Abstract
MicroRNA (miR)-613 has been reported to function as a tumor suppressor in several types of cancer. However, the biological function and underlying mechanism in gastric cancer (GC) has remained elusive. Therefore, the aim of the present study was to assess the expression and biological role of miR-613 in GC tissues and cell lines. miR-613 expression was found to be downregulated in 38 GC tissue samples compared to that in their adjacent non-cancerous tissues, and low expression of miR-613 was associated with lymph node metastasis and advanced tumor-nodes-metastasis stage. A gain-of-function assay demonstrated that miR-613 overexpression reduced tumor cell proliferation, migration and invasion of SGC-7901 cells, as determined by MTT and Transwell assays. Furthermore, brain-derived neutrophic factor (BDNF) was identified as a direct target of miR-613 in GC cells by a luciferase reporter assay. BDNF expression was upregulated and inversely correlated with miR-613 levels in GC tissues. In addition, knockdown of BDNF expression mimicked the tumor suppressive effect of miR-613 in GC cells. In conclusion, these findings demonstrated that miR-613 functions as a tumor suppressor in GC by targeting BDNF. Thus, miR-613 is a potential therapeutic target for GC.
Keywords: gastric cancer, miR-613, brain-derived neurotrophic factor, proliferation, invasion
Introduction
Gastric cancer (GC) is the is the fourth most commonly diagnosed cancer type and the second most frequent cause of cancer-associated mortalities worldwide, leading to >720,000 deaths annually (1,2). While advances have been made in the early detection of GC and therapy, and in part prevention, enhance the survival rate of patients with early GC, the disease remains incurable at the advanced stage with a poor 5-year survival rate of 4–5% (3), mainly due to unlimited tumor-cell proliferation as well as strong invasive and metastatic capacities (3). Therefore, further elucidation of the underlying molecular mechanisms involved in GC progression and metastasis is urgently required to improve the treatment and prolong the survival of patients with advanced GC.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs of 18–25 nucleotides in length that are important negative regulators of gene expression by directly targeting the three prime untranslated region (3′-UTR) of mRNAs, either promoting the degradation of target mRNAs or preventing their translation (4,5). Over the past few years, accumulating evidence suggested that miRNAs participate in the regulation of diverse biological processes, such as cell proliferation, cell cycle, apoptosis and cell differentiation, as well as metastasis, angiogenesis and immune responses (6). miRNAs have been reported to have important roles in tumorigenesis and function as tumor suppressors or oncogenes, depending on the function of their target genes (7,8). Recent studies also indicated that certain miRNAs are involved in GC development, including oncogenic miRNAs (such as miR-21, miR-210, miR-222 and miR-221) and tumor suppressor miRNAs (such as miR-124, miR145, miR-34 and miR-206) (9–11), suggesting miRNAs may act as diagnostic markers and therapeutic agents/targets for GC.
miR-613, a newly discovered miRNA, has been identified to act as a tumor suppressor in several types of cancer, including papillary thyroid carcinoma (12), prostate cancer (13), ovarian cancer (14), hepatocellular carcinoma (15,16), non-small cell lung cancer (17) and esophageal squamous cell carcinoma (18). However, the function of miR-613 in GC and the underlying molecular mechanisms have remained to be fully elucidated. The present study therefore investigated the association between miR-613 expression levels in GC tissues and clinicopathological parameters. The role of miR-613 in gastric carcinogenesis and metastasis as well as the underlying mechanisms were also investigated. The present study confirmed that miR-613 inhibits GC cell growth and metastasis through directly targeting brain-derived neurotrophic factor (BDNF), suggesting that miR-613 may be a potential therapeutic agent for GC.
Materials and methods
Patients and tissue samples
All clinical GC tissue samples and adjacent non-cancerous tissues were collected following obtainment of written informed consent from 38 patients who underwent gastric resection at the Department of Gastrointestinal Surgery of the China-Japan Union Hospital (Jilin University, Changchun, China) from September 2014 to September 2016. None of the patients had received any adjuvant chemotherapy prior to surgery. The pathological tumor type was diagnosed by two independent pathologists. Matched adjacent non-cancerous gastric epithelial tissues were collected >5 cm away from the tumors and were verified at the same time. The present study was approved by the Ethics Committee of the China-Japan Union Hospital, Jilin University (Changchun, China).
