing interest in targeting genes that are essential solely during infection2–4.
The advent of high-throughput sequencing technologies greatly enhanced essential gene discovery both in vitro and in vivo. One such technology, transposon insertion site sequencing (Tn-seq), allows simultaneous assessment of the relative frequencies of tens of thousands of individual mutants following growth in specific environments5. This technique has allowed our laboratory and others to determine the essential genomes of microbes both in vitro and during infection3,6–8. However, these studies have focused on mono-culture infections in single infection sites3,5–8, despite the fact that many bacterial pathogens cause disease in multiple tissue types and often as part of diverse, multispecies communities9. In this study, we set out to determine the role of both infection site and co-infecting microbes on the in vivo essential genome of the pathogenic bacterium Staphylococcus aureus.
S. aureus is an excellent bacterium for these studies because it is well-adapted to survival in humans, persistently colonizing 30% of the human population10, and capable of infecting nearly every tissue of the human body11. Types of infections caused by S. aureus range from primarily mono-species infections, such as skin and soft-tissue abscesses11,12, to polymicrobial infections such as chronic wounds, where on average 6 species are isolated from an infection13. S. aureus is the most common organism isolated from chronic wounds and is frequently found with the opportunistic pathogenic bacterium Pseudomonas aeruginosa13. Notably, chronic wounds harboring both S. aureus and P. aeruginosa are associated with increased wound severity and increased healthcare costs13,14.
Here, we used Tn-seq and a Monte Carlo-based analysis to determine what we refer to as the ‘essential genome’ of S. aureus in three murine infection models and during co-infection with Pseudomonas aeruginosa. Our results reveal that co-infection has a larger effect on the S. aureus essential genome than infection site with ~200 genes showing altered essentiality during co-infection. A similar number of Community Dependent Essential (CoDE) genes were also found in a periodontal pathogen using an unrelated polymicrobial infection model. Collectively, this work illustrates the importance of studying pathogens in complex polymicrobial communities.
Results and Discussion
First, we used a Monte Carlo-based analysis3,15 of a transposon mutant pool containing ~72,000 insertions7 in S. aureus strain HG003 to determine the in vitro S. aureus essential genome in rich laboratory media. Briefly, this analysis compares transposon abundances in the library (observed dataset) to an expected dataset where transposons are randomly redistributed in silico across the genome (see Methods). For this study, “essential genes” are defined as genes that in a condition: 1) have a reduced fold change in the insertion frequency compared to the expected dataset; and 2) cluster with the lower mode in the characteristic16 bimodal distribution of fold changes comparing the observed to expected datasets. This analysis identified 512 S. aureus genes that are essential for S. aureus growth in vitro (Supplementary Table 1). As expected, these genes are enriched for central cellular processes including central metabolism and ribosome activity (Fisher’s Exact Test p-value < 0.05).
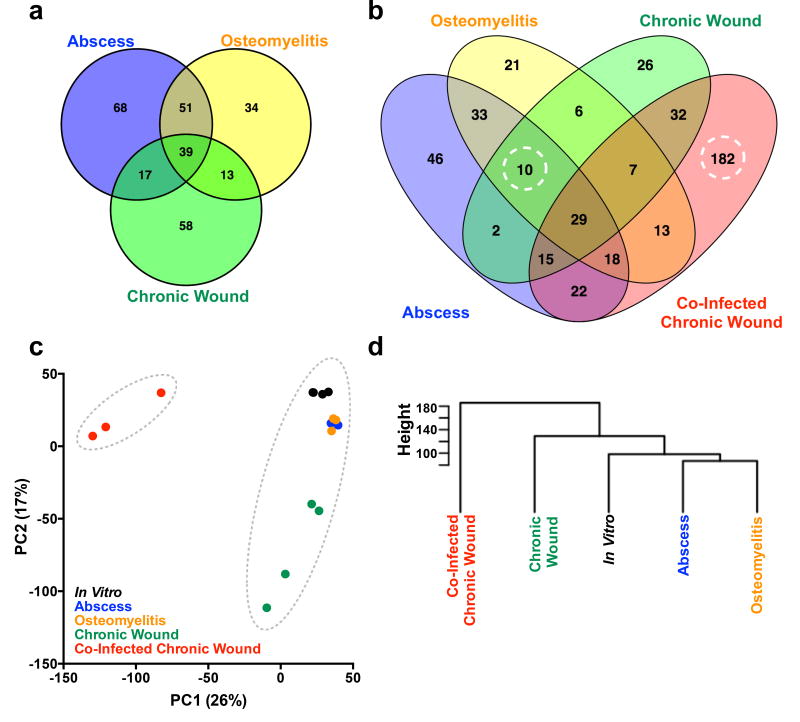
We then used the Monte Carlo approach to investigate how infection site affects the S. aureus essential genome during mono-culture infection. We analyzed previously generated Tn-seq data from murine abscess7 and osteomyelitis8 models, as well as data generated in this study from a murine chronic surgical wound model. This latter model is well established for studying polymicrobial S. aureus wound infections and requires over a week for resolution17,18. To specifically focus on the in vivo essential genome, we removed the 512 in vitro essential genes from all subsequent analyses. We identified 280 genes that are essential in at least one infection model (Figure 1a), but not essential in vitro. Of these genes, 39 (14%) were essential in all three mono-infection models, and 120 (43%) were essential in at least two models. However, 68 (24%) were unique to the abscess, 34 (12%) were unique to osteomyelitis, and 58 (21%) were unique to the chronic surgical wound. These data reveal infection site can have a substantial impact on the in vivo essential genome, and that S. aureus possesses a core set of in vivo essential genes across all infection models.
