Abstract
The bacterial flagellum is a supramolecular motility machine consisting of the basal body as a rotary motor, the hook as a universal joint, and the filament as a helical propeller. Intact structures of the bacterial flagella have been observed for different bacterial species by electron cryotomography and subtomogram averaging. The core structures of the basal body consisting of the C ring, the MS ring, the rod and the protein export apparatus, and their organization are well conserved, but novel and divergent structures have also been visualized to surround the conserved structure of the basal body. This suggests that the flagellar motors have adapted to function in various environments where bacteria live and survive. In this review, we will summarize our current findings on the divergent structures of the bacterial flagellar motor.
Keywords: bacterial flagellum, electron cryotomography, motility, rotary motor, structural diversity
Significance.
The bacterial flagellum is a supramolecular motility machine consisting of a rotary motor, a universal joint and a helical propeller. Component proteins of the flagellar motor are highly conserved among bacterial species. Recent structural analyses of intact flagellar motors derived from different bacterial species by electron cryotomography and subtomogram averaging have shown that novel and divergent structures surround the conserved structure in different species, suggesting that the flagellar motors have adapted to function in their living environment. In this review article, we will describe the conserved structure of the flagellar motor and its structural diversity.
A huge variety of bacteria live on our planet. Bacteria adapt and evolve in their living environment. Many bacteria swim in liquid environments and move on solid surfaces towards more favorable conditions and escape from undesirable ones for their survival. The bacterial flagellum is a supramolecular motility machine consisting of about 30 different proteins. These component proteins are highly conserved among bacterial species. The bacterial flagellum can be divided into at least three parts: the basal body, the hook and the filament. The basal body is embedded within the cell membranes and acts as a rotary motor. The hook and filament extend outwards in the cell exterior. The filament works as a helical propeller. The hook connects the basal body and filament and functions as a universal joint to smoothly transmit torque produced by the motor to the filament [1–4].
The flagellar basal body of Escherichia coli and Salmonella enterica serovar Typhimurium (hereafter referred to as Salmonella) consists of the C ring (FliG, FliM, FliN), the MS ring (FliF), the P ring (FlgI), the L ring (FlgH), the rod (FliE, FlgB, FlgC, FlgF, FlgG) and the protein export apparatus (FlhA, FlhB, FliH, FliI, FliJ, FliP, FliQ, FliR). In addition, a dozen stator units (MotA, MotB) surround the basal body and couple the proton flow through a proton channel with torque generation. These structures and their organization are highly conserved among bacterial species.
Electron cryotomography (ECT) is an imaging technique that directly provides 3D structure of cells and molecular complexes in their cellular environment at nanometer resolution. To obtain the structures of specimens, the specimen grid is tilted incrementally around an axis perpendicular to the electron beam, e.g. from −60° to +60° with 2° increments, and images are taken at each tilt angle. The images of a tilt series are aligned and are back-projected to generate a 3D image (tomogram) of the specimen. However, each tilt image is recorded at low electron doses to minimize radiation damage of biological samples. This leads to extremely low signal to noise ratio in each tomogram, making it necessary to align and average hundreds of particles images to increase the signal to noise ratio and to reconstruct the 3D structure at high resolution. Using this method, in situ structural analyses of the flagellar motors derived from different bacterial species have shown novel and divergent structures around the conserved core structure in different species (Fig. 1) [5,6].
Figure 1.
Structural differences in the flagellar basal body among bacterial species. in situ structures of the flagellar basal bodies are visualized by electron cryotomography and subtomogram averaging. The central section maps of the basal bodies of Salmonella enterica (EMDataBank ID: EMD-2521), Escherichia coli (EMDataBank ID: EMD-5311), Vibrio fischeri (EMDataBank ID: EMD-3155), Helicobacter pylori (EMDataBank ID: EMD-8459), Campylobacter jejuni (EMDataBank ID: EMD-3150), Acetonema longum (EMDataBank ID: EMD-5297), Caulobacter crescentus (EMDataBank ID: EMD-5312), Hyphomonas neptunium (EMDataBank ID: EMD-5313), Hylemonella gracilis (EMDataBank ID: EMD-5309), Leptospira interrogans (EMDataBank ID: EMD-5913) and Borrelia burgdorferi (EMDataBank ID: EMD-5627).
