Abstract
The study aim was to evaluate the association of the expression of serum microribonucleic acid-590 (miR-590) with the risk of lung squamous cell carcinoma (LUSC), clinicopathological staging and prognosis. A total of 237 patients with LUSC and 100 healthy volunteers (control group) were included in the study. Total RNA was extracted from the peripheral blood serum of the subjects, and the expression level of miR-590 was detected by reverse transcription real-time quantitative polymerase chain reaction. The baseline clinicopathological information of LUSC patients was evaluated, and the patients were followed up with the median follow-up of 47 months. Compared with that in the control group, the expression level of serum miR-590 in LUSC patients was significantly decreased [0.532 (0.367- 0.821) vs. 1.63 (0.893–1.347), P<0.001]. The receiver operating characteristic (ROC) curve showed that the value of predicting LUSC risk using miR-590 was high, the area under curve (AUC) was 0.883, and 95% confidence interval (CI) was 0.829–0.934. In addition, the expression level of serum miR-590 was correlated with pathological staging (P=0.022), lymph node metastasis (P=0.012), distant metastasis (P<0.001) and tumor, node and metastasis (TNM) staging (P=0.044). The overall survival (OS) of patients in the serum miR-590 low expression group was significantly lower than that of the serum miR-590 high expression group (P=0.012), and the low expression of miR-590 was an independent risk factor for the prognosis of patients [hazard ratio (HR)=2.152, 95% CI=1.285–3.233, P=0.004]. The results suggested that the expression level of miR-590 can be used as a biomarker for the risk of disease, disease staging and prognosis of LUSC patients.
Keywords: miR-590, lung cancer, prognosis, lung squamous cell carcinoma
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. In China, approximately 2.5 million individuals succumb to the disease each year. In addition, lung cancer is the main cause of cancer mortality in males and the third major cause of mortality in females. Approximately 80% of lung cancer is classified as non-small cell lung cancer (NSCLC), including four major histological subtypes with different pathological characteristics: adenocarcinoma, lung squamous cell carcinoma (LUSC), large cell carcinoma and neuroendocrine cancer (1,2). According to treatment results, only patients in the early phase are willing to undergo surgery, leading to a 5-year overall survival (OS) rate of 50–60% (1,2). Notably, even when patients in the late stage undergo chemotherapy, molecular-targeted therapy or radiotherapy and chemotherapy, they do not survive for more than 5 years (3,4). In addition, cisplatin-based combination chemotherapy and targeted drugs also greatly improve the survival time and quality of life of patients with NSCLC. However, the efficacy of new drugs, such as tyrosine kinase inhibitors, pemetrexed and bevacizumab, in the treatment of LUSC is significantly lower than that of drugs for the treatment of adenocarcinoma. Therefore, identification of innovative and reliable biomarkers and treatment strategies for the diagnosis or prognosis is very imperative.
Microribonucleic acid (miRNA) is a small (19–22 nucleotides) non-coding single-chain RNA molecule. It is also a class of important non-protein-encoding RNA and target messenger RNA (mRNA) regulating gene expression, which triggers, translates or inhibits mRNA cleavage. Bioinformatics data suggest that a single miRNA can bind to hundreds of target mRNAs, thus playing increasingly important roles in various biological processes (5). Increasing evidence revealed that miRNA is associated with the occurrence of tumors (6). The lethal-7 (let-7) family and miR-155 are reportedly associated with the clinical outcome of lung adenocarcinoma (7,8). Therefore, the behavior of Ras, the negative regulatory oncogene of the let-7 family, shows that it can be regarded as a tumor suppressor gene (9). Although the biological function of miRNAs is not fully understood, miRNA analysis revealed that a wide range of tumor types exhibit significantly different expression profiles compared with normal tissues.
In order to better understand the involvement of specific miRNAs in multi-step canceration and progression, the association of clinicopathological factors with clinical outcomes of LUSC was further analyzed.
Materials and methods
General data
A total of 237 LUSC patients admitted to the Shaoxing People's Hospital (Zhejiang, China) from July 2005 to May 2011 were collected. The patients had not received radiation therapy, chemotherapy, surgery or other anti-cancer treatments before blood sampling. At the same time, 100 normal healthy volunteers were included as controls.
The study was approved by the Ethics Committee of the Shaoxing People's Hospital and all participants signed informed consent.