Cell culture
Four human GC cell lines (BGC-823, SGC-7901, MKN-45, AGS) and an immortalized gastric epithelial cell line (GES-1), were obtained from the Cell Bank of the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China), and grown in RPM1640 medium (HyClone; GE Healthcare, Little Chalfont, UK) supplemented with 10% fetal bovine serum (FBS; HyClone; GE Healthcare) in a humidified atmosphere containing 5% CO2 at 37°C.
RNA isolation and reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the tissue samples and cultured cells using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The RNA concentration and purity were determined using a Nanodrop 2000 (Thermo Fisher Scientific, Inc.). For detection of miR-613 expression, RT-qPCR was performed by using TaqMan miRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) in an ABI7500 sequence detector (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 small nuclear RNA was used as an endogenous control for data normalization. For detection of BDNF, 100 ng mRNA was reverse-transcribed to complementary DNA using a PrimeScript® RT reagent kit (Takara Bio, Inc., Otsu, Japan). RT-qPCR was performed in an ABI7500 sequence detector (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR Premix ExTaq kit (Takara Bio, Inc.) with primers for BDNF and β-actin as described previously (19). β-actin was used as endogenous control for data normalization. All qPCR reactions were performed in triplicate and relative quantification was performed using the 2−∆∆Cq method (95% confidence interval) with calibration to the corresponding endogenous control (20).
Cell transfection
SGC-7901 cells (1×105 per well) were inoculated in six-well plates and cultured for 24 h. The cells were transfected with miR-613 mimics or the corresponding negative control (miR-NC), or the small interfering (si)RNAs targeting human BDNF (si-BDNF) or the corresponding negative control (si-NC) (all from Ribo Bio, Guangzhou, China) using Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions.
Cell proliferation assay
An MTT assay was used to determine the cell proliferative capacity after transfection with miR-613 mimics or si-BDNF. In brief, transfected cells (2×104 cells/well) were seeded into 96-well culture plates and cultured in RPM1640 medium containing 10% FBS. After transfection for 24, 48 or 72 h, MTT reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well, followed by incubation at 37°C for an additional 4 h. Subsequently, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to dissolve the crystals for 10 min at 37°C. The spectrometric absorbance at 490 nm was measured by an EnSpire Multimode Plate Reader (PerkinElmer, Inc., Waltham, MA, USA).
Cell migration and invasion assays
The migration assay was performed using Transwell plates (cat. no. 3422; Corning Inc., Corning, NY, USA) containing membranes with 8-µm pores. Cell invasion assays were performed with invasion chambers pre-coated with Matrigel (cat. no. 354480; BD Biosciences, Franklin Lakes, NJ, USA). Cells (5×104 for migration assays and 2×105 for invasion assays) were seeded into the upper chamber in serum-free medium. Culture medium containing 10% FBS was added to the lower chamber as the chemoattractant. After the cells were incubated in a humidified incubator at 37°C for 24 h (migration assay) or 36 h (invasion assay), cells on the upper surface of the membrane were removed by scraping with a cotton swab, whereas cells attached to the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 10 min. Five randomly selected fields of the fixed cells were imaged and counted using a X71 light invert microscope (magnification, ×200; Olympus, Tokyo, Japan).
Luciferase reporter assay
Based on various miRNA databases [microRNA.org; miRDB (miRDB) and TargetScan (targetscan.org)], BDNF was predicted as a target of miR-613 in humans. To experimentally verify this prediction, a fragment (AAA AUU UGA ACC AAA ACA UUC CG) of the BDNF 3′UTR containing the predicted binding site was synthesized by GeneCopoeia, Inc. (Rockville, MD, USA), and inserted into the plasmid pGL3 (Promega Corp., Madison, WI, USA) downstream of the luciferase reporter gene, and named as wild-type (WT)-BDNF. A BDNF 3′UTR fragment (AAAAUUUGAACCAAAAGGTTGGG) with mutant (Mut) sequence was synthesized by GeneCopoeia, Inc., also inserted into the pGL3 luciferase reporter vector and referred to as Mut-BDNF. For the luciferase assay, SGC-7901 cells were co-transfected with the reporter plasmid (WT-/Mut-BDNF) and miR-613 mimics or miR-NC using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After 48 h of transfection, cells were harvested and the luciferase activity was determined using the Dual-Luciferase Reporter Assay system (Promega Corp.).
