ABSTRACT
Objective: It is unknown whether bacteria play a role in the collagen matrix degradation that occurs during caries progression. Our aim was to characterize the expression level of genes involved in bacterial collagenolytic proteases in root biofilms with and without caries. Method: we collected samples from active cavitated root caries lesions (RC, n = 30) and from sound root surfaces (SRS, n = 10). Total microbial RNA was isolated and cDNA sequenced on the Illumina Hi-Seq2500. Reads were mapped to 162 oral bacterial reference genomes. Genes encoding putative bacterial collagenolytic proteases were identified. Normalization and differential expression analysis was performed on all metatranscriptomes (FDR<10-3).
Result: Genes encoding collagenases were identified in 113 bacterial species the majority were peptidase U32. In RC, Streptococcus mutans and Veillonella parvula expressed the most collagenases. Organisms that overexpressed collagenolytic protease genes in RC (Log2FoldChange>8) but none in SRS were Pseudoramibacter alactolyticus [HMPREF0721_RS02020; HMPREF0721_RS04640], Scardovia inopinata [SCIP_RS02440] and Olsenella uli DSM7084 [OLSU_RS02990].
Conclusion: Our findings suggest that the U32 proteases may be related to carious dentine. The contribution of a small number of species to dentine degradation should be further investigated. These proteases may have potential in future biotechnological and medical applications, serving as targets for the development of therapeutic agents.
KEYWORDS: Root caries, gene expression, collagen, microbial collagenase, sequence analysis, RNA, biofilms
Introduction
Root hard tissues (cementum and dentine) become vulnerable to demineralization once root surfaces are exposed. These tissues are less mineralized than enamel and are composed of high proportions of organic materials such as collagen [1,2]. From a clinical point of view, the development of caries in root hard tissues may be considered a two-stage process: the first stage is characterized by mineral dissolution and the second by the degradation of the organic matrix of the root surface [3]. Microbial invasion of cementum and dentine tissues has been reported even in the first stage of the caries process, whereas in enamel caries, dentine is invaded only once enamel is destroyed [4,5]. This fact has an impact on the microbiome associated with the caries process in root hard tissues.
The function of bacteria in the demineralization stage of caries development is well known. Root hard tissue demineralization may develop in the presence of a rich and diverse microbiota, and the acidification of the microenvironment selects some species that are able to survive at low pH and produce high amounts of organic acids [6]. Root dentine biofilms are composed of a variety of saccharolytic, aciduric, and acidogenic organisms, as well as proteolytic bacteria, which can produce acids or ammonia from the catabolism of nitrogenous substrates that are available exogenously or from the dentine organic matrix [3,7]; thus, they can affect the biofilm pH in several ways. In addition to demineralization, bacteria may be involved in matrix degradation. Collagen is resistant to most common proteases and can be degraded by only a few types of proteases from mammals or bacteria [8], including some metalloproteases and serine proteases. It has been suggested that host collagenases from dentine are associated with collagen matrix degradation during caries progression [9,10], representing a response of the host tissues to caries attack under acidic conditions. These proteases, which include matrix metalloproteinases (MMP-2, 3, 8, 9, and 20) and cysteine cathepsins (B and K), are present in the dentinal organic matrix and become activated once the cementum is degraded [3,9–13].
Recently, a tissue-dependent hypothesis for dental caries suggested that some bacteria could promote dentine degradation and caries development [14]. This hypothesis is based on the discovery of overexpression of genes related to proteolytic activity, as well as bacterial collagenases in dentinal caries from coronal lesions [14,15]. These studies showed for the first time that microbial proteolytic activity might contribute to dentinal protein degradation. Microbial collagenolytic activity has been demonstrated in a few oral bacteria [3]; however, a real contribution of bacteria to the degradation of the organic part of root dentine remains questionable. Protease PrtC from Porphyromonas gingivalis ATCC 53977 is one of the most reported microbial collagenolytic proteases produced by oral bacteria. It is part of the U32 protease family and contains 1,002 bp encoding a 333-residue PrtC protein. It can degrade soluble and reconstituted fibrillar type I collagen (the most common in root hard tissues) at body temperature or below [8,16]. Due to the relationship between the periodontal biofilm and the biofilm that cause root caries (RC), this protein could be involved in root dentine degradation.
The collagenase activity-dependent ability to degrade the dentinal collagen matrix could be an important virulence trait of plaque biofilms. In this study, we evaluated bacterial collagenolytic protease gene expression within natural biofilms from RC compared with supragingival biofilms of RC-free individuals by RNA-seq data analysis. The terminology ‘bacterial collagenolytic proteases’ was used to refer to all proteases that can degrade at least one type of collagen according to Zhang et al., including true collagenases and other proteases with collagenolytic activity [8]. These data may help clarify the role of bacteria in collagen matrix degradation in RC.
Materials and methods
Sample collection was carried out as described by Damé-Teixeira et al. [17]. Briefly, 10 volunteers with an exposed root surface and no RC lesion were included in the sound root surface group (SRS). Supragingival biofilms were collected from all exposed root surfaces. All participants recruited for the RC group had one primary cavitated root lesion in need of restorative treatment. All lesions presented characteristics of activity (soft and yellow dentine). Biofilm and carious dentine samples (soft and infected tissue) were collected from 30 patients during the restorative treatment.
Upon collection, samples were placed in a nuclease-free microtube containing 1 mL of RNAprotect reagent (Qiagen Inc.). Total RNA was extracted from all samples using the UltraClean® Microbial RNA Isolation (Mo-bio, San Diego, CA, USA) using on-column DNAse digestion (Qiagen, Inc.). The extracted RNA was quantified using the Quant-iT™ RiboGreen® RNA Assay Kit (Invitrogen), and samples with total RNA concentration <30 ng/RNA were pooled, leading to a final sample count of 10 SRS and 9 RC. The Ribo-Zero™ Meta-Bacteria Kit (Epicentre, Illumina) was used for mRNA enrichment, and Illumina®TruSeq™ library prep protocols (Illumina, SD) were used for library preparation and paired-end sequencing with the Illumina HiSeq2500.
Read sequences for each sample were quality trimmed using cutadapt and imported into the CLC Genomics Workbench v8 software (CLC Bio, Qiagen). The genomes of 162 bacteria and their associated information were downloaded from the DNA Data Bank of Japan, NCBI, the Broad Institute, and the HOMD database and mapped against the short-reads sequences (for the list of genomes, see [17]). The data produced are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive, under the accession numbers SRS779973 and SRS796739. Read count data for all potential collagenases were manually extracted from the 162 genomes, with particular focus on the U32 family proteases [8] due to the implication of this family as virulence factors in oral bacteria and its abundance. However, peptidolytic or gelatinolytic proteases were not included in this study’s analysis.
The number of genes with no activity was stated as ‘number of reads = 0’. The relative median expression level for genes from bacterial collagenolytic proteases was calculated for each of the sample groups, as described previously [18] within the R package ‘DESeq’ [19], and considered as the ‘gene expression value’. Graphs were generated within the R package ‘plotly’ [20].
Statistical analysis for inferring differential gene expression between sample groups was also carried out using the R package DESeq2 [21]. The cut-off for designating a gene as being differentially expressed was a change in transcript levels of at least 1-log fold change (two times difference, negative values = up-regulated in SRS and down-regulated in RC and positive values = down-regulated in RC and up-regulated in SRS) and Benjamini–Hochberg adjusted p-value (padj) of less than 10− 3 [22]. This high cut-off was chosen in order to avoid false-positive results and identify only true differences.
This study was approved by the ethics committee of the Federal University of Rio Grande do Sul (process n° 427.168) and by the Yorkshire & The Humber – Leeds West National Research Ethics Service Committee (protocol n° 2012002DD). Volunteers to the study were patients who attended dental clinics for any dental treatment in two centres: Faculty of Dentistry, Federal University of Rio Grande do Sul, Porto Alegre, Brazil; and the School of Dentistry, Dental Translational Research Unit, University of Leeds, Leeds, UK. All volunteers consented to participate and donate samples after receiving the information about the study.
