Abstract
Ankylosing spondylitis (AS) is a chronic inflammatory arthritis and autoimmune disease, the etiology and pathogenesis of which remain largely unknown. In the present study, blood samples were harvested from patients with AS and from healthy volunteers as a normal control (NC) for RNA-sequencing. Differentially expressed genes (DEGs) in the AS group compared with the NC group were identified, and gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were subsequently performed. Protein-protein interaction (PPI) network and AS-specific transcriptional regulatory network construction was performed for the DEGs. A total of 503 DEGs, including 338 upregulated and 165 downregulated DEGs, were identified in patients with AS compared with the NC group. Three upregulated DEGs identified, interferon-induced protein with tetratricopeptide repeats (IFIT)1, IFIT3 and radical S-adenosyl methionine domain containing (RSAD)2, are interferon (IFN)-stimulated genes that serve a role in the IFN signaling pathway. The most significantly enriched GO term was response to other organisms. Osteoclast differentiation was a significantly enriched pathway for eight DEGs [High affinity immunoglobulin gamma Fc receptor (FCGR)1A, FCGR2B, four and a half LIM domains 2, integrin β3, signal transducer and activator of transcription 2 (STAT2), suppressor of cytokine signaling 3 (SOCS3), leukocyte immunoglobulin like receptor (LILR)A4 and LILRA6]. The six hub genes in the PPI network constructed were interferon-stimulated gene 15, heat shock protein β1, microtubule-associated proteins 1A/1B light chain 3A, IFIT1, IFIT3 and SOCS3. POU domain class 2 transcription factor 1 (1-Oct) and ecotropic virus integration site-1 (Evi-1) were identified as two important transcription factors (TFs) in AS according to the AS-specific transcriptional regulatory network constructed. In addition, IFIT1 and IFIT3 were identified as targets of 1-Oct. The results of the present study indicate that osteoclast differentiation, the IFN signaling pathway and genes associated with these two signaling pathways, particularly FCGR2B, STAT2, SOCS3, IFIT1 and IFIT3, may serve a role in AS. In addition, Evi-1 and 1-Oct may be two important TFs associated with AS. These results may provide a basis for elucidating the underlying mechanisms of and developing novel treatments for AS.
Keywords: ankylosing spondylitis, differentially expressed genes, RNA-sequencing, protein-protein interaction networks, transcription factors
Introduction
Ankylosing spondylitis (AS) is a chronic inflammatory arthritis and autoimmune disease, which can develop into spondyloarthritis (SpA), with an estimated prevalence of 2–5% worldwide (1,2). AS primarily affects the sacroiliac joints and spine (2), and is characterized by inflammation of the spine and progressive spinal ankylosis (3–5). Back pain and spinal stiffness are two of the main symptoms of AS (5,6). AS is multifactorial and highly heritable; however, the etiology and pathogenesis of AS remain largely unknown (7,8). It has previously been reported that human leukocyte antigen (HLA)-B27 may be associated with the pathogenesis of AS (2). As a gene located near HLA-B on chromosome 6, MICA confers strong susceptibility to AS in US white and Chinese Han populations (9). Although ~90% of patients with AS have the HLA-B27 genotype, only 8% of HLA-B27 carriers develop AS (10–16), which indicates that other mechanisms are associated with the development of AS. Elucidating these mechanisms may increase understanding of the pathogenesis of and risk factors for AS. A total of 34 loci that affect disease susceptibility to AS have been identified (17). RUNX3, TBKBP1 and PPARGC1B were reported to be associated with the AS susceptibility in patients with Western European descent (18). Among them, RUNX3 was associated with the severity of AS and the function of daily life of Chinese Han patients with AS (18). The population-attributable AS risk associated with ERAP1 and IL-23R has been reported to be ~26 and 1%, respectively (19). Despite considerable research efforts, no effective therapeutic strategy for AS has been developed.
With rapid advances in next generation DNA sequencing technologies (20), RNA sequencing (RNA-Seq), which is a comprehensive and accurate method of transcriptome analysis, has become increasingly feasible and affordable (21,22). RNA-Seq can be performed to identify differentially expressed genes (DEGs), and quantify exons and isoforms accurately (23). Previous studies have used RNA-Seq to identify disease-associated genes; however, to the best of our knowledge, RNA-Seq has not previously been used to analyze the transcriptomes of patients with AS (21,24–26).
In the present study, a comprehensive transcriptome analysis of the blood of patients with AS and healthy controls was performed using RNA-Seq. DEGs in the AS group compared with the normal control (NC) group were identified. Functional annotation and protein-protein interaction (PPI) network construction of the DEGs was performed. In addition, a transcriptional regulatory network of the DEGs was constructed. The results of the present study may improve understanding of the pathogenesis of AS, and thus contribute to the development of novel and effective treatments for AS.
