Abstract
We previously showed that the chimeric proteins of microbial rhodopsins, such as light-driven proton pump bacteriorhodopsin (BR) and Gloeobacter rhodopsin (GR) that contain cytoplasmic loops of bovine rhodopsin, are able to activate Gt protein upon light absorption. These facts suggest similar protein structural changes in both the light-driven proton pump and animal rhodopsin. Here we report two trials to engineer chimeric rhodopsins, one for the inserted loop, and another for the microbial rhodopsin template. For the former, we successfully activated Gs protein by light through the incorporation of the cytoplasmic loop of β2-adrenergic receptor (β2AR). For the latter, we did not observe any G-protein activation for the light-driven sodium pump from Indibacter alkaliphilus (IndiR2) or a light-driven chloride pump halorhodopsin from Natronomonas pharaonis (NpHR), whereas the light-driven proton pump GR showed light-dependent G-protein activation. This fact suggests that a helix opening motion is common to G protein coupled receptor (GPCR) and GR, but not to IndiR2 and NpHR. Light-induced difference FTIR spectroscopy revealed similar structural changes between WT and the third loop chimera for each light-driven pump. A helical structural perturbation, which was largest for GR, was further enhanced in the chimera. We conclude that similar structural dynamics that occur on the cytoplasmic side of GPCR are needed to design chimeric microbial rhodopsins.
Keywords: microbial rhodopsin, GPCR, G-protein activation, retinal, FTIR
Significance.
Chimeric proteins of a light-driven proton pump GR containing the cytoplasmic loop of β2-adrenergic receptor (β2AR) activate Gs protein by light. In contrast, chimeric proteins of light-driven sodium pump IndiR2 or chloride pump NpHR containing the same loop of β2AR do not activate Gs protein at all. Light-induced difference FTIR spectroscopy showed largest helical structural perturbation for GR, which was further enhanced in the chimera. Similar structural dynamics that occur on the cytoplasmic side of GPCR are needed to design chimeric microbial rhodopsins.
Animal and microbial rhodopsins convert light into signals and energy by employing the photochemical reaction of retinal [1]. Animal rhodopsins contain 11-cis retinal as the chromophore, and photoisomerization from the 11-cis to the all-trans form initiates protein structural changes, leading to activation of the trimeric G protein transducin (Gt) [1–4]. Microbial rhodopsins contain all-trans retinal as the chromophore, and the initiation of protein structural changes, which are caused by the photoisomerization from the alltrans to the 13-cis form, lead to various functions such as light-driven pumps, light-gated channels, photosensors and light-activated enzymes [1,5–9]. There are no sequence homologies between animal and microbial rhodopsins, but both possess similar chromophore (retinal) and protein (7-transmembrane helices) structures.
Microbial rhodopsins have been used as tools in optogenetics, a field of study in which animal behavior is controlled by light [10–12]. In optogenetics, animal brain functions are studied by incorporating microbial rhodopsins, but not animal rhodopsins, into the animal brain. There are two reasons for this. One is the isomeric structure of the chromophore. Whereas 11-cis retinal is not abundant in animal cells, endogenous all-trans retinal is sufficient for optogenetics in animal cells. The second reason is the cyclic behavior of the chromophore in the photoreaction. In animal rhodopsins, isomerized all-trans retinal does not return to the 11-cis form, a process that is termed “photobleaching”. This is not a problem in visual cells because enzymatically isomerized 11-cis retinal is newly supplied, which is not the case in other cells. In contrast, the 13-cis form is thermally reisomerized into the all-trans form, and the spontaneous return leads to the “photocycle” in microbial rhodopsins. This is highly advantageous in optogenetics.
For these reasons, animal rhodopsins have not been actively used in optogenetics. Although Arian et al. engineered ‘optoXRs’ [13], in which a bovine rhodopsin chimera containing the cytoplasmic loop of other G-protein coupled receptors (GPCRs) was used to respond to light, problems with 11-cis retinal and photobleaching limit broad applications. Thus, the optogenetic application of GPCR signaling requires that these two problems be resolved. One approach is to use bistable animal rhodopsins whose photointermediate does not bleach and is thermally stable [14,15]. By photoconverting the intermediate (normally in an active state) into the original state (normally in an inactive state), activation of GPCR signaling is switchable by light. Some bistable rhodopsins can bind a 13-cis retinal, which exists in normal cells in thermal equilibrium with an all-trans form. In addition, it was recently reported that a ciliary opsin from Platynereis dumerilii can bind all-trans retinal directly and exhibit bistability [16].
