Abstract
Introduction
Mitomycin C is an anticancer antibiotic agent that has the potential for broad-spectrum use against several cancers, including mammary cancers. Because its half-life is 17 min after a 30 mg intravenous bolus administration, the suitability of mitomycin C for wide use in the clinical setting is limited. Based on tumor pathophysiology, pH-sensitive liposomes could provide better tumor-targeted effects. The aim of this study was to investigate the possibility of diminishing the side effect of mitomycin C by using pH-sensitive liposomes.
Materials and methods
pH-sensitive liposomes was employed to deliver mitomycin C and evaluate the characterization, release behaviors, cytotoxicity, in vivo pharmacokinetics and biochemical assay.
Results
The results demonstrated that mitomycin C-loaded pH-sensitive liposomes had a particle diameter of 144.5±2.8 nm and an entrapment efficiency of 66.5%. The in vitro release study showed that the pH-sensitive liposome release percentages at pH 7.4 and pH 5.5 were approximately 47% and 93%, respectively. The cell viability of MCF-7 cells showed that both the solution and liposome group exhibited a concentration-dependent effect on cell viability. The MCF-7 cell uptake of pH-sensitive liposomes with a folate modification was higher which was indicated by an increased fluorescence intensity compared to that without a folate modification. The area under the concentration–time curve of mitomycin C-loaded pH-sensitive liposomes (18.82±0.51 µg·h/L) was significantly higher than that of the mitomycin C solution group (10.07±0.31 µg·h/L). The mean residence times of the mitomycin C-loaded and mitomycin C solution groups were 1.53±0.16 and 0.05 h, respectively. In addition, there was no significant difference in terms of Vss (p>0.05). Moreover, the half-life of pH-sensitive liposomes and the mitomycin C solution was 1.35±0.15 and 1.60±0.04 h, respectively. In terms of safety, mitomycin C-loaded pH-sensitive liposomes did not affect the platelet count and the levels of blood urea nitrogen and aspartate aminotransferase.
Conclusion
The positive results of pH-sensitive liposomes demonstrated maintained the cytotoxicity and decrease the side effect.
Keywords: mitomycin C, liposomes, pH-triggered release, pharmacokinetics, side effect
Introduction
Mitomycin C, an anticancer antibiotic agent, is a fermentation product of Streptomyces caespitosus which was discovered over half a century ago.1 Mitomycin C has the potential for broad-spectrum use against several cancers, including mammary cancers, such as breast cancer, as well as stomach, colon, anal, and bladder cancers. Most of the chemotherapeutic agents are obstructed by multidrug resistance, which is a cause of chemotherapeutic treatment failure.2 Unlike general chemotherapeutic agents, mitomycin C is not associated with multidrug resistance by a common mechanism, and this allows the possibility of using mitomycin C in other clinical applications.3,4
Mitomycin C (C15H18N4O5) has a molecular weight of 334.33 Da, appears as a blue to purple crystalline powder, and is readily soluble in water (0.5 mg/mL). A clinical mitomycin C package has been commercialized as a 2, 10, 20, or 40 mg powder which can be injected as a solution using sodium chloride as dissolving medium; however, the half-life of mitomycin C is 17 min after a 30 mg intravenous bolus administration.5 Moreover, due to high toxicity, clinical intravenous administration of mitomycin C should be carried out avoiding extravascular leakage at the injection site, and the catheter location should be carefully checked to avoid possible necrosis.6 A serious drawback of mitomycin C is its dose-limiting toxicity, and its nonspecific antineoplastic effect must be taken into account when it is considered for broad-spectrum use.
Clinically, a mitomycin C dose of 0.58–1.2 mg/kg can achieve a satisfactory therapeutic response. The treatment course, however, which depends on the patient’s condition, particularly with respect to cumulative myelosuppression and bone marrow recovery, should be carefully reevaluated after treatment. Increasing the dose intervals or decreasing the dose administered is a common strategy to reduce adverse effects on patients; however, patient dysfunction is observed in laboratory setting within the therapeutic course.7,8
In clinical settings, more than 80% of all cancers are solid tumors.9 Based on tumor pathophysiology, there is a pH gradient between the extracellular pH (pHe 7.2) and intracellular pH (pHi 5.7–7.8), which exists as a gradient gap. By contrast, normal tissue maintains a constant pH of 7.4, and the pHi remains at 7.2.10 Moreover, many studies have noted that the extracellular environment of solid tumors is acidic due to cancer cells’ high rate of glycolysis.11 Based on tumor pathophysiology, pH-sensitive biomedical materials can be applied to a tumor-targeted drug delivery system. pH-sensitive liposomes have been widely investigated as they exhibit higher targeting and accumulating effect in cancer cells while also decreasing the toxicity of the therapeutic agent.12 The objective of this study was to design a suitable pH-sensitive liposomes formulation and understand the relative characteristics. The study investigated the in vitro characteristics, release behavior, cytotoxicity, and in vivo pharmacokinetic behavior and biochemical influence of mitomycin C-encapsulated pH-sensitive liposomes.
Materials and methods
Chemicals and reagents
Mitomycin C, 4-aminoacetophenone, dimethylsulfoxide, cholesteryl hemisuccinate (CHEMS), and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) were purchased from Sigma-Aldrich (St Louis, MO, USA). 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[folate(polyethylene glycol)-2000] (DSPE-PEG2000 folate), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) were supplied by Avanti Polar Lipid (Alabaster, AL, USA). Sodium hydroxide, phosphoric acid, disodium hydrogen phosphate dihydrate, and sodium dihydrogen phosphate monohydrate were obtained from Merck (Darmstadt, Germany). Triton X-100 was obtained from TCI (Tokyo, Japan). Methanol, ethanol, and analytical reagent-grade acetonitrile were obtained from Mallinckrodt (Staines-upon-Thames, UK). Human breast adenocarcinoma cell line (MCF-7) was purchased from the Culture Collection and Research Center (CCRC) of the Food Industry Research and Development Institute (FIRDI, Hsinchu, Taiwan).