Materials and methods
Patients
A total of 3 patients with AS and 3 NCs were recruited from Jining No. 1 People's Hospital (Shandong, China) between February 2016 and April 2016. The inclusion criteria of patients were as follows: i) Patients diagnosed with AS who were in compliance with the 1984 revised New York AS diagnostic criteria; ii) patients with the duration of AS for >1 month; and iii) patients over 18 years old. The inclusion criteria of NCs were as follows: i) Subjects without clinical manifestations of AS; ii) subjects without abnormalities of the sacroiliac joint and spinal column; iii) subjects who were HLA-B27 negative; and iv) subjects over 18 years old. Subjects with other immune disorders, pregnant women and people who were unable to draw blood samples were excluded from the present study. Of the 3 patients with AS, 1 patient was a 38-year-old male who had been admitted to the hospital due to hip pain that had lasted for a year and worsened in the past 40 days. Patient 2 was a 73-year-old female who was hospitalized for systemic pain that had lasted for >20 years and worsened in the prior month. Patient 3 was a 43-year-old male who had been admitted to hospital due to pain and stiffness of the waist that had lasted for 5 years. All of the patients with AS were HLA-B27 positive. The 3 NCs consisted of a 35-year-old male, 44-year-old female and 75-year-old female, respectively. None of the sujects in the AS or NC groups had any other autoimmune diseases. All participants provided written informed consent, and this was reviewed by the Ethics Committee of Jining No. 1 People's Hospital (Shandong, China).
RNA isolation, polymerase chain reaction (PCR) amplification and sequencing
Blood samples were obtained from 3 NCs and 3 patients with AS between February 2016 and April 2016. Total RNA was isolated from blood samples using TRIizol reagent (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions (27). The quantity and integrity of RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA; RNA integrity number >7). RNA libraries were constructed with TruSeq RNA Sample Prep Kit v2 (Illumina, Inc., San Diego, CA, USA) according to the manufacturer's instructions (28). mRNA was purified using Oligo (dT) magnetic beads and fragmented at 95°C for 8 min with fragmentation buffer in TruSeq RNA Sample Prep Kit v2 (Illumina, Inc., San Diego, CA, USA). Using mRNA as a template, the first cDNA strand was synthesized with random oligonucleotides and SuperScript III reverse transcriptase (Invitrogen; Thermo Fisher Scientific Inc., Waltham, MA, USA) under the following conditions: 5 min at 65°C, 2 min at 4°C, 1 h at 42°C and 10 min at 70°C as described previously (29). The second cDNA strand was then synthesized by the addition of the buffer, dNTPs, RNase H (Invitrogen; Thermo Fisher Scientific Inc.) and DNA polymerase I (Invitrogen; Thermo Fisher Scientific Inc.) under the following conditions: 2.5 h at 16°C and 10 min at 70°C. After purification using the QIAQuick PCR Purification kit (Qiagen, Inc., Valencia, CA, USA), end repair and ligation of the sequencing adapter were performed with using the NEB Next End Repair Module (New England Biolabs, Ipswich, MA, USA) according to the manufacturer's protocol (30).
The PCR products were analyzed on 2% agarose gel (Certified Low Range Ultra Agarose) and stained with ethidium bromide. The 300 nucleotide-long fragments were isolated and purified by using QIAQuick PCR Purification kit (Qiagen, Inc., Valencia, CA, USA). Cluster generation (bridge PCR) amplification was performed using an Illumina cBot system (Illumina, Inc., San Diego, CA, USA) as described (29). Sequencing was performed using a HiSeq 2500 system (Illumina, Inc.).
Identification of DEGs
The quality of the sequencing data obtained was evaluated using FastQC software (version 0.11.4, www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cutadapt software (version 1.9.1, cutadapt.readthedocs.io/en/stable/changes.html#v1-9-1-2015-12-02) was used to remove low quality reads, including sequences with a quality score <20 and sequences with an N base rate of raw reads >10%. The cleaned reads and human genome (GRCh38.p7 assembly) were then aligned using TopHat version 2.1.1 (www.ccb.jhu.edu/software/tophat/index.shtml). Cuffquant version 2.2.1 (www.cole-trapnell-lab.github.io/cufflinks/) was used to obtain the count and fragments per kilobase of transcript per million fragments mapped (FPKM) of each gene. Cuffdiff version 2.2.1 (www.cole-trapnell-lab.github.io/cufflinks/) was used to calculate the differential expression of genes. The thresholds for DEGs were as follows: An FDR <0.05; an absolute value of FPKM of 1; and a coefficient of variation <1. The heat map of the top 50 upregulated and downregulated DEGs was obtained using pheatmap package in R version 3.1.3 (www.r-project.org/).
Functional annotation
Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using Metascape (metascape.org/gp/index.html#/main/step1) to further investigate the biological function of each DEG. P<0.01 was considered to indicate a significantly enriched DEG.
PPI network construction
Using the Biological General Repository for Interaction Datasets (BioGrid, www.uniprot.org/database/DB-0184), the 100 most upregulated and downregulated DEGs were scanned. A PPI network was then constructed using Cytoscape software (version 3.3.0, www.cytoscape.org) in order to explore of the functions of the DEGs.
Construction of AS-specific transcriptional regulatory network
The 2 kb upstream promoter sequence of the 10 most upregulated and downregulated DEGs was downloaded from UCSC Genome Browser (www.genome.ucsc.edu). The transcription factors (TFs) involved in regulating these DEGs were derived from the match tools using the transcription factor database TRANSFAC (http://gene-regulation.com/pub/databases.html), which is a database of TFs (31). Cytoscape software (version 3.3.0, www.cytoscape.org) was used to construct the AS-specific transcriptional regulatory network.