In this review article, we describe the conserved structure of the flagellar basal body and its structural diversity.
Conserved structure of the flagellar basal body
Intact structures of the bacterial flagella derived from different bacterial species have been visualized by ECT (Fig. 1). The MS ring, the C ring, the rod and the protein export apparatus show similar structures to those of the Salmonella flagellar basal body (Figs. 1 and 2) [7,8]. The transmembrane protein, FliF, self-assembles into the MS ring in the cytoplasmic membrane [9]. Three cytoplasmic proteins, namely FliG, FliM and FliN, form the C ring on the cytoplasmic face of the MS ring [10]. The MS and C rings together act as a rotor of the flagellar motor. The rod is composed of three distal rod proteins, FlgB, FlgC, FlgF and the distal rod protein, FlgG. The rod is a rigid, tubular structure composed of 11 protofilaments and acts as a drive shaft [11,12]. A basal body protein, FliE, which is highly conserved among bacterial species, assembles at the periplasmic surface of the MS ring to form a MS ring/rod junction zone to presumably overcome the mismatch between the rotational symmetry of the MS ring and the helical symmetry of the rod [13].
Figure 2.
Cartoons of the flagellar motor derived from (a) Salmonella, (b) B. subtilis, (c) V. alginolyticus, (d) C. jejuni and (e) B. burgdorferi. The schematic diagrams of the Salmonella, V. alginolytics, C. jejuni and B. burgdorfer flagellar motors are shown based on their in situ structures whereas the B. subtilis motor is drawn based on purified basal body structure with some speculations. The common architectures are indicated by following colors: MS ring, green; C ring, light green; export apparatus, orange; rod, cyan; hook, navy; LP ring, purple; stator, wine red. The position of the stator unit in the Salmonella and B. subtilis motors remains unclear. The additional structures are showed in following colors: H-ring, yellow; T-ring, brown; O-ring, dark gray; basal disk, blue; median disk, light blue; proximal disk, red; P-collar, pink. IM, inner membrane; PG, peptidoglycan layer; OM, outer membrane.
The protein export apparatus is composed of a transmembrane export gate complex made of FlhA, FlhB, FliP, FliQ, and FliR, and a cytoplasmic ATPase ring complex consisting of FliH, FliI, and FliJ [14–16]. The transmembrane protein, FliO, is required for efficient assembly of the core structure of the export gate complex made of FliP, FliQ and FliR although it is not essential for flagellar protein export [16,17]. The cytoplasmic ATPase ring complex is associated with the basal body through interactions of the extreme N-terminal region of FliH with FlhA and FliN [18–22]. The protein export apparatus utilizes ATP hydrolysis by the FliI ATPase and proton motive force across the cytoplasmic membrane as the energy sources to transport flagellar component proteins from the cytoplasm to the distal end of the growing structure where their assembly occurs [23–25].
The stator unit is composed of two transmembrane proteins, commonly referred to as MotA and MotB in the H+-driven motor, PomA and PomB in the Na+-driven motor of marine Vibrio and MotP and MotS in the Na+-driven motor of alkalophilic Bacillus species. The stator unit acts as an ion channel to couple the ion flow with torque generation [26]. The MotAB and PomAB complexes are anchored to the peptidoglycan layer through a highly conserved peptidoglycan binding motif in the C-terminal periplasmic domains of MotB and PomB, respectively, thereby allowing the MotAB and PomAB complexes to act as an active H+-type and Na+-type stator units in the motor, respectively [27,28]. A dozen MotAB stator units surround the rotor in the E. coli and Salmonella flagellar motors. However, these stator units have not been visualized by ECT because they are not permanently fixed in place around the rotor (Fig. 1). Interestingly, the flagellar motor itself regulates the number of active stator units around the rotor in response to changes in external load and ion motive force across the cytoplasmic membrane [29–34].