Western blot analysis
Protein was extracted from cells or tissues using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China), and the total protein concentration was measured with a bicinchoninic acid kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 30 µg protein/lane was separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat dry milk in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.1% Tween-20 (TBST), the membranes were incubated with antibody against BDNF (1:1,000 dilution, cat. no. sc-20981) or β-actin (1:5,000 dilution; cat. no. sc-70931; both from Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. The membranes were then washed with TBST and incubated with peroxidase-conjugated goat anti-mouse immunoglobulin G (1:3,000 dilution; cat. no. sc-516102; Santa Cruz Biotechnology, Inc.) for 2 h at 37°C. Protein bands were visualized using Pierce® enhanced chemiluminescence reagents (Thermo Fisher Scientific, Inc.) and exposed on an X-ray film(Thermo Fisher Scientific, Inc.). β-actin was used as an internal reference for relative quantification. Relative protein expression was evaluated using Image J software 3.1 (NIH, Bethesda, MD, USA).
Statistical analysis
Values are expressed as the mean ± standard deviation from at least three separate experiments. SPSS 19.0 software (International Business Machines, Corp., Armonk, NY, USA) was used for statistical analysis. Differences between groups were analyzed using Student's t-test and one-way analysis of variance followed by the Bonferroni post hoc test. The correlation between miR-613 and BDNF expression was tested using Pearson's correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-613 expression is downregulated in GC cells and tissues
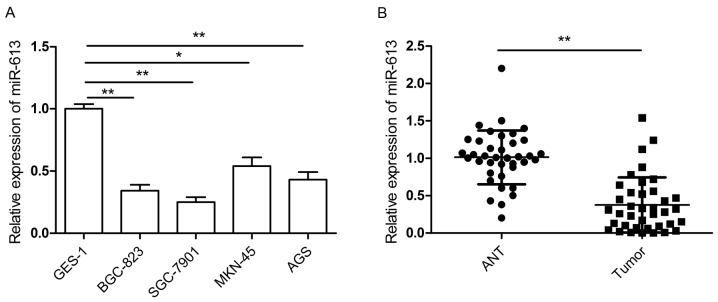
The present study assessed the expression of miR-613 in four human GC cell lines (BGC-823, SGC-7901, MKN-45 and AGS) as well as in a normal gastric epithelial cell line (GES-1) as a control. The RT-qPCR results demonstrated that miR-613 was downregulated in GC cell lines compared to the human normal gastric cell line (Fig. 1A). The expression of miR-613 in 38 pairs of GC tissue specimens and adjacent non-cancerous tissues was also examined. The results showed that miR-613 expression was significantly decreased in GC tissues compared with that in paired adjacent non-cancerous tissues (P<0.01; Fig. 1B). To examine the association between miR-613 expression and clinicopathological features, GC patients were categorized into two groups (low and high expression group) based on the median expression level of miR-613 (0.34) relative to U6 RUA. As shown in Table I, decreased expression of miR-613 was significantly associated with lymph node metastasis and advanced tumor-nodes-metastasis (TNM) stage. However, there was no significant association of miR-613 expression with any other factors, including age, sex and tumor size. These results indicated that downregulation of miR-613 may be involved in GC development and progression.
Figure 1.
miR-613 is downregulated in GC cell lines and tissues specimens. (A) Relative expression of miR-613 was detected in four GC cell lines and normal gastric cells using RT-qPCR. (B) Relative expression of miR-613 was measured in 38 pairs of GC tissues and ANT using RT-qPCR. Data are expressed as the mean ± standard deviation. *P<0.05, **P<0.01. miR, microRNA; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; GC, gastric cancer; ANT, adjacent noncancerous tissues.