Figure 1. The S. aureus in vivo essential genome in three mono-culture infections amd during co-infection with P. aeruginosa.
a) Venn diagram of the S. aureus in vivo essential genome in mono-culture murine abscess (blue, n=2), osteomyelitis (yellow, n=3), and chronic surgical wound (green, n=4) models of infection. b) Venn diagram of the S. aureus in vivo essential genome during abscess (blue), osteomyelitis (yellow), chronic surgical wound mono-culture infection (green) and chronic surgical wound co-culture infection with P. aeruginosa (red). Dashed circles highlight the 10 S. aureus CoDE genes that were essential in all mono-culture infections but non-essential in co-culture, and the 182 S. aureus CoDE genes that were unique to co-infection in the chronic surgical wound. False positive rates for the essential gene analysis were determined as outlined in Methods (‘Essential Gene Analysis’) and yielded 0 genes for abscess, 4 genes for osteomyelitis, 1 gene for mono-infection chronic surgical wound, and 18 genes for co-infection chronic surgical wound. c) Plot of the first two principal components (PC) generated by Principal Component Analysis of the normalized read counts per S. aureus gene in 5 conditions: in vitro BHI growth7 (black, n=4, only three points are distinguishable due to overlap), murine abscess (blue, n=2), murine osteomyelitis (orange, n=3), murine chronic surgical wound mono-infection (green, n=4), and murine chronic surgical wound co-infection with P. aeruginosa (red, n=3). Dotted grey circles indicate the two clusters that are generated by k-means clustering analysis. d) Hierarchal clustering (Ward method) of the average normalized counts per gene in each of the conditions described above. Height indicates the Euclidean distance between clusters. Similar clustering results were obtained with individual replicates (Supplementary Figure 3).
Chronic wound infections are predominantly polymicrobial, with S. aureus and P. aeruginosa the most commonly isolated microbes13,14,19,20. Therefore, we next assessed how the presence of P. aeruginosa impacts the S. aureus in vivo essential genome in the murine chronic surgical wound model. For these experiments, 10-fold less P. aeruginosa than S. aureus was used in the inoculum to ensure that the effects observed were not due to increased bacterial inoculum. We compared the S. aureus Tn-seq data generated in all 4 infections and in vitro through k-means clustering of the principal component analysis and hierarchal clustering, and found that the chronic surgical wound co-infection data clustered independently from mono-culture infections by both methods (Figure 1c&d). These findings indicate the essential genome of S. aureus is distinct during co-infection compared to mono-infection.
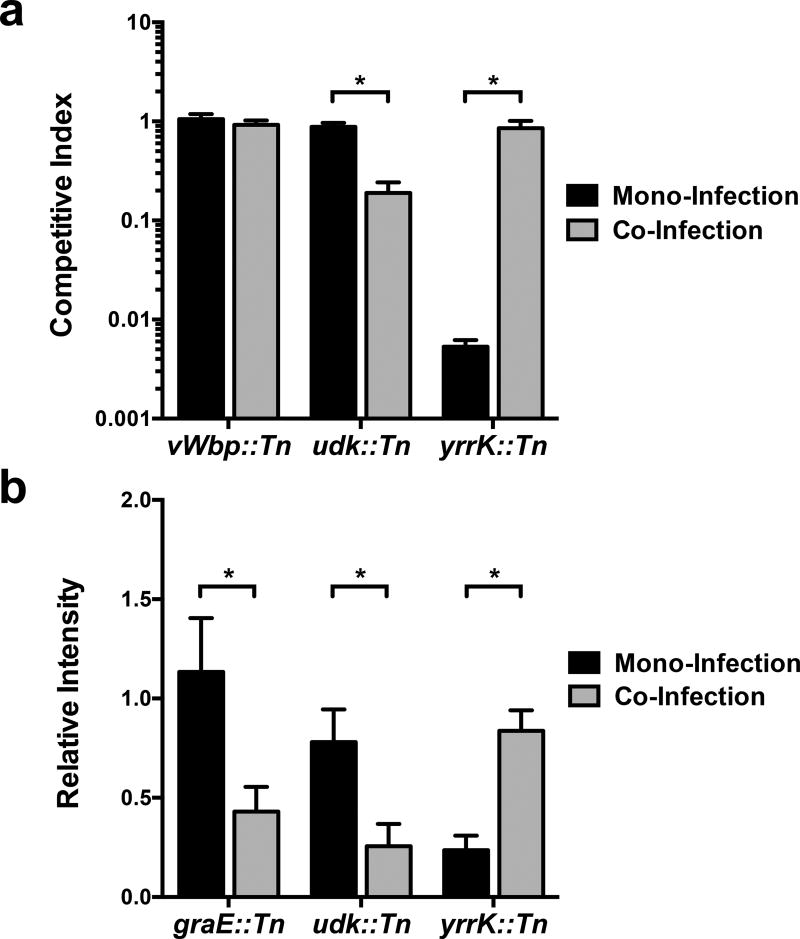
Additional examination revealed that 182 genes, representing over 6% of the S. aureus genome, are uniquely essential to S. aureus during chronic surgical wound co-infection (Figure 1b). Furthermore, these genes were significantly enriched (Fisher’s Exact Test, P-value < 0.05) in the Clusters of Orthologous Groups (COG) functional categories for energy production and conversion as well as amino acid, nucleotide, carbohydrate, lipid, and inorganic ion metabolism (Supplementary Table 1). These data suggest that P. aeruginosa induces a metabolic stress on S. aureus in the chronic surgical wound. Of note, these genes also included the global regulator Repressor of Toxins (rot, SAOUHSC_01879), a phenol-soluble modulin (psmβ1, SAOUHSC_1135), and multiple components of the S. aureus Type VII secretion system21,22 (SAOUHSC_00260, SAOUHSC_00264, SAOUHSC_00265). In addition, we discovered 10 genes, representing 26% of the genes essential in all three mono-infections, which were no longer essential during co-infection with P. aeruginosa (Figure 1b and Supplementary Table 1). These included genes coding for menaquinol oxidase (qoxC, SAOUHSC_01000) and ATP synthase (atpD, SAOUHSC_02341). To ensure the accuracy of our population scale Tn-seq analyses, we competed three individual transposon mutants (the mono-infection-specific essential gene mutant yrrK, the