Specimen collection
Peripheral blood (5 ml) of LUSC patients and volunteers in the normal control group was placed into an ethylenediaminetetraacetic acid (EDTA)-free test tube before surgery. After standing for 45 min at 25°C, the centrifugation was conducted at 5,000 × g for 5 min, and the supernatant was removed, followed by another centrifugation at 12,000 × g at 4°C for 10 min. The supernatant was discarded and preserved in a refrigerator at −80°C for preparation.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Serum total RNA was extracted using TRIzol reagent according to the instructions of the RNA extraction kit (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA). The concentration and purity of RNA were determined using an ultraviolet spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sample quality requirements: A240/A300 of the total RNA solution was within the range of 1.8–2.1. If it failed to meet the standard, the total RNA was re-extracted. The quality of the extracted total RNA was analyzed by 1% denatured agarose gel electrophoresis. Reverse transcription of RNA was performed according to the instructions of the One Step PrimeScript miRNA cDNA Synthesis Kit (Takara Bio, Inc., Dalian, China), and quantitative PCR was performed using the SYBR® Premix Ex Taq™ II Kit (Takara Bio, Inc.). The relative expression of miR-590 was expressed as 2−ΔΔt, and the experiment was repeated three times for each sample. Primer Premier 5.0 software was used for miR-590 and U6 primer design: miR-590 (forward: 5′-ACACTCCAGCTGGGTATGGCTTTTTATTCCT-3′; reverse: 5′-GGTGTCGTGGAGTCGGCAA-3′); U6 (forward: 5′-CTCGCTTCGGCAGCACA-3′; reverse: 5′-AACGCTTCACGAATTTGCGT-3′).
Follow-up
All 237 patients were followed up by telephone or outpatient. The cut-off date of follow-up was June 2016 with the median follow up of 47 months.
Statistical analysis
Statistical analysis was performed using Statistical Product and Service Solutions (SPSS, Inc., Chicago, IL, USA) 21.0 software. Micro RNA-590 (miR-590) expression data were expressed as median (quartile spacing) [M (P 25, P 75)], and the Wilcoxon rank-sum test was used to compare the differences between the two groups. Differences in qualitative data between two groups were analyzed using the χ2 test or the Fisher's exact test. The receiver operating characteristic (ROC) curve was used to analyze the onset risk of lung cancer. The association between miR-590 and the OS of patients with lung cancer was analyzed by the Kaplan-Meier curve and log-rank test, and the univariate and multivariate Cox regression analysis. P<0.05 indicated that the difference was statistically significant.
Results
Basic information of tested subjects
Clinicopathological characteristics of the 237 LUSC patients are shown in Table I.
Table I.
Clinicopathological characteristics of LUSC patients.
| Characteristics | No. (%) | miR-590 [M (P 25-P 75)] | P-value |
|---|---|---|---|
| Sex | |||
| Male | 189 (79.7) | 0.587 (0.392–0.871) | 0.671 |
| Female | 48 (20.3) | 0.499 (0.357–0.776) | |
| Age | |||
| <60 | 167 (70) | 0.533 (0.362–0.859) | 0.897 |
| ≥60 | 70 (30) | 0.539 (0.370–0.810) | |
| Pathological staging | |||
| Stage I | 52 (21.9) | 0.420 (0.256–0.621) | 0.022 |
| Stage II | 42 (17.7) | 0.487 (0.327–0.688) | |
| Stage III | 105 (44.3) | 0.551 (0.387–0.751) | |
| Stage IV | 38 (16.1) | 0.620 (0.499–0.849) | |
| Tumor size | |||
| ≤50 mm | 187 (78.9) | 0.607 (0.492–0.891) | 0.152 |
| >50 mm | 50 (21.1) | 0.475 (0.320–0.722) | |
| Tumor, node and metastasis (TNM) staging | |||
| I/II | 154 (64.9) | 0.611 (0.501–0.913) | 0.044 |
| III/IV | 83 (35.1) | 0.399 (0.267–0.661) | |
| Lymph node metastasis | |||
| Positive | 72 (30.3) | 0.388 (0.255–0.620) | 0.022 |
| Negative | 165 (69.7) | 0.622 (0.513–0.855) | |
| Distant metastasis | |||
| Positive | 32 (13.6) | 0.384 (0.259–0.601) | <0.001 |
| Negative | 205 (86.4) | 0.633 (0.489–0.872) | |
| Smoking status | |||
| Yes | 198 (83.5) | 0.684 (0.487–0.874) | 0.042 |
| No | 39 (16.5) | 0.348 (0.247–0.583) |
LUSC, lung squamous cell carcinoma; miR-590, micro ribonucleic acid-590; TNM, tumor, node and metastasis.