Results
A total of 201 genes coding for bacterial collagenolytic proteases were identified in 113 bacterial species; 24 from Prevotella spp. and 20 from Streptococcus spp. Table 1 describes genes encoding bacterial collagenolytic proteases identified in the metatranscriptome analysis of root biofilms, showing that a majority expressed genes for the peptidase U32 family (basically protease PrtC).
Table 1.
List of genes coding for bacterial collagenolytic proteases.
| Genome | Locus tag | Protein product | Protein annotation |
|---|---|---|---|
| Aggregatibacter actinomycetemcomitans D11S-1 | D11S_1802 | ACX83163.1 | Peptidase U32 |
| Aggregatibacter aphrophilus NJ8700 [23] | NT05HA_RS01075 | WP_005701395.1 | Collagenase-like protease, PrtC family |
| Alloprevotella tannerae ATCC 51259 | GCWU000325_RS05925 | WP_006255366.1 | Collagenase |
| Alloprevotella tannerae ATCC 51259 | GCWU000325_RS08705 | WP_006256078.1 | Collagenase-like protease, PrtC family |
| Alloprevotella tannerae ATCC 51259 | GCWU000325_RS05925 | WP_006255366.1 | Collagenase |
| Atopobium rimae ATCC 49626 | ATORI0001_RS02210 | WP_003148443.1 | Peptidase U32 |
| Bifidobacterium breve UCC2003 [24] | BBR_RS19280 | WP_015439232.1 | Peptidase U32 |
| Bifidobacterium dentium Bd1 | BDP_RS01815 | WP_012901869.1 | Collagenase |
| Bifidobacterium kashiwanohense PV20-2 | AH68_RS01480 | WP_039196994.1 | Collagenase |
| Bifidobacterium thermophilum RBL67 | D805_RS06990 | WP_044282489.1 | Collagenase |
| Campylobacter concisus 13826 | CCC13826_RS05820 | WP_048809830.1 | Collagenase-like protease, PrtC family |
| Campylobacter curvus 525.92 | CCV52592_RS06400 | WP_011992484.1 | Collagenase-like protease, PrtC family |
| Campylobacter gracilis strain ATCC 33236 | CGRAC_RS08900 | WP_005873169.1 | Collagenase-like protease, PrtC family |
| Campylobacter rectus RM3267 | CAMRE0001_RS04590 | WP_004318907.1 | Collagenase-like protease, PrtC family |
| Candidate division SR1 bacterium Aalborg_AAW-1 | XF24_00476 | AKH32809.1 | Collagenase |
| Capnocytophaga gingivalis ATCC 33624 | CAPGI0001_RS09340 | WP_002669179.1 | Collagenase |
| Capnocytophaga sputigena ATCC 33612 | CAPSP0001_RS09700 | WP_002680903.1 | Collagenase |
| Cardiobacterium hominis ATCC 15826 | HMPREF0198_RS02295 | WP_004139642.1 | Collagenase-like protease, PrtC family |
| Catonella morbi ATCC 51271 | GCWU000282_RS01655 | WP_035039351.1 | Peptidase U32 |
| Catonella morbi ATCC 51271 | GCWU000282_RS01780 | WP_023353272.1 | Peptidase U32 |
| Clostridium saccharolyticum WM1WM1 | CLOSA_RS11275 | WP_013272899.1 | Peptidase U32 |
| Clostridium saccharolyticum WM1WM1 | CLOSA_RS04645 | WP_013271613.1 | Peptidase U32 |
| Dialister invisus DSM 15470 | GCWU000321_RS07150 | WP_007070508.1 | Peptidase U32 |
| Dichelobacter nodosus VCS1703A | DNO_RS01910 | WP_012030735.1 | Collagenase-like protease, PrtC family |
| Eikenella corrodens ATCC 23834 | EIKCOROL_RS00310 | WP_003821765.1 | Collagenase-like protease, PrtC family |
| Eikenella corrodens ATCC 23834 | EIKCOROL_RS00745 | WP_035579870.1 | Collagenase-like protease, PrtC family |
| Eubacterium eligens ATCC 27750 | EUBELI_RS04560 | WP_012739182.1 | Collagenase-like protease, PrtC family |
| Eubacterium eligens ATCC 27750 | EUBELI_RS03735 | WP_041688528.1 | Peptidase U32 |
| Eubacterium saphenum ATCC 49989 | GCWU000322_RS00740 | WP_005837827.1 | Peptidase U32 |
| Filifactor alocis ATCC 35896 | HMPREF0389_RS02570 | WP_014262170.1 | Protease |
| Filifactor alocis ATCC 35896 | HMPREF0389_RS00640 | WP_014261808.1 | Peptidase U32 |
| Fusobacterium nucleatum subsp. animalis 7_1 | FSDG_RS09345 | WP_008702184.1 | Collagenase-like protease, PrtC family |
| Fusobacterium nucleatum subsp. animalis 7_1 | FSDG_RS08285 | WP_008702419.1 | Collagenase-like protease, PrtC family |
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | FN1931 | NP_602731.1 | Collagenase-like protease, PrtC family |
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | FN1826 | NP_602626.1 | Collagenase-like protease, PrtC family |
| Fusobacterium nucleatum subsp. vincentii 3_1_36A2 | HMPREF0946_RS00995 | WP_008800359.1 | Collagenase-like protease, PrtC family |
| Fusobacterium nucleatum subsp. vincentii 3_1_36A2 | HMPREF0946_RS02055 | WP_008796619.1 | Collagenase-like protease, PrtC family |
| Fusobacterium periodonticum ATCC 33693 | FUSPEROL_RS01940 | WP_039984117.1 | Collagenase-like protease, PrtC family |
| Fusobacterium periodonticum ATCC 33693 | FUSPEROL_RS01460 | WP_005970981.1 | Collagenase-like protease, PrtC family |
| Gemella haemolysans M341 | HMPREF0428_RS03755 | WP_003146785.1 | Collagenase-like protease, PrtC family |
| Gemella haemolysans M341 | HMPREF0428_RS03760 | WP_003146787.1 | Peptidase U32 |
| Gemella moribillum M424 | HMPREF0432_RS03545 | WP_004632787.1 | Collagenase-like protease, PrtC family |
| Gemella moribillum M424 | HMPREF0432_RS03550 | WP_004632788.1 | Peptidase U32 |
| Granulicatella adiacens ATCC 49175 | HMPREF0444_RS08360 | WP_005607759.1 | Collagenase-like protease, PrtC family |
| Granulicatella adiacens ATCC 49175 | HMPREF0444_RS08365 | WP_005607760.1 | Peptidase U32 |
| Granulicatella elegans ATCC 700633 | HMPREF0446_RS01120 | WP_006703604.1 | Peptidase U32 |
| Granulicatella elegans ATCC 700633 | HMPREF0446_RS01125 | WP_006703603.1 | Collagenase-like protease, PrtC family |
| Haemophilus influenzae F3047 | HICON_RS03890 | WP_013526492.1 | Collagenase-like protease, PrtC family |
| Haemophilus parainfluenzae T3T1 | PARA_RS00685 | WP_005695474.1 | Collagenase-like protease, PrtC family |
| Kingella oralis ATCC 51147 | GCWU000324_RS07250 | WP_003796734.1 | Collagenase-like protease, PrtC family |
| Lachnoanaerobaculum saburreum DSM 3986 | HMPREF0381_RS09405 | WP_040461351.1 | Peptidase U32 |
| Lachnoanaerobaculum saburreum DSM 3986 | HMPREF0381_RS11935 | WP_008752244.1 | Peptidase U32 |