Table I.
Association between clinicopathological features and miR-613 expression in 38 patients with gastric cancer.
| miR-613 expression | ||||
|---|---|---|---|---|
| Variables | Cases (n) | Low, n (%) | High, n (%) | P-value |
| Age (years) | 0.41 | |||
| <60 | 20 | 13 (65.0) | 7 (35.0) | |
| ≥60 | 18 | 10 (55.6) | 8 (44.4) | |
| Sex | 0.64 | |||
| Male | 21 | 13 (62.0) | 8 (38.0) | |
| Female | 17 | 10 (58.8) | 7 (41.2) | |
| Tumor size (cm) | 0.57 | |||
| <5 | 24 | 14 (58.3) | 8 (41.2) | |
| ≥5 | 14 | 9 (64.3) | 6 (35.6) | |
| Tumor-nodes-metastasis stage | <0.01 | |||
| I–II | 26 | 12 (46.2) | 13 (53.5) | |
| III–IV | 12 | 11 (91.7) | 1 (8.3) | |
| Lymph node metastasis | <0.01 | |||
| No | 25 | 11 (44.0) | 13 (56.0) | |
| Yes | 13 | 12 (92.3) | 1 (7.7) | |
miR, microRNA.
miR-613 inhibits cell proliferation, invasion and migration of GC cells
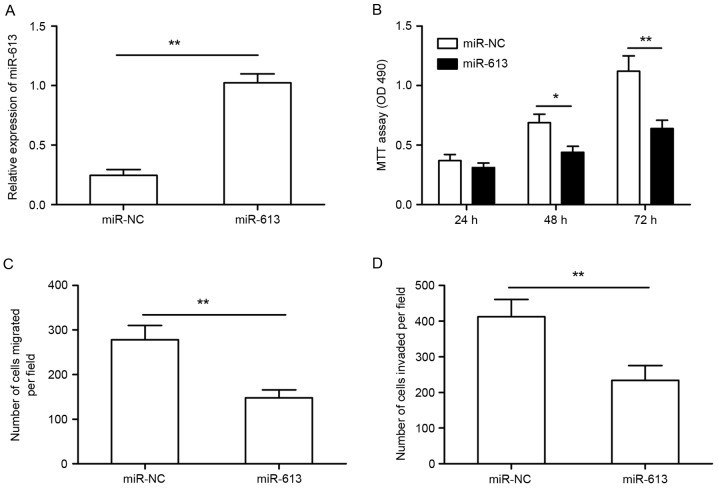
To explore the possible biological functions of miR-613 in GC cells, miR-613 mimics or miR-NC were transfected into SGC-7901 cells, and the transfection efficiency was determined by RT-qPCR. As shown Fig. 2A, miR-613 expression was significantly higher in SGC-7901 cells transfected with miR-613 mimic compared to that in cells transfected with miR-NC. An MTT assay demonstrated that cell proliferation was significantly impaired in SGC-7901 cells transfected with miR-613 mimics compared to that of cells transfected with miR-NC (Fig. 2B). To assess the effect of miR-613 on cellular motility, we Transwell assays were used to measure the migratory and invasive capacity of SGC-7901 cells. As shown in Fig. 2C and D, upregulation of miR-613 significantly suppressed cell migration and invasion, respectively, of SGC-7901 cells.
Figure 2.
miR-613 inhibits cell proliferation, invasion and migration in gastric cancer cells. (A) Reverse-transcription quantitative polymerase chain reaction was used to detect the transfection efficiency of miR-613 mimics in SGC-7901 cells. (B) The proliferation of SGC-7901 cells after transfection with miR-613 or miR-NC was determined by an MTT assay. (C and D) The cell migration and invasion, respectively, were determined in SGC-7901 cells after transfection with miR-613 or miR-NC by Transwell assays. *P<0.05, **P<0.01. miR-NC, negative control microRNA; OD, optical density.