Expression of serum miR-590 in LUSC patients
The expression of serum miR-590 in LUSC patients was significantly decreased compared with that in the control group [0.513 (0.387, 0.834) vs. 1.187 (0.901, 1.355), P<0.001]. The ROC curve was further applied, which showed that the risk of predicting LUSC using miR-590 was high, the area under curve (AUC) was 0.882, and the 95% confidence interval (CI) was 0.829–0.9334 (Fig. 1).
Figure 1.
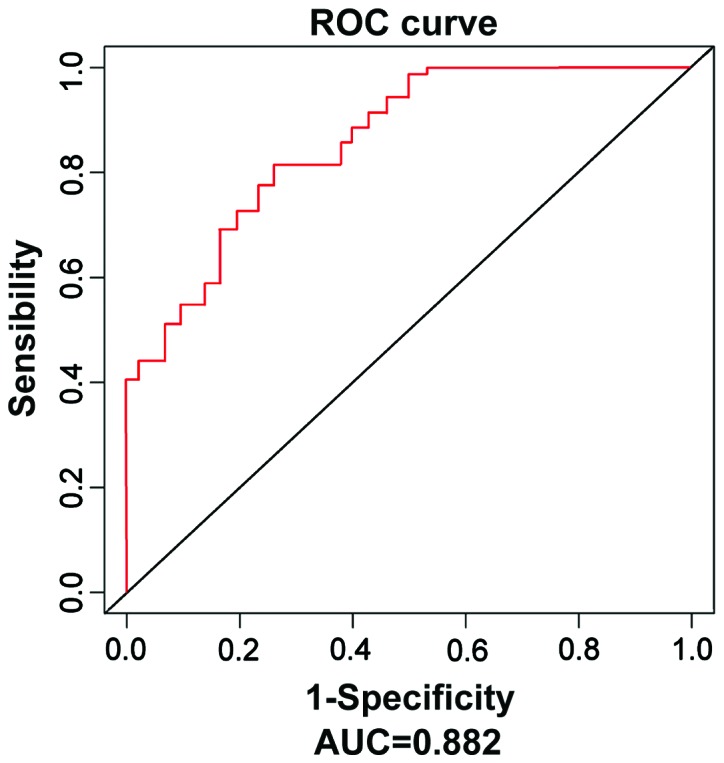
ROC curves of miRNA-195 in LUSC serum and serum in normal control group. miR-590 has a high value in predicting the LUSC risk (AUC=0.882, 95% CI=0.829–0.9334). ROC, receiver operating characteristic; miR-590, micro ribonucleic acid-590; LUSC, lung squamous cell carcinoma; AUC, area under curve; CI, confidence interval.
Association of the expression level of miR-590 with clinicopathological characteristics of LUSC patients
The expression level of miR-590 in LUSC patients was correlated with pathological staging (P=0.022), lymph node metastasis (P=0.012), distant metastasis (P<0.001), TNM staging (P=0.044) and smoking status (P=0.042), but not correlated with the age, sex or tumor size (Table I).
Association between the expression level of serum miR-590 and the OS of LUSC patients
Based on the median value (0.513) of the expression level of miR-590 in all 237 LUSC patients, these patients were divided into the high expression group (miR-590>0.513) and the low expression group (miR-590<0.513). The Kaplan-Meier curve and log-rank tests revealed that the OS of miR-590 low expression group was significantly lower than that of the high expression group (P=0.012) (Fig. 2).
Figure 2.