Another approach is to use chimeric proteins of animal and microbial rhodopsins. We have engineered chimeric proteins of microbial rhodopsins containing cytoplasmic loops of animal rhodopsin [17,18]. These chimera contain alltrans retinal, display a photocycle (no bleaching), and activate Gt. So far, the second and third cytoplasmic loops of bovine rhodopsin have been used, in which the third loop is essential for the activation of Gt [18]. As templates of microbial rhodopsin, we attempted light-driven proton pumps bacteriorhodopsin (BR), Gloeobacter rhodopsin (GR), proteorhodopsin (PR), and sensory rhodopsin II (SRII). Among these, BR, GR and SRII chimera activated Gt, but PR chimera did not.
Gt activation by these chimera suggest a common activation mechanism between animal and microbial rhodopsins, in which helix opening occurs at the cytoplasmic surface [18]. These chimera are potential candidates of new optogenetic tools for GPCR signaling. In this paper, we report two trials that advance the engineering of chimeric rhodopsins, one for the inserted loop, and another for the microbial rhodopsin template. Regarding the first trial, we have so far only tested the activation of Gt, which is localized in the retina. Here we examined the activation of Gs protein by light, for which we incorporated the cytoplasmic loop of β2-adrenergic receptor (β2AR). Similar structural changes for G-protein activation have been suggested for bovine rhodopsin and β2AR [19–21], and indeed we successfully activated Gs-protein by using a microbial rhodopsin chimera with β2AR. Regarding the second trial, we have so far only tested proton-pump proteins as the template of microbial rhodopsins. SRII is a phototaxis sensor but functions as a light-driven proton pump without its transducer protein [22]. Here we examined two light-driven pumps, the sodium pump [23,24] and the chloride pump [24–26]. Interestingly, we did not observe any G-protein activation for the light-driven sodium pump from Indibacter alkaliphilus ( IndiR2) and the light-driven chloride pump halorhodopsin from Natronomonas pharaonis ( NpHR), although a light-driven proton pump GR showed light-dependent G-protein activation. The molecular mechanism of G-protein activation by chimeric proteins will be discussed.
Materials and Methods
Sample Preparation
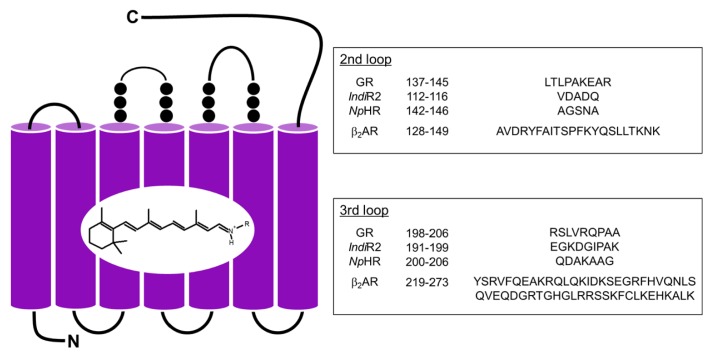
The chimeric constructions were designed based on the wild-type (WT) GR (GenBank accession number: BAC88139), IndiR2 (BAV92787) and NpHR (P15647) (Fig. 1), and the DNA template of the human β2AR loop was exchanged by the following three-step PCR. First, three PCR products were constructed and purified: the front side of microbial rhodopsins before the exchange region and adding the top 15 mer β2AR loop by a primer to the end (f-mRh); the last side of microbial rhodopsins were constructed in the same way (l-mRh). The loop region of β2AR was designed by transforming the sequence of a rare codon in Escherichia coli into one with a high codon usage, and it was total synthesized and amplified by PCR. The products of the first PCR were used to amplify a second round PCR product. Then, f-mRh was extended to the loop region by PCR with a β2AR loop and l-mRh as same. Finally, the resulting full-length chimera fragment was used to amplify the former two products which were cloned into pMS by insertion after XbaI/NotI digestion (in GR), or into the pET21 vector by insertion after NdeI/XhoI digestion (in IndiR2 and NpHR). After ligation, the plasmids were transformed into E. coli strain JM109. All of the chimeras were confirmed by DNA sequencing. The WT and chimeric proteins possessing a six histidine tag at the C-terminus were expressed in E. coli strain BL21 for GR or in strain C41(DE3) for IndiR2 and NpHR, solubilized with 1% n-dodecyl-β-D-maltoside (DDM), and purified by Co2+-column chromatography as described previously [27]. Absorption spectra of the solubilized proteins (300 mM NaCl, 300 mM imidazole, 50 mM Tris-HCl, pH 7.0 and 0.1% DDM) were measured at 20°C using a UV-visible spectrophotometer (UV-2400PC, Shimadzu, Japan). The bovine Gs-protein α-subunit was expressed in E. coli strain BL21 (DE3) using the pQE60 vector containing Gsα cDNA (M13006) and a six histidine tag at the N-terminus and purified using Ni-affinity chromatography. Purified Gs-protein α-subunit was mixed with an equal amount of Gt βγ-subunits purified from bovine retina.
Figure 1.