co-infection-specific essential gene mutant udk, and mutant vWbp (a.k.a. graB) that did not change in abundance) against the wildtype strain in mono- and co-infection in the murine chronic surgical wound (Figure 2a, Supplementary Table 7). In addition, we assessed a small pool of transposon mutants in a small-scale murine chronic surgical wound Tn-seq experiment (Figure 2b). In all cases, these experiments confirmed the findings of the original genome scale Tn-seq experiments. Together, these results indicate that the S. aureus in vivo essential genome is dramatically altered during co-infection with P. aeruginosa both through the emergence of essential genes (182 genes) and the alleviation of the requirement of mono-culture essential genes (10 genes), which in sum, we term “Community Dependent Essential” (CoDE) genes. The fact that the number of CoDE genes (192) is twice as large as the total number of genes unique to all mono-culture infection sites (93) suggests that the presence of a co-infecting bacterium impacts the in vivo essential genome more significantly than infection site.
Figure 2. Confirmation of S. aureus mutant Tn-Seq phenotypes.
a) Three S. aureus transposon mutants were competed with the wildtype S. aureus strain HG003 in mono- and co-infection with P. aeruginosa PAO1 in the murine chronic surgical wound. Mutations in the CoDE gene udk::TnMariner (SAOUHSC_01715) was predicted to be essential in co-infection but not mono-infection, while yrrK::TnMariner (SAOUHSC_01720) was predicted to be essential in mono-infection but not in co-infection. An S. aureus mutant whose relative abundance did not change (vWbp::TnMariner, SAOUHSC_00814) in the initial Tn-seq experiments (Supplementary Table 1) was used as a control. For each condition, three biological replicates were used. b) A sub-set of S. aureus transposon mutants were pooled and used to infect the murine chronic surgical wound alone and in co-infection with P. aeruginosa (3 mice each). DNA was extracted four days post-infection, and PCR used to quantify relative abundance of each S. aureus mutant. As a control, an S. aureus mutant whose relative abundance did not change (vWbp::TnMariner, SAOUHSC_00814) in the initial Tn-seq experiments (Supplementary Table 1) was used for normalization. Mutations in the CoDE genes graE::TnMariner (SAOUHSC_01600) and udk::TnMariner were predicted to be essential in co-infection but not mono-infection, while yrrK::TnMariner was predicted to be essential in mono-infection but not in co-infection. Intensity of PCR amplicons was calculated using ImageJ (FIJI), and relative intensity calculated by dividing CoDE gene amplicon intensity by vWbp::TnMariner amplicon intensity. Statistical analysis was performed using a Student’s t-test (* = p < 0.05). Error bars represent the standard error of the mean (SEM). Bars represent three biological replicates, and three technical replicates (total of 9 replicates) for udk::TnMariner and graE::TnMariner, and three biological, two technical replicates (6 total replicates) for yrrK::TnMariner. The average total number of S. aureus (± standard error of the mean) recovered from mono-culture wounds was 3 × 108 ± 5 × 107 CFU/g and from co-culture wounds was 6.0 ×108 ± 3 × 107 CFU/g.
An important question is whether S. aureus CoDE genes are restricted to the murine chronic surgical wound infection. To investigate this, we assessed the phenotypes of multiple CoDE gene mutants in an in vitro wound model23. This well-characterized23,24 model allows P. aeruginosa and S. aureus to spatially segregate in a coagulated matrix and thus promotes synergistic interactions, such as enhanced resistance to antibiotics and formation of distinct aggregates24. From our Tn-seq analysis (Supplementary Table 1), we predicted graE, udk, and tcyA would be required in co-culture but not in mono-culture, and conversely yrrK and atpD would be required in mono-culture but not co-culture. Surprisingly, four of the five S. aureus CoDE gene mutants showed the predicted phenotypes in the in vitro wound model (Supplementary Figure 1) supporting the importance of CoDE genes for S. aureus fitness beyond the murine wound environment.