BDNF is a direct target of miR-613 in GC cells
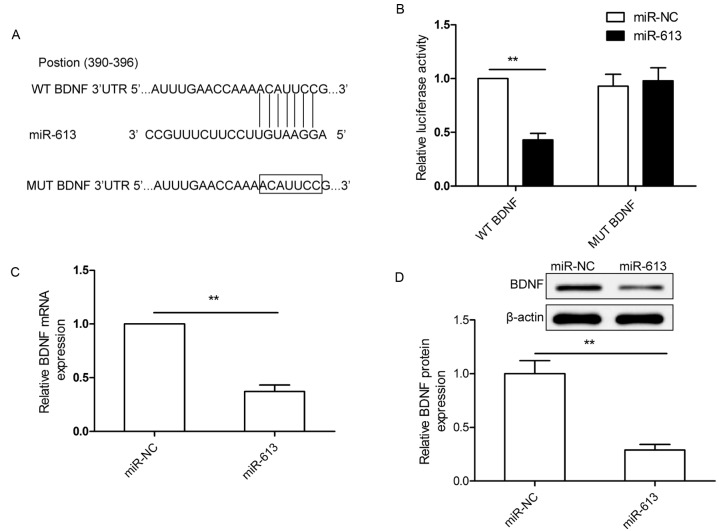
To investigate the mechanism by which miR-613 suppresses GC cell growth and metastasis, potential mRNA targets of miR-613 were screened by using three bioinformatics online tools for miRNA target prediction (microRNA.org, miRDB and TargetScan databases). BDNF was predicted to be a target of miR-613, since the seed sequence of miR-613 was complementary to the 3′-UTR of BDNF at position 390–396 (Fig. 3A). To further confirm that BDNF is a direct target of miR-613 in GC cells, a luciferase reporter assay was performed, which revealed that miR-613 had an obvious inhibitory effect on the luciferase activity in SGC-7901 cells transfected with the plasmid containing the WT, but not the Mut 3′-UTR fragment of BDNF (Fig. 3B). In addition, BDNF expression in SGC-7901 cells transfected with miR-613 mimics was decreased compared with that in cells transfected with miR-NC at the mRNA and the protein level (Fig. 3C and D). These results suggested that BDNF is a direct target of miR-613 in GC cells.
Figure 3.
BDNF is a direct target of miR-613 in GC cells. (A) miR-613 binding sites in the BDNF 3′UTR. Mut BDNF indicates the BDNF 3′UTR with a mutation in the miR-613 binding site. (B) Luciferase assay in SGC-7901 cells co-transfected with miR-613 mimic or miR-NC and luciferase reporter plasmid (WT/Mut 3′UTR BDNF). (C and D) BDNF expression in SGC-7901 cells after transfection with miR-613 mimics or miR-NC was determined at (C) the mRNA level by reverse-transcription quantitative polymerase chain reaction and (D) the protein level by western blot analysis. β-actin was used as an internal control. **P<0.01. WT, wild-type; Mut, mutant-type; UTR, untranslated region; miR-NC, negative control microRNA; BDNF, brain-derived neurotrophic factor.
BDNF is inversely correlated with miR-613 in GC tissues
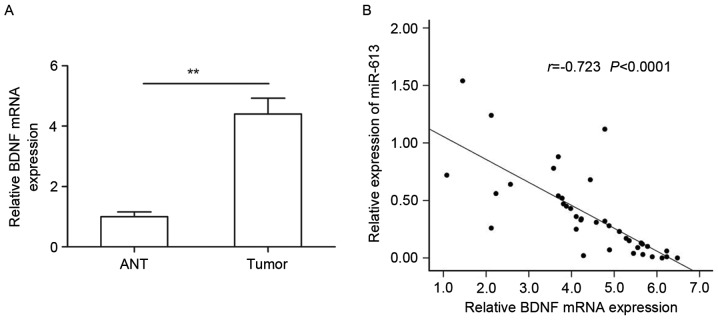
Next, BDNF mRNA expression in 38 pairs of GC tissue specimens and adjacent non-cancerous tissues was examined by RT-qPCR. The results revealed that BDNF expression in GC specimens was higher than that in the corresponding non-cancerous tissues (Fig. 4A). The inverse correlation between miR-613 and BDNF mRNA levels was further confirmed by a Pearson correlation analysis in 38 GC tissues (r=−0.738 P<0.0001; Fig. 4B).