Results
Identification of DEGs
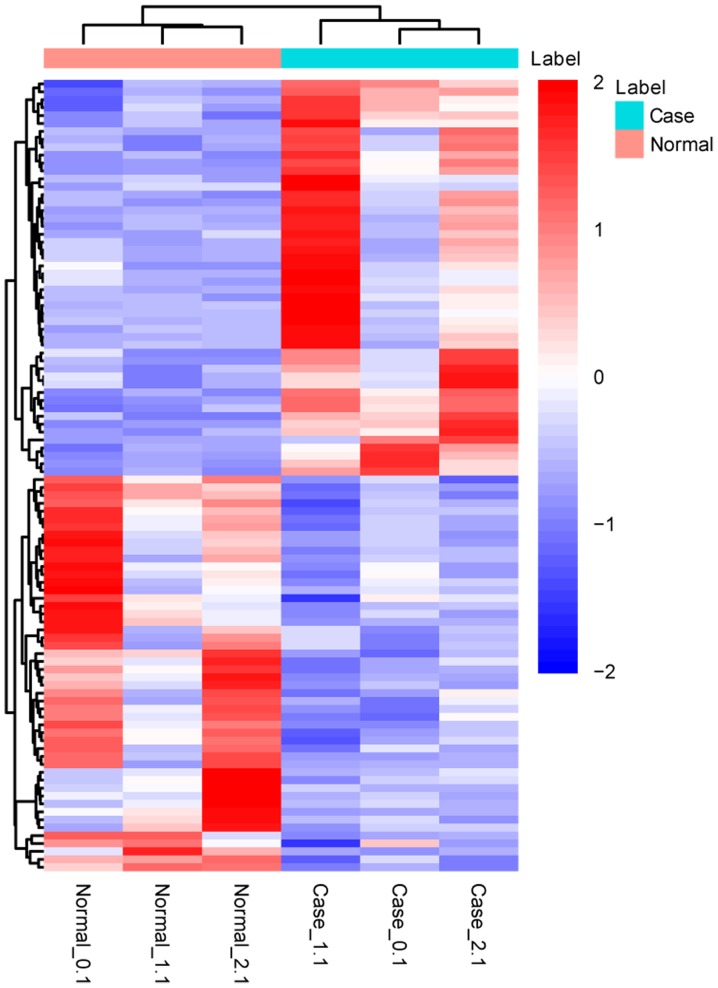
A total of 503 DEGs, including 338 upregulated and 165 downregulated DEGs, were identified in patients with AS compared with the NC group (Fig. 1). The 10 most upregulated and downregulated DEGs are displayed in Table I.
Figure 1.
Heatmap of the 50 most upregulated and downregulated differentially expressed genes in patients with ankylosing spondylitis compared with the normal controls.
Table I.
Top 10 most upregulated and downregulated DEGs in patients with ankylosing spondylitis.
| A, Upregulated DEGs | |||
|---|---|---|---|
| Gene | log2 fold change | t-value | P-value |
| SIGLEC1 | 3.25845 | 4.26359 | 5.0×10−5 |
| VSIG4 | 4.02414 | 4.19381 | 5.0×10−5 |
| BATF2 | 2.55908 | 3.45345 | 5.0×10−5 |
| IFIT1 | 2.09875 | 3.29697 | 5.0×10−5 |
| IFIT3 | 2.14216 | 3.1677 | 5.0×10−5 |
| SERPING1 | 2.00997 | 3.03775 | 5.0×10−5 |
| RSAD2 | 2.45907 | 2.96724 | 5.0×10−5 |
| CASP5 | 3.04181 | 2.93592 | 5.0×10−5 |
| MCEMP1 | 2.20974 | 2.92794 | 5.0×10−5 |
| LOC441081 | 2.63115 | 2.79881 | 5.0×10−5 |
| B, Downregulated DEGs | |||
| Gene | log2 fold change | t-value | P-value |
| GPR162 | −2.24236 | 2.24236 | 5.0×10−5 |
| HAPLN3 | −2.47863 | −2.47863 | 5.0×10−5 |
| PTGDS | −2.64354 | −2.64354 | 5.0×10−5 |
| FCGBP | −2.32069 | −2.32069 | 2.5×10−4 |
| GZMM | −2.06931 | −2.06931 | 5.5×10−4 |
| SH2D1B | −1.91722 | −1.91722 | 6.0×10−4 |
| TIGD3 | −2.05767 | −2.05767 | 6.0×10−4 |
| LGALS9C | −1.91957 | −1.91957 | 6.5×10−4 |
| KIR2DL3 | −2.24251 | −2.24251 | 9.5×10−4 |
| COL6A2 | −1.87265 | −1.8726 | 1.4×10−3 |
Functional annotation
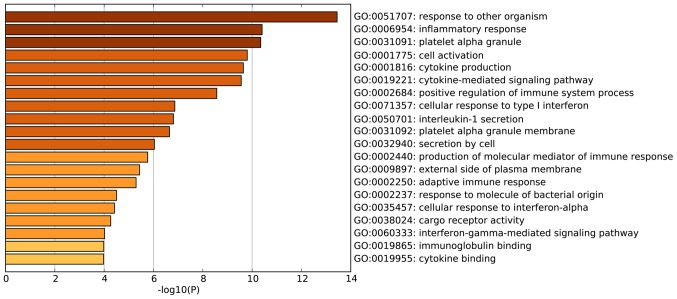
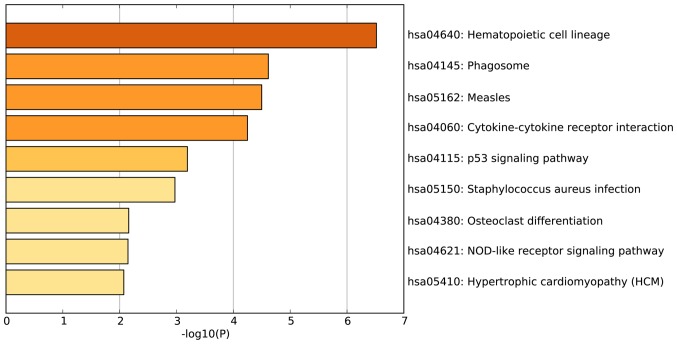
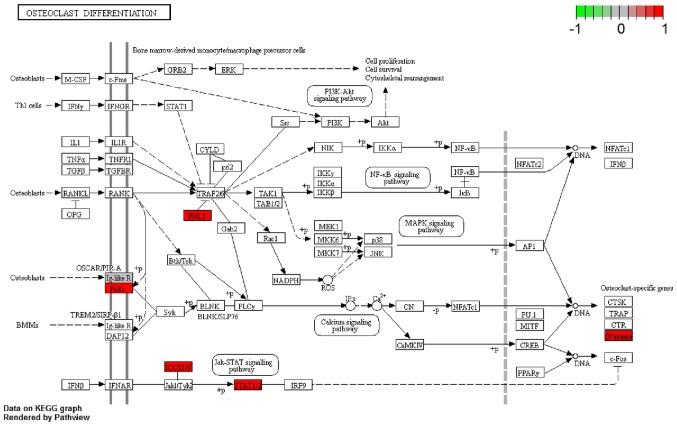
According to the GO enrichment analysis, the DEGs were significantly enriched in the following GO terms: Response to other organisms (P<0.001); inflammatory response (P<0.001); platelet a granule (P<0.001); cell activation (P<0.001); and cytokine production (P<0.001). The 20 most enriched GO terms of the DEGs in patients with AS are presented in Fig. 2. KEGG pathway enrichment analysis demonstrated that the DEGs identified