Design of chimeric proteins from GR, IndiR2 and NpHR. Each loop of microbial rhodopsins was replaced by that of β2AR.
G-Protein Activation Assays
A radionucleotide filter-binding assay, which measures a light-dependent GDP/GTPγS exchange by Gs-protein, was carried out with slight modifications of our previous method using other G-protein subtypes [28]. All procedures were carried out at 20°C. The assay mixture consisted of 50 mM HEPES (pH 7.0), 140 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.05% DDM, 1 μM [35S]GTPγS and 2 μM GDP. The mixture of target protein (final concentration: 2 μM WT or chimera) and Gs-protein (final concentration: 100 nM) was constantly irradiated with white light or was kept in the dark. After incubation for a selected period of time, an aliquot (20 μl) was removed from the sample and placed into 200 μl of stop solution (20 mM Tris/Cl (pH 7.4), 100 mM NaCl, 25 mM MgCl2, 1 μM GTPγS and 2 μM GDP), and it was immediately filtered through a nitrocellulose membrane to trap [35S]GTPγS bound to Gs-protein. The amount of bound [35S]GTPγS was quantified by assaying the membrane with a liquid scintillation counter (Tri-Carb 2910 TR, Perkin Elmer).
Light-induced Difference FTIR Spectroscopy
Light-induced difference FTIR spectroscopy of GR, IndiR2, NpHR and their chimeras was performed as described previously [18,29–31]. Each protein was reconstituted into L-α-phosphatidylcholine liposomes by removing the detergent with Bio-beads, in which the molar ratio of the added lipid to protein was 30:1. The samples in PC liposomes were washed twice in pH 7.5 buffers (2 mM phosphate for GR, or 2 mM phosphate, 2 mM NaCl for IndiR2 and NpHR). An 80 μl aliquot of the sample was deposited on a BaF2 window of 18 mm diameter and dried in a glass vessel that was evacuated by an aspirator. The film sample was hydrated with 1 μL of H2O before measurements. Although the salt concentration cannot be precisely measured for hydrated IndiR2 and NpHR films, we roughly estimated it to be >100 mM. Then, the sample was placed in a cryostat (Oxford DN-1704, UK) mounted in the FTIR spectrometer (Bio-Rad FTS-7000, USA). The cryostat was equipped with a temperature controller (Oxford ITC-4, UK), and the temperature was regulated with 0.1 K precision.
Long-lived intermediates must be responsible for Gprotein activation of chimeric microbial rhodopsins, such as metarhodopsin-II in the case of bovine rhodopsin. Therefore, we attempted to capture late intermediates that accumulate at the last stage of the photocycle. In GR [18] and IndiR2 [32], the late intermediate that forms is the red-shifted O intermediate. We illuminated GR, IndiR2 and their chimeras with 520±5 nm light (an interference filter) at 250 K for 2 min. On the other hand, the O intermediate does not accumulate in NpHR under high salt conditions [33], and our previous FTIR study showed that the L2 (or N) intermediate is formed at 250 K [34]. Thus, we illuminated NpHR and its chimeras with >500 nm light at 250 K for 2 min. The difference spectra were obtained with 2 cm−1 resolution. We averaged 3–4 independent measurements with 128 scans.
Results
Absorption Properties of the GR, IndiR2 and NpHR Chimera
In the present study, we replaced the second or third cytoplasmic loop of microbial rhodopsins into those of β2AR. The schematic structure of the second and third loop of β2AR inserted into chimeras is shown in Figure 1, together with the removed amino-acid sequences in GR, IndiR2 and NpHR. The crystal structures of NpHR [35] and β2AR [36] are known, and we designed the amino acids to replace them based on these structures. Although the structures of GR and IndiR2 have not been reported, we used the structures of homologous proteins, xanthorhodopsin (XR) [37] and Krokinobacter eikastus rhodopsin 2 (KR2) [38], respectively, for amino acid design.
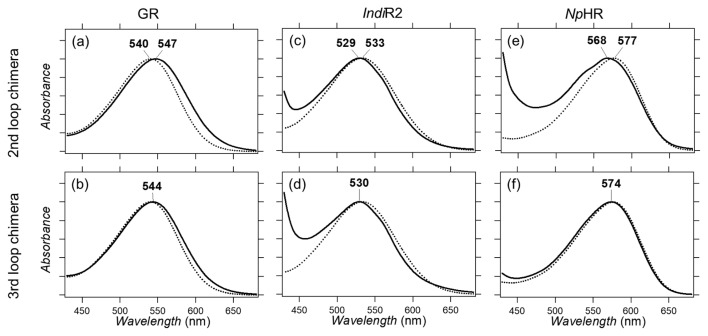
We expressed the GR, IndiR2 and NpHR chimeras in E. coli, followed by solubilization with DDM and purification through a Co2+:NTA column. Figure 2 compares the absorption spectra of the GR, IndiR2 and NpHR loop chimeras with WT. Figure 2a and b show that the λmax of GR WT, GR/second and GR/third chimeras are 540 nm, 547 nm, and 544 nm, respectively. Previously, we designed bovine rhodopsin chimeras, whose λmax was 546 nm and 543 nm for the second and third loops, respectively [18]. In the case of GR, both loop replacements resulted in a spectral red-shift during which the second loop is more influential. On the other hand, there are no differences between bovine rhodopsin and β2AR.