HPLC analysis of mitomycin C
Mitomycin C analysis was performed using high-performance liquid chromatography (HPLC). The HPLC system consisted of a Hitachi model L-2130 pump (Tokyo, Japan), a Hitachi model L-2200 autosampler, a Hitachi model L-2450 diode array detector at 365 nm, and a Lichrocart® RP-18 column (125×4 mm, internal diameters I.D., 5 µm) (Merck). The mobile phase was a mixture of 0.01 M diammonium hydrogen phosphate (adjusted to pH 3.0 by phosphoric acid) and methanol (76:24, v/v), and the flow rate was 1.0 mL/min. 4-Aminoacetophenone was prepared as the internal standard (IS). Mitomycin C’s limit of detection and limit of quantitation were determined by dissolving mitomycin C at decreasing concentrations in distilled-deionized water until the signal-to-noise ratios were 3 and 10, respectively. The linearity of the standard curves, intraday and inter day precision, and accuracy were determined using six mitomycin C concentrations of 5, 10, 50, 100, 150, and 200 µg/mL.
Stability of mitomycin C at different pH values
Mitomycin C undergoes rapid decomposition in an acidic medium; hence, we tested different pH values to determine the stability of mitomycin C. A mitomycin C solution was dissolved in phosphate buffer saline (PBS) adjusted with 0.1 M NaOH and H3PO4 to different pH values at 25°C and analyzed by HPLC.
Preparation of pH-sensitive liposomes
Mitomycin C-loaded pH-sensitive liposomes were prepared using the thin-film method. The formulation was composed of DOPE, CHEMS, and DSPE-PEG2000 folate as the lipid phase, which was dissolved in 99.5% ethanol. The mixture was evaporated by a rotary evaporator at 60°C, and residual solvents were removed under nitrogen for 15 min. The film was hydrated with a 15 mM HEPES buffer solution with or without mitomycin C, and coarse colloids were dispersed by a probe sonicator (Branson Digital Sonifier® Model 250; Branson Ultrasonics Corp., Danbury, CT, USA) for 10 min at 5 W.
Particle size and zeta potential
The particle diameter (d) and zeta potential (ζ) of pH-sensitive liposomes were measured by laser-light scattering with a helium–neon laser at 630 nm (Zetasizer 3000HSA; Malvern Instruments, Malvern, UK). The polydispersity index (PdI) was used to measure the size distribution. All vesicles were diluted 80-fold with double-distilled water to record the count rate for the measurements. The determination was repeated three times for each of the three samples.
Encapsulation efficiency
The entrapment efficiency of mitomycin C in pH-sensitive liposomes was determined using an ultracentrifugation method. Fresh liposomal colloids were centrifuged at 36,000 rpm for 30 min at 4°C in a Hitachi CS150 GXL ultracentrifuge. The supernatant was removed and measured by HPLC. The pellets were dissolved with Triton X-100, passed through a PVDF filter, and then analyzed to determine the mitomycin C content.
The drug entrapment efficiency was calculated using the following equation:
Morphology
The morphology of the pH-sensitive liposome was observed by transmission electron microscopy (TEM) to characterize their microstructure. The liposomal colloids were dropped on a formvar/carbon film on a 200-mesh copper grid (FCF-200-Cu; Electron Microscopy Sciences, Hatfield, PA, USA) and stained with 1% phosphotungstic acid. This was adjusted to pH 6.5 by 0.1 M NaOH within 20 min of the negative staining process. The sample was washed for 8 h with distilled-deionized water, which was vacuumed out, and then observed using a transmission electron microscope (JEM-2000EX II; JEOL, Tokyo, Japan).
Storage stability
The liposomal colloids were stored at 4°C for 1 month. An aliquot of a sample was taken at predetermined time intervals to investigate d, PdI, and ζ. The concentration of mitomycin C was also determined at a final time point to ensure the changes in the drug encapsulation efficiency.
In vitro release study
An in vitro release study was carried out using dialysis method. Dialysis bags with a molecular weight cutoff of 6,000–8,000 Da (pore size 1.8 nm; Orange Scientific, Braine-l’Alleud, Belgium) were soaked in double-distilled water for 12 h before the experiment. Dialysis bags, containing mitomycin C with or without liposomes, were subsequently incubated in flasks containing 15 mL of medium (pH 7.4 or 5.5 PBS) at 100 rpm in a 37°C water bath. At appropriate intervals, 5,000 µL aliquots of the medium were withdrawn for analysis and immediately replaced with an equal volume of fresh medium.
Cytotoxicity assay
MCF-7 cells were incubated in minimum essential medium with 10% fetal bovine serum and 1% antibiotics in a humidified atmosphere containing 5% CO2/95% air at 37°C. The cells were seeded on 96-well culture plates at a density of 5×105 cells/well. After incubation for overnight, fresh medium containing mitomycin C with or without liposomes at final concentrations of 0.5, 5, 10, and 15 µM were added to the cells. After incubation for 48 h, the plates were washed with PBS and reacted with 10 µL of Cell Counting Kit-8 for 3 h. The number of surviving cells was measured by an enzyme-linked immunosorbent assay reader (Epoch™; Bio-Tek, Winooski, VT, USA) at 450 nm.
Flow cytometry
The uptake of mitomycin C-loaded liposomes by MCF-7 cells was evaluated using a flow cytometric assay (FACScan™; BD Biosciences, San Jose, CA, USA). DOPE-rhodamine B was used as a fluorescence indicator. MCF-7 cells were seeded on a 10 cm dish at a density of 1×106 cells and incubated for 24 h. The medium was removed and replaced with fresh medium containing mitomycin C-loaded liposomes at final concentrations of 0.5, 5, and 15 µM and incubated for 24 h. MCF-7 cells were lysed with Trype LE™ (Thermo Fisher Scientific, Waltham, MA, USA) and neutralized with medium. The mixture was centrifuged at 800 rpm for 5 min, and the supernatants were removed. Then, the cell pellets were washed with PBS twice, after which the final cells were suspended in PBS and analyzed by flow cytometry. The fluorescence signal was read with a 488 nm laser using an FACScan flow cytometer.
In vivo pharmacokinetics
In vivo pharmacokinetic studies were performed in healthy male Sprague Dawley (SD) rats that were 6–8 weeks old and weighed 220–250 g (the rats were obtained from the Laboratory Animal Centre of the National Science Council, Taipei, Taiwan). All animal experiments were conducted in accordance with institutional guidelines and approved by the Animal Care and Use Committee of Kaohsiung Medical University, Kaohsiung, Taiwan. All animals were starved overnight prior to the experiments. Rats were anesthetized by an intraperitoneal injection of 25% (w/v) urethane (3 mL/kg) before the experiment. Mitomycin C with or without liposomes was administered via the jugular vein at dosages of 5 mg/kg. The polyethylene tube (PE50) was implanted into the jugular vein for blood sampling. The tube was maintained patency by flushing with heparinized saline in each sampling time. At specified time intervals of 0.083, 0.167, 0.333, 0.5, 1, 2, 4, 6, and 8 h, 300 µL of blood was collected from the jugular vein in a heparin-containing tube (BD Vacutainer™; BD, Franklin Lakes, NJ, USA) and was immediately centrifuged at 12,000 rpm for 10 min. Plasma was collected and stored at −20°C. Rats were sacrificed at the end of time point, and the heart, liver, spleen, lungs, and kidneys were harvested and washed with PBS and then stored at −20°C.