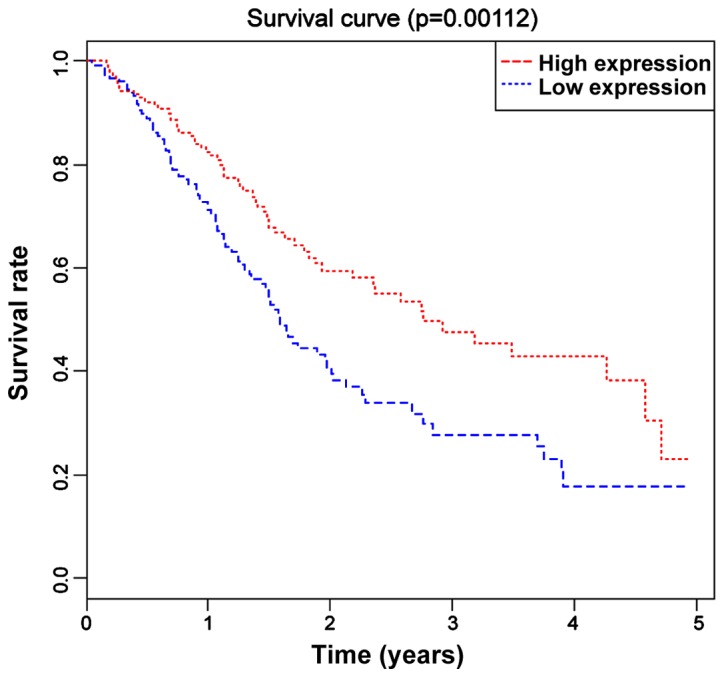
K-M survival curves of LUSC patients in high/low expression miR-590 groups. OS in the low expression miR-590 group is significantly lower than that in the high expression group (P=0.008, P<0.05). LUSC, lung squamous cell carcinoma; miR-590, microribonucleic acid-590; OS, overall survival.
Low expression of serum miR-590 is an independent risk factor for the prognosis of LUSC patients
The expression level of miR-590 and the clinicopathological characteristics of patients with LUSC were analyzed by the univariate Cox regression model. The low expression of miR-590 was significantly correlated with the prognosis of patients [hazard ratio (HR)=1.647, 95% CI=1.172–3.006, P=0.009]. In addition, pathological staging, lymph node metastasis, distant metastasis and TNM staging were also predictive factors for the prognosis of patients (Table II).
Table II.
Independent risk factors for the prognosis of LUSC patients.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Factor | HR (95% CI) | P-value | HR (95% CI) | P-value |
| miR-590 | 1.647 | 0.009 | 2.152 | 0.004 |
| (low vs. high) | (1.122–2.896) | (1.285–3.233) | ||
| Age | 1.014 | 0.062 | ||
| (>60 vs. ≤60) | (0.999–1.029) | |||
| Sex | 0.819 | 0.788 | ||
| (male vs. female) | (0.277–2.424) | |||
| Pathological staging | 2.143 | 0.013 | 1.268 | 0.113 |
| (low vs. high/medium differentiation) | (1.288–2.715) | (0.918–2.471) | ||
| Maximum diameter of tumor | 0.812 | 0.682 | ||
| (>50 mm vs. ≤50 mm) | (0.357–1.847) | |||
| Lymph node metastasis | 3.064 | 0.012 | 1.097 | 0.013 |
| (positive vs. negative) | (1.282–7.323) | (0.336–3.582) | ||
| Distant metastasis | 2.124 | 0.001 | 1.984 | 0.005 |
| (positive vs. negative) | (1.557–4.575) | (1.137–3.754) | ||
| TNM staging | 2.112 | |||
| (III/IV vs. I/II) | (1.231–3.625) | 0.036 | ||
LUSC, lung squamous cell carcinoma; HR, hazard ratio; CI, confidence interval; miR-590, micro ribonucleic acid-590; TNM, tumor, node and metastasis.
Furthermore, the multivariate Cox regression model was used to analyze the predictive factor for OS in patients with LUSC. The results indicated that the low expression of miR-590 was an independent risk factor for the prognosis of patients (HR=2.152, 95% CI=1.285–3.233, P=0.004). In addition, lymph node metastasis and distant metastasis were independent predictive factors for OS in patients with LUSC (Table II).
Discussion
Lung cancer remains the most common cause of cancer-associated mortalities in developed countries, and approximately 1.8 million new cases of lung cancer occurred in 2012 (10). Approximately 80% of lung cancer is classified as NSCLC, including four major histological subtypes: adenocarcinoma, LUSC, large cell carcinoma and neuroendocrine cancer (11). Compared with small cell lung cancer, NSCLC is less sensitive to anticancer drugs and radiation therapy. Recently, molecular-targeted therapy for adenocarcinoma (gefitinib, erlotinib and crizotinib) has shown a significant therapeutic effect. However, no targeted therapy has been approved for the treatment of LUSC (12). Therefore, a new treatment program for LUSC is required.
The present findings have shown that the expression of non-coding single-chain miR-590 was used to predict the risk of LUSC as well as the patient's pathological and TNM staging, and distant and lymph node metastasis. The most important finding was that the low expression of miR-590 was correlated with the low OS of patients, which was an independent prognostic factor for LUSC patients.