in patients with AS were the most significantly enriched in the following signaling pathways: Hematopoietic cell lineage (P<0.001); phagosome (P<0.001); and measles (P<0.001) (Fig. 3). Another significantly enriched signaling pathway, osteoclast differentiation (P=0.007; Fig. 3) consisted of DEGs including high affinity immunoglobulin gamma Fc receptor (FCGR)1A, FCGR2B, four and a half LIM domains 2 (FHL2), integrin β3 (ITGB3), signal transducer and activator of transcription 2 (STAT2), suppressor of cytokine signaling 3 (SOCS3), leukocyte immunoglobulin like receptor (LILR)A4 and LILRA6 (Fig. 4).
Figure 2.
Top 20 most enriched gene ontology terms for the differentially expressed genes identified in patients with ankylosing spondylitis.
Figure 3.
Enriched Kyoto Encyclopedia of Genes and Genomes signaling pathways for the differentially expressed genes identified in patients with ankylosing spondylitis.
Figure 4.
Osteoclast differentiation signaling pathway. The DEGs identified in patients with ankylosing spondylitis were significantly enriched in the osteoclast differentiation signaling pathway. Red rectangles represent downregulated DEGs. DEG, differentially expressed gene.
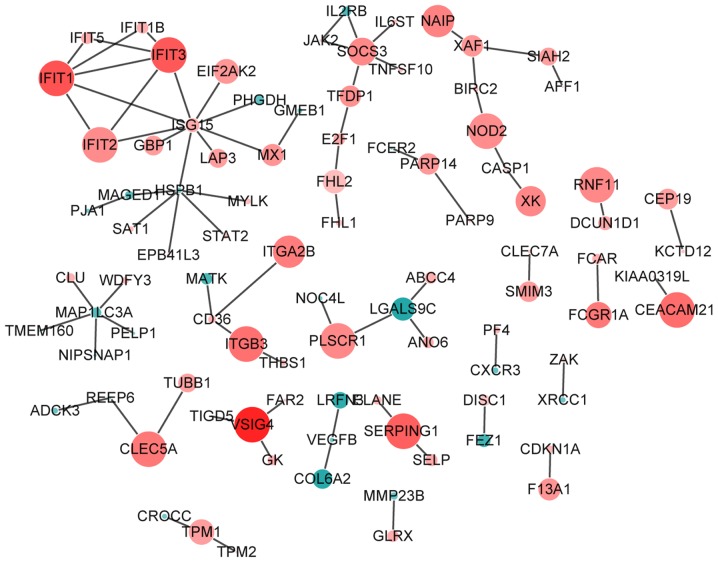
PPI network
Based on the BioGrid, the proteins interacted with the proteins encoded by the top 100 most upregulated and downregulated DEGs were searched. After removal of the proteins that were not encoded by DEGs in AS, a AS-specific PPI network was constructed, consisting of 92 nodes and 76 edges (Fig. 5). The proteins which were integrated with at least five proteins encoded by DEGs in AS (degree ≥5) were defined as hub proteins in the AS-specific PPI network in the study. The PPI network revealed that interferon-stimulated gene (ISG)15 (degree, 9), heat shock protein family B member 1 (degree, 6), microtubule-associated proteins 1A/1B light chain 3A (MAPILC3A; degree, 5), interferon-induced protein with tetratricopeptide repeats, IFIT1 (degree, 5), IFIT3 (degree, 5) and SOCS3 (degree, 5) were hub proteins. In addition, the clustering coefficients of IFIT3 and SOCS3 were 0.5 and 0.1 respectively.