LP ring complex of the flagellar basal body
FlgH and FlgI assemble around the rod in the outer membrane and the peptidoglycan layer, respectively, to form the LP ring complex, which acts as a molecular bushing [35]. In contrast to component proteins of the rod, hook and filament, FlgH and FlgI are synthesized as precursor forms with their secretion signal sequence in the cytoplasm, translocated across the cytoplasmic membrane by the Sec translocon, and subjected to signal peptide cleavage [36,37]. The LP ring complex is missing in gram-positive bacteria such as Bacillus subtilis (Fig. 2b) [38], suggesting that much thicker peptidoglycan layer can act as a bushing of the Bacillus flagellar motor instead of the LP ring complex.
The flagellar motor of marine Vibrio
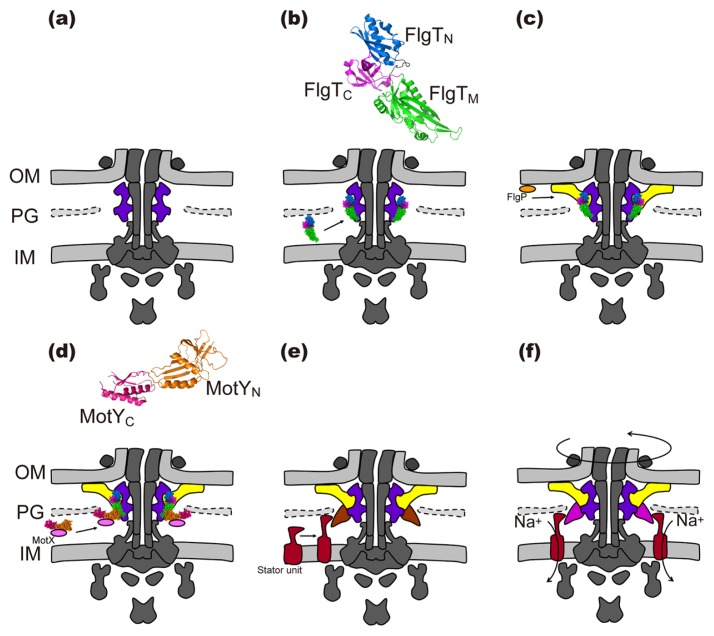
The flagellar motor of Vibrio alginolyticus can spin at the maximum speed of 1,700 revolutions per second compared to the Salmonella motor, which rotates at 300 revolutions per second [39]. The marine Vibrio motor has two additional H and T ring structures surrounding the LP ring complex [40–43] (Fig. 2c). The H and T rings are responsible for stable anchoring of the flagellum to the cell for such a high-speed rotation [41,44]. FlgT directly binds to the basal body and promotes the formation of the T and H rings around the LP ring complex (Fig. 3) [41]. FlgT consists of three domains: FlgTN, FlgTM and FlgTC (Fig. 3b) [42]. FlgTN forms an outer rim of the H ring structure made of FlgP in the outer membrane (Fig. 3c) [42,45]. Inter-subunit interactions between FlgTC domains stabilize the FlgT ring structure [42]. FlgTM is crucial for H ring formation as well as T ring formation. MotX and MotY together form the T ring with the 13-fold rotational symmetry [40,45,46]. MotY directly binds to FlgTM, thereby inducing the formation of the T ring structure beneath the P ring (Fig. 3d) [40,41]. MotY consists of two domains: MotYN and MotYC (Fig. 3d) [47]. MotYC associates with the peptidoglycan layer through its peptidoglycan binding motif [47]. MotYN is responsible not only for the interaction with FlgTM but also for the interaction with MotX [40,47]. In contrast to the MotAB stator unit in the E. coli and Salmonella motors, clearly defined electron density corresponding to the PomAB stator unit has been observed by ECT [45,46]. MotX allows thirteen Na+-type PomAB stator units to assemble into the basal body through an interaction between PomB and MotX (Fig. 3d). Since the Na+-type PomAB stator complex cannot assemble into the Vibrio motor in the absence of the T ring, the T ring is responsible not only for efficient stator assembly around the rotor but also for strongly anchoring the PomAB stator to the motor, allowing the motor to spin at much higher speed up to 1,700 revolutions per second (Fig. 3e) [40,45].
