Abstract
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy of the head and neck with the highest incidence rate in southern China. The aim of the present study was to understand the molecular mechanisms that underlie the progression of NPC. The relative expression of miR-93 and CDKN1A was detected by the reverse-transcription quantitative PCR. Western blot analysis was applied to detect the protein levels of genes. Luciferase activity report was applied to verify the target of miRNA. Cell growth was assayed by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. miR-93 was upregulated in NPC tissues and cell lines compared with normal samples. Re-expression of miR-93 promoted cell growth in vitro as determined by the MTT assay. CDKN1A was identified by luciferase reporter as a direct target of miR-93. Its expression was downregulated by miR-93. Furthermore, the results showed that the expression of miR-93 was inversely correlated with the expression of CDKN1A protein. miR-93 enhanced cell proliferation in NPC by directly targeting CDKN1A. It is suggested that miR-93/CDKN1A axis may present a new target for the treatment of NPC.
Keywords: nasopharyngeal carcinoma, CDKN1A, microRNA-93, proliferation
Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy of the head and neck with the highest incidence rate in southern China, with a high prevalence of 20–50 cases per 100,000 individuals (1). Several factors are involved in the progression and development of NPC, including Epstein-Barr virus (EBV) infection, genetic components, environmental factors and interactions between these factors (2). Although the application of intensity-modulated radiotherapy and chemoradiotherapy has increased the 5-year overall survival rate to approximately 70%, the prognosis remains poor because of distant metastasis and recurrence (3,4). Thus, it is critical to understand the molecular mechanisms that underlie the progression of NPC in order to promote the development of novel therapeutic strategy.
MicroRNAs (miRNAs) are small non-coding RNA sequences, 19–25 nucleotides long, which negatively regulate gene expression by targeting the 3-untranslated region (3′-UTR) of their target mRNA transcripts at the post-transcriptional level (5,6). Increasing evidence showed that miRNAs regulate a wide range of cell functions, including proliferation, migration, differentiation and apoptosis (5). Emerging evidence indicates that miRNAs are abnormally expressed in human disease, especially cancer, and they function as either tumor suppressors or oncogenes in the progression of tumors (7,8). Multiple miRNAs have been found to be dysregulated in NPC, including miR-124 (9), miR-29c (10), miR-34c (11) and miR-16 (12). These findings indicated that the abnormally expressed miRNAs may contribute to the progression and development of NPC. miR-93, derived from a paralogue (miR-106b-25), is dysregulated in several cancer types, such as epithelial ovarian carcinoma (13), ischemic heart disease (14) and bone cancer pain mouse model (15). The evidence indicates that miR-93 plays a key role in cancer progression.
CDKN1A, also known as p21, is a cyclin-dependent kinase inhibitor that inhibits the complexes of CDK2 and CDK1, and regulates the cell cycle progression at G1 and S phase (16). Its expression is tightly controlled by the tumor suppressor protein p53, through which CDKN1A mediates the p53-dependent cell cycle G1 phase arrest in response to a variety of stress stimuli (17). p21 can also interact with proliferating cell nuclear antigen (PCNA) and plays a regulatory role in DNA damage repair and S-phase DNA replication (18).
In the present study, we demonstrated that miR-93 was upregulated in NPC tissues and cells. Re-expression of miR-93 promoted cell growth in vitro. CDKN1A was downregulated by miR-93 by directly targeting its 3′-UTR. The newly identified miR-93/CDKN1A axis may provide new evidence to understand the molecular mechanism of NPC and present a new target for the treatment of NPC.
Materials and methods
Tissue specimens and cell lines
A collection of 23 freshly frozen NPC biopsy specimens and 13 normal nasopharyngeal epithelial tissue specimens were obtained from the Yidu Central Hospital. The specimens were confirmed by histopathological examination. None of the patients with NPC had received chemotherapy or radiotherapy before biopsy. Written informed consent was obtained from the patients. This study was approved by the Ethics Review Committee of the Yidu Central Hospital (Shandong, China).
The human NPC cell line SUNE-1 and the human immortalized nasopharyngeal epithelial cell line NP69 were used in this study. All cells were cultured in a humidified atmosphere at 37°C with 5% CO2.