Figure 5.
AS-specific protein-protein interaction network. Nodes (circles) represent the proteins encoded by DEGs. The radius of the circle indicates the significance of enrichment, a red color indicates that the DEG is upregulated and a green color indicates that the DEG is downregulated. DEG, differentially expressed gene.
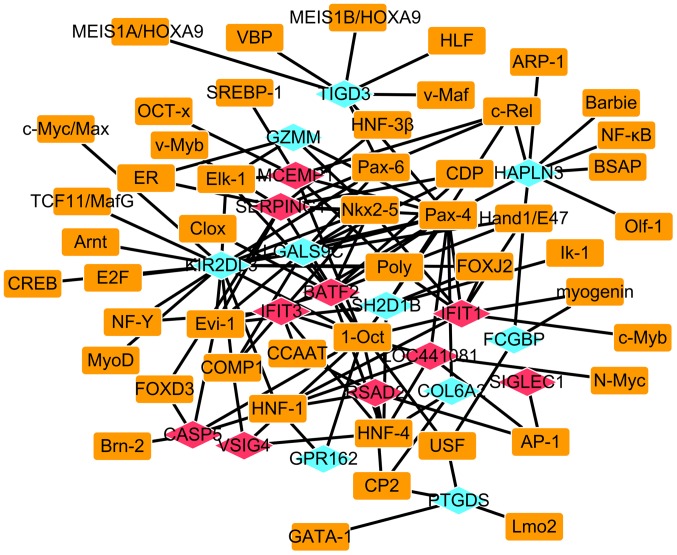
AS-specific transcriptional regulatory network
A total of 49 TFs which regulated the expression of the top 10 most upregulated and downregulated DEGs in AS were identified and the AS-specific transcriptional regulatory network of the 10 most upregulated and downregulated DEGs was constructed, which consisted of 69 nodes and 127 edges (Fig. 6). In this network, POU domain class 2 transcription factor 1 (1-Oct), paired box gene 4, hepatocyte nuclear factor (HNF)-4, natural killer 2 homeobox 5, ecotropic virus integration site-1 (Evi-1) and HNF-1 regulated the majority of the 10 most upregulated and downregulated DEGs (Table II).
Figure 6.
Ankylosing spondylitis-specific transcriptional regulatory network. Blue rhombuses represent downregulated DEGs, and red rhombuses represent upregulated DEGs. Rectangles represent TFs. The lines indicate TF-DEG pairs. DEG, differentially expressed gene; TF, transcription factor,.
Table II.
Top six transcription factors regulating the majority of differentially expressed genes identified in patients with ankylosing spondylitis.
| TF | No. of DEGs regulated | DEG |
|---|---|---|
| 1-Oct | 13 | SH2D1B, RSAD2, LOC441081, VSIG4, IFIT1, IFIT3, SERPING1, CASP5, BATF2, GPR162, LGALS9C, KIR2DL3, MCEMP1 |
| Pax-4 | 13 | SH2D1B, LOC441081, IFIT3, IFIT1, SERPING1, BATF2, TIGD3, HAPLN3, LGALS9C, GZMM, MCEMP1, COL6A2, KIR2DL3 |
| HNF-4 | 7 | SH2D1B, RSAD2, LOC441081, VSIG4, IFIT3, BATF2, COL6A2 |
| Nkx2-5 | 7 | IFIT3, IFIT1, SERPING1, BATF2, LGALS9C, GZMM, KIR2DL3 |
| Evi-1 | 6 | SH2D1B, SERPING1, CASP5, BATF2, LGALS9C, KIR2DL3 |
| HNF-1 | 6 | RSAD2, LOC441081, IFIT1, CASP5, GPR162, KIR2DL3 |
DEG, differentially expressed gene; TF, transcription factor.
Discussion
In the present study, RNA-Seq was used to identify 503 DEGs (338 upregulated and 165 downregulated) in patients with AS compared with the NC group. To further investigate the pathogenesis of AS at a molecular level, functional annotation, PPI network construction and AS-specific transcriptional regulatory network construction were performed.
IFIT1, IFIT3 and radical S-adenosyl methionine domain containing (RSAD2) were in the 10 most upregulated DEGs identified in patients with AS. In addition, the patients with AS included in the present study were all positive for HLA-B27. The expression of HLA-B27 is able to cause deficiencies in the interferon (IFN) signaling pathway, suggesting that the IFN signaling pathway may be associated with the pathogenesis of AS (32). IFIT1, IFIT3 and RSAD2 are ISGs (33) that are associated with the IFN signaling pathway. It has previously been demonstrated that IFIT3 is downregulated in disease-prone HLA-B27 transgenic rats and patients with SpA (32). In the present study IFIT1, IFIT3 and RSAD2 were upregulated in patients with AS. Furthermore, the PPI network identified IFIT1 and IFIT3 as hub genes. These results suggest that these three DEGs may affect the progression of AS through the IFN signaling pathway.