Figure 4.
BDNF was upregulated and inversely correlated with miR-613 in GC tissues. (A) BDNF mRNA levels were measured in 38 pairs of GC tissues and ANT using reverse-transcription quantitative polymerase chain reaction. (B) Correlation of miR-613 and BDNF levels in human GC tissues was explored by Pearson correlation analysis (n=38). **P<0.01. ANT, adjacent noncancerous tissues; BDNF, brain-derived neurotrophic factor; GC, gastric cancer; miR, microRNA.
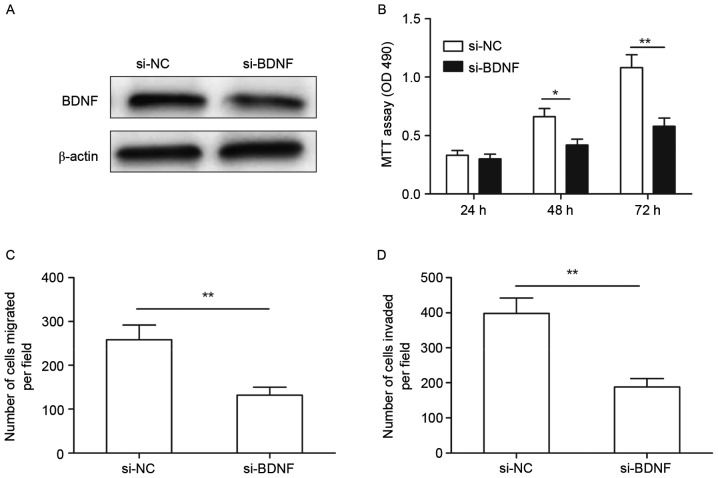
Knockdown of BDNF has a similar effect to miR-613 overexpression in GC cells
To investigate the function of BDNF in GC cells, si-BDNF or si-NC were individually transfected into SGC-7901 cells. As shown in Fig. 5A, knockdown BDNF by si-BDNF significantly decreased BDNF protein expression in SGC-7901 cells. In addition, downregulation of BDNF significantly inhibited cell proliferation (Fig. 5B), migration (Fig. 5C) and invasion (Fig. 5D) in SGC-7901 cells, which was a similar effect to that of miR-613 overexpression in SGC-7901 cells.
Figure 5.
Knockdown of BDNF has a similar effect to that of microRNA-613 overexpression in gastric cancer cells. (A) BDNF protein expression was determined in SGC-7901 cells transfected with si-BDNF or si-NC. β-actin was used for as an internal control. (B) Proliferation, (C) migration and (D) invasion of MG63 cells were determined after transfection with si-BDNF or si-NC. *P<0.05, **P<0.01. si-BDNF, small interfering RNA targeting brain-derived neurotrophic factor; si-NC, scrambled control small interfering RNA.
Discussion
Accumulating evidence has suggested that miRNAs regulate the expression of a variety of genes involved in various processes associated with the progression and genesis of various cancer types, including GC (9–11). The present study observed that miR-613 was frequently downregulated in GC cell lines and tissue samples compared to a normal gastric cell line and adjacent non-cancerous tissues, respectively. Furthermore, low expression of miR-613 was associated with lymph node metastasis and advanced TNM stage. A gain-of-function assay suggested that miR-613 suppressed GC cell proliferation, migration and invasion by repressing BDNF. These results suggested that miR-613 has crucial roles in GC initiation and progression.