| Lautropia mirabilis | HMPREF0551_RS01810 | WP_040529625.1 | Collagenase-like protease, PrtC family |
| Leptotrichia buccalis [25] | LEBU_RS05040 | WP_041760865.1 | Peptidase U32 |
| Leptotrichia buccalis [25] | LEBU_RS10190 | WP_015770252.1 | Collagenase-like protease, PrtC family |
| Neisseria bacilliformis | HMPREF9123_RS07480 | WP_007342950.1 | Collagenase-like protease, PrtC family |
| Neisseria bacilliformis | HMPREF9123_RS08480 | WP_007343222.1 | Collagenase-like protease, PrtC family |
| Neisseria elongata subsp. glycolytica ATCC 29315 [26] | NELON_RS10680 | WP_003769563.1 | Collagenase-like protease, PrtC family |
| Neisseria elongata subsp. glycolytica ATCC 29315 [26] | NELON_RS07015 | WP_003771571.1 | Collagenase-like protease, PrtC family |
| Neisseria flavescens SK114 | NEIFL0001_RS03385 | WP_003684307.1 | Collagenase-like protease, PrtC family |
| Neisseria flavescens SK114 | NEIFL0001_RS01600 | WP_003683417.1 | Collagenase-like protease, PrtC family |
| Neisseria lactamica 020-06 [27] | NLA_RS03260 | WP_013448613.1 | Collagenase-like protease, PrtC family |
| Neisseria mucosa C102 | HMPREF0604_RS08420 | WP_003748766.1 | Collagenase-like protease, PrtC family |
| Neisseria mucosa C102 | HMPREF0604_RS07970 | WP_003748589.1 | Collagenase-like protease, PrtC family |
| Neisseria subflava | NEISUBOT_RS03855 | WP_004519592.1 | Collagenase-like protease, PrtC family |
| Neisseria subflava | NEISUBOT_RS04305 | WP_004519683.1 | Collagenase-like protease, PrtC family |
| Olsenella uli DSM7084 [28] | OLSU_RS02990 | WP_041549197.1 | Peptidase U32 |
| Oribacterium sp. oral taxon 078 str. F0262 | GCWU000341_RS02740 | WP_009214193.1 | Peptidase U32 |
| Parvimonas micra ATCC 33270 | PEPMIC_RS07615 | WP_004833569.1 | Peptidase U32 |
| Parvimonas micra ATCC 33270 | PEPMIC_RS08350 | WP_041954953.1 | Peptidase U32 |
| Peptostreptococcus stomatis | HMPREF0634_RS06810 | WP_007790248.1 | Collagenase-like protease, PrtC family |
| Peptostreptococcus stomatis | HMPREF0634_RS00830 | WP_007788149.1 | Peptidase U32 |
| Porphyromonas asaccharolytica DSM 20707 | PORAS_RS01075 | WP_013759854.1 | Peptidase U32 |
| Porphyromonas asaccharolytica DSM 20707 | PORAS_RS04355 | WP_013760316.1 | Collagenase |
| Porphyromonas endodontalis | POREN0001_RS06205 | WP_004334244.1 | Peptidase U32 |
| Porphyromonas endodontalis | POREN0001_RS08830 | WP_052296722.1 | Collagenase |
| Porphyromonas gingivalis ATCC 33277 [29] | PGN_RS02685 | WP_039416961.1 | Collagenase |
| Porphyromonas gingivalis ATCC 33277 [29] | PGN_RS03720 | WP_012457772.1 | Collagenase |
| Porphyromonas gingivalis ATCC 33277 [29] | PGN_RS02685 | WP_039416961.1 | Collagenase |
| Porphyromonas gingivalis ATCC 33277 [29] | PGN_RS03720 | WP_012457772.1 | Collagenase |
| Prevotella amnii DSM 23384 = JCM 14753 | F596_RS0106960 | WP_026302377.1 | Collagenase-like protease, PrtC family |
| Prevotella amnii DSM 23384 = JCM 14753 | F596_RS0104625 | WP_019036032.1 | Collagenase |
| Prevotella bergensis | HMPREF0645_RS06070 | WP_007174036.1 | Collagenase-like protease, PrtC family |
| Prevotella bergensis | HMPREF0645_RS09130 | WP_007174691.1 | Collagenase |
| Prevotella bivia DSM 20514 | PREBIDRAFT_RS05630 | WP_004336717.1 | Collagenase |
| Prevotella bryantii C21a | G638_RS0101200 | WP_027452885.1 | Collagenase-like protease, PrtC family |
| Prevotella bryantii C21a | G638_RS0104370 | WP_027453233.1 | Collagenase |
| Prevotella buccae ATCC 33574 | HMPREF6485_RS08010 | WP_044125959.1 | Collagenase-like protease, PrtC family |
| Prevotella buccae ATCC 33574 | HMPREF6485_RS03410 | WP_004341966.1 | Collagenase |
| Prevotella buccalis ATCC 35310 | HMPREF0650_RS10745 | WP_004350712.1 | Collagenase-like protease, PrtC family |
| Prevotella buccalis ATCC 35310 | HMPREF0650_RS05375 | WP_004348964.1 | Collagenase |
| Prevotella copri | PREVCOP_RS02145 | WP_006846714.1 | Collagenase-like protease, PrtC family |
| Prevotella copri | PREVCOP_RS08960 | WP_006848256.1 | Collagenase |
| Prevotella dentalis DSM 3688 | PREDE_RS10275 | WP_005847167.1 | Collagenase-like protease, PrtC family |
| Prevotella dentalis DSM 3688 | PREDE_RS07105 | WP_005845431.1 | Collagenase |
| Prevotella denticola F0289 | HMPREF9137_RS07010 | WP_013671465.1 | Collagenase-like protease, PrtC family |
| Prevotella denticola F0289 | HMPREF9137_RS02430 | WP_013670722.1 | Collagenase |
| Prevotella disiens JCM 6334 = ATCC 29426 | HMPREF0653_RS10015 | WP_021670023.1 | Peptidase U32 |
| Prevotella disiens JCM 6334 = ATCC 29426 | HMPREF0653_RS06040 | WP_021669257.1 | Collagenase |
| Prevotella intermedia 17 chr1 and 2 | PIN17_RS08560 | WP_014709923.1 | Collagenase-like protease, PrtC family |
| Prevotella intermedia 17 chr1 and 2 | PIN17_RS04505 | WP_014709153.1 | Collagenase |
| Prevotella marshii DSM 16,973 = JCM 13450 | HMPREF0658_RS00735 | WP_006947833.1 | Collagenase-like protease, PrtC family |
| Prevotella marshii DSM 16,973 = JCM 13450 | HMPREF0658_RS07640 | WP_006949885.1 | Collagenase |
| Prevotella melaninogenica ATCC 25845 | HMPREF0659_RS08000 | WP_013265267.1 | Collagenase-like protease, PrtC family |
| Prevotella melaninogenica ATCC 25845 | HMPREF0659_RS07035 | WP_044045939.1 | Collagenase |
| Prevotella multiformis | HMPREF9141_RS08830 | WP_007368357.1 | Collagenase-like protease, PrtC family |
| Prevotella multiformis | HMPREF9141_RS06410 | WP_007367797.1 | Collagenase |
| Prevotella nigrescens ATCC 33563 | HMPREF9419_RS03665 | WP_004366273.1 | Collagenase-like protease, PrtC family |
| Prevotella nigrescens ATCC 33563 | HMPREF9419_RS06820 | WP_004366953.1 | Collagenase |
| Prevotella oralis ATCC 33269 | HMPREF0663_RS01445 | WP_004368289.1 | Collagenase-like protease, PrtC family |
| Prevotella oralis ATCC 33269 | HMPREF0663_RS04675 | WP_004369102.1 | Collagenase |
| Prevotella oris F0302 | HMPREF0971_RS00090 | WP_004370849.1 | Collagenase-like protease, PrtC family |
| Prevotella oris F0302 | HMPREF0971_RS12160 | WP_004375331.1 | Collagenase |