Oligonucleotide and plasmid transfection
Cells were cultured on 6-well plates 24 h prior to transfection. The oligonucleotide was transfected into NPC cells using Lipofectamine™ 2000 reagent (Invitrogen, Carlsbad, CA, USA). The transfected cells were incubated at 37°C in complete medium and harvested at the indicated time-points.
Quantitative real-time PCR
qRT-PCR was used to detect miR-93 and CDKN1A expression levels. Total RNA was extracted from cells or tissues by using TRIzol reagent (Invitrogen) according to the manufacturers instruction. qRT-PCR was performed using agents from Takara Bio (Dalian, China) with the Stratagene Mx3000P real-time PCR system (Agilent Technologies, Santa Clara, CA, USA). For miR-93 detection, U6 was used as an internal control. For CDKN1A detection, GAPDH was used as an endogenous control. The PCR primers for CDKN1A were as follows: 5-GTGGGGTTATCTCTGTGTTAGGG-3 and 5-CCCTGT CCATAGCCTCTACTGC-3. The primers for GAPDH were as follows: 5-CTCCTCCTGTTCGACAGTCAGC-3 and 5-CCCAATACGACCAAATCCGTT-3. The relative expression was calculated with the 2−∆∆CT. All reactions were performed in triplicate.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
MTT assay was used to detect the proliferation of NPC cells. After transfection, cells were plated in 96-well plates at 1,000 cells/well and cultured for 1–5 days. Cell viability was examined once daily. On the indicated days, the MTT dye were added to the well and incubated for 4 h at 37°C. Then the medium were removed and dimethyl sulfoxide (DMSO) at 150 µl/well (Sigma Corp., Ronkonkoma, NY, USA) was added to dissolve the formazan crystals. The absorbance at 490 nm was measured on a spectrophotometer (Perkin-Elmer, Waltham, MA, USA).
Luciferase reporter assay
293 cells were used for luciferase reporter assay. 3′-UTR of CDKN1A sequences were cloned from mRNA and inserted into the downstream of luciferase expression gene in psiCHECK-2 plasmids (psi-CDKN1A-3′-UTR).
Cells were co-transfected with miR-93 mimic or control, and psi-CDKN1A-3′-UTR-WT or psi-CDKN1A-3′-UTR-MUT using Lipofectamine 2000 (Invitrogen). The mutation (MUT) of 3′-UTR was generated using QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technologies). After 24-h incubation, cells were collected and assayed for luciferase activity using the Dual-Luciferase reporter assay system (Promega Bio Systems, Sunnyvale, CA, USA). The Renilla luciferase activity was used as internal control.
Western blotting
Cells were cultutred, lysed and processed for western blotting according to the standard protocols. The primary antibodies include anti-CDKN1 (1:1,000; Cell Signaling Technology, Danvers, MA, USA), anti-GAPDH (1:1,000; Bioworld Technology, Inc., Nanjing, China). Then the membrane was incubated with appropriate secondary antibody at room temperature for 1 h. Protein bands were detected using enhanced chemiluminescence system (GE Healthcare, Chicago, IL, USA).
Statistical analysis
All statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). All data are presented as mean ± SD. Differences between groups were assessed using the Students t-test or Tukey's post hoc test after one-way analysis of variance in SPSS. Differences were considered significant at P<0.05.