Figure 2.
Absorption spectra of WT GR (dotted line in a and b), and the second (solid line in a) and third (solid line in b) loop chimeras. Absorption spectra of WT IndiR2 (dotted line in c and d), and the second (solid line in c) and third (solid line in d) loop chimeras. Absorption spectra of WT NpHR (dotted line in e and f), and the second (solid line in e) and third (solid line in f) loop chimeras.
Figure 2c and d show that IndiR2 chimeras exhibit a λmax of 533 nm for WT, which is blue-shifted by 3 nm for both chimeras (λmax: 530 nm). Compared to WT, the loop chimera of IndiR2 shows a strong absorption at <450 nm, and less purified samples suggest that the IndiR2 chimera is thermally less stable than WT. Figure 2e and f show that the NpHR chimera exhibits a λmax of 577 nm for WT, and 568 nm and 574 nm for the second and third loop chimeras, respectively. In the case of NpHR, only the second loop chimera has strong absorption at <450 nm, suggesting that the third loop chimera is as thermally stable as WT.
Gs-Protein Activation Properties of the GR, IndiR2 and NpHR Chimera
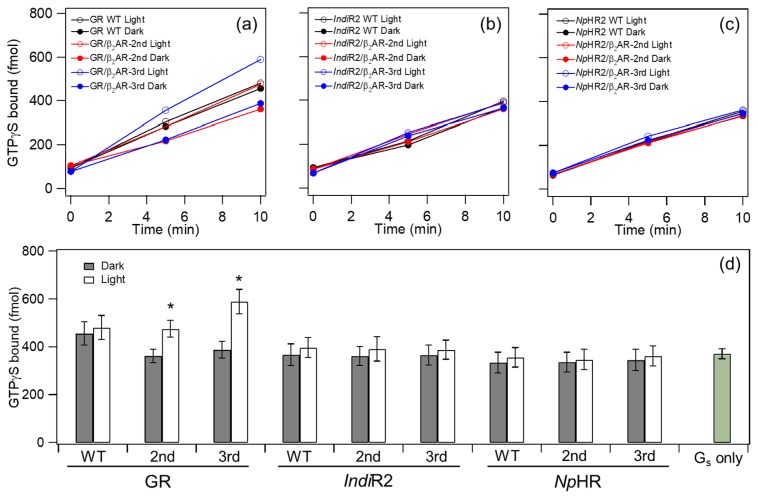
We next tested the Gs-protein activation of GR, IndiR2 and NpHR chimeras. Figure 3a–c shows the time-course of the binding of GTPγS to Gs-protein, where the light-dependent GDP/GTPγS exchange was monitored by using [35S]GTPγS. At least 200 DDM molecules are needed to solubilize one microbial rhodopsin [39], thus 0.05% DDM was used in our study to fully solubilize the chimeric proteins whose concentration was ca. 2 μM (molecular ratio of chimera: DDM = 1:500). In the case of IndiR2 (Fig. 3b) and NpHR (Fig. 3c) chimeras, all time-courses looked similar. This fact indicates that the amount of light-induced time-dependent GTPγS binding is similar to the level of each chimera in the dark, regardless of whether it is a second or third loop chimera, and this is also the case for WT (dotted lines). This feature is clearly seen in Figure 3d, where the amount of GTPγS binding is similar between dark and light conditions within current experimental accuracy, whose level coincides with the spontaneous incorporation of GTPγS to trimeric Gs without receptors (Gs only in Fig. 3d). Thus, we conclude that chimeric proteins of light-driven sodium (IndiR2) and chloride (NpHR) pumps do not activate G-protein.
Figure 3.
G-protein activation by GR chimeras (a), IndiR2 chimeras (b), and NpHR chimeras (c). Time-dependent GTPγS-binding to Gs-protein was monitored in the light (open circle) and dark (filled circle). Black, red and blue circles/lines represent the results of WT, the second and third loop chimera of β2AR, respectively. (d) Comparison of G-protein activation ability by WT and chimeras. GTPγS-binding to Gs-protein was monitored at 10 min in the light (open bar) and dark (filled bar). It should be noted that the spontaneous incorporation of GTPγS of Gs (Gs only) was about 40 times higher than that of Gt [17,18]. Data are presented as the means±S.D. of more than three independent experiments and the marked chimeras (*) exhibit a significant difference between light-dependent and dark activations (p<0.05; Student’s t-test, one-tailed).