| Prevotella pallens ATCC 700821 | HMPREF9144_RS05520 | WP_040595396.1 | Collagenase-like protease, PrtC family |
| Prevotella pallens ATCC 700821 | HMPREF9144_RS01260 | WP_006043979.1 | Collagenase |
| Prevotella ruminicola 23 | PRU_RS03090 | WP_013063209.1 | Collagenase-like protease, PrtC family |
| Prevotella ruminicola 23 | PRU_RS14605 | WP_013065324.1 | Collagenase |
| Prevotella salivae DSM 15606 | HMPREF9420_RS01830 | WP_007133684.1 | Collagenase-like protease, PrtC family |
| Prevotella salivae DSM 15606 | HMPREF9420_RS07750 | WP_044079174.1 | Collagenase |
| Prevotella sp. oral taxon 299 str. F0039 | HMPREF0669_RS01095 | WP_009228902.1 | Collagenase-like protease, PrtC family |
| Prevotella sp. oral taxon 299 str. F0039 | HMPREF0669_RS08220 | WP_009227931.1 | Collagenase |
| Prevotella sp. oral taxon 472 str. F0295 | HMPREF6745_RS04325 | WP_009235927.1 | Collagenase-like protease, PrtC family |
| Prevotella sp. oral taxon 472 str. F0295 | HMPREF6745_RS08625 | WP_009236867.1 | Collagenase |
| Prevotella timonensis 4401737 | BN35_RS03705 | WP_025071979.1 | Collagenase-like protease, PrtC family |
| Prevotella timonensis 4401737 | BN35_RS06480 | WP_028900923.1 | Collagenase |
| Prevotella veroralis F0319 | HMPREF0973_RS06810 | WP_004383383.1 | Collagenase-like protease, PrtC family |
| Prevotella veroralis F0319 | HMPREF0973_RS05650 | WP_039851216.1 | Collagenase |
| Pseudoramibacter alactolyticus | HMPREF0721_RS02020 | WP_050786939.1 | Collagenase-like protease, PrtC family |
| Pseudoramibacter alactolyticus | HMPREF0721_RS04640 | WP_006598435.1 | Peptidase U32 |
| Pyramidobacter piscolens | HMPREF7215_RS04740 | WP_040550474.1 | Peptidase U32 |
| Scardovia inopinata JCM 12537 [30] | SCIP_RS02440 | WP_006292938.1 | Peptidase U32 |
| Selenomonas infelix ATCC 43532 | HMPREF9334_RS00910 | WP_006691631.1 | Peptidase U32 |
| Selenomonas infelix ATCC 43532 | HMPREF9334_RS02810 | WP_006691984.1 | Peptidase U32 |
| Selenomonas infelix ATCC 43532 | HMPREF9334_RS06450 | WP_006692674.1 | Peptidase U32 |
| Selenomonas noxia ATCC 43541 | HMPREF7545_RS02205 | WP_006694041.1 | Peptidase U32 |
| Selenomonas noxia ATCC 43541 | HMPREF7545_RS03310 | WP_040571168.1 | Peptidase U32 |
| Selenomonas noxia ATCC 43541 | HMPREF7545_RS04210 | WP_006694441.1 | Peptidase U32 |
| Selenomonas ruminantium subsp. ruminantium ATCC 12561 | G598_RS0108180 | WP_026766556.1 | Peptidase U32 |
| Selenomonas sp. oral taxon 478 | ADJ74_RS10000 | WP_050343958.1 | Peptidase U32 |
| Selenomonas sp. oral taxon 478 | ADJ74_RS10520 | WP_050344100.1 | Peptidase U32 |
| Selenomonas sp. oral taxon 478 | ADJ74_RS07050 | WP_050343039.1 | Peptidase U32 |
| Selenomonas sputigena ATCC 35185 | SELSP_RS05470 | WP_006192437.1 | Peptidase U32 |
| Shuttleworthia satelles DSM 14600 | GCWU000342_RS00125 | WP_006905076.1 | Peptidase U32 |
| Shuttleworthia satelles DSM 14600 | GCWU000342_RS05060 | WP_006906224.1 | Collagenase-like protease, PrtC family |
| Solobacterium moorei DSM 22971 | H345_RS12725 | WP_051240871.1 | Collagenase-like protease, PrtC family |
| Solobacterium moorei DSM 22971 | H345_RS0101730 | WP_028077445.1 | Collagenase-like protease, PrtC family |
| Streptococcus anginosus C238 [31] | SANR_RS03650 | WP_003035012.1 | Peptidase U32 |
| Streptococcus anginosus C238 [31] | SANR_RS03655 | WP_020999544.1 | Collagenase-like protease, PrtC family |
| Streptococcus australis ATCC 700641 | HMPREF9421_RS05000 | WP_006597381.1 | Peptidase U32 |
| Streptococcus australis ATCC 700641 | HMPREF9421_RS05005 | WP_006597406.1 | Collagenase-like protease, PrtC family |
| Streptococcus constellatus subsp. constellatus SK53 | HMPREF1044_RS08695 | WP_006270660.1 | Collagenase-like protease, PrtC family |
| Streptococcus constellatus subsp. constellatus SK53 | HMPREF1044_RS08700 | WP_037566276.1 | Peptidase U32 |
| Streptococcus constellatus subsp. pharyngis C232 [31] | SCRE_RS05335 | WP_006267751.1 | Collagenase-like protease, PrtC family |
| Streptococcus constellatus subsp. pharyngis C232 [31] | SCRE_RS05340 | WP_006267951.1 | Peptidase U32 |
| Streptococcus cristatus ATCC 51100 | HMPREF9422_RS00190 | WP_005589468.1 | Peptidase U32 |
| Streptococcus cristatus ATCC 51100 | HMPREF9422_RS04040 | WP_005590706.1 | Collagenase-like protease, PrtC family |
| Streptococcus gordonii str. Challis substr. CH1 [32] | SGO_RS03645 | WP_012000207.1 | Peptidase U32 |
| Streptococcus gordonii str. Challis substr. CH1 [32] | SGO_RS03650 | WP_012000208.1 | Collagenase-like protease, PrtC family |
| Streptococcus infantis ATCC 700779 | HMPREF9423_RS07500 | WP_006148729.1 | Peptidase U32 |
| Streptococcus infantis ATCC 700779 | HMPREF9423_RS07175 | WP_006148665.1 | Collagenase-like protease, PrtC family |
| Streptococcus intermedius B196 [31] | SIR_RS12835 | WP_021002602.1 | Collagenase-like protease, PrtC family |
| Streptococcus intermedius B196 [31] | SIR_RS12840 | WP_021002603.1 | Collagenase-like protease, PrtC family |
| Streptococcus mitis B6 [33] | smi_1316 | YP_003446424.1 | Collagenase-like protease, PrtC family |
| Streptococcus mitis B6 [33] | smi_0854 | YP_003445970.1 | Collagenase-like protease, PrtC family |
| Streptococcus mutans UA159 [48] | SMU_759 | NP_721176.1 | Collagenase-like protease, PrtC family |
| Streptococcus mutans UA159 [48] | SMU_761 | NP_721177.1 | Collagenase-like protease, PrtC family |
| Streptococcus oligofermentans AS 1.3089 [34] | I872_RS05755 | WP_015605207.1 | U32 family peptidase |
| Streptococcus oligofermentans AS 1.3089 [34] | I872_RS06980 | WP_015605435.1 | Peptidase U32 |
| Streptococcus oralis Uo5 [35] | SOR_RS05810 | WP_000411175.1 | Peptidase U32 |
| Streptococcus oralis Uo5 [35] | SOR_RS05510 | WP_000169101.1 | Collagenase-like protease, PrtC family |
| Streptococcus parasanguinis ATCC 15912 | HMPREF0833_RS04350 | WP_003002878.1 | Collagenase-like protease, PrtC family |