KEGG pathway enrichment analysis demonstrated that osteoclast differentiation was a significantly enriched pathway of the DEGs identified in patients with AS. Excessive and ectopic bone formation with concomitant systemic bone loss occurs in patients with AS, suggesting that osteoclasts are important in the progression of AS (3,34–37). Osteoclast differentiation has also been reported to serve a crucial role in the development of AS (2,3). Taken together, these reports suggest that osteoclast differentiation-associated DEGs (FCGR1A, FCGR2B, FHL2, ITGB3, STAT2, SOCS3, LILRA4 and LILRA6) may be associated with the pathogenesis of AS. Of these DEGs, FCGR2B and STAT2 have previously been reported to be correlated with AS (7,38–41).
FCGR2B belongs to the immunoglobulin super family (42). FCGRs have been reported to serve an important role in immune responses (38,39) and FCGR2B deficiency may contribute to autoimmune diseases (40,41). The number of FCGR2B-postive B cells and the expression of FCGR2B have been reported to be downregulated in patients with rheumatoid arthritis (40). Furthermore, FCGR2B gene polymorphisms have previously been demonstrated to be associated with autoimmune diseases, including AS (8). In the present study, FCGR2B was identified to be upregulated in AS, which suggests that the dysregulation of FCGR2B may be associated with the progression of AS, potentially through regulating autoimmunity and osteoclast differentiation. However, the precise role of FCGR2B in the development and progression of AS requires further research.
STAT2, a member of the STAT gene family, has been reported to be associated with AS (7). A previous study reported that STAT2 mRNA and protein expression were significantly reduced in patients with AS cases compared with NCs (7). However, the RNA-Seq results in the present study demonstrated that STAT2 was upregulated in patients with AS. The results of the current study suggest that STAT2 may serve a role in AS via regulating the osteoclast differentiation signaling pathway, although large scale studies are required to investigate this further.
With the exception of FCGR2B and STAT2, the results of the present study indicate that the other six osteoclast differentiation-associated DEGs (FCGR1A, FHL2, ITGB3, SOCS3, LILRA4 and LILRA6) may be associated with the development of AS. In particular, overexpression of SOCS3 was identified in B27-transgenic rats and patients with spondyloarthritis, which may be induced by HLA-B27 and involve in the pathogenesis of spondyloarthritis (32). In the present study, SOCS3 was identified to be a hub gene in the PPI network, and was upregulated in the AS group compared with the NC group. These results suggest that SOCS3 dysregulation may cause susceptibility to the development of AS, which is due to its function in osteoclast differentiation.
Based on the AS-specific transcriptional regulatory network constructed in the present study, two TFs, Evi-1 and 1-Oct, were identified as being particularly important in AS. Evi-1 is a TF that regulates numerous osteoclast differentiation-associated genes and may affect skeletal health (43). Deletion of Evi-1 in hematopoietic cells in adult mice has been demonstrated to induce an increase in osteoclast precursors (44,45). Bone loss due to an increased number and activity of osteoclasts has also been reported in mice with Evi-1 deletion (43). Evi-1 is therefore thought to inhibit osteoclast precursor cell abundance, and osteoclastic activity and formation, which contributes to overall bone mass (43). In the present study, Evi-1 was one of the six TFs that regulated the majority of DEGs identified in patients with AS. These results suggest that Evi-1 may be associated with AS via regulating osteoclastogenesis, bone resorption and osteoclast differentiation.
Another TF, 1-Oct was also identified to be one of the six TFs that regulated the majority of DEGs identified in patients with AS. 1-Oct regulates the induction of activation-induced cytidine deaminase, which in turn regulates the balance between efficient immunity and autoimmunity (46). Dysregulation of 1-Oct may therefore be associated with autoantibody-mediated autoimmune disease by altering this balance. Systemic lupus has been reported to be correlated with an upregulation of 1-Oct in B cells (47,48). Notably, IFIT1 and IFIT3 are targets of 1-Oct. These results suggest that 1-Oct is associated with the pathogenesis of AS.
In conclusion, the results of the present study indicate that osteoclast differentiation, the IFN signaling pathway and genes associated with these two signaling pathways are associated with the pathogenesis of AS. Evi-1 and 1-Oct may be crucial TFs that are associated with AS via regulating bone mass, and the balance between efficient immunity and autoimmunity. These findings may provide a basis for elucidating the pathogenesis of, and developing novel diagnostic and therapeutic strategies for AS. However, the small sample size for RNA-sequencing is a limitation of the present study, therefore studies with a larger sample size are required to further elucidate the present findings.