Figure 3.
Assembly process of the marine Vibrio motor. (a) The common architecture is formed. (b) FlgT (PDB ID: 3W1E) assembles around the LP ring. (c) FlgP, which is shown by an orange ellipse, associates with FlgTN to form the H ring in the outer membrane (OM). (d) MotY (PDB ID: 2ZF8) binds to FlgTM and the PG layer, and then MotX, which is shown by a pink ellipse, interacts with MotY to form the T ring. (e) The stator units (wine red) assemble around the basal body through interactions of PomB with the PG layer and MotX. (f) The flagellar motor utilizes Na+ as a coupling ion to drive motor rotation at the maximum speed of 1,700 revolutions per second.
The Vibrio flagellum is wrapped by a sheath, which is derived from the outer membrane. The outer membrane surrounds the hook and filament, thereby causing a 90° bend of the outer membrane. Recently, a new ring structure, named the O ring, has been identified around the basal body [46]. The O ring is located at the 90° bend end of the outer membrane (Fig. 2c). Since unsheathed Vibrio flagellum does not possess the O ring structure, the O ring may contribute to remodeling of the outer membrane for the sheath formation surrounding the hook and filament [46]. Interestingly, the LPHT quaternary ring complex shows a sliding motion along the axial rod structure, suggesting that the friction between the rod and the inner surface of the LP ring complex is extremely small [46].
The flagellar motor of Campylobacter jejuni
Many of pathogenic ɛ-proteobacteria prefer to live in digestive tracts and hence can powerfully swim through mucous layer of the digestive tracts. In fact, Helicobacter pylori cells can keep swimming even under very high viscosity condition where E. coli cells cannot [48]. Torque generated by the flagellar motor of H. pylori has been estimated to be 3,600 pN nm, which is about two times higher than that by the E. coli motor [49]. This suggests that the flagellar motor of ɛ-proteobacteria has adapted to function in high viscous environments. The flagellar motor derived from Campylobacter and Helicobacter spp. possesses extensive disk structures in the periplasmic space (Fig. 1) [7]. The C. jejuni motor has three distinct disk-like structures: a basal disk beneath the outer membrane, a proximal disk located at a position adjacent to the cytoplasm membrane and a medial disk connecting the proximal and basal disks (Fig. 2d) [45]. Seventeen MotAB stator units have been visualized to surround the basal body [45]. FlgP forms the basal disk structure with the help of FlgQ [50]. Interestingly, there is no FlgT homologue in C. jejuni, suggesting that FlgP itself directly binds to the basal body. The basal disk adopts a funnel shape, and hence the outer membrane is distorted [45]. Lack of the basal disk leads to a loss of the medial and proximal disks, which are formed by PflA and PflB, respectively, suggesting that the basal disk is required for the assembly of the medial and proximal disks [45]. PflA is required for the assembly of PflB into the proximal disk. Since PflB is required for stator assembly around the rotor, these two disks presumably allow seventeen stator units to tightly associate with the motor to generate much larger torque than that of E. coli [45].
The flagellar motor of Spirochete
Spirochetes cause some serious diseases in human and animals, including leptospirosis (Leptospira interrogans), Lyme disease (Borrelia burgdorferi) and syphilis (Treponema pallidum). Their morphology and motility is markedly different from other bacteria. The cell shape is an elongated corkscrew that is suitable for swimming in highly viscous environments. The flagella are elongated between the outer membrane and the peptidoglycan layer and contribute to such a unique morphology of spirochetes. The flagellar motor has a bowl-like structure called “collar” that surrounds the MS ring and the rod (Figs. 1 and 2e). The collar is easily dissociated from the basal body during the purification of the hook-basal body, suggesting that it is not tightly associated with the basal body. Recently, flbB has been identified as a gene responsible for collar assembly [51]. FlbB is a transmembrane protein with a single N-terminal transmembrane helix and is located in the basal part of the collar structure, suggesting that FlbB anchors the collar structure to the cell membrane.