The procedure of analysis of plasma mitomycin C was as follows. Plasma samples (180 µL) were mixed with 20 µL of a 300 µL/mL IS solution, and 360 µL of acetonitrile was added for protein precipitation. The mixture was centrifuged at 12,000 for 10 min, and the supernatants were evaporated under vacuum. The final dry residue, resuspended in mobile phase, was analyzed by HPLC. The organs were homogenized with 1 mL of PBS at 17,800 rpm for 30 s. They were then mixed with IS solution and acetonitrile and analyzed with HPLC.
Pharmacokinetic analysis
The pharmacokinetic compartment model was evaluated using the correlation coefficient from the observed and predicted data by using WinNonLin software (V5.3; Pharsight Software, Mountain View, CA, USA). The area under the concentration–time curve (AUC0–∞) was calculated by the trapezoidal rule. The half-life of the distribution phase (t1/2α), half-life of the elimination phase (t1/2β), and clearance (Cl) were also calculated. The Cl value was calculated as the dose/AUC0–∞, and the mean residence time (MRT) and volume of distribution at a steady state (Vss) were obtained by summation of the central and tissue compartments.
Biochemical analysis
Blood was collected in a test tube containing a heparin anti-coagulant (BD Vacutainer™; BD) for complete blood count analysis. Blood was analyzed to determine the number of erythrocytes, platelets, and total white blood cells (WBCs) by an automated blood analyzer (Sysmex XT-1800i; Sysmex, Kobe, Japan). Hemoglobin (Hb), red blood cells (RBCs), and hepatic function were also analyzed.
Statistical analysis
Statistical analyses were carried out utilizing unpaired Student’s t-test with Winks SDA 6.0 software (TexaSoft, Cedar Hill, TX, USA). An analysis of variance test was also used. Subgroup comparisons were carried out using Newman–Keuls multiple comparisons. A 0.05 level of probability was used as the level of significance. All data are expressed as mean ± standard deviation.
Results and discussion
Stability of mitomycin C at various pH values
Mitomycin C is unstable in an acidic environment.13 To confirm the reality of changes at different pH values, we investigated the mitomycin C content after incubation in mediums with different pH values. The results showed that at pH 3 mitomycin C significantly decreased to approximately 10% (p<0.05), after incubation for 6 or 24 h (Figure 1). At pH 5, however, mitomycin C remained at 30% after incubation for 24 h. An incubation period longer than 6 h did not significantly decrease the mitomycin C content (p>0.05). Over the past few decades, several studies have been carried out to evaluate the influence of pH on mitomycin C.14 Underberg and Beijnen indicated that mitomycin C mainly degraded in acidic solutions according to the protonation status of the aziridine nitrogen in the molecules as well as the protonation degree of the opened aziridine in key intermediate species.15 In the present study, to choose a suitable aqueous medium and confirm that pH had no influence on mitomycin C, a brief pH stability test was carried out. The results clearly showed that mitomycin C was unstable, with rapid degradation at pH 3. Moreover, mitomycin C was stable at both neutral pH 7.4 and alkali pH 8.5. When incubated at pH 5.5, mitomycin C decreased by only approximately 10% within 6 h; however, when mitomycin C was incubated for 24 h, the decrease in its content was greater, approximately 30%. This implies that the stability of mitomycin C in a pH 5.5 environment is relative to exposure time. The primary screen provided evidence for choosing a suitable aqueous medium as the hydration medium for liposome production.
Figure 1.
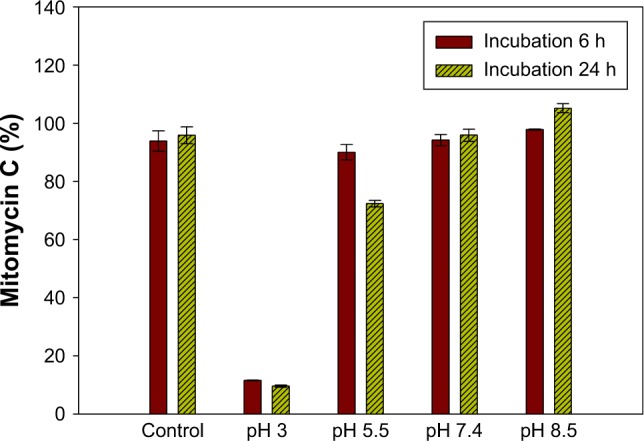
Mitomycin C contents after incubation at a range of pH 3–8.5. Water was the dissolved medium of the control group.
Note: Data are presented as mean ± standard deviation (n=3).
Characterization of mitomycin C-loaded pH-sensitive liposomes
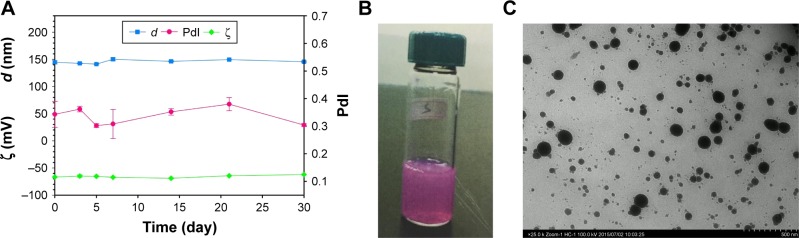
The diameter of the particles (d), PdI, and zeta potential (ζ) of mitomycin C-loaded pH-sensitive liposomes were examined (Figure 2A). Liposomal colloids were prepared by the thin-film method, using empty liposomes with a small size of 136.8±1.3 nm, PdI of 0.18±0.02, and negative charge of −44.64±0.658 mV. Mitomycin C-loaded liposomes consisted of spherical nanoparticles of 144.5±2.8 nm diameter with a PdI of 0.343 and a negative charge of approximately −64.9±5.1 mV. The formulation could carry hydrophilic mitomycin C in pH-sensitive liposomes and achieved an encapsulation efficiency of 66.5% when freshly prepared. Moreover, mitomycin C-loaded pH-sensitive liposomes appeared purple in color which was contributed by mitomycin C itself (Figure 2B). In addition, the microstructure of mitomycin C-loaded pH-sensitive liposomes was analyzed by TEM which showed nanometer-sized spherical outlines (Figure 2C).