| Streptococcus parasanguinis ATCC 15912 | HMPREF0833_RS04355 | WP_013903889.1 | Peptidase U32 |
| Streptococcus peroris | HMPREF9180_RS06815 | WP_006145781.1 | Peptidase U32 |
| Streptococcus peroris | HMPREF9180_RS06495 | WP_006145710.1 | Collagenase-like protease, PrtC family |
| Streptococcus salivarius CCHSS3 | SALIVB_RS06560 | WP_002886038.1 | Peptidase U32 |
| Streptococcus salivarius CCHSS3 | SALIVB_RS06555 | WP_004182776.1 | Collagenase-like protease, PrtC family |
| Streptococcus sanguinis SK36 [36] | SSA_1541 | YP_001035482.1 | U32 family peptidase |
| Streptococcus sanguinis SK36 [36] | SSA_1542 | YP_001035483.1 | U32 family peptidase |
| Streptococcus sobrinus DSM 20742 = ATCC 33478 | BS63_RS0108710 | WP_028798546.1 | Collagenase-like protease, PrtC family |
| Streptococcus sobrinus DSM 20742 = ATCC 33478 | BS63_RS0100290 | WP_002962408.1 | Collagenase-like protease, PrtC family |
| Streptococcus sp. VT 162 | V470_RS05500 | WP_044020909.1 | Peptidase U32 |
| Streptococcus sp. VT 162 | V470_RS05200 | WP_000169101.1 | Collagenase-like protease, PrtC family |
| Streptococcus thermophilus LMG 18311 [37] | STU_RS12905 | WP_002952720.1 | Peptidase U32 |
| Streptococcus thermophilus LMG 18311 [37] | STU_RS12910 | WP_002945995.1 | Collagenase-like protease, PrtC family |
| Streptococcus vestibularis ATCC 49124 | HMPREF9425_RS05965 | WP_003097482.1 | Peptidase U32 |
| Streptococcus vestibularis ATCC 49124 | HMPREF9425_RS05960 | WP_003094356.1 | Collagenase-like protease, PrtC family |
| Tannerella forsythia 92A2 | BFO_RS03710 | WP_014224272.1 | Collagenase-like protease, PrtC family |
| Tannerella forsythia 92A2 | BFO_RS05860 | WP_014224717.1 | Collagenase |
| Treponema brennaborense | TREBR_RS02850 | WP_013757721.1 | Peptidase U32 |
| Treponema brennaborense | TREBR_RS10885 | WP_013759230.1 | Peptidase U32 |
| Treponema denticola ATCC 35405 [38] | TDE0071 | NP_970688.1 | U32 family peptidase |
| Treponema denticola ATCC 35405 [38] | TDE2262 | NP_972862.1 | U32 family peptidase |
| Treponema putidum [39] | JO40_RS07200 | WP_044978748.1 | Peptidase U32 |
| Treponema putidum [39] | JO40_RS05960 | WP_044978548.1 | Peptidase U32 |
| Treponema vincentii F0403 | HMPREF1222_RS11455 | WP_016519531.1 | Collagenase-like protease, PrtC family |
| Veillonella atypica KON | HMPREF0870_RS02545 | WP_005382667.1 | Peptidase U32 |
| Veillonella atypica KON | HMPREF0870_RS05775 | WP_005376158.1 | Peptidase U32 |
| Veillonella dispar ATCC 17748 | VEIDISOL_RS04770 | WP_005386324.1 | Peptidase U32 |
| Veillonella dispar ATCC 17748 | VEIDISOL_RS04185 | WP_005386127.1 | Peptidase U32 |
| Veillonella parvula DSM 2008 [40] | VPAR_RS05935 | WP_012864557.1 | Peptidase U32 |
| Veillonella parvula DSM 2008 [40] | VPAR_RS05390 | WP_012864475.1 | Peptidase U32 |
The microorganism associated with each gene annotation is indicated in the first column along with the corresponding reference when available. Taxonomical and protein assignments were identified in the metatranscriptome analysis of root biofilms.
Overall, bacterial collagenolytic proteases showed low levels of expression. The higher proportion of reads assigned to the bacterial collagenolytic proteases was 0.1% of total reads (RC_7). The other samples had an average of proportion of reads assigned to the bacterial collagenolytic proteases of 0.04% for SRS and 0.05% for the RC group, and no statistically significant differences were found (t test; p = 0.2) (Figure 1(a,b)). However, the number of collagenase genes with no expression (number of reads = 0) was SRS = 73.1 ± 9.6 (36.4%) and RC = 109.1 ± 23.7 (54.3%) (t test; p = 0.000). Thus, in spite of similar number of reads in RC and SRS, the number of genes encoding collagenases in RC was lower than in SRS.
Figure 1.
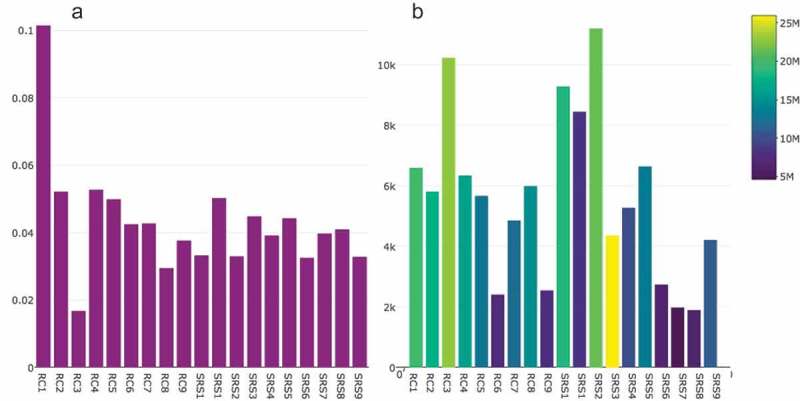
Bacterial collagenolytic proteases present in samples from sound root surfaces (SRS) and root caries (RC). (a) Proportion (%) of bacterial collagenolytic proteases based on the total read count per sample; (b) Number of reads mapped to bacterial collagenolytic protease genes (yellow = sample with more total reads per sample; blue = sample with less total reads per sample).
The heatmap showing the distances between the samples (n = 19) is represented in Figure 2. It takes into account the level of expression of the genes that code bacterial collagenolytic proteases within the sample for each group. There was less sample-to-sample variation between the SRS samples than the RC samples (RC_8, RC_D and RC_E differ from the other RC samples). The diversity of gene expression patterns in the RC samples could represent differences in the lesion characteristics, such as caries stages and lesion sizes.
Figure 2.
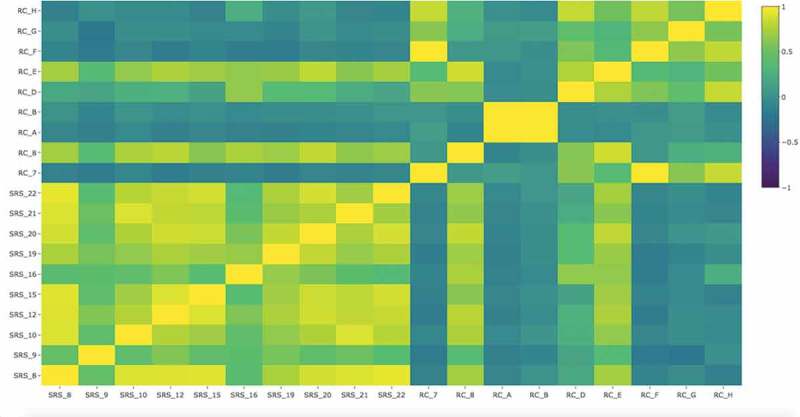
Heatmap showing the distances between the samples as calculated from the normalized count data of the gene expression of bacterial collagenolytic proteases. RC = root caries samples; SRS = sound root surfaces samples.
Figure 3 shows the median expression value of collagenolytic proteases in RC lesions, i.e. the median of the normalized read numbers. Eight collagenolytic proteases had a median of expression value higher than 100, including those from S. mutans, Veillonella parvula, V. dispar, Leptotrichia buccalis, Olsenella uli, and Scardovia inopinata. It is important to point out that in two RC samples, S. inopinata had the highest collagenolytic protease expression value (RC_A = 14,838.83 and RC_B = 3,305.65), although the median was lower than other species. Three collagenolytic proteases had expression values higher than 200, meaning that these were very highly expressed in RC: SMU_761 and SMU_759 from S. mutans and RS05935 from V. parvula. S. mutans possessed collagenolytic proteases with the highest gene expression in RC, while L. buccalis possessed collagenolytic proteases with the highest gene expression in SRS.
Figure 3.