The distal rod length in spirochetes is different from that of other bacteria, such as E. coli and Salmonella. The rod length of B. burgdorferi is ~4 nm whereas the proximal and distal rods of Salmonella is 6.9 nm and 17.7 nm, respectively [12,52,53]. Since the periplasmic filament does not penetrate the outer membrane, such a shorter rod may be long enough to function as a drive shaft of the flagellar motor in B. burgdorferi.
The flagellar motor derived from L. interrogans has both the L and P rings [54] whereas the T. pallidum motor does not [55]. Interestingly, B. burgdorferi possesses only the P ring [56]. Thus, there is a clear difference in existence of the LP ring complex among the spirochetes.
The flagellar motor of marine magnetotactic bacterium MO-1
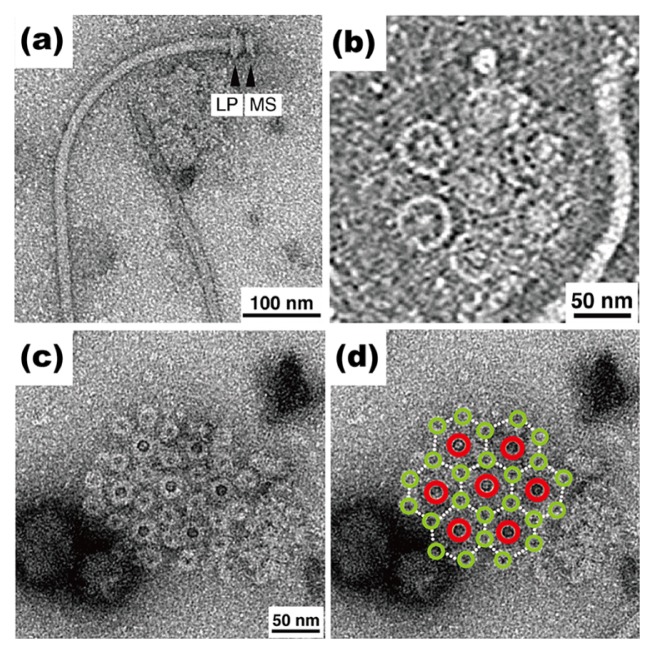
Marine magnetotactic bacterium MO-1 senses the geomagnetic direction by a unique organelle containing nanoscale magnets, namely magnetosome, and hence can orient its cell body along the geomagnetic field line. The swimming speed of MO-1 cells is up to 300 μm/sec, which is about 10-fold faster than those of E. coli and Salmonella cells. The entire architecture of the flagella isolated form the MO-1 cells look similar to that of Salmonella (Fig. 4a). However, the diameter of the LP ring complex is somehow larger than that of the Salmonella one, raising the possibility that additional structures surround the LP ring complex in a way similar to the Vibrio and Campylobacter motors. The MO-1 cell has two highly organized flagellar bundle structures on the cell surface. Each flagellar bundle structure is composed of seven flagella and 24 fibrils in a sheath [57]. The seven flagellar filaments, which are arranged in a hexagonal array (Fig. 4b), are enveloped with 24 fibrils in the sheath (Fig. 4c, d) [57]. The basal bodies of seven flagella and 24 fibrils form intertwined hexagonal array in the cell membranes (Fig. 4d). One unit lattice is located in the center of the hexagonal array, and the other six lattices surround the center unit. Each flagellum is located at the center of each unit lattice, and six fibrils surround each flagellum in the hexagonal array. As a result, the 24 fibrils are packed together with the seven flagella in the sheath, and hence the formation of a much tighter and stronger bundle structure allows for very first swimming of MO-1 cells [57].
Figure 4.
Flagellar structure of magnetotactic bacterium MO-1. (a) Electron micrograph of isolated flagellum with negative staining. The MS ring and the LP ring complex are shown. (b) ECT image of the flagellar basal body around the cytoplasmic membrane. (c) Negatively stained EM image of a detergent-solubilized base platform isolated from MO-1 cells. Seven flagella and twenty four fibrils together form an intertwined hexagonal array. (d) The same image as (c) with large red and small light green circles overlaid on the flagellar and fibril basal bodies, respectively.