In contrast, different features were observed for the GR chimera. Figure 3a shows light-dependent Gs-protein activations of GR, where G-protein activation was almost identical between light and dark conditions for WT (black circles in Fig. 3a). Unlike WT, clear light-dependent Gs-protein activation was observed for the second (red circles in Fig. 3a) and third (blue circles in Fig. 3a) loop chimeras, where light-dependent activation was more enhanced in the latter. These features are obvious from Figure 3d. Dark activation was higher for WT GR than those for GR chimera and other proteins, but the reason is unclear. However, similar results for light activation of WT GR suggest no difference between light and dark, nor between IndiR2 and NpHR chimeras.
Only the GR chimera activated G-protein, in which the third loop chimera exhibited stronger activation than the second loop chimera. We also designed a double mutant containing both second and third cytoplasmic loops, but we were not able to obtain such protein, presumably because of the structural instability. In the previous study, light-dependent Gt activation by the GR chimera was quantitatively compared with those of bovine rhodopsin [18]. Here we also measured the Gs activation ability by β2AR as a positive control. However, we utilized membrane-embedded β2AR, not purified samples, and we could not estimate the amount of β2AR in the sample. Thus, we could not compare the Gs activation ability between β2AR and our chimeras.
We found that the proton pump chimera (GR) possessed the ability to activate Gs-protein and Gt-protein, whereas the sodium pump (IndiR2) and the chloride pump (NpHR) were unable to activate Gs-protein. Similar absorption spectra for all chimeras in Figure 2 show a retained protein structure around their retinal chromophore. To further characterize the molecular properties of these chimeras, we applied light-induced difference FTIR spectroscopy.
Light-Induced Difference FTIR Spectroscopy of the GR, IndiR2 and NpHR Chimera
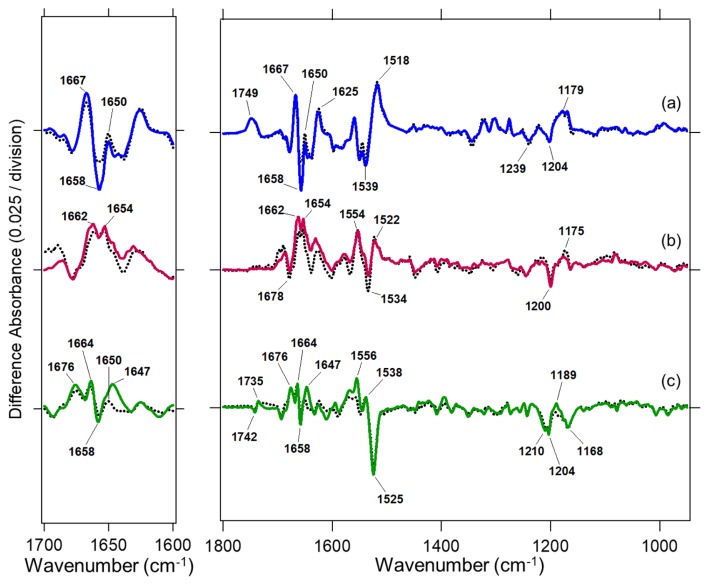
Figure 4 compares structural changes of WT and the third loop chimeras of GR (Fig. 4a), IndiR2 (Fig. 4b) and NpHR (Fig. 4c). All difference spectra were measured at 250 K, where late intermediates accumulated during their photocycles. The spectra of WT and the chimera in each rhodopsin were very similar, particularly the frequency region of the C=C (1550–1500 cm−1) and C–C (1250–1150 cm−1) stretches of the retinal chromophore. This indicates that similar intermediates formed in both WT and the loop chimera of each rhodopsin. In other words, different properties among ionpump rhodopsins such as G-protein activation by chimeric proteins essentially originate from the structural dynamics of each ion-pump protein.
Figure 4.
Light-induced difference FTIR spectra of WT GR (black dotted line in a), the third loop chimera of GR (blue line in a), WT IndiR2 (black dotted line in b), the third loop chimera of IndiR2 (red line in b), WT NpHR (black dotted line in c), and the third loop chimera of NpHR (green line in c). Positive and negative bands originate from the photointermediate and unphotolyzed states, respectively.
The left panel of Figure 4 highlights an amide-I vibration that appears at 1700–1600 cm−1. The frequency of that vibration strongly depends on the secondary structure of the protein, where the frequency of the α-helix appears at 1660–1650 cm−1. Figure 4a shows the results for GR, where difference spectra correspond to those between the O intermediate and the resting state [18]. In the case of GR, the bands at 1667 (+)/1659 (−)/1650 (+) cm−1 can be interpreted as the structural perturbation of α-helices. These bands were observed for WT GR, and their amplitude for the third loop chimera was 1.42 times larger. This was also the case for the loop chimera of bovine rhodopsin, and we interpreted that structural perturbation of the α-helix as being related to the helix opening at the cytoplasmic side, which is larger in the chimera than in WT [18]. The present study shows that this is also the case for the β2AR chimera.