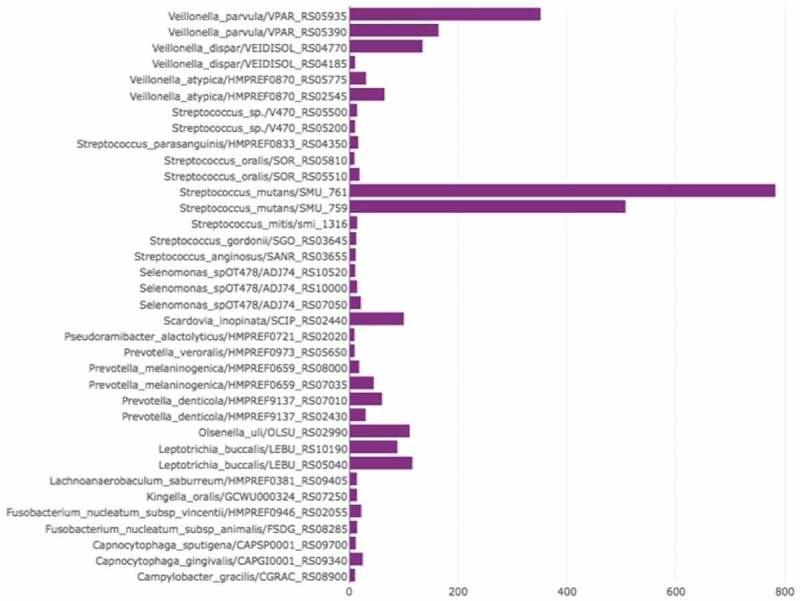
Gene expression level (median of expression value) of putative bacterial collagenolytic proteases (presented as ‘bacterial species name/gene locus tag’) in root caries. Only genes that had gene expression level >10 are displayed.
Using a very high cut-off (FDR <10− 3) for considering differential expression between sound and carious biofilms, we observed 42 bacterial collagenolytic proteases with significant differential expression: 24 were overexpressed in SRS and 18 in RC (Figure 4). P. alactolyticus [HMPREF0721_RS02020], S. inopinata JCM 12537 [SCIP_RS02440], P. alactolyticus [HMPREF0721_RS04640], and O. uli DSM7084 [OLSU_RS02990] were the organisms with highly overexpressed proteases in RC (Log2FC>8), but no expression in SRS.
Figure 4.
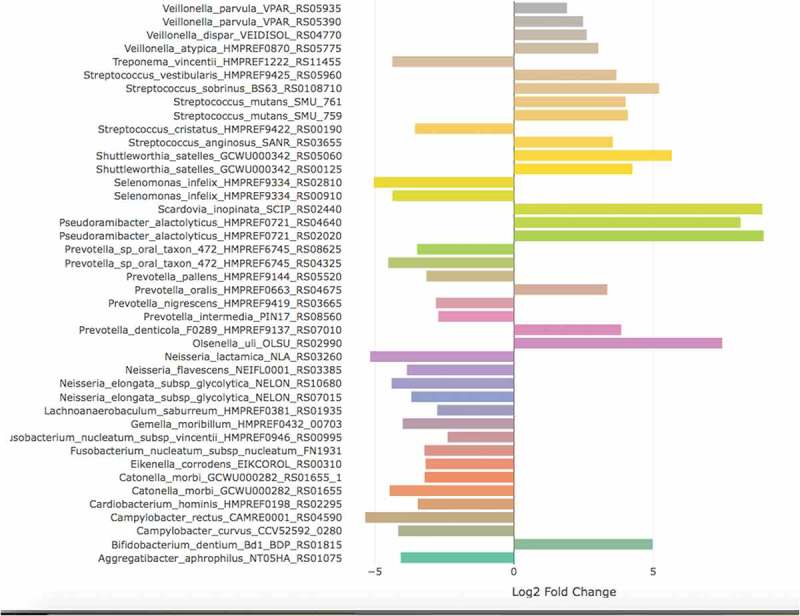
Genes with significant differential expression coding for bacterial collagenolytic proteases (presented as ‘bacterial species name/gene locus tag’) in the metatranscriptome analysis of root biofilms. Positive log2FoldChange means up-regulated genes in root caries, while negative log2FoldChange means up-regulated in sound root surfaces.
Discussion
The current understanding of the microbial functions in RC and dentine caries remains limited compared with enamel caries. In a recent review of caries ecological hypotheses, it was proposed that bacteria play a role in the degradation of the organic components of teeth [3]. Although a lot of bacteria are found to secrete collagenolytic proteases, their roles and the mechanisms involved in cariogenic processes are still largely unknown [8]. This is the first study showing bacterial collagenolytic proteases gene expression within the metatranscriptome of clinical dental biofilms with and without RC. Our findings show that a few species were responsible for high expression of genes that code for bacterial collagenolytic proteases in RC, namely S. mutans, V. parvula, V. dispar, and S. inopinata.
The progression of caries lesions involves the degradation of the collagen matrix in the root hard tissues. The collagen protein family is characterized by the presence of the proline-rich tripeptide ‘Gly-X-Y’, forming a triple helix of polypeptide chains in which the glycine residue is positioned in the centre [41]. Collagen type I, the most common in dentine, has a heterotrimer structure. The collagen structure contributes to the molecular stabilization and mechanical properties of dentine. Only a specific group of proteases, the collagenases, are able to degrade collagen. The triple-helix is interrupted in its internal structure by digesting the triple-helix three-quarters of the way from the terminal amino group ‘Gly-Leu’ bond. This may cause intramolecular flexibility and allow specific proteolytic cleavage [41]. Bacterial collagenolytic proteases include some metalloproteases of the M9 family and some serine proteases. These are distributed in the S1, S8, and S53 families and also some members of the U32 family, mainly from pathogenic bacteria [8]. In this study, protease PrtC was detected to have a relatively low gene expression levels. Other protease families were not detected in the genomes annotation, and these still remain to be investigated (i.e. the M9, S8, and S53 families).
Dental caries occurs not by continuous demineralization but by alternating demineralization and remineralization. According to a recent theory proposed by Takahashi and Nyvad (2016), the exposed collagen is broken down and the collagen content may be denatured during a second stage of RC. The theory suggests that collagen matrix degradation could only be possible after demineralization because the substrate is not accessible by collagenases in the mineralized tissue. Some endogenous collagenases have been shown to be involved in this process [9,10,42]. MMPs, zinc-dependent endopeptidases, are able to cleave denatured collagen. They function in tissue development and repair and in pathological processes as well [43]. It has been found that bacterial collagenases have no activity during demineralization in an acid environment (pH 4.3) [43,44], and it was shown that collagenase works during the remineralizing phase and predominantly attacks the organic matrix of the root after demineralization [44]. However, collagen degradation products are known to be released from dentine when treated with lactic acid and bacterial collagenase or trypsin [45]. Therefore, acids from bacterial metabolism may render dentinal collagen more susceptible to host and microbial proteases such as those of the U32 family.
It has been reported that S. mutans is not associated with collagen matrix degradation in cavitated RC [46,47]. However, in this study, we detected high expression of genes SMU_761 and SMU_759 (S. mutans UA159). Both genes encode collagenase-like protease, PrtC family (peptidase U32 family) [48]. SMU_761 codes for a 428 aa protein, while SMU_759 encodes a 308 aa protein. S. mutans is widely known as an important aetiological agent of dental caries, due to its involvement in biofilm formation and its aciduricity and acidogenicity. Furthermore, most culture-based studies have shown a strong relationship between RC and these bacteria, which have higher isolation frequencies and/or higher proportions on carious root surfaces [49–53]. Our results suggest that the collagenase activity could also be an important virulence factor of S. mutans in RC. These proteases were also elevated under conditions of glucose excess in another in vitro transcriptome study [54].
Along with S. mutans, two species of Veillonella (V. parvula and V. dispar) showed high collagenase gene expression levels in RC. These species have been implicated in dentinal caries due to their overexpressed functions in caries lesions, inferring a role in disease [18]. Other species such as P. alactolyticus, S. inopinata, and O. uli had high differential expression in RC when compared to SRS. These species have been included in the complex microbial community of coronal caries [15] and RC [53,55–58], but their roles and functions have been underexplored.
A higher level of gene expression of some bacterial collagenases was observed in samples from the control group of this study (supragingival biofilm – SRS). Periodontopathogens, such as Prevotella intermedia, showed high differential expression in SRS. The SRS group included patients in preventive periodic maintenance for periodontal disease: the U32 proteases explored here have been previously related to periodontal disease [16]. So this result could be linked to collagen degradation of periodontal tissues.