A remaining question is whether other pathogens exhibit a similarly high prevalence of CoDE genes during polymicrobial infection? Therefore, we tested for CoDE genes in the oral pathogenic bacterium Aggregatibacter actinomycetemcomitans during mono- and co-infection with the oral commensal bacterium Streptococcus gordonii. This model is for assessing CoDE genes as A. actinomycetemcomitans and S. gordonii co-localize in the human oral cavity25 and synergistically interact in a murine abscess infection model26. As with S. aureus, genes that were essential in vitro were removed from our analysis to focus on A. actinomycetemcomitans in vivo essential genes (Supplementary Table 1). A. actinomycetemcomitans Tn-seq data from mono- and co-infected abscesses revealed that ~47% (155 genes) of its in vivo essential genome are CoDE genes (Supplementary Figure 2). Similar to our findings with S. aureus, co-infection with S. gordonii places unique metabolic stresses on A. actinomycetemcomitans as multiple nutrient transporters and biosynthesis pathways were solely required during co-infection (Supplementary Table 1). Additionally, this analysis confirmed a previous observation that while A. actinomycetemcomitans requires atpB in mono-infection, the presence of S. gordonii alleviates this requirement27. Collectively, these data indicate CoDE genes are not limited to S. aureus/P. aeruginosa wound infections.
For over 100 years, it has been recognized that many human infections are caused by microbial communities, not individual microbes. However, microbial pathogens are frequently studied in isolation in animal infection models. Our discovery of CoDE genes reveals it is critical to identify a pathogen’s essential genes in complex communities that reflect native infections. This is particularly relevant when designing antimicrobials since co-infection-specific genes could serve as new therapeutic targets for multi-species infections, while CoDE genes that are no longer essential in co-infection may only be effective targets in mono-culture infections. As with all essential gene analyses, there are limitations to these findings. Due to the nature of Tn-seq, mutants can cross-complement each other, and therefore secreted factors are often not identified. Additionally, it is possible some mutations will not disrupt protein function or some genes may have few transposon insertions, limitations we have addressed in our essential gene analysis (see Methods). Finally it should be noted we have used a precise definition for ‘essential genes’ that may differ from other studies, although all CoDE genes can clearly be defined as in vivo fitness determinants. Overall, we believe these findings have broad implications for polymicrobial infections, and it will be interesting to explore the exact mechanisms by which the presence of co-infecting microbes leads to CoDE genes.
Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animal protocols were approved by the Institutional Animal Care and Use Committees of The University of Texas at Austin (protocol number 00136) and Texas Tech University Health Sciences Center (protocol numbers 07044).
Bacterial Strains and Culture Conditions
Previously generated transposon mutant libraries of S. aureus strain HG0037 and P. aeruginosa strain PAO128 were used in this study. S. aureus in vitro cultures were grown in Brain-Heart Infusion broth (BHI) at 37°C with shaking at 225 rpm and a flask to media volume ratio of 5:1. All of the transposon mutants used in this study were provided by The Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) and distributed by BEI Resources, NIAID, NIH. Mutations were moved to the S. aureus HG003 strain background by transduction using staphylococcal phage Φ1129, and confirmed by PCR. All primer sequences are listed in Supplementary Table 3. The strains of oral bacteria used were A. actinomycetemcomitans 624 (a clinical isolate) and S. gordonii Challis DL1.1 (ATCC 49818). A. actinomycetemcomitans was grown as lawns on tryptic soy agar + 0.5% (w/vol) yeast extract (TSBYE agar medium) under aerobic (5% CO2 atmosphere) conditions. S. gordonii was grown in filter-sterilized TSBYE liquid medium under aerobic conditions without shaking.
Creation of A. actinomycetemcomitans Transposon Library
Plasmid pMR361-K was used to create the A. actinomycetemcomitans transposon library. Plasmid pMR361-K was created by first digesting the plasmid pMRKO26 with SalI and NotI to excise mCherry. Then, the mariner transposase C9 gene from pZXL530 was PCR-amplified with primers Tnp-F-SalI and Tnp-R-NotI (Supplementary Table 3). These primers include SalI and NotI restriction sites at the 5’ ends, respectively. The transposase amplicon was then digested with SalI and NotI, and ligated into the SalI/NotI-digested pMRKO fragment with mCherry removed. These steps replaced mCherry with the transposase now positioned downstream of the lac promoter. This intermediate vector was then transformed into E. coli DH5α, purified, and digested with NotI. Next, the kanamycin resistance (KanR) cassette from pCGL024331 was PCR-amplified with primers KanR-UP-F and KanR-DN-R (Supplementary Table 3). These primers included flanking sequences that will base pair with the up- and downstream fragments of the mariner inverted repeats. The upstream fragment was PCR-amplified from pZXL5 with primers mar-IR-NotI and mar-UP-R, and the downstream fragment was amplified from pZXL5 with primers mar-DN-F and mar-IR-NotI (Supplementary Table 3). These fragments were then assembled on each end of the KanR cassette by overlap extension PCR, digested with NotI, and ligated into the NotI-digested intermediate vector, generating pMR361-K. pMR361-K was then transformed into E. coli DH5α and purified to check the mariner inverted repeats by Sanger sequencing. Finally, pMR361-K was moved into the conjugative E. coli strain MFDpir32. A transposon mutant pool in A. actinomycetemcomitans 624 was generated as described33 by conjugation with E. coli MFDpir (a diaminopimelate auxotroph)32 containing pMR361-K. Conjugations were performed under aerobic and anaerobic conditions on TSBYE agar + 0.3 mM diaminopimelate, and pooled prior to counter-selection under aerobic and anaerobic conditions on TSBYE agar + 40 µg/ml kanamycin. Anaerobic conditions were maintained in a vinyl chamber (Coy Lab Products) with the following atmosphere: 85% N2, 10% CO2, 5% H2. Independent conjugations were combined then aliquoted to generate the final mutant pool.