Results
miR-93 is upregulated in NPC tissues and cell lines
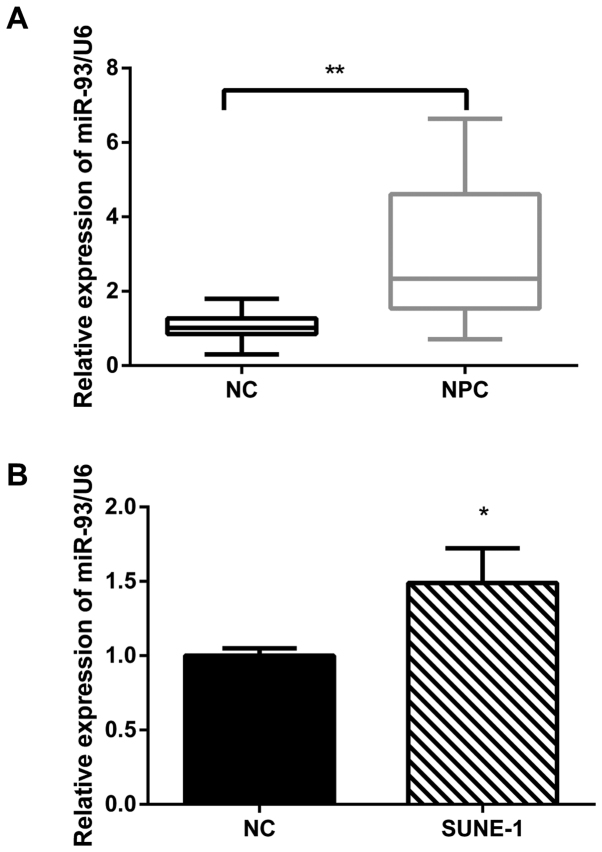
To study the expression level of miR-93 in NPC, 23 freshly frozen NPC biopsy specimens and 13 normal nasopharyngeal epithelial tissue specimens were analyzed by the qRT-PCR. The expression of miR-93 was increased by ~2-fold in 23 NPC tissues compared with 13 normal tissues (Fig. 1A). Furthermore, we detected the expression of miR-93 in SUNE-1 NPC cell lines. The qRT-PCR results revealed that miR-93 was significantly upregulated in SUNE-1 cell line compared with the normal nasopharyngeal epithelial cell line NP69 (Fig. 1B). Taken together, miR-93 was aberrantly upregulated in NPC tissues and cell lines.
Figure 1.
miR-93 is upregulated in NPC tissues and cell lines. (A) The relative expression of miR-93 in 23 NPC tissues and 13 normal tissues. U6 was used as internal control. (B) miR-93 was significantly upregulated in SUNE-1 cell line compared with the normal nasopharyngeal epithelial cell line NP69. *P<0.05, **P<0.01.
miR-93 promotes NPC cell proliferation in vitro
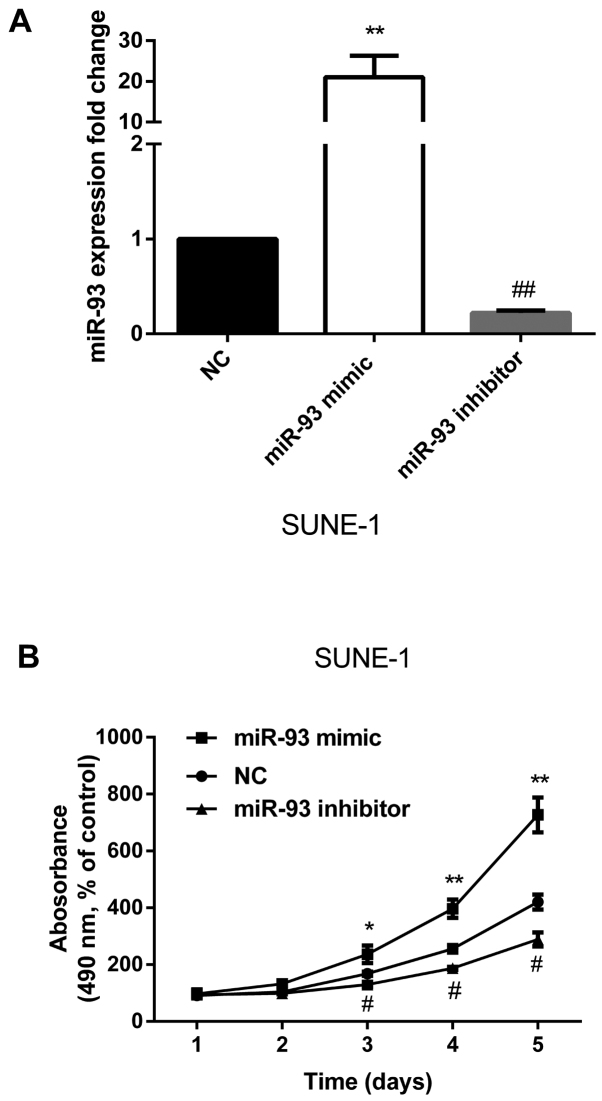
To further understand the biological functions of miR-93 in the development of NPC, we re-expressed miR-93 or inhibited miR-93 expression separately. The successful re-expression or inhibition of miR-93 was confirmed by qRT-PCR (Fig. 2A). MTT assay showed that re-expression of miR-93 significantly promoted cell proliferation and miR-93 inhibitor significantly decreased cell proliferation compared with control (Fig. 2B). Collectively, these results indicate that miR-93 enhances the proliferation of NPC.