It is important to acknowledge that we cannot state that there is activity of bacterial collagenolytic proteases in the degradation of dentine because our data are based on gene expression and the enzymes could be inactive in vivo. It is also important to note that other organisms not included as reference genomes in this analysis could be expressing collagenases, as the analysis presented here relies on the current reference databases and other not yet identified collagenases (for example, those currently identified as hypothetical proteins) may play an important role in collagen degradation. This work represents a preliminary screening of transcripts coding for collagenases using clinical data and the validation is being planned in further investigations. However, it is important to point out that the level of protease transcripts observed in this study may indicate the importance of this function within the RC biofilm communities, considering that the transcription of irrelevant genes would be a waste of energy to the microorganisms.
The results suggest that the U32 proteases could be related to RC lesions (carious dentine). The contribution of some species in dentine degradation should be further investigated, such as S. mutans, V. parvula, and V. dispar (high gene expression level in RC), as well as P. alactolyticus, S. inopinata, and O. uli (high differential expression in RC when compared to SRS). Our results provide novel insights into the collagenase activity of some bacterial species in RC. These studies lay the foundations for further investigations involving the use of proteomic tools, to better understand the aetiology of RC, and microbial metabolic activities leading to disease progression. These proteases may have potential for future biotechnological and medical applications serving as targets for the development of therapeutic agents.
Acknowledgements
This study was developed in partnership with the University of Leeds and the Federal University of Rio Grande do Sul (UFRGS). Financial support was provided by the Leeds Hospital Charitable Foundation (R&D/PP/12011), the Dunhill Medical Trust (R245/0212), the Brazilian National Counsel of Technological and Scientific Development (CNPQ) (process no. 482504/2013-7), the Coordination for the Improvement of Higher Level Education (CAPES) (process no 18097-12-0), and the Rio Grande do Sul State Foundation for Research Support (FAPERGS) (process no. 001/2013 PQG). We gratefully thank A. Tugnait, V. Yorke, and J. Rowbotham (School of Dentistry, University of Leeds) for their help in sample collection.
Funding Statement
This work was supported by the Dunhill Medical Trust [R245/0212]; Brazilian National Counsel of Technological and Scientific Development [482504/2013-7]; The Leeds Hospital Charitable Foundation [1170369]; Coordination for the Improvement of Higher Level Education [18097-12-0]; and Rio Grande do Sul State Foundation for Research Support [001/2013 PQG].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1]. Bosshardt DD, Selvig KA.. Dental cementum: the dynamic tissue covering of the root. Periodontol 2000. 1997;13: 41–11. PubMed PMID: 9567923. [DOI] [PubMed] [Google Scholar]
- [2]. Goldberg M, Kulkarni AB, Young M, et al. Dentin: structure, composition and mineralization. Front Biosci (Elite Ed). 2011;3:711–735, PubMed PMID: 21196346; PubMed Central PMCID: PMCPMC3360947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Takahashi N, Nyvad B. Ecological hypothesis of dentin and root caries. Caries Res. 2016;50(4):422–431. PubMed PMID: 27458979 [DOI] [PubMed] [Google Scholar]
- [4]. Schupbach P, Guggenheim B, Lutz F. Human root caries: histopathology of advanced lesions. Caries Res. 1990;24(3): 145–158. PubMed PMID: 2364398. [DOI] [PubMed] [Google Scholar]
- [5]. Parolo CC, Maltz M. Microbial contamination of noncavitated caries lesions: a scanning electron microscopic study. Caries Res. 2006;40(6):536–541. 95654 [pii]. [DOI] [PubMed] [Google Scholar]
- [6]. Marsh PD. Microbiologic aspects of dental plaque and dental caries. Dent Clin North Am. 1999;43(4): 599-614, v-vi PubMed PMID: 10553246; eng. [PubMed] [Google Scholar]
- [7]. Takahashi N. Oral microbiome metabolism: from “who are they?” to “what are they doing? J Dent Res. 2015;94 12:1628–1637. PubMed PMID: 26377570. [DOI] [PubMed] [Google Scholar]
- [8]. Zhang YZ, Ran LY, Li CY, et al. Diversity, structures, and collagen-degrading mechanisms of bacterial collagenolytic proteases. Appl Environ Microbiol. 2015. September;81(18):6098–6107. PubMed PMID: 26150451; PubMed Central PMCID: PMCPMC4542243. eng DOI: 10.1128/AEM.00883-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Tjäderhane L, Buzalaf MA, Carrilho M, et al. Matrix metalloproteinases and other matrix proteinases in relation to cardiology: the era of ‘dentin degradomics’. Caries Res. 2015;49(3):193–208. PubMed PMID: 25661522; eng. [DOI] [PubMed] [Google Scholar]
- [10]. Toledano M, Nieto-Aguilar R, Osorio R, et al. Differential expression of matrix metalloproteinase-2 in human coronal and radicular sound and carious dentine. J Dent. 2010;38(8):635–640. PubMed PMID: 20452393; eng. [DOI] [PubMed] [Google Scholar]
- [11]. Boushell LW, Kaku M, Mochida Y, et al. Distribution and relative activity of matrix metalloproteinase-2 in human coronal dentin. Int J Oral Sci. 2011;3(4):192–199. PubMed PMID: 22010577; PubMed Central PMCID: PMC3469976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Mazzoni A, Pashley DH, Tay FR, et al. Immunohistochemical identification of MMP-2 and MMP-9 in human dentin: correlative FEI-SEM/TEM analysis. J Biomed Mat Res Part A. 2009;88(3):697–703. PubMed PMID: 18335530. [DOI] [PubMed] [Google Scholar]
- [13]. Sulkala M, Larmas M, Sorsa T, et al. The localization of matrix metalloproteinase-20 (MMP-20, enamelysin) in mature human teeth. J Dent Res. 2002;81(9):603–607. PubMed PMID: 12202640. [DOI] [PubMed] [Google Scholar]
- [14]. Simón-Soro A, Belda-Ferre P, Cabrera-Rubio R, et al. A tissue-dependent hypothesis of dental caries. Caries Res. 2013;47(6):591–600. PubMed PMID: 24080530; eng. [DOI] [PubMed] [Google Scholar]
- [15]. Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, et al. The oral metagenome in health and disease. Isme J. 2012;6(1):46–56. PubMed PMID: 21716308; PubMed Central PMCID: PMCPMC3246241. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Kato T, Takahashi N, Kuramitsu HK. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J Bacteriol. 1992;174(12): 3889–3895. PubMed PMID: 1317840; PubMed Central PMCID: PMCPMC206096. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Dame-Teixeira N, Parolo CCF, Maltz M, et al. Actinomyces spp. gene expression in root caries lesions. J Oral Microbiol. 2016;8:32383 PubMed PMID: MEDLINE:27640531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Do T, Sheehy EC, Mulli T, et al. Transcriptomic analysis of three Veillonella spp. present in carious dentine and in the saliva of caries-free individuals. Front Cell Infect Microbiol. 2015;5 DOI: 10.3389/fcimb.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106 PubMed PMID: 20979621; PubMed Central PMCID: PMCPMC3218662. eng [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Sievert C, Parmer C, Hocking T, et al. Plotly:create interactive web graphics via ‘plotly.js’. https://plot.ly/r,https://cpsievert.github.io/plotly_book/, https://github.com/ropensci/plotly.