miR-613 has been reported to be downregulated and to exert suppressive roles in several types of cancer by regulating oncogenes (12–18). For instance, Qiu et al (12) reported that miR-613 functions as a tumor suppressor in papillary thyroid carcinoma and its suppressive effect is mediated by repressing sphingosine kinase 2 expression. Wang et al (16) found that miR-613 suppresses hepatocellular carcinoma growth and invasiveness through targeting doublecortin-like kinase 1. Li et al (17) demonstrated that miR-613 inhibited cell proliferation and cell cycle progression in non-small cell lung cancer via direct binding to cyclin D kinase 4. Ren et al (13) reported that miR-613 suppressed cell proliferation and invasion in prostate cancer trough by downregulating the Frizzled 7/Wnt signaling pathway. However, to date, the biological function of miR-613 in GC and the underlying mechanisms have remained to be determined. The present study confirmed that miR-613 was downregulated in GC tissues and cell lines. To better understand the function of miR-613 in GC, the effects of miR-613 on GC cell proliferation, migration and invasion were examined in vitro using MTT and Transwell assays. The results demonstrated that miR-613 overexpression significantly inhibited GC cell proliferation, migration and invasion. These results suggested that miR-613 exerted a tumor suppressor role in GC.
To further investigate the molecular mechanisms underlying the ability of miR-613 to inhibit cell proliferation, migration and invasion in GC, BDNF, which was identified as a potential target gene of miR-613 according to a bioinformatics analysis, was selected for further study, as BDNF is known to have an oncogenic role in multiple cancer-associated processes (21–25). BDNF has been reported to be upregulated and contribute to the pathogenesis of multiple types of human cancer (20–24). A previous study showed that BDNF was upregulated in GC tissues compared to adjacent normal tissues, and overexpression of BDNF in GC cells promoted proliferation, migration, invasion and inhibition of anoikis (26), which suggested that BDNF was an oncogene in GC (26). In the present study, BDNF was further confirmed to be a direct target of miR-613 through a luciferase activity assay, RT-qPCR and western blot analysis. In addition, BDNF expression was found to be upregulated and to be inversely correlated with miR-613 in GC tissues. Downregulation of BDNF had similar anti-tumor effects on GC as overexpression of miR-613. These results suggested that miR-613 has a tumor suppressor role in GC, at least in part, by targeting BDNF.
In conclusion, the present study provided evidence that miR-613 expression was significantly decreased in GC cell lines and tissues, and its expression was significantly associated with advanced TNM stage and lymph node metastasis. Furthermore, miR-613 inhibited GC proliferation, migration and invasion by targeting BDNF. These findings may contribute to the further elucidation of the molecular mechanisms underlying GC progression and provide candidate targets for the prevention and treatment of GC.
Acknowledgements
This study was supported by Industrial Technology Research and Development Projects of Jilin Province (no. 2013C014-2) and the Provincial Economic Structure Strategic Adjustment Projects of Jilin Province (no. 2014Y083) from the Jilin Province Development and Reform Commission.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Yang CS, Varelas X, Monti S. Altered RNA editing in 3′ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci Rep. 2016;6:23226. doi: 10.1038/srep23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: From functions to targets. PLoS One. 2010;5:pii: e13067. doi: 10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JL, Hu Y, Kong X, Wang ZH, Chen HY, Xu J, Fang JY. Candidate microRNA biomarkers in human gastric cancer: A systematic review and validation study. PLoS One. 2013;8:e73683. doi: 10.1371/journal.pone.0073683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrestha S, Hsu SD, Huang WY, Huang HY, Chen W, Weng SL, Huang HD. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3:878–888. doi: 10.1002/cam4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, Lv M, Wang H, Guan W. Identification of circulating microRNAs as novel potential biomarkers for gastric cancer detection: A systematic review and meta-analysis. Dig Dis Sci. 2014;59:911–919. doi: 10.1007/s10620-013-2970-9. [DOI] [PubMed] [Google Scholar]