The abnormal expression of miRNAs is closely related to tumour formation, and certain key factors can promote or inhibit cell proliferation by regulating the pathways thereof (13,14). In addition, miRNAs are involved in regulating migration, metastasis and apoptosis of cells (15–17).
miR-590 is mainly composed of two genes, miR-590-3p and miR-590-5p. The present study revealed that the low expression of miR-590 was an independent prognostic factor for LUSC patients, which is consistent with the findings by Keller et al (18). Those authors identified that miR-590 expression is significantly downregulated in lung cancer tissues and cells compared with those in normal controls.
In the present study, the expression of miR-590 was associated with pathological TNM staging and lymph node and distant metastasis, which may be due to its inhibition of tumor cell migration and metastasis. In a related study, it was shown that decreased miR-590 upregulates transforming growth factor-β1 (TGF-β1) and TGF-TGF-βRII proteins removed, translated and inhibited by these miRNAs. Increased TGF-β1 and TGF-βRII result in enhanced collagen production and fibrosis (19).
Finally, findings of the present study indicated that the low expression of serum miR-590 was associated with low OS of patients with LUSC, which was an independent risk factor for prognosis. Possible causes were that, the low expression of miR-590 was related to the severity of disease of patients with LUSC (pathology and metastasis), and the more severe the disease, the worse the prognosis (20). There were also shortcomings to the study. The sample size was relatively small, and miR-590 affected LUSC clinicopathological staging as well as prognosis by regulating which target gene has yet to be explored. Future investigations should focus on the evaluation of the relationship between serum miR-590 and target gene expression levels as well as their association with clinicopathology and prognosis by including a large sample size, thereby evaluating the relationship of serum miR-590 with the efficacy and prognosis of patients receiving different treatments.
In summary, this study further showed that miRNAs can be used as an independent prognostic factor for tumors, and the low expression of miR-590 in LUSC patients is a risk factor of patients, which is correlated with pathological staging, lymph node and distant metastasis and TNM staging. In addition, the downregulation of miR-590 is associated with the OS of LUSC patients, which is an independent prognostic factor for LUSC patients. The aforementioned results demonstrate that the miR-590 expression level can be used as a biomarker for onset risk, disease staging and prognosis of LUSC patients.
References
- 1.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P, International Staging Committee and Participating Institutions The International Association for the Study of Lung Cancer Staging Project. Prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. Zhongguo Fei Ai Za Zhi Zhi stage in surgically managed non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2010;13:9–18. doi: 10.3779/j.issn.1009-3419.2010.01.02. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawabata N, Asamura H, Goya T, Mori M, Nakanishi Y, Eguchi K, Koshiishi Y, Okumura M, Miyaoka E, Fujii Y, Japanese Joint Committee for Lung Cancer Registry Japanese Lung Cancer Registry Study: First prospective enrollment of a large number of surgical and nonsurgical cases in 2002. J Thorac Oncol. 2010;5:1369–1375. doi: 10.1097/JTO.0b013e3181e452b9. [DOI] [PubMed] [Google Scholar]
- 3.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH, Eastern Cooperative Oncology Group Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 4.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 8.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 11.Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32:669–692. doi: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: Recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, Hu J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai J, Zhu X, Ma J, Wang W. miR-205 regulates A549 cells proliferation by targeting PTEN. Int J Clin Exp Pathol. 2015;8:1175–1183. [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H, Ding C, Zhang H, Gao J. Let 7 miRNAs sensitize breast cancer stem cells to radiation induced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep. 2016;14:3285–3292. doi: 10.3892/mmr.2016.5656. [DOI] [PubMed] [Google Scholar]
- 16.Bracken CP, Gregory PA, Khew-Goodall Y, Goodall GJ. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci. 2009;66:1682–1699. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang BB, Xie H, Wu T, Xie N, Wu J, Gu Y, Tang F, Liu J. Controlled-release mitomycin C-polylactic acid film prevents epidural scar hyperplasia after laminectomy by inducing fibroblast autophagy and regulating the expression of miRNAs. Eur Rev Med Pharmacol Sci. 2017;21:2526–2537. [PubMed] [Google Scholar]
- 18.Keller A, Backes C, Leidinger P, Kefer N, Boisguerin V, Barbacioru C, Vogel B, Matzas M, Huwer H, Katus HA, et al. Next-generation sequencing identifies novel microRNAs in peripheral blood of lung cancer patients. Mol Biosyst. 2011;7:3187–3199. doi: 10.1039/c1mb05353a. [DOI] [PubMed] [Google Scholar]
- 19.Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y, et al. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]