- [21]. Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014;15:550 DOI: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed]
- [22]. Zeng L, Choi SC, Danko CG, et al. Gene regulation by CcpA and catabolite repression explored by RNA-Seq in Streptococcus mutans . PLoS One. 2013;8(3):e60465 PubMed PMID: 23555977; PubMed Central PMCID: PMCPMC3610829. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Di Bonaventura MP, DeSalle R, Pop M, et al. Complete genome sequence of Aggregatibacter (Haemophilus) aphrophilus NJ8700. J Bacteriol. 2009;191(14):4693–4694. PubMed PMID: 19447908; PubMed Central PMCID: PMCPMC2704734. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. O’Connell Motherway M, Zomer A, Leahy SC, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A. 2011;108(27):11217–11222. PubMed PMID: 21690406; PubMed Central PMCID: PMCPMC3131351. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Ivanova N, Gronow S, Lapidus A, et al. Complete genome sequence of Leptotrichia buccalis type strain (C-1013-b). Stand Genomic Sci. 2009;1(2):126–132. PubMed PMID: 21304648; PubMed Central PMCID: PMCPMC3035221. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Veyrier FJ, Biais N, Morales P, et al. Common cell shape evolution of two nasopharyngeal pathogens. PLoS Genet. 2015;11(7):e1005338 PubMed PMID: 26162030; PubMed Central PMCID: PMCPMC4498754. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Bennett JS, Bentley SD, Vernikos GS, et al. Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics. 2010;11:652 PubMed PMID: 21092259; PubMed Central PMCID: PMCPMC3091772. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Göker M, Held B, Lucas S, et al. Complete genome sequence of Olsenella uli type strain (VPI D76D-27C). Stand Genomic Sci. 2010;3(1):76–84. PubMed PMID: 21304694; PubMed Central PMCID: PMCPMC3035265. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Naito M, Hirakawa H, Yamashita A, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis . DNA Res. 2008;15(4):215–225. PubMed PMID: 18524787; PubMed Central PMCID: PMCPMC2575886. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Oshima K, Hayashi J, Toh H, et al. Complete Genome sequence of Scardovia inopinata JCM 12537T, isolated from human dental caries. Genome Announc. 2015;3(3). PubMed PMID: 25977411; PubMed Central PMCID: PMCPMC4432351. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Olson AB, Kent H, Sibley CD, et al. Phylogenetic relationship and virulence inference of Streptococcus anginosus group: curated annotation and whole-genome comparative analysis support distinct species designation. BMC Genomics. 2013;14:895 PubMed PMID: 24341328; PubMed Central PMCID: PMCPMC3897883. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Vickerman MM, Iobst S, Jesionowski AM, et al. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol. 2007. November;189(21):7799–7807. PubMed PMID: 17720781; PubMed Central PMCID: PMCPMC2168715. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Denapaite D, Brückner R, Nuhn M, et al. The genome of Streptococcus mitis B6 – what is a commensal? PLoS One. 2010;5(2):e9426 PubMed PMID: 20195536; PubMed Central PMCID: PMCPMC2828477. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Tong H, Shang N, Liu L, et al. Complete genome sequence of an oral commensal, Streptococcus oligofermentans strain AS 1.3089. Genome Announc. 2013;1(3). PubMed PMID: 23788543; PubMed Central PMCID: PMCPMC3707592. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Reichmann P, Nuhn M, Denapaite D, et al. Genome of Streptococcus oralis strain Uo5. J Bacteriol. 2011;193(11):2888–2889. PubMed PMID: 21460080; PubMed Central PMCID: PMCPMC3133139. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Xu P, Alves JM, Kitten T, et al. Genome of the opportunistic pathogen Streptococcus sanguinis . J Bacteriol. 2007;189(8):3166–3175. PubMed PMID: 17277061; PubMed Central PMCID: PMCPMC1855836. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Bolotin A, Quinquis B, Renault P, et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus . Nat Biotechnol. 2004;22(12):1554–1558. PubMed PMID: 15543133; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Seshadri R, Myers GS, Tettelin H, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101(15):5646–5651. PubMed PMID: 15064399; PubMed Central PMCID: PMCPMC397461. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Lacap-Bugler DC, Jiang J, Huo YB, et al. Complete genome sequence of the oral spirochete bacterium Treponema putidum strain OMZ 758T (ATCC 700334T). Genome Announc. 2014;2(5). PubMed PMID: 25342686; PubMed Central PMCID: PMCPMC4208330. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Gronow S, Welnitz S, Lapidus A, et al. Complete genome sequence of Veillonella parvula type strain (Te3). Stand Genomic Sci. 2010;2(1):57–65. PubMed PMID: 21304678; PubMed Central PMCID: PMCPMC3035260. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Gelse K, Pöschl E, Aigner T. Collagens – structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55(12): 1531–1546.. [DOI] [PubMed] [Google Scholar]
- [42]. Tjäderhane L, Larmas M. A high sucrose diet decreases the mechanical strength of bones in growing rats. J Nutr. 1998;128(10): 1807–1810.. [DOI] [PubMed] [Google Scholar]
- [43]. Ricard-Blum S. The collagen family. PubMed PMID: 21421911; PubMed Central PMCID: PMCPMC3003457. eng Cold Spring Harb Perspect Biol. 2011;3 1:a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Kawasaki K, Featherstone JD. Effects of collagenase on root demineralization. J Dent Res. 1997;76(1): 588–595. PubMed PMID: 9042082; eng. [DOI] [PubMed] [Google Scholar]
- [45]. Dung SZ, Gregory RL, Li Y, et al. Effect of lactic acid and proteolytic enzymes on the release of organic matrix components from human root dentin. Caries Res. 1995;29(6):483–489. PubMed PMID: 8556753; eng. [DOI] [PubMed] [Google Scholar]
- [46]. Dung TZ, Liu AH. Molecular pathogenesis of root dentin caries. Oral Dis. 1999;5(2): 92–99. PubMed PMID: 10522203; eng. [DOI] [PubMed] [Google Scholar]
- [47]. Argimón S, Caufield PW. Distribution of putative virulence genes in Streptococcus mutans strains does not correlate with caries experience. J Clin Microbiol. 2011;49(3:984–992. PubMed PMID: 21209168; PubMed Central PMCID: PMCPMC3067729. eng [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Ajdić D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99(22):14434–14439. PubMed PMID: 12397186; PubMed Central PMCID: PMCPMC137901. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Van Houte J, Jordan HV, Laraway R, et al. Association of the microbial flora of dental plaque and saliva with human root-surface caries. J Dent Res. 1990;69(8):1463–1468. PubMed PMID: 2384622; eng. [DOI] [PubMed] [Google Scholar]
- [50]. Syed SA, Loesche WJ, Pape HL, et al. Predominant cultivable flora isolated from human root surface caries plaque. Infect Immun. 1975;11(4):727–731. PubMed PMID: 1091550; PubMed Central PMCID: PMCPMC415128. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Ravald N, Birkhed D. Factors associated with active and inactive root caries in patients with periodontal disease. Caries Res. 1991;25(5): 377–384. PubMed PMID: 1747889; eng. [DOI] [PubMed] [Google Scholar]
- [52]. Beighton D, Lynch E, Heath MR. A microbiological study of primary root-caries lesions with different treatment needs. J Dent Res. 1993;72(3): 623–629. PubMed PMID: 8450122; eng. [DOI] [PubMed] [Google Scholar]
- [53]. Mantzourani M, Fenlon M, Beighton D. Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol Immunol. 2009;24(1):32–37. PubMed PMID: 19121067; eng. [DOI] [PubMed] [Google Scholar]
- [54]. Moye ZD, Zeng L, Burne RA. Modification of gene expression and virulence traits in Streptococcus mutans in response to carbohydrate availability. Appl Environ Microbiol. 2014;80(3):972–985.PubMed PMID: 24271168; PubMed Central PMCID: PMCPMC3911228. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Bizhang M, Ellerbrock B, Preza D, et al. Detection of nine microorganisms from the initial carious root lesions using a TaqMan-based real-time PCR. Oral Dis. 2011;17(7):642–652. PubMed PMID: 21605286; eng. [DOI] [PubMed] [Google Scholar]
- [56]. Preza D, Olsen I, Aas JA, et al. Bacterial profiles of root caries in elderly patients. J Clin Microbiol. 2008;46(6):2015–2021. PubMed PMID: 18385433; PubMed Central PMCID: PMCPMC2446847. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Chen L, Qin B, Du M, et al. Extensive description and comparison of human supra-gingival microbiome in root caries and health. PLoS One. 2015;10(2):e0117064 PubMed PMID: 25658087; PubMed Central PMCID: PMCPMC4319720. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Do T, Damé-Teixeira N, Naginyte M, et al. Root surface biofilms and caries. Monogr Oral Sci. 2017;26:26–34. PubMed PMID: 29050018; eng. [DOI] [PubMed] [Google Scholar]