Figure 2.
(A) The characterization of mitomycin C-loaded pH-sensitive liposomes by evaluation of d, PdI, ζ and its storage for a 1-month period. (B) Observational view. Mitomycin C contributed to the purple color. (C) Observation of morphology using TEM (original magnification ×25,000).
Note: Data are presented as mean ± standard deviation (n=3).
Abbreviations: d, diameter of particle; PdI, polydispersity index; ζ, zeta potential; TEM, transmission electron microscopy.
We also attempted to prepare the same formulation using ethanol injection technique. This produced smaller-sized particles of approximately 60 nm with a smaller distribution in PdI; however, the encapsulation efficiency was only approximately 10% (data not shown). Therefore, we utilized the thin-film method. In the art of drug loading, generating a hydrophilic composition has proven to be difficult for liposome loading and depends on the small volume of the internal aqueous compartment of the liposome.16 The ethanol injection method generally forms smaller internal aqueous areas than other preparation methods.17
Choosing FDA-approved materials can accelerate the research and development of new drugs for clinical use. Phosphatidylethanolamine (PE) derivatives are commonly used components of a series of phospholipid materials. PE derivatives have a smaller head group (ethanolamine) than phosphatidylcholine (PC).18 Therefore, they have a lower volume compared to hydrocarbon chains and are cone shaped, in contrast to cylinder-shaped PC.19 Moreover, due to the ethanolamine head group, PE derivatives have a more fluid membrane property than PC.20 DOPE, which has a dioleoyl carbon side chain, has a self-formed hexagonal (HII) structure in the physiological environment.21 When incorporated with CHEMS as a stabilizer, DOPE is assembled into a bilayer that is stable in the physiological environment. pH-sensitive liposomes are destabilized in an acidic environment (pH <6). The Warburg effect, pertaining to tumor pathology, is a theory which states that tumors produce energy due to a high rate of glycolysis followed by lactic acid fermentation in the cytosol, rather than by a comparatively low rate of glycolysis followed by the oxidation of pyruvate in the mitochondria, as occurs in most normal cells.18 Tumor growth progresses in an acidic microenvironment, which occasionally triggers mitomycin C release from pH-sensitive liposomes. Moreover, to increase the half-life of mitomycin C, PEGylation was required.
In the hydration process, which is important in this case, pH 7.4 PBS was initially selected as the aqueous phase; there was a caked layer that was not hydrated with the thin film. For easy hydration, we chose pH 8.5 HEPES buffer as the hydration medium. Past evidence, obtained by employing the same formulation loaded with another hydrophobic compound, showed the same results. We also used a formulation that only included DOPE and CHEMS, which again provided the same result. The results depended on the composition of DOPE and CHEMS. First, with DOPE at a physiological pH (pH 7.4), the vesicles did not form a bilayer, but instead formed an inverted hexagonal (HII). At a higher pH of approximately 8.5–9.0, the amine group of DOPE was deprotonated, leading to easy self-assembly with CHEMS. According to the afore-mentioned evidence, the hydration medium also plays an important role in the preparation of pH-sensitive liposomes.
Storage stability
The results also showed that there were changes in the particle size (d), PdI, and zeta potential (ζ) within 1 month of storage at 4°C (Figure 2A). However, the changes were not significant (p>0.05). Moreover, the encapsulation efficiency was maintained at 64%, with no significant decrease at the end point at 4°C storage (p>0.05). A stability evaluation showed that after storage for 1 month, mitomycin C-loaded pH-sensitive liposomes retained all of their initial parameters, including particle size, zeta potential, and entrapment efficiency. The liposomes that had a negative charge of approximately −65 mV in zeta potential were stable after 1 month of storage; this mitomycin C-loaded pH-sensitive liposomal colloid existed in an environment with a negative charge, which provided electric repulsion to help prevent particle aggregation.22 On the other hand, pH-sensitive liposomes released their contents in an acidic environment.23 This concept was observed as detailed in the following in vitro release study.
In vitro release
We examined the pattern of release of mitomycin C from pH-sensitive liposomes under physiological and acidic pH conditions (Figure 3). Mitomycin C release from pH-sensitive liposomes at pH 7.4 and pH 5.5 was approximately 47% and 93%, respectively. There was a significant difference in mitomycin C release from pH-sensitive liposomes at pH 7.4 and 5.5 (p<0.05). In addition, mitomycin C was barely released from pH-sensitive liposomes at pH 7.4 after 1 h. By contrast, mitomycin C was released from pH-sensitive liposomes in a pH 5.5 environment. In an acidic environment, such as tumor sites, CHEMS played a predominant role in pH-sensitive liposomes. CHEMS is protonated in an acidic environment, triggering liposome release.24 Moreover, a rapid release of mitomycin C from pH-sensitive liposomes was observed at pH 5.5, with a relatively low pH-induced destabilization of liposomes.25 At pH 6.0–6.5 in early endosomes and pH 4.5–5.5 in late endosomes, the rapid-release behavior could potentially lead to enhanced cytoplasmic delivery through passive diffusion and/or endosomal escape mechanisms.26
Figure 3.
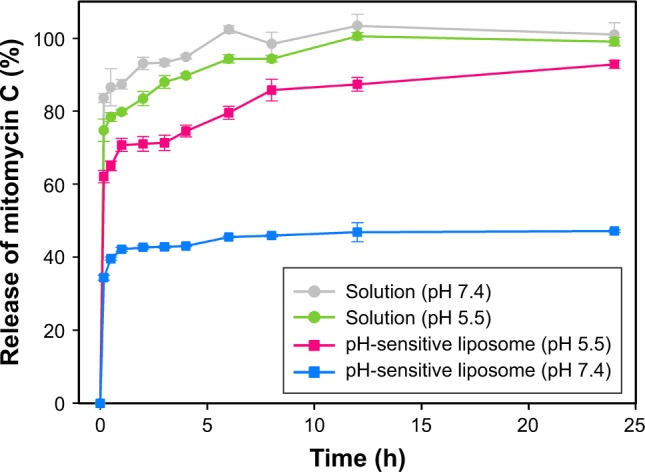
In vitro release behavior of mitomycin C from pH-sensitive liposomes in a physiological (pH 7.4) and a tumor acidic environment (pH 5.5).
Note: Data are presented as mean ± standard deviation (n=3).