Conclusion
Recent structural analyses of the flagellar motors derived from different bacterial species by ECT have revealed a considerable diversity in the flagellar motor structures among bacterial species, suggesting that flagellar motors have adapted to function in the context of phylogenetically diverse bacteria. Functional and structural characterizations of the flagellar motors of different bacteria will provide insights into the evolution and function of the flagellar motor.
Acknowledgements
We gratefully thank Keiichi Namba, Michio Homma and Masahiro Ueda for continuous support and encouragement. Our research is supported in part by JSPS KAKENHI Grant Numbers JP15K14498 and JP15H05593 (to Y. V. M), JP15H02386 (to K. I.) and JP26293097 (to T. M.) and MEXT KAKENHI Grant Numbers JP26115720 and JP15H01335 (to Y. V. M) and JP24117004 and JP15H01640 (to T. M.).
Footnotes
Conflict of Interest
The authors declare no conflict of interests
Author Contributions
H. T., A. K., Y. V. M., K. I. and T. M. wrote the paper.
References
- 1.Pallen MJ, Matzke NJ. From the origin of species to the origin of bacterial flagella. Nat Rev Microbiol. 2006;4:789–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 3.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Terashima H, Kojima S, Homma M. Flagellar motility in bacteria: Structure and function of flagellar motor. Int Rev Cell Mol Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhao X, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry. 2014;53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minamino T, Imada K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015;23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamoto A, Morimoto YV, Miyata T, Minamino T, Hughes KT, Kato T, et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno T, Oosawa K, Aizawa S. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992;227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 10.Khan IH, Reese TS, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 12.Fujii T, Kato T, Hiraoka D, Miyata T, Minamino T, Chevance F, et al. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun. 2017;8:14276. doi: 10.1038/ncomms14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minamino T, Yamaguchi S, Macnab RM. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol. 2000;182:3029–3036. doi: 10.1128/jb.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minamino T, Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamino T, Macnab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 16.Fukumura T, Makino F, Dietsche T, Kinoshita M, Kato T, Wagner S, et al. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 2017;15:e2002281. doi: 10.1371/journal.pbio.2002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabiani FD, Renault TT, Peters B, Dietsche T, Gálvez EJC, Guse A, et al. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol. 2017;15:e2002267. doi: 10.1371/journal.pbio.2002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Pedrajo B, Minamino T, Kihara M, Namba K. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol Microbiol. 2006;60:984–998. doi: 10.1111/j.1365-2958.2006.05149.x. [DOI] [PubMed] [Google Scholar]
- 19.Minamino T, Yoshimura SDJ, Morimoto YV, González-Pedrajo B, Kami-ike N, Namba K. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol Microbiol. 2009;74:1471–1483. doi: 10.1111/j.1365-2958.2009.06946.x. [DOI] [PubMed] [Google Scholar]
- 20.Hara N, Morimoto YV, Kawamoto A, Namba K, Minamino T. Interaction of the extreme N-terminal region of FliH with FlhA is required for efficient bacterial flagellar protein export. J Bacteriol. 2012;194:5353–5360. doi: 10.1128/JB.01028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai F, Morimoto YV, Yoshimura SDJ, Hara N, Kami-ike N, Namba K, et al. Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Sci Rep. 2014;4:6528. doi: 10.1038/srep06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notti RQ, Bhattacharya S, Lilic M, Stebbins CE. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun. 2015;6:7125. doi: 10.1038/ncomms8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 24.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of the flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 25.Minamino T, Morimoto YV, Hara N, Namba K. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat Commun. 2011;2:475. doi: 10.1038/ncomms1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morimoto YV, Minamino T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules. 2014;4:217–234. doi: 10.3390/biom4010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, et al. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol Microbiol. 2009;73:710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhu S, Takao M, Li N, Sakuma M, Nishino Y, Homma M, et al. Conformational change in the periplasmic region of the flagellar stator complex coupled with the assembly around the rotor. Proc Natl Acad Sci USA. 2014;111:13523–13528. doi: 10.1073/pnas.1324201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 30.Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tipping MJ, Steel BC, Delalez NJ, Berry RM, Armitage JP. Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol Microbiol. 2013;87:338–347. doi: 10.1111/mmi.12098. [DOI] [PubMed] [Google Scholar]