Figure 2.
Re-expression of miR-93 promotes NPC cell growth in vitro. (A) The successful re-expression of miR-93 and inhibition of miR-93 in SUNE-1 cells were confirmed by qRT-PCR. (B) MTT assay showed that re-expression of miR-93 significantly promoted cell growth and miR-93 inhibitor significantly decreased cell growth. #P<0.05, ##P<0.01, *P<0.05, **P<0.01.
CDKN1A is a direct target of miR-93
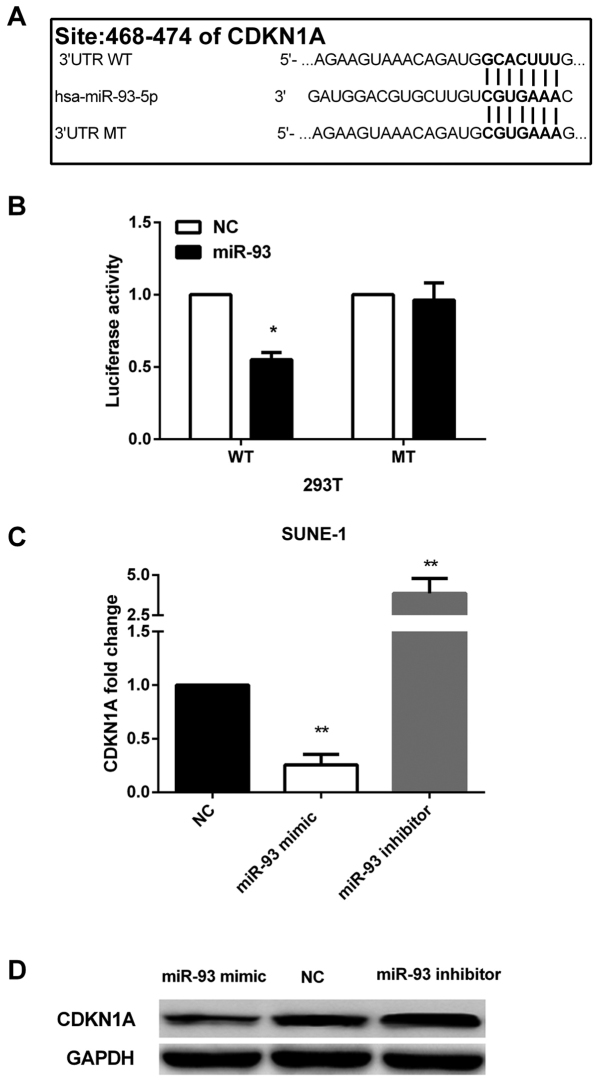
To explore the downstream of miR-93, we performed bioinformatics analysis by using two online algorithms, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do) to predict its putative mRNA targets. CDKN1A was identified as a direct target, and a highly conserved putative binding site was found at 468–474 bp of CDKN1A 3′-UTR (Fig. 3A). Thus, we further performed Dual-Luciferase reporter assay to confirm the prediction. In cells co-transfected with psi-CDKN1A-3UTR-WT and miR-93 mimic, luciferase activity was dramatically decreased compared with cells co-transfected with miR-control and psi-CDKN1A-3UTR-WT. However, the inhibition of luciferase activity was abolished when cells were co-transfected with miR-93 mimic and psi-CDKN1A-3UTR-MT (Fig. 3B).
Figure 3.
CDKN1A is a direct target of miR-93. (A) The putative binding site of miR-93 was at 468–474 bp of CDKN1A 3′-UTR. (B) Luciferase activity assay. Firefly luciferase activity was decreased in 293T cells co-tranfected with miR-93 mimic and WT 3′-UTR reporter. The inhibition of miR-93 was abolished in cells cotransfected with miR-93 mimic and MUT 3′-UTR reporter. Luciferase activity was normalized to Renilla. (C and D) CDKN1A expression fold change was detected by qRT-PCR and western blot analysis. Re-expression of miR-93 decreased CDKN1A expression and miR-93 inhibitor increased CDKN1A expression. *P<0.05, **P<0.01.