Cell viability and uptake of MCF-7 cells
The cell viability of MCF-7 cells was examined as follows (Figure 4). An aqueous solution was used as the medium in the mitomycin C solution group. The addition of 0.5, 5, 10, and 15 µM of mitomycin C solution reduced the viability of MCF-7 cells from 64.8% to 38.9%. There was also a concentration-dependent trend with pH-sensitive liposomes; however, the cell viability of MCF-7 cells slightly increased at each concentration. The results showed that the cytotoxic activity of mitomycin C was substantially decreased when it was incorporated into pH-sensitive liposomes. This may have been due to the binding of mitomycin C to phospholipids, limiting its cytotoxicity. Past work has indicated that the lipids bind to the drug itself, subsequently reducing its cytotoxicity.27 A cell viability assay was performed using a physiological pH culture system, which may have limited the cytotoxicity of mitomycin C-loaded pH-sensitive liposomes.
Figure 4.
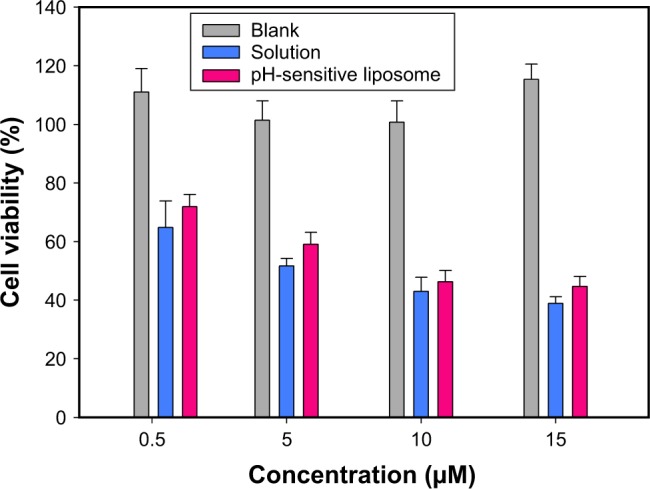
Cellular viability of MCF-7 cells after a 48-h treatment with different concentrations of mitomycin C solution and mitomycin C-loaded pH-sensitive liposomes. Blank indicates cells treated with pH-sensitive liposomes without mitomycin.
Note: Data are presented as mean ± standard deviation (n=3).
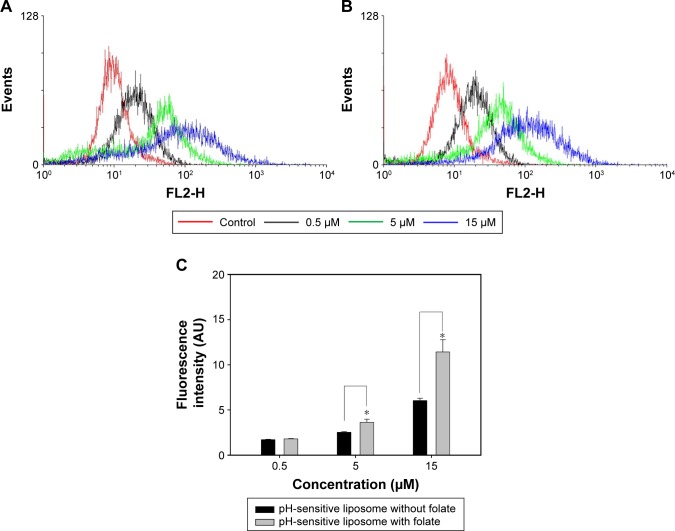
Flow cytometry was performed to understand MCF-7 cell uptake of mitomycin C-loaded pH-sensitive liposomes (Figure 5). Due to the high affinity of folic acid for folate receptors, we prepared two pH-sensitive liposomes with and without a folate modification. The quantification results showed that for pH-sensitive liposomes with the folate modification, the fluorescence intensity increased (to the right side). In addition, the quantification data showed a significantly increased uptake of liposomes with the addition of 15 µM mitomycin C-loaded pH-sensitive liposomes (p<0.05). This phenomenon demonstrated that the folate modification increased uptake by MCF-7 cells. The folate receptor-targeting concept has been evaluated for its ability to enhance tumor cell-selective delivery of a wide variety of therapeutic agents;28 however, this strategy does not always work. Previous research also discussed folate receptor-targeting treatment as being successful relative to the folate receptor expression levels on patients’ tumor cells.29 To understand the influence of the folate modification on uptake of MCF-7 cells, mitomycin C-loaded pH-sensitive liposomes with or without the folate modification were examined. We found that mitomycin C-loaded pH-sensitive liposomes with the folate modification showed a significantly higher uptake behavior, despite the lack of folate receptors on MCF-7 cells.30 This indicates that MCF-7 cells might have folate receptors; however, the levels of folate receptor on MCF-7 cells are insufficient.
Figure 5.
For flow cytometric analyses, MCF-7 cells were treated for 24 h with 0.5, 5, and 15 µM of pH-sensitive liposomes (A) with or (B) without a folate modification. (C) Fluorescence intensities were quantified as cellular uptake. Control was a cell blank without any treatment.
Notes: Data are presented as mean ± standard deviation (n=3). *p<0.05 was considered statistically significant.
In vivo pharmacokinetics
The plasma concentration–time profile of mitomycin C was evaluated with or without loading in pH-sensitive liposomes after intravenous administration in SD rats at a 5 mg/kg dose (Figure 6). There was a significant difference in the plasma concentration of mitomycin C between loaded and unloaded pH-sensitive liposomes. The mitomycin C concentration in plasma dramatically decreased from 12.78 to 2.18 µg/mL within 1 h. In addition, mitomycin C was almost not detected at 6 and 8 h after administration. To understand this pharmacokinetic behavior, WinNonLin software was used to calculate the data. Table 1 summarizes the pharmacokinetic parameters of mitomycin C with or without loading in pH-sensitive liposomes according to the intravenous bolus two-compartment model analysis. There was a marked difference in the AUC of mitomycin C with (18.82±0.51) and without (10.07±0.31) pH-sensitive liposomes. The Cl value of mitomycin C-loaded pH-sensitive liposomes was significantly lower than that of the mitomycin C solution (p<0.05). Moreover, mitomycin C-loaded pH-sensitive liposomes had higher AUC and MRT than the mitomycin C solution (p<0.05). In addition, there was no significant difference in terms of Vss (p>0.05).
Figure 6.