- 32.Che YS, Nakamura S, Morimoto YV, Kami-ike N, Namba K, Minamino T. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol Microbiol. 2014;91:175–184. doi: 10.1111/mmi.12453. [DOI] [PubMed] [Google Scholar]
- 33.Castillo DJ, Nakamura S, Morimoto YV, Che YS, Kami-ike N, Kudo S, et al. The C-terminal periplasmic domain of MotB is responsible for load-dependent control of the number of stators of the bacterial flagellar motor. Biophysics. 2013;9:173–181. doi: 10.2142/biophysics.9.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terahara N, Noguchi Y, Nakamura S, Kami-ike N, Ito M, Namba K, et al. Load- and polysaccharide-dependent activation of the Na+-type MotPS stator in the Bacillus subtilis flagellar motor. Sci Rep. 2017;7:46081. doi: 10.1038/srep46081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiba T, Yoshimura H, Namba K. Monolayer crystallization of flagellar L-P rings by sequential addition and depletion of lipid. Science. 1991;252:1544–1546. doi: 10.1126/science.2047860. [DOI] [PubMed] [Google Scholar]
- 36.Homma M, Komeda Y, Iino T, Macnab RM. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987;169:1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones CJ, Homma M, Macnab RM. Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli. J Bacteriol. 1987;169:1489–1492. doi: 10.1128/jb.169.4.1489-1492.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubori T, Okumura M, Kobayashi N, Nakamura D, Iwakura M, Aizawa S. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol Microbiol. 1997;24:399–410. doi: 10.1046/j.1365-2958.1997.3341714.x. [DOI] [PubMed] [Google Scholar]
- 39.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, et al. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 40.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol Microbiol. 2006;62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 41.Terashima H, Koike M, Kojima S, Homma M. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol. 2010;192:5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terashima H, Li N, Sakuma M, Koike M, Kojima S, Homma M, et al. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc Natl Acad Sci USA. 2013;110:6133–6138. doi: 10.1073/pnas.1222655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosogi N, Shigematsu H, Terashima H, Homma M, Nagayama K. Zernike phase contrast cryo-electron tomography of sodium-driven flagellar hook-basal bodies from Vibrio alginolyticus. J Struct Biol. 2011;173:67–76. doi: 10.1016/j.jsb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Martinez RM, Jude BA, Kirn TJ, Skorupski K, Taylor RK. Role of FlgT in anchoring the flagellum of Vibrio cholerae. J Bacteriol. 2010;192:2085–2092. doi: 10.1128/JB.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci USA. 2016;113:E1917–E1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu S, Nishikino T, Hu B, Kojima S, Homma M, Liu J. Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc Natl Acad Sci USA. 2017;114:10966–10971. doi: 10.1073/pnas.1712489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kojima S, Shinohara A, Terashima H, Yakushi T, Sakuma M, Homma M, et al. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc Natl Acad Sci USA. 2008;105:7696–7701. doi: 10.1073/pnas.0800308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hazell SL, Lee A, Brady L, Hennessy W. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153:658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 49.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sommerlad SM, Hendrixson DR. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol. 2007;189:179–186. doi: 10.1128/JB.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moon KH, Zhao X, Manne A, Wang J, Yu Z, Liu J, et al. Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol. 2016;102:336–348. doi: 10.1111/mmi.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2013;110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones CJ, Macnab RM, Okino H, Aizawa S. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 54.Raddi G, Morado DR, Yan J, Haake DA, Yang XF, Liu J. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol. 2012;194:1299–1306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruan J, Kato T, Santini CL, Miyata T, Kawamoto A, Zhang WJ, et al. Architecture of a flagellar apparatus in the fast-swimming magnetotactic bacterium MO-1. Proc Natl Acad Sci USA. 2012;109:20643–20648. doi: 10.1073/pnas.1215274109. [DOI] [PMC free article] [PubMed] [Google Scholar]