Besides, re-expression of miR-93 in SUNE-1 cells could reduce the expression of CDKN1A at both miRNA and protein levels and silencing of miR-93 could increase endogenous CDKN1A at both mRNA and protein levels in SUNE-1 cells (Fig. 3C and D). The evidence indicated that miR-93 downregulated CDKN1A by directly targeting its 3′-UTR.
The expression of miR-93 is inversely correlated with the expression of CDKN1A
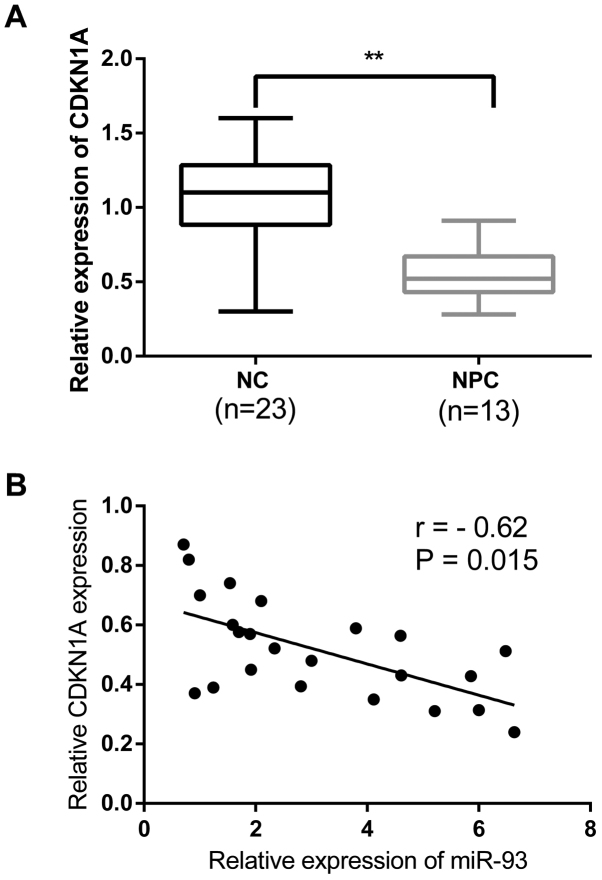
To further investigate the expression of CDKN1A, 23 freshly frozen NPC tissues and 13 normal nasopharyngeal epithelial tissue specimens were detected to analyze the clinicopathological significance. The relative expression of CDKN1A in NPC tissues were significantly decreased compared with that in normal tissues as assayed by qRT-PCR (Fig. 4A). Furthermore, we detected the relationship between the endogenous expression levels of CDKN1A and miR-93 in the same NPC tissues. The results showed a significant inverse correlation between the expression of miR-93 and CDKN1A in NPC tissues (r=−0.62; P<0.05) (Fig. 4B).
Figure 4.
The expression of miR-93 is inversely correlated with the expression of CDKN1A. (A) CDKN1A expression was significantly downregulated in NPC tissues compared with normal samples. (B) The expression of miR-93 was inversely correlated with the expression of CDKN1A. **P<0.01.
Discussion
Dysregulation of miRNAs has been reported in many types of cancer. Increasing evidence indicate that miRNAs function as oncogenes or tumor suppressors. Although the tumorigenesis of NPC is a multistage process relying on the control of the gene expression, several dysregulated miRNA expression profiles and dysregulated genes of NPC have been identified in some studies (19–21). Several miRNAs are identified upregulated, including miR-206, miR-99a, miR-30a and some are down-regulated, such as let-7i, miR-7 and miR-222 (21). The capability to regulate proliferation, invasion and migration is considered as an important determinant in the process of tumor progression. In the present study, we demonstrated that the expression of miR-93 was significantly increased in NPC cell lines and tumor tissues. The evidence indicates that miR-93 was upregulated in NPC, which is consistent with the results obtained from bone cancer (15), myocardial ischemia/reperfusion (I/R) injury (14) and polycystic ovarian syndrome (22). The enhanced proliferation of NPC cell lines indicated that miR-93 was an oncogene in NPC. To date, the ectopic expression of miR-93 is shown as a frequent epigenetic tumor promoter in various types of cancer (23–25). Furthermore, aberrant expression of miR-93 has been demonstrated to regulate tumor migration and invasion (26). The evidence indicates that miR-93 exerts pivotal biological and pathological functions, which may contribute to the development and progression of NPC.