Murine Chronic Surgical Wound Infection
Murine chronic surgical wound infections were performed with 6–8 week old female Swiss Webster mice as previously described6,34. For Tn-seq experiments, 4 × 105 CFU of the S. aureus HG003 transposon library was used for mono-infection (n=4 mice), and 2 × 105 S. aureus and 2 × 104 P. aeruginosa were used for co-infection (n=3 mice). Wound tissue was harvested 4 days post-infection and stored in RNAlater (Ambion). No blinding was used in any of the chronic surgical wound animal experiments and animals were not randomized. At least three biological replicates were used per condition in this model, which was determined from previous6,34 and preliminary data to be sufficient to yield statistically significant differences.
Murine Abscess Infection
Murine abscess infections with the A. actinomycetemcomitans transposon mutant pool were performed with 9–11 week old female Swiss Webster mice as described26,27 with minor modifications. No blinding was used in these experiments, and sample size was determined from previous Tn-seq experiments in this model27 to be sufficient to yield statistically significant differences. Briefly, an aliquot of the mutant pool was grown overnight as lawns under aerobic and anaerobic conditions. The lawns were then pooled in phosphate buffered saline (pH 7.4), adjusted to an OD600 of 2, and mixed with an equal amount of either wild-type A. actinomycetemcomitans (for mono-infection inoculum) or S. gordonii (for co-infection inoculum). For each mono-infected abscess, 100 µl containing ~108 of the A. actinomycetemcomitans mutant pool + ~108 of wild-type A. actinomycetemcomitans was used (n = 2 mice, with 2 abscesses per mouse). For each co-infected abscess, 100 µl containing ~108 of the A. actinomycetemcomitans mutant pool + ~108 of S. gordonii was used (n = 2 mice, with 2 abscesses per mouse). At 3 days post-infection, each abscess was harvested, suspended in 0.9 ml phosphate buffered saline (pH 7.4) in BeadBug™ tubes prefilled with 2.8 mm steel beads (Sigma), and homogenized for 30 seconds in a Mini-Beadbeater (Biospec). 500 µl of each homogenate was then grown in 4.5 ml of TSBYE + 40 µg/ml kanamycin for 6 hours under aerobic conditions without shaking prior to being frozen and stored until preparation for sequencing. The left thigh abscess of each mouse was used for Tn-seq library preparation.
Preparation of S. aureus Tn-seq Libraries
Sequencing data for the S. aureus murine abscess and osteomyelitis models were previously generated7,8. DNA from murine chronic surgical wounds stored in RNAlater was extracted by bead beating as previously described6,35, with the exception that wound tissue was resuspended in Goodman Buffer A + 0.1% SDS + 0.1% sodium deoxycholate. Extracted wound DNA was size selected for fragments between 100 and 700 bp with Agencourt AMPure XP beads (Beckman Coulter, Inc.), and prepared for Tn-seq analysis as previously described6,35. Cytidine tails were added with terminal deoxynucleotidyltransferase (TdT), followed by 2 PCRs. The primers olj37636 and the transposon-specific 5’-biotinylated primer PCR1-Ba-Bio were used for the first PCR for S. aureus Tn-seq. An Illumina barcoded primer3 and the transposon-specific primer PCR2-Ba were used for the second PCR for S. aureus Tn-seq. The libraries were sequenced at the Genome Sequencing and Analysis Facility at the University of Texas at Austin on an Illumina NextSeq 500 using a 75-bp single-end run.
Preparation of A. actinomycetemcomitans Tn-seq Libraries
DNA from abscess outgrowths was extracted by vortexing for 1 minute with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, pH 8) and then purified, beginning at the isopropanol precipitation step2, as previously described. Extracted DNA from abscess outgrowths was prepared for Tn-seq analysis as previously described6,27. Cytidine tails were added with terminal deoxynucleotidyltransferase (TdT), followed by 2 PCRs. The primers olj37636 and the transposon-specific 5’-biotinylated primer mariner-1 were used for the first PCR27 for A. actinomycetemcomitans Tn-seq. An Illumina barcoded primer3 and the transposon-specific primer mariner-2 were used for the second PCR27 for A. actinomycetemcomitans Tn-seq. The libraries were sequenced at the Genome Sequencing and Analysis Facility at the University of Texas at Austin on an Illumina NextSeq 500 using a 75-bp single-end run.
Essential Gene Analysis
Reads were trimmed of adapter sequences and mapped to the reference genomes (strain NCTC8325 for S. aureus and strain 624 for A. actinomycetemcomitans) using the TnSeq2.sh script (available at https://github.com/spleonard1/Tn-seq) with bowtie2 v2.2.5. To minimize the effect of insertions that may not abolish gene function, modified open reading frame assignments were generated for each genome where the 3’ 10% of every gene was removed. The following minor modifications were made to the TnSeq2.sh script for A. actinomycetemcomitans Tn-Seq analysis: (1) cutadapt (v1.12)37 was used to trim reads of 3’ low-quality bases and 3’ poly(C) tail sequences, and to remove reads less than 20 bases long; (2) after mapping, reads that were filtered for high mapping quality (MAPQ > 39) were used to determine the abundance of unique insertion sites while accounting for the 2 bp duplication associated with mariner insertion events; and (3) to correct for DNA polymerse slippage28, the read counts of adjacent sites were collapsed onto the site with the highest count (the local maximum) using a custom Python script (available upon request).