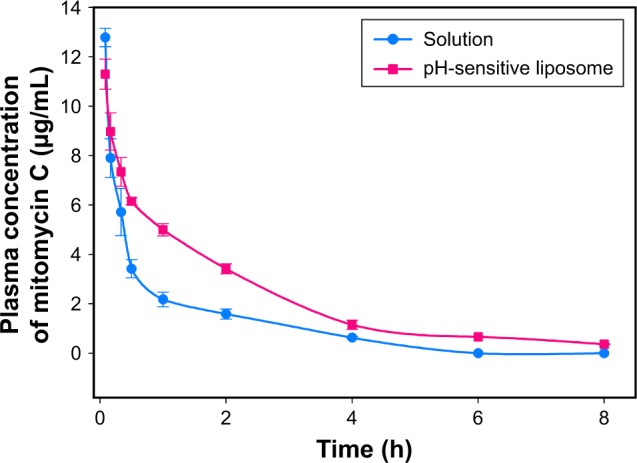
Plasma concentration–time curves of mitomycin C in the control solution and pH-sensitive liposomes after intravenous administration of 5 mg/kg in Sprague Dawley rats.
Note: Each value is presented as mean ± SD (n=3).
Table 1.
Pharmacokinetic parameters after 8 h of intravenous administration of mitomycin C or mitomycin C-loaded pH-sensitive liposomes to rats at a dose of 5 mg/kg
| PK parameters | Solution | pH-sensitive liposomes |
|---|---|---|
| AUC0–∞ (µg·h/L) | 10.07±0.31 | 18.82±0.51* |
| t1/2α (h) | 0.10±0.04 | 0.08±0.02 |
| t1/2β (h) | 1.35±0.15 | 1.60±0.04* |
| Cl (L/h/kg) | 0.10±0.03 | 0.05±0.01* |
| MRT (h) | 1.53±0.16 | 2.21±0.05* |
| Vss (L/kg) | 0.13±0.01 | 0.12±0.01 |
Notes: Data are presented as mean ± standard deviation (n=3).
p<0.05 was considered statistically significant.
Abbreviations: PK, pharmacokinetic; AUC0–∞, area under the concentration–time curve; t1/2α, half-life of the distribution phase; t1/2β, half-life of the elimination phase; Cl, clearance; MRT, mean residence time; Vss, volume of distribution at a steady state.
According to the pharmacokinetic results, the AUC of the mitomycin C solution in plasma was significantly lower than that of the pH-sensitive liposomes group. This result was in spite of the fact that the initial sampling time was longer than that of the pH-sensitive liposomes group (solution 12.78±0.37 µg/mL; pH-sensitive liposomes 11.30±0.61 µg/mL). Moreover, the Cl value was also significantly higher in mitomycin C solution group compared to the pH-sensitive liposomes group. This suggested that the mitomycin C solution was rapidly eliminated from blood circulation. On the other hand, mitomycin C-loaded pH-sensitive liposomes had increased AUC and MRT values; they demonstrated not only better performance with regard to the AUC but also increased mitomycin C in plasma circulation. Moreover, the t1/2 of the pH-sensitive liposomes group was slightly higher than that of the mitomycin C solution, with mitomycin C exhibiting extended retention time in plasma due to encapsulation in the pH-sensitive liposomes. When mitomycin C was encapsulated in pH-sensitive liposomes, the elimination phase was prolonged, although there was a significant difference. Based on pharmacokinetic analysis, mitomycin C-loaded pH-sensitive liposomes exhibited enhanced plasma circulation times and a higher concentration compared with the mitomycin C solution group.
The results concluded that the following contributed to the increase in the half-life of mitomycin C. In general, larger particles are eliminated from blood circulation more rapidly than smaller ones. In particular, phagocytes can distinguish between different-sized foreign particles. The mononuclear phagocyte system (MPS) primarily takes up foreign particles that are between 500 and 5,000 nm in size.31,32 Moreover, polyethylene glycol modification contributes to the increased half-life and decreased Cl of mitomycin C-loaded liposomes.33 PEGylation also altered pharmacokinetics, reduced MPS uptake, and enhanced the stability of pH-sensitive liposomes, decreasing vesicle aggregation in our study.34
Biochemical assay
pH-sensitive liposomes could increase the short half-life of mitomycin C; however, the safety of mitomycin C-loaded pH-sensitive liposomes remains a matter of concern. A common side effect of mitomycin C was low blood counts including temporary decrease of WBCs, RBCs, and platelets. When mitomycin C is continuously used, the side effect may occur after 7–10 days.35,36 Hence, the biochemical parameters of mitomycin C-loaded pH-sensitive liposomes were tested after intravenous injections for 7 days (Table 2). Mitomycin C solution induced significant increases in blood urea nitrogen and aspartate aminotransferase compared with the control (p<0.05); however, the effect of pH-sensitive liposomes was similar to that of control. Moreover, the mitomycin C solution also caused a significant decrease in platelets (p<0.05). The mitomycin C-loaded pH-sensitive liposomes did not cause significant changes compared to unloaded liposomes (p>0.05). In addition, WBCs, lymphocytes, Hb, hematocrit, creatinine, and alanine aminotransferase were not significantly changed in the presence of mitomycin C solution and mitomycin C-loaded pH-sensitive liposomes.
Table 2.
Complete blood counts after 7 days of administration of a mitomycin C solution or mitomycin C-loaded pH-sensitive liposomes to rats at a dose of 5 mg/kg
| Assay | Blank | Solution | pH-sensitive liposomes | Reference values |
|---|---|---|---|---|
| White blood cells (×103/mm3) | 6.2±2.4 | 4±0.7 | 4.6±0.2 | 6–17 |
| Lymphocyte (%) | 58.8±15.6 | 70±6.2 | 53.1±19.6 | 65–85 |
| Hemoglobin (mg/dL) | 16.1±1.7 | 15.45±1.1 | 12.4±1.6 | 11–18 |
| Hematocrit (%) | 43.8±4.5 | 43.45±2.6 | 34.3±5.5 | 36–48 |
| Red blood cells (×106/mm3) | 7.71±1.14 | 7.36±0.8 | 5.67±0.97 | 7–10 |
| Platelets (×103/mm3) | 690±174 | 356±144.2* | 645±295 | 500–1,300 |
| Blood urea nitrogen (mg/dL) | 16.3±0.3 | 24.2±3.7* | 14.1±1.9 | 15–21 |
| Creatinine (mg/dL) | 0.1±0.1 | 0.25±0.2 | 0.2±0.1 | 0.2–0.8 |
| Aspartate aminotransferase (IU/L) | 127±10 | 203.5±17.7* | 114±25 | 63–175 |
| Alanine aminotransferase (IU/L) | 24±0 | 21.5±3.5 | 20±3 | 20–92 |
Notes: Data are presented as mean ± standard deviation (n=3).
p<0.05 was considered statistically significant.