miRNAs exert their function by regulating their target genes by binding to the 3′-UTR. Several studies have identified the target genes of miR-93, including RAB11 family interacting protein 1 (27), PTEN (14), PDCD4 (24), Sirt7 and Tbx3 (28). In the present study, we identified CDKN1A as a direct target of miR-93 by bioinformatics analysis and luciferase activity report, which is consistent with the findings in polycystic ovarian syndrome (22). Re-expression of miR-93 significantly reduced CDKN1A expression and silencing of miR-93 increased CDKN1A expression. Furthermore, we demonstrated that the expression of miR-93 was inversely correlated with the expression of CDKN1A protein. Recently, it has been reported that CDKN1A is directly regulated by multiple miRNAs, including miR-663 (29), miR-106b (30), miR-221 and miR-222 (31). In NPC, p21, functioning as a critical cell cycle regulator, is regulated by miR-663 to promote the G1/S transition (29). miR-106b family members contribute to tumor cell proliferation partially by regulating cell cycle progression via p21/CDKN1A and by modulating checkpoint functions (30). Abnormal expression of miR-221 and miR-222 promotes cancer growth by inhibiting the expression of p27Kip1 (31). Thus, emerging evidence presents that miRNAs induce the aberrant G1/S transition, promote cell proliferation and tumorigenicity in cancer by targeting CDKN1A.
In conclusion, this study found that miR-93 was upregulated in NPC cell lines and tissues. Re-expression of miR-93 promotes NPC proliferation, functioning as an oncogene. CDKN1A was identified as a direct target of miR-93. It is suggested that miR-93/CDKN1A axis may present a new target for the treatment of NPC.
References
- 1.Jia WH, Luo XY, Feng BJ, Ruan HL, Bei JX, Liu WS, Qin HD, Feng QS, Chen LZ, Yao SY, et al. Traditional Cantonese diet and nasopharyngeal carcinoma risk: A large-scale case-control study in Guangdong, China. BMC Cancer. 2010;10:446. doi: 10.1186/1471-2407-10-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu FH, Xiong D, Xu YF, Cao SM, Xue WQ, Qin HD, Liu WS, Cao JY, Zhang Y, Feng QS, et al. An epidemiological and molecular study of the relationship between smoking, risk of nasopharyngeal carcinoma, and Epstein-Barr virus activation. J Natl Cancer Inst. 2012;104:1396–1410. doi: 10.1093/jnci/djs320. [DOI] [PubMed] [Google Scholar]
- 3.Yoshizaki T, Ito M, Murono S, Wakisaka N, Kondo S, Endo K. Current understanding and management of nasopharyngeal carcinoma. Auris Nasus Larynx. 2012;39:137–144. doi: 10.1016/j.anl.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu LZ, Sun Y, Lin AH, Liu MZ, Ma J. How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? Int J Radiat Oncol Biol Phys. 2011;80:661–668. doi: 10.1016/j.ijrobp.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Bandres E, Agirre X, Ramirez N, Zarate R, Garcia-Foncillas J. MicroRNAs as cancer players: Potential clinical and biological effects. DNA Cell Biol. 2007;26:273–282. doi: 10.1089/dna.2006.0544. [DOI] [PubMed] [Google Scholar]
- 6.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Peng XH, Huang HR, Lu J, Liu X, Zhao FP, Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer. 2014;13:186. doi: 10.1186/1476-4598-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N, Tang LL, Sun Y, Cui RX, Wang HY, Huang BJ, He QM, Jiang W, Ma J. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013;329:181–188. doi: 10.1016/j.canlet.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 11.Rietzschel N, Pierik AJ, Bill E, Lill R, Mühlenhoff U. The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol Cell Biol. 2015;35:370–378. doi: 10.1128/MCB.01033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Q, Ren X, Chen J, Li Y, Tang X, Wen X, Yang X, Zhang J, Wang Y, Ma J, et al. miR-16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signaling pathways. Oncotarget. 