The S. aureus and A. actinomycetemcomitans essential genomes were determined using the following custom scripts with DESeq2 v1.10.1: TnSeqEssential.sh, TnGeneBin.pl, and TnSeqDESeq2Essential.R (available at https://github.com/spleonard1/Tn-seq), as previously described3,15. Briefly, the mapped reads were tallied by insertion site, and the 50 most abundant insertion sites were removed to correct for bias resulting from PCR amplification. Only sites that were identified in at least two replicates were considered for downstream analysis. Insertion read counts were smoothed locally by weighted LOESS regression to correct for genomic position-dependent effects on perceived mutant abundance16. The data were normalized for differences in sequencing depth with estimateSizeFactors() in DESeq238, and the number of unique insertions and their associated read counts were tallied per gene using the modified open reading frame assignments. Next, 400 pseudodatasets (“expected data”) were generated where for each Tn-seq dataset the unique insertions and their associated reads were randomly redistributed among the TA sites present in the reference genomes (see Supplementary Tables 5 and 6 for a list of the TA sites in the S. aureus and A. actinomycetemcomitans reference genomes, respectively). Differential mutant abundance between the observed data and the “expected” pseudodatasets was calculated with a negative binomial test using DESeq238, with the normalization factors set to one (since normalization was already performed based on the insertions), with Cook’s distance cutoff for outlier removal turned off, and with the p-values adjusted for multiple testing using the Benjamini-Hochberg method38. A parameterized bimodal Gaussian mixture model was fit to the log2-transformed fold changes in mutant abundance to determine whether a gene was “reduced” or “unchanged” using the mclust39 package in R. Genes were called “essential” if they: 1) had a reduced fold change compared to the expected dataset (Benjamini-Hochberg adjusted p-value < 0.01, negative binomial Wald test in DESeq238) and 2) clustered with the lower mode in the characteristic17 bimodal distribution of fold changes comparing the observed to “expected” datasets (mclust39 p-value < 0.01). To assess the false positive rate, we used this analysis method to randomly generate datasets from the input library reads that contained the same number of replicates, number of reads, and number of insertion sites as the actual data for each condition and determine the number of falsely identified “essential genes” using fitness neutral data. Venn diagrams were generated using Venny (v2.1)40.
Clustering Analysis
Read counts for each condition were binned by gene and then used to perform Principal Component Analysis (PCA) in R. Differences in sequencing depth between each replicate were normalized using the estimateSizeFactors() function and transformed using the rlog function in DESeq2 (v1.10.1) prior to analysis. Principal components were then generated from this data using the R prcomp function, and Principal Component 1 (PC1) and Principal Component 2 (PC2) were plotted using GraphPad Prism 6 (La Jolla, CA). Clustering analysis of the PCA plot was performed in R using k-means in the stats package (v3.2.2). For hierarchal clustering, Euclidean distances were calculated from the average normalized read counts for each condition prior to determining clusters using the Ward method in hclust in the stats package (v3.2.2).
Comparison of Mutant Fitness in the In Vivo Chronic Surgical Wound Model
S. aureus transposon mutants were constructed by phage transduction (described in Bacterial Strains and Culture Conditions section) in the HG003 background from the S. aureus JE2 Nebraska Transposon Mutant Library homologues (listed in Supplementary Table 6). For in vivo competition experiments, S. aureus wildtype strain HG003 or the S. aureus HG003 transposon mutants were grown overnight, normalized to an OD600 reading of 1.0 in phosphate buffered saline (pH 7.4), and mixed at a ratio of 1:1 mutant to wildtype. Mice (n=3 per condition) were inoculated at 7 × 105 for mono-infection, and for co-infection at 5 × 105 S. aureus and 5 × 105 P. aeruginosa PAO1. Wound tissue and associated bandage was harvested (~200–400 mg per wound) 4 days post-infection and added to 0.9 ml phosphate buffered saline (pH 7.4) in BeadBug™ tubes prefilled with 2.8 mm steel beads (Sigma), and homogenized for 1 minute in a Mini-Beadbeater (Biospec). The homogenate was serially diluted in PBS and plated on BHI agar (to quantify all S. aureus) and BHI agar + 10 µg/ml erythromycin (to quantify S. aureus mutant numbers). S. aureus colonies were enumerated and competitive indices for each mutant in each condition were calculated as (# mutant S. aureus CFU/g / # wildtype S. aureus CFU/g). For co-culture experiments, both S. aureus and P. aeruginosa formed colonies on BHI but were easily distinguished by colony color and morphology (small, circular, smooth, yellow colonies for S. aureus).