In the case of IndiR2, the difference spectra correspond to the O intermediate and the resting state [32], similar to GR. Positive peaks appear at 1662 and 1654 cm−1, and these spectral features and amplitudes are similar in both WT and loop chimera (Fig. 4b). It should be noted that changes in amplitude at 1660–1650 cm−1 were much smaller in IndiR2 than in GR, suggesting smaller helical structural changes in IndiR2.
In the case of NpHR, the difference spectra correspond to the L2 (or N) intermediate and the resting state [34]. The bands at 1664 (+)/1658 (−)/1650 (+) cm−1 are assigned as amide-I vibrations of the α-helix (Fig. 4b). Unlike IndiR2, structural changes of the α-helix are obvious for NpHR, which is also the case for GR. In NpHR, the relative amplitude of the amide-I vibration to that of the C=C (1550–1500 cm−1) and C–C (1250–1150 cm−1) stretches of the retinal chromophore was smaller in NpHR (Fig. 4c) than in GR (Fig. 4a). This suggests a smaller structural perturbation in the α-helix of NpHR than in the late intermediates of GR. These observations of the structural dynamics in light-driven proton, sodium and chloride pumps may be related to the fact that proton-pump chimera can activate G-protein, while sodium and chloride pumps cannot.
Discussion
Microbial rhodopsin chimeras that contain the cytoplasmic loop of GPCR offer potential as optogenetic tools. We previously reported that the chimera of bovine rhodopsin are able to activate Gt-protein [17,18]. It is important to extend this ability to more general G-proteins such as Gs, Gi, Go, and others, and there is interest in studying if these chimeras are able to activate various G-proteins, or not. In this paper, we showed that GR chimeras containing the second and third loops of β2AR are able to activate Gs-protein. Gs-activating rhodopsin in jellyfish was reported as a potential optogenetic tool for light-dependent Gs activation [40,41], and the present study provides different kind of Gs activation tool by light using microbial rhodopsin. Previously we suggested similar protein structural changes between bovine rhodopsin and microbial rhodopsins such as BR, SRII and GR [17,18]. The present study further generalizes such structural changes for another GPCR, β2AR. This generalization is reasonable because the outward motion of helix 6 was reported upon activation of bovine rhodopsin and β2AR, together with microbial rhodopsins [1,3,6,19–21].
GR chimeras show no G-protein activation in the dark, suggesting that the interaction surface of the receptor is hidden in the dark and is only exposed by light. This concept is also common for GPCR activation [19–21]. A weakened hydrogen bond of the amide-I vibration of the α-helix was observed upon formation of the O intermediate of WT GR, and we infer that this α-helical perturbation is caused by opening of helix 6 on the cytoplasmic side. The GR template was identical for the GR chimera of the third loop of bovine rhodopsin [18] and β2AR (Fig. 1). In addition, the amplitude of the amide-I signal for the positive 1667-cm−1 and negative 1659-cm−1 bands was enhanced 1.67-times for bovine rhodopsin chimera [18] and 1.42-times for β2AR chimera (Fig. 4a) relative to WT. This suggests that inserted third loop enlarges opening motion of helix 6, which is larger in bovine rhodopsin chimeras.
The present study shows that light-driven proton pump (GR) chimeras contain cytoplasmic loops that activate Gs-protein, whereas light-driven sodium (IndiR2) and chloride (NpHR) pump chimeras do not. Thus, we are able to classify a new template of microbial rhodopsins into two classes: BR (proton pump), SRII (sensor, but acts as proton pump without transducer) and GR (proton pump) chimeras that can activate G-proteins (class I), and PR (proton pump), IndiR2 (sodium pump) and NpHR (chloride pump) chimeras that cannot (class II). A remaining question involves the possible mechanism to distinguish the activation of G-proteins. G-protein activation is caused by helix opening at the cytoplasmic side. Thus, no G-protein activation by the chimeras of light-driven sodium and chloride pumps suggests a small helix opening on the cytoplasmic side. This hypothesis is strongly supported by the present FTIR observation, in which the helical structural perturbation was smaller in the sodium-pump IndiR2 and the chloride-pump NpHR than in the proton-pump GR (Fig. 4).