References
- 1.Baeten D, Baraliakos X, Braun J, Sieper J, Emery P, van der Heijde D, McInnes I, van Laar JM, Landewé R, Wordsworth P, et al. Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: A randomised, double-blind, placebo-controlled trial. Lancet. 2013;382:1705–1713. doi: 10.1016/S0140-6736(13)61134-4. [DOI] [PubMed] [Google Scholar]
- 2.Yang C, Ding P, Wang Q, Zhang L, Zhang X, Zhao J, Xu E, Wang N, Chen J, Yang G, et al. Inhibition of complement retards ankylosing spondylitis progression. Sci Rep. 2016;6:34643. doi: 10.1038/srep34643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin S, Qiu M, Chen J. IL-4 modulates macrophage polarization in ankylosing spondylitis. Cell Physiol Biochem. 2015;35:2213–2222. doi: 10.1159/000374026. [DOI] [PubMed] [Google Scholar]
- 4.Ronneberger M, Schett G. Pathophysiology of spondyloarthritis. Curr Rheumatol Rep. 2011;13:416–420. doi: 10.1007/s11926-011-0202-x. [DOI] [PubMed] [Google Scholar]
- 5.Dakwar E, Reddy J, Vale FL, Uribe JS. A review of the pathogenesis of ankylosing spondylitis. Neurosurg Focus. 2008;24:E2. doi: 10.3171/FOC/2008/24/1/E2. [DOI] [PubMed] [Google Scholar]
- 6.Dean LE, Jones GT, Macdonald AG, Downham C, Sturrock RD, Macfarlane GJ. Global prevalence of ankylosing spondylitis. Rheumatology (Oxford) 2014;53:650–657. doi: 10.1093/rheumatology/ket387. [DOI] [PubMed] [Google Scholar]
- 7.Uddin M, Codner D, Hasan SM, Scherer SW, O'Rielly DD, Rahman P. Integrated genomics identifies convergence of ankylosing spondylitis with global immune mediated disease pathways. Sci Rep. 2015;5:10314. doi: 10.1038/srep10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan ZH, Pan FM, Zeng Z, Zhang TC, Wang S, Li GX, Mei Y, Gao J, Ge R, Ye DQ, et al. The FCGR2B rs10917661 polymorphism may confer susceptibility to ankylosing spondylitis in Han Chinese: A case-control study. Scand J Rheumatol. 2012;41:219–222. doi: 10.3109/03009742.2011.625972. [DOI] [PubMed] [Google Scholar]
- 9.Zhou X, Wang J, Zou H, Ward MM, Weisman MH, Espitia MG, Xiao X, Petersdorf E, Mignot E, Martin J, et al. MICA, a gene contributing strong susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2014;73:1552–1557. doi: 10.1136/annrheumdis-2013-203352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reveille JD. Major histocompatibility genes and ankylosing spondylitis. Best Pract Res Clin Rheumatol. 2006;20:601–609. doi: 10.1016/j.berh.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Brown MA, Laval SH, Brophy S, Calin A. Recurrence risk modelling of the genetic susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2000;59:883–886. doi: 10.1136/ard.59.11.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi CB, Kim TH, Jun JB, Lee HS, Shim SC, Lee B, Pope A, Uddin M, Rahman P, Inman RD. ARTS1 polymorphisms are associated with ankylosing spondylitis in Koreans. Ann Rheum Dis. 2010;69:582–584. doi: 10.1136/ard.2008.105296. [DOI] [PubMed] [Google Scholar]
- 13.Danoy P, Pryce K, Hadler J, Bradbury LA, Farrar C, Pointon J, Australo-Anglo-American Spondyloarthritis Consortium. Ward M, Weisman M, Reveille JD, et al. Association of variants at 1q32 and STAT3 with ankylosing spondylitis suggests genetic overlap with Crohn's disease. PLoS Genet. 2010;6:e1001195. doi: 10.1371/journal.pgen.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Genetics of Ankylosing Spondylitis Consortium (IGAS) Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, Cremin K, Pryce K, Harris J, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uddin M, Maksymowych WP, Inman R, Gladman D, Munn A, Yazdani R, Pellett F, Hamilton S, O'Rielly DD, Rahman P. UGT2B17 copy number gain in a large ankylosing spondylitis multiplex family. BMC Genet. 2013;14:67. doi: 10.1186/1471-2156-14-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortes A, Maksymowych WP, Wordsworth BP, Inman RD, Danoy P, Rahman P, Stone MA, Corr M, Gensler LS, Gladman D, et al. Association study of genes related to bone formation and resorption and the extent of radiographic change in ankylosing spondylitis. Ann Rheum Dis. 2015;74:1387–1393. doi: 10.1136/annrheumdis-2013-204835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Lian Z, Xiao Y, Shi LL, Chai W, Wang Y. Analysis of clinical indexes and RUNX3, TBKBP1, PPARGC1B polymorphisms in Chinese Han patients with ankylosing spondylitis. Genet Test Mol Biomarkers. 2015;19:37–43. doi: 10.1089/gtmb.2014.0194. [DOI] [PubMed] [Google Scholar]
- 19.Tam LS, Gu J, Yu D. Pathogenesis of ankylosing spondylitis. Nat Rev Rheumatol. 2010;6:399–405. doi: 10.1038/nrrheum.2010.79. [DOI] [PubMed] [Google Scholar]