For small-scale Tn-seq experiments using 7 mutants, overnight cultures of the S. aureus HG003 transposon mutants were normalized to an OD600 reading of 1.0 in phosphate buffered saline (pH 7.4) and mixed in equal proportions. An inoculum consisting of 3 × 105 of the S. aureus mixture was used for mono-infection (n=3 mice), and co-infection consisted of 1 × 105 S. aureus and 6 × 106 PAO1 (n=3 mice). Wound tissue was harvested (~100–300 mg of tissue per wound) 4 days post-infection and added to 0.9 ml phosphate buffered saline (pH 7.4) in BeadBug™ tubes prefilled with 2.8 mm steel beads (Sigma), and homogenized three times for 1 minute in a Mini-Beadbeater (Biospec). 500 µl of the homogenate was added to 4.5 ml of BHI broth containing 10 µg/ml erythromycin, and cells were outgrown for 2 hours prior to preparation of DNA for semi-quantitative PCR. Cells were pelleted by centrifugation at 10,000 × g and resuspended in 200 mM Tris (pH 8.0). Lysostaphin (Sigma) was added at 1 mg/ml, and S. aureus cells were lysed for 1 hour at 37°C. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, pH 8) was added, and samples were vortexed for 30 seconds prior to centrifugation. DNA was purified from the aqueous phase by ethanol precipitation, and resuspended in 50 µl H2O. DNA concentrations were quantified using a NanoDrop, and samples were adjusted to 5 ng/µl. Semi-quantitative PCRs (see Supplementary Table 3 for primer sequences) were performed using Phusion High Fidelity DNA Polymerase (NEB) per manufacturer recommendations for 35 cycles. Densitometry measurements were performed using ImageJ FIJI software (v2.0.0)41, the average of 10 background measurements was subtracted from each gel, and bands were normalized to PCRs from the S. aureus mutant SAOUHSC_00814 (vWbp, secreted von Willebrand factor-binding protein) that did not display a fitness defect in mono- or co-infection.
Comparison of Mutant Fitness in the In Vitro Lubbock Chronic Wound Model
Wound-like media was prepared as previously described23 and 600 µl was added to tubes (glass, 6 × 50 mm). For mono-culture experiments wound-like media was inoculated with individual transposon mutants (construction described in Bacterial Strains and Culture Conditions) or the S. aureus WT strain HG003 at 5 × 104 S. aureus. For co-culture experiments wound-like media was inoculated with 2.5 × 104 CFU of the S. aureus WT or individual S. aureus transposon mutants and 2.5 × 104 CFU P. aeruginosa PAO1. Cultures were incubated statically at 37°C for 6 days, and 10 µl samples removed for bacterial counts on days 2, 4, and 6. S. aureus cells were enumerated by plate counts on Mannitol Salt Phenol Red Agar (Sigma) as this medium does not permit growth of P. aeruginosa.
Data Availability
The raw sequencing files that support this study have been deposited into the NCBI Sequence Read Archive under accession SRP093229 for S. aureus Tn-seq data and accession SRP095181 for A. actinomycetemcomitans Tn-seq data. The sequenced genome for S. aureus reference strain NCTC8325 is available from under RefSeq accession number NC_007795.1. The sequenced genome for A. actinomycetemcomitans Strain 624 is available under RefSeq accession number NZ_CP012959.1. The remaining data that support the findings of this study are available from the corresponding author upon request.
All material requests and correspondences should be addressed to Marvin Whiteley, mwhiteley@austin.utexas.edu
Supplementary Material
Supplementary Figure 1. Some S. aureus CoDE genes are not in vivo specific. Comparison of wildtype S. aureus and S. aureus CoDE gene mutant cell numbers in an in vitro chronic wound model during mono-culture and P. aeruginosa co-culture. graE::TnMariner (b, green), udk (c, orange), and tcyA::TnMariner (d, purple) were predicted to be required in co-culture but not mono-culture, while while yrrK::TnMariner (a, red) and atpD::TnMariner (e, blue) were predicted to be required in mono-infection but not in co-infection. All mutants are reported as % wildtype cell number (S. aureus HG003) in the same condition. Statistical analysis was performed by arcsine transforming the data prior to analysis with a Student’s t-test (* = p < 0.05). For each condition, 4 biological replicates were used. Error bars represent the standard error of the mean (SEM).
Supplementary Figure 2. Identification of CoDE genes in A. actinomycetemcomitans during co-infection with S. gordonii. Venn diagram of the A. actinomycetemcomitans in vivo essential genome in mono-culture (blue, n=2) and co-culture (yellow, n=2) infection with S. gordonii. 72 CoDE genes were identified that were essential in mono-culture infection but non-essential in co-culture, and 83 CoDE genes were uniquely essential in co-infection. Data are the result of two replicates per condition.
Supplementary Figure 3. Experimental replicates cluster by condition. Hierarchal clustering (Ward method) of the average normalized read counts per gene in each of the replicates (numbered and color coded) for all experimental conditions. Height indicates the Euclidean distance between clusters.
Acknowledgments
This work was supported by National Institutes of Health Grants R01GM116547-01A1, 1R01DE023193-01 (to M.W.) and a grant from Human Frontiers Science (to M.W.). C.B.I is supported by postdoctoral fellowship IBBERS16F0 from the Cystic Fibrosis Foundation. Contributions by M.S.G were supported by PHS grant AI107248 and the Harvard-wide Program on Antibiotic Resistance AI083214. A.S. is supported by a predoctoral fellowship from the NIH (F31DE024931). We would like to acknowledge Dr. Matthew Ramsey for generating pMR361-K, Kelly Michie for assistance with the chronic surgical wound experiments, Sean Leonard for computational assistance, and Dr. Daniel Cornforth for discussion of this manuscript.
Footnotes
Author Contributions
C.B.I., A.S., K.P.R and M.W. designed experiments; C.B.I., A.S., and M.W. analyzed data; C.B.I., A.S., and D.F. performed experiments; J.L.D. prepared sequencing libraries; M.S.G. provided S. aureus HG003 transposon library and provided the raw data from previous studies7,8 included in the analysis; C.B.I., A.S., J.L.D, M.S.G., K.P.R., M.W. wrote the paper.