Information on the structural dynamics of light-driven sodium pumps is limited, whereas X-ray structure and computational studies of KR2 suggested a more polar environment at the cytoplasmic region than proton pumps [38,42,43]. Therefore, large conformational changes on the cytoplasmic side might not be needed for the uptake of sodium ions. In the case of NpHR, detailed structural dynamics have been performed based on the crystal structures of intermediates [44]. Although the authors detected a large deformation of helix 6, the motion did not accompany helix opening at the cytoplasmic surface. These structural dynamics in the literature for light-driven sodium and chloride pumps are consistent with the mechanism of G-protein activation by chimeric proteins.
Conclusion
In the present study, chimeric proteins of a light-driven proton pump GR containing the cytoplasmic loop of β2AR successfully activated Gs protein when exposed to light. In contrast, no G-protein activation was observed in chimeric proteins of a light-driven sodium pump IndiR2 and a light-driven chloride pump NpHR carrying the cytoplasmic loop of β2AR. This fact suggests that the helix opening motion, which is common to GPCR and GR, is different for IndiR2 and NpHR. Thus, GR chimera can serve as a potential tool in optogenetics, where the activation of various G-proteins can be initiated by light.
Acknowledgments
This work was financially supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology to K. I. (26708001, 26115706, 26620005) and to H. K. (25104009, 15H02391).
Footnotes
Conflicts of Interest
All authors declare that they have no conflicts of interest.
Author Contributions
H. K. directed the research, and wrote the manuscript. K. Y. prepared samples with the help of K. S. and K. I. T. Y. performed the G-protein activation assay with the help of K. Y. and Y. S. K. Y. measured light-induced difference FTIR spectra. All authors discussed and commented on the manuscript.
References
- 1.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haupts U, Tittor J, Oesterhelt D. Closing in on bacteriorhodopsin: progress in understanding the molecule. Annu Rev Biophys Biomol Struct. 1999;28:367–399. doi: 10.1146/annurev.biophys.28.1.367. [DOI] [PubMed] [Google Scholar]
- 6.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 7.Grote M, Engelhard M, Hegemann P. Of ion pumps, sensors and channels—perspectives on microbial rhodopsins between science and history. Biochim Biophys Acta. 2014;1837:533–545. doi: 10.1016/j.bbabio.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Inoue K, Kato Y, Kandori H. Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 2015;23:91–98. doi: 10.1016/j.tim.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Govorunova EG, Sineshchekov OA, Li H, Spudich JL. Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. Annu Rev Biochem. 2017;86:845–872. doi: 10.1146/annurev-biochem-101910-144233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 11.Zhang F, Wang L-P, Brauner M, Liewald JF, Kay K, Watzke N, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 12.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 14.Koyanagi M, Kawano E, Kinugawa Y, Oishi T, Shichida Y, Tamotsu S, et al. Bistable UV pigment in the lamprey pineal. Proc Natl Acad Sci USA. 2004;101:6687–6691. doi: 10.1073/pnas.0400819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyanagi M, Takada E, Nagata T, Tsukamoto H, Terakita A. Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc Natl Acad Sci USA. 2013;110:4998–5003. doi: 10.1073/pnas.1219416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto H, Chen IS, Kubo Y, Furutani Y. A ciliary opsin in the brain of a marine annelid zooplankton is ultraviolet-sensitive, and the sensitivity is tuned by a single amino acid residue. J Biol Chem. 2017;292:12971–12980. doi: 10.1074/jbc.M117.793539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatsuma A, Yamashita T, Sasaki K, Kawanabe A, Inoue K, Furutani Y, et al. Chimeric microbial rhodopsins containing the third cytoplasmic loop of bovine rhodopsin. Biophys J. 2011;100:1874–1882. doi: 10.1016/j.bpj.2011.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki K, Yamashita T, Yoshida K, Inoue K, Shichida Y, Kandori H. Chimeric proton-pumping rhodopsins containing the cytoplasmic loop of bovine rhodopsin. PLoS ONE. 2014;9:e91323. doi: 10.1371/journal.pone.0091323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deupi X, Standfuss J, Schertler G. Conserved activation pathways in G-protein-coupled receptors. Biochem Soc Trans. 2012;40:383–388. doi: 10.1042/BST20120001. [DOI] [PubMed] [Google Scholar]