2016;7:3047–3058. doi: 10.18632/oncotarget.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Chen S, Xiu YL, Sun KX, Zong ZH, Zhao Y. RhoC is a major target of microRNA-93-5P in epithelial ovarian carcinoma tumorigenesis and progression. Mol Cancer. 2015;14:31. doi: 10.1186/s12943-015-0304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke ZP, Xu P, Shi Y, Gao AM. MicroRNA-93 inhibits ischemia-reperfusion induced cardiomyocyte apoptosis by targeting PTEN. Oncotarget. 2016;7:28796–28805. doi: 10.18632/oncotarget.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao WF, Li YS, Lou W, Cai T, Zhang S, Hu XY, Zhang XW, Luo W. MicroRNA-93-5p may participate in the formation of morphine tolerance in bone cancer pain mouse model by targeting Smad5. Oncotarget. 2016;7:52104–52114. doi: 10.18632/oncotarget.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol Biol Cell. 2006;17:402–412. doi: 10.1091/mbc.E05-07-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-H. [DOI] [PubMed] [Google Scholar]
- 19.Ho SL, Chan HM, Ha AW, Wong RN, Li HW. Direct quantification of circulating miRNAs in different stages of nasopharyngeal cancerous serum samples in single molecule level with total internal reflection fluorescence microscopy. Anal Chem. 2014;86:9880–9886. doi: 10.1021/ac5025182. [DOI] [PubMed] [Google Scholar]
- 20.Bruce JP, Yip K, Bratman SV, Ito E, Liu FF. Nasopharyngeal Cancer: Molecular Landscape. J Clin Oncol. 2015;33:3346–3355. doi: 10.1200/JCO.2015.60.7846. [DOI] [PubMed] [Google Scholar]
- 21.Wang HY, Yan LX, Shao Q, Fu S, Zhang ZC, Ye W, Zeng YX, Shao JY. Profiling plasma microRNA in nasopharyngeal carcinoma with deep sequencing. Clin Chem. 2014;60:773–782. doi: 10.1373/clinchem.2013.214213. [DOI] [PubMed] [Google Scholar]
- 22.Jiang L, Huang J, Li L, Chen Y, Chen X, Zhao X, Yang D. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. J Clin Endocrinol Metab. 2015;100:E729–E738. doi: 10.1210/jc.2014-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Wang H, Wang Y, Li Z, Pan Y, Liu Q, Yang M, Wang J. Repression of the miR-93-enhanced sensitivity of bladder carcinoma to chemotherapy involves the regulation of LASS2. Onco Targets Ther. 2016;9:1813–1822. doi: 10.2147/OTT.S97399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang H, Wang F, Chu D, Zhang W, Liao Z, Fu Z, Yan X, Zhu H, Guo W, Zhang Y, et al. miR-93 functions as an oncomiR for the downregulation of PDCD4 in gastric carcinoma. Sci Rep. 2016;6:23772. doi: 10.1038/srep23772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu MH, Han C, Srivastava AK, Cui T, Zou N, Gao ZQ, Wang QE. miR-93 promotes TGF-β-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37:5645–5651. doi: 10.1007/s13277-015-4328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu YF, Mao YP, Li YQ, Ren XY, He QM, Tang XR, Sun Y, Liu N, Ma J. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett. 2015;363:146–155. doi: 10.1016/j.canlet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Wang F, Xu J, Wang X, Ye F, Xie X. Micro ribonucleic acid-93 promotes oncogenesis of cervical cancer by targeting RAB11 family interacting protein 1. J Obstet Gynaecol Res. 2016;42:1168–1179. doi: 10.1111/jog.13027. [DOI] [PubMed] [Google Scholar]
- 28.Cioffi M, Vallespinos-Serrano M, Trabulo SM, Fernandez-Marcos PJ, Firment AN, Vazquez BN, Vieira CR, Mulero F, Camara JA, Cronin UP, et al. MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell R. 2015;12:1594–1605. doi: 10.1016/j.celrep.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Yi C, Wang Q, Wang L, Huang Y, Li L, Liu L, Zhou X, Xie G, Kang T, Wang H, et al. MiR-663, a microRNA targeting p21WAF1/CIP1, promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31:4421–4433. doi: 10.1038/onc.2011.629. [DOI] [PubMed] [Google Scholar]
- 30.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]