The major side effect of mitomycin C is bone marrow suppression, which may cause severe blood problems, such as low platelet levels and low WBC levels (leukopenia).37 Moreover, mitomycin C also increases the risk of developing homolytic uremic syndrome.38 In our study, mitomycin C caused a decrease in platelet count, which was dependent on its concentration. In addition, mitomycin C solution led to a low WBC count. However, mitomycin C-loaded pH-sensitive liposomes did not affect the platelet or WBC count, and there was no significant difference from unloaded liposomes. A previous study found that liposomes lead to platelet count fluctuations that depend on the deposition, dose, and charge of the liposomes.39 In addition, we observed that RBCs and hematocrit mildly decreased below normal range in mitomycin C-loaded liposome treatment group. Stoll et al demonstrated that changes in acyl chain length of phospholipid and cholesterol-to-phospholipid ratio led to conformational disorder in the RBC membranes.40 Mui et al also pointed out that the phospholipids composition is an important factor, and phospholipids containing DOPE are much more hemolytic than those containing DOPC, and destabilization is induced by excess DOPE.41,42 A recent research also provides a viewpoint that RBCs treated with uncharged liposomes underwent lower hemolysis compared with those treated with charged liposomes despite the presence of PC.43 We presumed that liposomes have a protective platelet membrane that might relate to their escape from the macrophage system; moreover, the interaction between liposomes and RBCs requires a stricter study design to be properly elucidated.
Conclusion
In this work, mitomycin C was successfully entrapped in pH-sensitive liposomes. The loaded liposomes were 144 nm in diameter, had an encapsulation efficiency of 66%, and could retain stability for 1 month when stored at 4°C. The cell uptake study demonstrated that folate modification of pH-sensitive liposomes increased their uptake by MCF-7 cells. Pharmacokinetic data indicated that mitomycin C-loaded pH-sensitive liposomes had increased half-life during the elimination phase, as well as higher AUC and MRT, and significantly decreased Cl value. Moreover, pH-sensitive liposomes diminished the hematological toxicity of mitomycin C. The positive results encourage us to continuously investigate how to modify the liposome structure to decrease the side effect of drugs and understand the underlying mechanism.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Szybalski W, Iyer VN. Crosslinking of DNA by enzymatic or chemically activated mitomycins and porfiromycins, bifunctionally “alkylating” antibiotics. Fed Proc. 1964;23:946–957. [PubMed] [Google Scholar]
- 2.Persidis A. Cancer multidrug resistance. Nat Biotechnol. 1999;17(1):94–95. doi: 10.1038/5289. [DOI] [PubMed] [Google Scholar]
- 3.Golan T, Grenader T, Ohana P, et al. Pegylated liposomal mitomycin C prodrug enhances tolerance of mitomycin C: a phase 1 study in advanced solid tumor patients. Cancer Med. 2015;4(10):1472–1483. doi: 10.1002/cam4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ihnat MA, Lariviere JP, Warren AJ, et al. Suppression of P-glycoprotein expression and multidrug resistance by DNA cross-linking agents. Clin Cancer Res. 1997;3(8):1339–1346. [PubMed] [Google Scholar]
- 5.Spratto GR, Woods AL. Delmar Healthcare Drug Handbook. 22nd ed. Clifton Park, NY: Delmar Cengage Learning; 2013. [Google Scholar]
- 6.Itano JK, Brant JM, Conde FA, Saria MG, editors. Core Curriculum for Oncology Nursing. 5th ed. St Louis, MO: Elsevier; 2015. [Google Scholar]
- 7.Mauch P, Constine L, Greenberger J, et al. Hematopoietic stem cell compartment: acute and late effects of radiation therapy and chemotherapy. Int J Radiat Oncol Biol Phys. 1995;31(5):1319–1339. doi: 10.1016/0360-3016(94)00430-S. [DOI] [PubMed] [Google Scholar]
- 8.Marsh JC. The effects of cancer chemotherapeutic agents on normal hematopoietic precursor cells: a review. Cancer Res. 1976;36(6):1853–1882. [PubMed] [Google Scholar]
- 9.Mauerer R, Gruber R. Monoclonal antibodies for the immunotherapy of solid tumours. Curr Pharm Biotechnol. 2012;13(8):1385–1398. doi: 10.2174/138920112800784844. [DOI] [PubMed] [Google Scholar]
- 10.Lee ES, Gao Z, Bae YH. Recent progress in tumor pH targeting nanotechnology. J Control Release. 2008;132(3):164–170. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stubbs M, McSheehy PM, Griffiths JR, Bashford CL. Causes and consequences of tumour acidity and implications for treatment. Mol Med Today. 2000;6(1):15–19. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 12.Balamuralidhara V, Pramodkumar TM, Srujana N, et al. pH sensitive drug delivery systems: a review. Am J Drug Discov Dev. 2011;1:24–48. [Google Scholar]
- 13.Underberg WJ, Lingeman H. Aspects of the chemical stability of mitomycin and porfiromycin in acidic solution. J Pharm Sci. 1983;72(5):549–553. doi: 10.1002/jps.2600720518. [DOI] [PubMed] [Google Scholar]
- 14.Beijnen JH, Underberg WJ. Degradation of mitomycin C in acidic solution. Int J Pharm. 1985;24:219–229. [Google Scholar]
- 15.Underberg WJ, Beijnen JH. Kinetics and mechanism of the degradation of 1a-acetylmitomycin C in aqueous solution. Pharm World Sci. 1993;15(3):123–127. doi: 10.1007/BF02113940. [DOI] [PubMed] [Google Scholar]
- 16.Eloy JO, Claro de Souza M, Petrilli R, Barcellos JP, Lee RJ, Marchetti JM. Liposomes as carriers of hydrophilic small molecule drugs: strategies to enhance encapsulation and delivery. Colloids Surf B Biointerfaces. 2014;123:345–363. doi: 10.1016/j.colsurfb.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 17.Jaafar-Maalej C, Diab R, Andrieu V, Elaissari A, Fessi H. Ethanol injection method for hydrophilic and lipophilic drug-loaded liposome preparation. J Liposome Res. 2010;20(3):228–243. doi: 10.3109/08982100903347923. [DOI] [PubMed] [Google Scholar]