- 12.Qiu W, Yang Z, Fan Y, Zheng Q. MicroRNA-613 inhibits cell growth, migration and invasion of papillary thyroid carcinoma by regulating SphK2. Oncotarget. 2016;28:39907–39915. doi: 10.18632/oncotarget.9530. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ren W, Li C, Duan W, Du S, Yang F, Zhou J, Xing J. MicroRNA-613 represses prostate cancer cell proliferation and invasion through targeting Frizzled7. Biochem Biophys Res Commun. 2016;469:633–638. doi: 10.1016/j.bbrc.2015.12.054. [DOI] [PubMed] [Google Scholar]
- 14.Fu X, Cui Y, Yang S, Xu Y, Zhang Z. MicroRNA-613 inhibited ovarian cancer cell proliferation and invasion by regulating KRAS. Tumour Biol. 2016;37:6477–6483. doi: 10.1007/s13277-015-4507-7. [DOI] [PubMed] [Google Scholar]
- 15.Zhong D, Zhang Y, Zeng YJ, Gao M, Wu GZ, Hu CJ, Huang G, He FT. MicroRNA-613 represses lipogenesis in HepG2 cells by downregulating LXRα. Lipids Health Dis. 2013;12:32. doi: 10.1186/1476-511X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Zhang H, Wang L, Zhang S, Tang M. miR-613 inhibits the growth and invasiveness of human hepatocellular carcinoma via targeting DCLK1. Biochem Biophys Res Commun. 2016;473:987–992. doi: 10.1016/j.bbrc.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Li DQ, Liu D, Tang XJ. MiR-613 induces cell cycle arrest by targeting CDK4 in non-small cell lung cancer. Cell Oncol (Dordr) 2016;39:139–147. doi: 10.1007/s13402-015-0262-4. [DOI] [PubMed] [Google Scholar]
- 18.Guan S, Wang C, Chen X, Liu B, Tan B, Liu F, Wang D, Han L, Wang L, Huang X, et al. MiR-613: A novel diagnostic and prognostic biomarker for patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37:4383–4391. doi: 10.1007/s13277-015-4271-8. [DOI] [PubMed] [Google Scholar]
- 19.Yan H, Wu W, Ge H, Li P, Wang Z. Up-Regulation of miR-204 enhances anoikis sensitivity in epithelial ovarian cancer cell line via brain-derived neurotrophic factor pathway in vitro. Int J Gynecol Cancer. 2015;25:944–952. doi: 10.1097/IGC.0000000000000456. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Kang HJ, Kim JM, Kim SY, Kim SW, Shin IS, Kim HR, Park MH, Shin MG, Yoon JH, Yoon JS. A Longitudinal Study of BDNF promoter methylation and depression in breast cancer. Psychiatry Investig. 2015;12:523–531. doi: 10.4306/pi.2015.12.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang SM, Lin C, Lin HY, Chiu CM, Fang CW, Liao KF, Chen DR, Yeh WL. Brain-derived neurotrophic factor regulates cell motility in human colon cancer. Endocr Relat Cancer. 2015;22:455–464. doi: 10.1530/ERC-15-0007. [DOI] [PubMed] [Google Scholar]
- 23.Kang HJ, Kim JM, Kim SY, Kim SW, Shin IS, Kim HR, Park MH, Shin MG, Yoon JH, Yoon JS. A Longitudinal Study of BDNF promoter methylation and depression in breast cancer. Psychiatry Investig. 2015;12:523–531. doi: 10.4306/pi.2015.12.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka K, Okugawa Y, Toiyama Y, Inoue Y, Saigusa S, Kawamura M, Araki T, Uchida K, Mohri Y, Kusunoki M. Brain-derived neurotrophic factor (BDNF)-induced tropomyosin-related kinase B (Trk B) signaling is a potential therapeutic target for peritoneal carcinomatosis arising from colorectal cancer. PLoS One. 2014;9:e96410. doi: 10.1371/journal.pone.0096410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan DR, Matsumoto K, Lucarelli E, Thiele CJ. Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Eukaryotic Signal Transduction Group. Neuron. 1993;11:321–331. doi: 10.1016/0896-6273(93)90187-V. [DOI] [PubMed] [Google Scholar]
- 26.Okugawa Y, Tanaka K, Inoue Y, Kawamura M, Kawamoto A, Hiro J, Saigusa S, Toiyama Y, Ohi M, Uchida K, et al. Brain-derived neurotrophic factor/tropomyosin-related kinase B pathway in gastric cancer. Br J Cancer. 2013;108:121–130. doi: 10.1038/bjc.2012.499. [DOI] [PMC free article] [PubMed] [Google Scholar]