References
- 1.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 2.Le Breton Y, et al. Essential genes in the core genome of the human pathogen Streptococcus pyogenes. Sci Rep. 2015;5:9838. doi: 10.1038/srep09838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umland TC, et al. In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. MBio. 2012;3 doi: 10.1128/mBio.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentino MD, et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. MBio. 2014;5:e01729–01714. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilde AD, et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015;11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nature Reviews Microbiology. 2015;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. doi:nrmicro2200 [pii]10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 12.Cheng AG, DeDent AC, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 2011;19:225–232. doi: 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gjødsbøl K, et al. Multiple bacterial species reside in chronic wounds: a longitudinal study. International Wound Journal. 2006;3:225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madsen SM, Westh H, Danielsen L, Rosdahl VT. Bacterial colonization and healing of venous leg ulcers. APMIS. 1996;104:895–899. doi: 10.1111/j.1699-0463.1996.tb04955.x. [DOI] [PubMed] [Google Scholar]
- 15.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:13887–13892. doi: 10.1073/pnas.1610856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One. 2012;7:e43012. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton T, et al. An in vivo polymicrobial biofilm wound infection model to studyinterspecies interactions. PLoS One. 2011;6(11):e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korgaonkar A, et al. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A. 2013;110(3):1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazli M, et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol. 2009;47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottrup F. Optimizing wound treatment through health care structuring and professional education. Wound Repair and Regeneration. 2004;12:129–133. doi: 10.1111/j.1067-1927.2004.012204.x. [DOI] [PubMed] [Google Scholar]
- 21.Anderson M, Chen YH, Butler EK, Missiakas DM. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J Bacteriol. 2011;193:1583–1589. doi: 10.1128/JB.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. 2016;2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008;16:805–813. doi: 10.1111/j.1524-475X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 24.DeLeon S, et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun. 2014;82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacy A, Fleming D, Lamont RJ, Rumbaugh KP, Whiteley M. A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. MBio. 2016;7 doi: 10.1128/mBio.00782-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2:e00315–00310. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson ME. Bacteriophage transduction in Staphylococcus aureus. The Genetic Manipulation of Staphylococci. 2015;1373:69–74. doi: 10.1007/7651_2014_186. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 2012;8:e1002804. doi: 10.1371/journal.pgen.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes O, et al. 'Integron'-bearing vectors: a method suitable for stable chromosomal integration in highly restrictive corynebacteria. Gene. 1991;107:61–68. doi: 10.1016/0378-1119(91)90297-o. [DOI] [PubMed] [Google Scholar]
- 32.Ferrieres L, et al. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol. 2010;192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mintz KP. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology. 2004;150:2677–2688. doi: 10.1099/mic.0.27110-0. [DOI] [PubMed] [Google Scholar]
- 34.Watters C, et al. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol. 2013;202:131–141. doi: 10.1007/s00430-012-0277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray JL, Kwon T, Marcotte EM, Whiteley M. Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa. MBio. 2015;6:e01603–01615. doi: 10.1128/mBio.01603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein BA, et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: http://dx.doi.org/10.14806/ej.17.1.200. [Google Scholar]
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. Journal of the American Statistical Association. 2002;97:611–631. [Google Scholar]
- 40.Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. 2007–2015 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 41.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Some S. aureus CoDE genes are not in vivo specific. Comparison of wildtype S. aureus and S. aureus CoDE gene mutant cell numbers in an in vitro chronic wound model during mono-culture and P. aeruginosa co-culture. graE::TnMariner (b, green), udk (c, orange), and tcyA::TnMariner (d, purple) were predicted to be required in co-culture but not mono-culture, while while yrrK::TnMariner (a, red) and atpD::TnMariner (e, blue) were predicted to be required in mono-infection but not in co-infection. All mutants are reported as % wildtype cell number (S. aureus HG003) in the same condition. Statistical analysis was performed by arcsine transforming the data prior to analysis with a Student’s t-test (* = p < 0.05). For each condition, 4 biological replicates were used. Error bars represent the standard error of the mean (SEM).
Supplementary Figure 2. Identification of CoDE genes in A. actinomycetemcomitans during co-infection with S. gordonii. Venn diagram of the A. actinomycetemcomitans in vivo essential genome in mono-culture (blue, n=2) and co-culture (yellow, n=2) infection with S. gordonii. 72 CoDE genes were identified that were essential in mono-culture infection but non-essential in co-culture, and 83 CoDE genes were uniquely essential in co-infection. Data are the result of two replicates per condition.
Supplementary Figure 3. Experimental replicates cluster by condition. Hierarchal clustering (Ward method) of the average normalized read counts per gene in each of the replicates (numbered and color coded) for all experimental conditions. Height indicates the Euclidean distance between clusters.
Data Availability Statement
The raw sequencing files that support this study have been deposited into the NCBI Sequence Read Archive under accession SRP093229 for S. aureus Tn-seq data and accession SRP095181 for A. actinomycetemcomitans Tn-seq data. The sequenced genome for S. aureus reference strain NCTC8325 is available from under RefSeq accession number NC_007795.1. The sequenced genome for A. actinomycetemcomitans Strain 624 is available under RefSeq accession number NZ_CP012959.1. The remaining data that support the findings of this study are available from the corresponding author upon request.
All material requests and correspondences should be addressed to Marvin Whiteley, mwhiteley@austin.utexas.edu