- 18.Li XB, Gu JD, Zhou QH. Review of aerobic glycolysis and its key enzymes – new targets for lung cancer therapy. Thorac Cancer. 2015;6(1):17–24. doi: 10.1111/1759-7714.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raja M. Do small headgroups of phosphatidylethanolamine and phosphatidic acid lead to a similar folding pattern of the K(+) channel? J Membr Biol. 2011;242(3):137–143. doi: 10.1007/s00232-011-9384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monteiro N, Martins A, Reis RL, Neves NM. Liposomes in tissue engineering and regenerative medicine. J R Soc Interface. 2014;11(101):20140459. doi: 10.1098/rsif.2014.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnsson M, Edwards K. Phase behavior and aggregate structure in mixtures of dioleoylphosphatidylethanolamine and poly(ethylene glycol)-lipids. Biophys J. 2001;80(1):313–323. doi: 10.1016/S0006-3495(01)76016-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volodkin D, Ball V, Schaaf P, Voegel JC, Mohwald H. Complexation of phosphocholine liposomes with polylysine. Stabilization by surface coverage versus aggregation. Biochim Biophys Acta. 2007;1768(2):280–290. doi: 10.1016/j.bbamem.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Venugopalan P, Jain S, Sankar S, Singh P, Rawat A, Vyas SP. pH-sensitive liposomes: mechanism of triggered release to drug and gene delivery prospects. Pharmazie. 2002;57(10):659–671. [PubMed] [Google Scholar]
- 24.Leite EA, Souza CM, Carvalho-Júnior AD, et al. Encapsulation of cisplatin in long-circulating and pH-sensitive liposomes improves its antitumor effect and reduces acute toxicity. Int J Nanomedicine. 2012;7:5259–5269. doi: 10.2147/IJN.S34652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, Paxton JW, Wu Z. Development of long-circulating pH-sensitive liposomes to circumvent gemcitabine resistance in pancreatic cancer cells. Pharm Res. 2016;33(7):1628–1637. doi: 10.1007/s11095-016-1902-8. [DOI] [PubMed] [Google Scholar]
- 26.Sorkin A, Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3(8):600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 27.Szoka FC, Jr, Milholland D, Barza M. Effect of lipid composition and liposome size on toxicity and in vitro fungicidal activity of liposome-intercalated amphotericin B. Antimicrob Agents Chemother. 1987;31(3):421–429. doi: 10.1128/aac.31.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Li H, Lee RJ. Targeted drug delivery via folate receptors. Expert Opin Drug Deliv. 2008;5(3):309–319. doi: 10.1517/17425247.5.3.309. [DOI] [PubMed] [Google Scholar]
- 29.Zhao XB, Lee RJ. Tumor-selective targeted delivery of genes and antisense oligodeoxyribonucleotides via the folate receptor. Adv Drug Deliv Rev. 2004;56(8):1193–1204. doi: 10.1016/j.addr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Ahn R, Van den Bossche J, Thompson DH, O’Halloran TV. Folate-mediated intracellular drug delivery increases the anticancer efficacy of nanoparticulate formulation of arsenic trioxide. Mol Cancer Ther. 2009;8(7):1955–1963. doi: 10.1158/1535-7163.MCT-09-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barza M, Stuart M, Szoka F., Jr Effect of size and lipid composition on the pharmacokinetics of intravitreal liposomes. Invest Ophthalmol Vis Sci. 1987;28(5):893–900. [PubMed] [Google Scholar]
- 32.Nag OK, Awasthi V. Surface engineering of liposomes for stealth behavior. Pharmaceutics. 2013;5(4):542–569. doi: 10.3390/pharmaceutics5040542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caliceti P, Veronese FM. Pharmacokinetic and biodistribution properties of poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev. 2003;55(10):1261–1277. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]
- 34.Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomedicine. 2006;1(3):297–315. [PMC free article] [PubMed] [Google Scholar]
- 35.Verwey J, de Vries J, Pinedo HM. Mitomycin C-induced renal toxicity, a dose-dependent side effect? Eur J Cancer Clin Oncol. 1987;23(2):195–199. doi: 10.1016/0277-5379(87)90014-9. [DOI] [PubMed] [Google Scholar]
- 36.Muhonen TT, Wiklund TA, Blomqvist CP, Pyrhönen SO. Unexpected prolonged myelosuppression after mitomycin, mitoxantrone and methotrexate. Eur J Cancer. 1992;28A(12):1974–1976. doi: 10.1016/0959-8049(92)90241-s. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto S, Shibata S, Kobayashi B. The effect of mitomycin C on platelet aggregation and adenosine 3′,5′-monophosphate metabolism. Thromb Haemost. 1978;39(1):177–185. [PubMed] [Google Scholar]
- 38.El-Ghazal R, Podoltsev N, Marks P, Chu E, Saif MW. Mitomycin-C-induced thrombotic thrombocytopenic purpura/hemolytic uremic syndrome: cumulative toxicity of an old drug in a new era. Clin Colorectal Cancer. 2011;10(2):142–145. doi: 10.1016/j.clcc.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Reinish LW, Bally MB, Loughrey HC, Cullis PR. Interactions of liposomes and platelets. Thromb Haemost. 1988;60(3):518–523. [PubMed] [Google Scholar]
- 40.Stoll C, Stadnick H, Kollas O, et al. Liposomes alter thermal phase behavior and composition of red blood cell membranes. Biochim Biophys Acta. 2011;1808(1):474–481. doi: 10.1016/j.bbamem.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Zelphati O, Szoka FC., Jr Intracellular distribution and mechanism of delivery of oligonucleotides mediated by cationic lipids. Pharm Res. 1996;13(9):1367–1372. doi: 10.1023/a:1016026101195. [DOI] [PubMed] [Google Scholar]
- 42.Mui B, Ahkong QF, Chow L, Hope MJ. Membrane perturbation and the mechanism of lipid-mediated transfer of DNA into cells. Biochim Biophys Acta. 2000;1467(2):281–292. doi: 10.1016/s0005-2736(00)00226-1. [DOI] [PubMed] [Google Scholar]
- 43.da Silveira Cavalcante L, Feng Q, Chin-Yee I, Acker JP, Holovati JL. Effect of liposome-treated red blood cells in an anemic rat model. J Liposome Res. 2017;27(1):56–63. doi: 10.3109/08982104.2016.1149867. [DOI] [PubMed] [Google Scholar]