- 21.Deupi X. Relevance of rhodopsin studies for GPCR activation. Biochem Biophys Acta. 2014;1837:674–682. doi: 10.1016/j.bbabio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Sudo Y, Iwamoto M, Shimono K, Sumi M, Kamo N. Photo-induced proton transport of pharaonis phoborhodopsin (sensory rhodopsin II) is ceased by association with the transducer. Biophys J. 2001;80:916–922. doi: 10.1016/S0006-3495(01)76070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, et al. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 24.Kandori H. Ion-pumping microbial rhodopsins. Front Mol Biosci. 2015;2:52. doi: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- 26.Essen LO. Halorhodopsin: light-driven ion pumping made simple? Curr Opin Struct Biol. 2002;12:516–522. doi: 10.1016/s0959-440x(02)00356-1. [DOI] [PubMed] [Google Scholar]
- 27.Kandori H, Shimono K, Sudo Y, Iwamoto M, Shichida Y, Kamo N. Structural changes of Pharaonis phoborhodopsin upon photoisomerization of the retinal chromophore: infrared spectral comparison with bacteriorhodopsin. Biochemistry. 2001;40:9238–9246. doi: 10.1021/bi0103819. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita T, Terakita A, Shichida Y. Distinct roles of the second and third cytoplasmic loops of bovine rhodopsin in G protein activation. J Biol Chem. 2000;275:34272–34279. doi: 10.1074/jbc.M002954200. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto K, Choi AR, Furutani Y, Jung KH, Kandori H. Low-temperature FTIR study of Gloeobacter rhodopsin: presence of strongly hydrogen-bonded water and long-range structural protein perturbation upon retinal photoisomerization. Biochemistry. 2010;49:3343–3350. doi: 10.1021/bi100184k. [DOI] [PubMed] [Google Scholar]
- 30.Kandori H, Yamazaki Y, Shichida Y, Raap J, Lugtenburg J, Belenky M, et al. Tight Asp-85--Thr-89 association during the pump switch of bacteriorhodopsin. Proc Natl Acad Sci USA. 2001;98:1571–1576. doi: 10.1073/pnas.98.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furutani Y, Kamada K, Sudo Y, Shimono K, Kamo N, Kandori H. Structural changes of the complex between pharaonis phoborhodopsin and its cognate transducer upon formation of the M photointermediate. Biochemistry. 2005;44:2909–2915. doi: 10.1021/bi047893i. [DOI] [PubMed] [Google Scholar]
- 32.Kajimoto K, Kikukawa T, Nakashima H, Yamaryo H, Saito Y, Fujisawa T, et al. Transient resonance Raman spectroscopy of a light-driven sodium-ion-pump rhodopsin from Indibacter alkaliphilus. J Phys Chem B. 2017;121:4431–4437. doi: 10.1021/acs.jpcb.7b02421. [DOI] [PubMed] [Google Scholar]
- 33.Váró G, Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, et al. Light-driven chloride ion transport by halorhodopsin from Natronobacterium pharaonis. 1. The photochemical cycle. Biochemistry. 1995;34:14490–14499. doi: 10.1021/bi00044a027. [DOI] [PubMed] [Google Scholar]
- 34.Shibata M, Muneda N, Sasaki T, Shimono K, Kamo N, Demura M, et al. Hydrogen-bonding alterations of the protonated Schiff base and water molecule in the chloride pump of Natronobacterium pharaonis. Biochemistry. 2005;44:12279–12286. doi: 10.1021/bi050726d. [DOI] [PubMed] [Google Scholar]
- 35.Kouyama T, Kanada S, Takeguchi Y, Narusawa A, Murakami M, Ihara K. Crystal structure of the light-driven chloride pump halorhodopsin from Natronomonas pharaonis. J Mol Biol. 2010;396:564–579. doi: 10.1016/j.jmb.2009.11.061. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 37.Luecke H, Schobert B, Stagno J, Imasheva ES, Wang JM, Balashov SP, et al. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc Natl Acad Sci USA. 2008;105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato HE, Inoue K, Abe-Yoshizumi R, Kato Y, Ono H, Konno M, et al. Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature. 2015;521:48–53. doi: 10.1038/nature14322. [DOI] [PubMed] [Google Scholar]
- 39.Møller JV, le Maire M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J Biol Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 40.Koyanagi M, Takano K, Tsukamoto H, Ohtsu K, Tokunaga F, Terakita A. Jellyfish vision starts with cAMP signaling mediated by opsin-Gs cascade. Proc Natl Acad Sci USA. 2008;105:15576–15580. doi: 10.1073/pnas.0806215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailes HJ, Zhuang LY, Lucas RJ. Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS ONE. 2012;7:e30774. doi: 10.1371/journal.pone.0030774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gushchin I, Shevchenko V, Polovinkin V, Kovalev K, Alekseev A, Round E, et al. Crystal structure of a light-driven sodium pump. Nat Struct Mol Biol. 2015;22:390–395. doi: 10.1038/nsmb.3002. [DOI] [PubMed] [Google Scholar]
- 43.Suomivuori CM, Gamiz-Hernandez AP, Sundholm D, Kaila VRI. Energetics and dynamics of a light-driven sodium-pumping rhodopsin. Proc Natl Acad Sci USA. 2017;114:7043–7048. doi: 10.1073/pnas.1703625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kouyama T, Kawaguchi H, Nakanishi T, Kubo H, Murakami M. Crystal structures of the L1, L2, N, and O states of pharaonis halorhodopsin. Biophys J. 2015;108:2680–2690. doi: 10.1016/j.bpj.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]