- 20.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 21.Heruth DP, Gibson M, Grigoryev DN, Li QZ, Shui QY. RNA-seq analysis of synovial fibroblasts brings new insights into rheumatoid arthritis. Cell Biosci. 2012;2:43. doi: 10.1186/2045-3701-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannopoulou EG, Elemento O, Ivashkiv LB. Use of RNA sequencing to evaluate rheumatic disease patients. Arthritis Res Ther. 2015;17:167. doi: 10.1186/s13075-015-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabbari A, Suárez-Fariñas M, Dewell S, Krueger JG. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol. 2012;132:246–249. doi: 10.1038/jid.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, Mcpherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips P, Progulske-Fox A, Grieshaber S, Grieshaber N. Expression of porphyromonas gingivalis small RNA in response to hemin availability identified using microarray and RNA-seq analysis. FEMS Microbiol Lett. 2014;351:202–208. doi: 10.1111/1574-6968.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Rong R, Zhao S, Zhu X, Zhang K, Xiong X, Yu X, Cui Q, Li S, Chen L, et al. Proteomic analysis of metabolic, cytoskeletal and stress response proteins in human heart failure. J Cell Mol Med. 2012;16:59–71. doi: 10.1111/j.1582-4934.2011.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du J, Aleff RA, Soragni E, Kalari K, Nie J, Tang X, Davila J, Kocher JP, Patel SV, Gottesfeld JM, et al. RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy. J Biol Chem. 2015;290:5979–5990. doi: 10.1074/jbc.M114.621607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z, Zhang H, Wang Z, Bin S, He H, Lin J. Discovery of chemosensory genes in the oriental fruit fly, bactrocera dorsalis. PLoS One. 2015;10:e0129794. doi: 10.1371/journal.pone.0129794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan SJ, Phan H, Gerry BM, Kuhn A, Hong LZ, Min Ong Y, Poon PS, Unger MA, Jones RC, Quake SR, Burkholder WF. A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps. PLoS One. 2013;8:e64084. doi: 10.1371/journal.pone.0064084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fert I, Cagnard N, Glatigny S, Letourneur F, Jacques S, Smith JA, Colbert RA, Taurog JD, Chiocchia G, Araujo LM, Breban M. Reverse interferon signature is characteristic of antigen-presenting cells in human and rat spondyloarthritis. Arthritis Rheumatol. 2014;66:841–851. doi: 10.1002/art.38318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Yang B, Zhang D. Activation of interferon signaling pathways in spinal cord astrocytes from an ALS mouse model. Glia. 2011;59:946–958. doi: 10.1002/glia.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perpétuo IP, Raposeiro R, Caetano-Lopes J, Vieira-Sousa E, Campanilho-Marques R, Ponte C, Canhão H, Ainola M, Fonseca JE. Effect of tumor necrosis factor inhibitor therapy on osteoclasts precursors in ankylosing spondylitis. PLoS One. 2015;10:e0144655. doi: 10.1371/journal.pone.0144655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miossec P. IL-17 and Th17 cells in human inflammatory diseases. Microbes Infect. 2009;11:625–630. doi: 10.1016/j.micinf.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Koenders MI, Marijnissen RJ, Devesa I, Lubberts E, Joosten LA, Roth J, van Lent PL, van de Loo FA, van den Berg WB. Tumor necrosis factor-interleukin-17 interplay induces S100A8, interleukin-1β, and matrix metalloproteinases, and drives irreversible cartilage destruction in murine arthritis: Rationale for combination treatment during arthritis. Arthritis Rheum. 2011;63:2329–2339. doi: 10.1002/art.30418. [DOI] [PubMed] [Google Scholar]
- 37.Lubberts E, Joosten LA, van de Loo FA, Schwarzenberger P, Kolls J, van den Berg WB. Overexpression of IL-17 in the knee joint of collagen type II immunized mice promotes collagen arthritis and aggravates joint destruction. Inflamm Res. 2002;51:102–104. doi: 10.1007/BF02684010. [DOI] [PubMed] [Google Scholar]
- 38.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 39.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 40.Prokopec KE, Rhodiner M, Matt P, Lindqvist U, Kleinau S. Down regulation of Fc and complement receptors on B cells in rheumatoid arthritis. Clin Immunol. 2010;137:322–329. doi: 10.1016/j.clim.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Wu J, Carter RH, Edberg JC, Su K, Cooper GS, Kimberly RP. A novel polymorphism in the Fcgamma receptor IIB (CD32B) transmembrane region alters receptor signaling. Arthritis Rheum. 2003;48:3242–3252. doi: 10.1002/art.11313. [DOI] [PubMed] [Google Scholar]
- 42.Sondermann P, Huber R, Jacob U. Crystal structure of the soluble form of the human Fcgamma-receptor IIb: A new member of the immunoglobulin superfamily at 1.7 Å resolution. EMBO J. 1999;18:1095–1103. doi: 10.1093/emboj/18.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soung do Y, Kalinowski J, Baniwal SK, Jacome-Galarza CE, Frenkel B, Lorenzo J, Drissi H. Runx1-mediated regulation of osteoclast differentiation and function. Mol Endocrinol. 2014;28:546–553. doi: 10.1210/me.2013-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacquin C, Gran DE, Sun KL, Lorenzo JA, Aguila HL. Identification of multiple osteoclast precursor populations in murine bone marrow. J Bone Miner Res. 2006;21:67–77. doi: 10.1359/JBMR.051007. [DOI] [PubMed] [Google Scholar]
- 46.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White AC, Hawkins JS, Pone EJ, Yu ES, Al-Qahtani A, Mai T. AID dysregulation in lupus-prone MRL/Faslpr/lpr mice increases class switch DNA recombination and promotes interchromosomal c-Myc/IgH loci translocations: Modulation by HoxC4. J Immunol. 2011;44:585–598. doi: 10.3109/08916934.2011.577128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White CA, Pone EJ, Lam T, Tat C, Hayama KL, Li G, Zan H, Casali P. Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses. J Immunol. 2014;193:5933–5950. doi: 10.4049/jimmunol.1401702. [DOI] [PMC free article] [PubMed] [Google Scholar]