Abstract
Background
Antheraea yamamai, also known as the Japanese oak silk moth, is a wild species of silk moth. Silk produced by A. yamamai, referred to as tensan silk, shows different characteristics such as thickness, compressive elasticity, and chemical resistance compared with common silk produced from the domesticated silkworm, Bombyx mori. Its unique characteristics have led to its use in many research fields including biotechnology and medical science, and the scientific as well as economic importance of the wild silk moth continues to gradually increase. However, no genomic information for the wild silk moth, including A. yamamai, is currently available.
Findings
In order to construct the A. yamamai genome, a total of 147G base pairs using Illumina and Pacbio sequencing platforms were generated, providing 210-fold coverage based on the 700-Mb estimated genome size of A. yamamai. The assembled genome of A. yamamai was 656 Mb (>2 kb) with 3675 scaffolds, and the N50 length of assembly was 739 Kb with a 34.07% GC ratio. Identified repeat elements covered 37.33% of the total genome, and the completeness of the constructed genome assembly was estimated to be 96.7% by Benchmarking Universal Single-Copy Orthologs v2 analysis. A total of 15 481 genes were identified using Evidence Modeler based on the gene prediction results obtained from 3 different methods (ab initio, RNA-seq-based, known-gene-based) and manual curation.
Conclusions
Here we present the genome sequence of A. yamamai, the first genome sequence of the wild silk moth. These results provide valuable genomic information, which will help enrich our understanding of the molecular mechanisms relating to not only specific phenotypes such as wild silk itself but also the genomic evolution of Saturniidae.
Keywords: Antheraea yamamai, genome assembly, Japanese silk moth, Japanese oak silk moth, wild silkworm
Data Description
Antheraea yamamai (NCBI Taxonomy ID: 7121), also known as the Japanese oak silk moth, is a wild silk moth species belonging to the Saturniidae family (Fig. 1). Silk moths can be categorized into 2 families—Bombycidae and Saturniidae. Saterniidae has been estimated to contain approximately 1861 species, with 162 genera [1], and is known as the largest family in the Lepidoptera. Among the many species in family Saturniidae, only a few species, including A. yamamai, can be utilized for silk production. Previous phylogenetic studies have shown that the family Saturniidae shares common ancestors with the family Sphingidae, including the hawk moth (Macroglossum stellatarum), and the Bombycidae family, including the most representative silkworm, Bombyx mori [2]. The estimated divergence time between A. yamamai and B. mori is 84 million years ago (MYA), making it similar to the 88 MYA estimated divergence time between the human and mouse [3, 4].
Figure 1:
Photograph of Antheraea Yamamai. From left: larva, cocoon, and adult A. yamamai, respectively. Green color is one of the representative characteristics of tensan silk.
A. yamamai produces a specific silk called tensan silk [5], which shows distinctive characteristics compared with common silk from B. mori, including characteristics such as thickness, bulkiness, compressive elasticity, and resistance to dyeing chemicals [6–8]. These characteristics have received the attention of researchers as a new biomaterial for use in various fields [9–11]. Additionally, tensan silk also has been studied for its applications to human health [12–15]. However, despite the potential importance of the wild silk moth in research and economic fields, no whole genomic information is currently available for this or any other species from the family Saturniidae.
In this study, we present the annotated genome sequence of A. yamamai, the first published genome in the family Saturniidae, with transcriptome datasets collected from 10 different body organ tissues. These data will be a fundamental resource for future studies and provide more insight into the genome evolution and molecular phylogeny of the family Saturniidae.
Sample preparation and sequencing
For whole-genome sequencing, we selected 1 male sample (Ay-7-male1) from a breeding line (Ay-7) of A. yamamai raised at the National Academy of Agricultural Science, Rural Development Administration, Korea. In lepidopterans, males are homogametic (ZZ), and selecting a male sample can reduce the complexity of assembly from excessive repeats on the W chromosome in females. For genomic library construction, we removed the guts of A. yamamai to prevent contamination of genomes from other organisms such as gut microbes and oak, the main food source of A. yamamai. Details of the sample preparation process used in this study are presented in the Supplementary Data. Genomic DNA was extracted using a DNeasy Animal Mini Kit (Qiagen, Hilden, Germany), and the quality of extracted DNA was checked using trenean, picogreen assay, and gel electrophoresis (1% agarose gel/40 ng loading). After quality control processing, we were left with a total of 61.5 ug of A. yamamai DNA for genome sequencing. Using standard Illumina whole-genome shotgun (WGS) sequencing protocol (paired-end and mate-pair), we added 2 long read sequencing platforms, Moleculo (Illumina synthetic long read) and RS II (Pacific Bioscience). Tables 1–3 show a summary of generated data for each library used in this study. RNA-seq libraries were also constructed for 10 different tissues (hemocyte, malpighian tube, midgut, fat body, anterior-middle/silk gland, posterior/silk gland, head, integument, testis, ovary) with 3 biological replicates following standard manufacturer protocol (Illumina, San Diego, CA, USA). For this, more than 100 individual A. yamamai samples in 5 instar stages from the same breeding line were used for tissue anatomy, and 3 samples from each tissue were selected based on the quality of extracted RNA. Details of transcriptome library construction are shown in the Supplementary Data. Information of libraries and generated data is provided in Table 4, and a total of 147 Gb of genomic data and 76 Gb of transcriptomic data was generated for this study.
Table 1:
Summary statistics of generated whole-genome shotgun sequencing data using Illumina Nextseq 500
| Library name | Library type | Insert size | Platform | Read length | No. of reads | Total base, bp | Reads retained after trimming |
|---|---|---|---|---|---|---|---|
| 350 bp | Paired-end | 350 bp | Nextseq500 | 151 | 293 176 268 | 44 269 616 468 | 291 070 362 |
| 700 bp | Paired-end | 700 bp | Nextseq500 | 151 | 246 945 900 | 37 288 830 900 | 244 698 580 |
| 3 Kbp | Mate-pair | 3 Kbp | Nextseq500 | 76 | 284 204 762 | 21 599 561 912 | 195 095 164 |
| 6 Kbp | Mate-pair | 6 Kbp | Nextseq500 | 76 | 246 238 370 | 18 714 116 120 | 152 496 372 |
| 9 Kbp | Mate-pair | 9 Kbp | Nextseq500 | 76 | 239 919 538 | 18 233 884 888 | 148 612 724 |
| Total | 1 310 484 838 | 140 106 010 288 | 1 031 973 202 |
Table 3:
Summary statistics of generated long reads data using Pacbio RS II system
| No. of reads | 1005,571 |
|---|---|
| Total bases | 5836 969 225 |
| Length of longest (shortest) read | 50 132 (50) |
| Average read length | 5804.63 |
Table 4:
Summary statistics of generated transcriptome data obtained from 6 organ tissues using Illumina platform
| Tissue | Sample name | Read length | Read count | Total base, bp |
|---|---|---|---|---|
| Hemocyte | Hemocyte_1 | 76 | 20 815 674 | 1 581 991 224 |
| Hemocyte_2 | 76 | 26 704 666 | 2 029 554 616 | |
| Hemocyte_2 | 76 | 53 068 562 | 4 033 210 712 | |
| Malpighian tube | Malpighi_1 | 76 | 22 635 428 | 1 720 292 528 |
| Malpighi_2 | 76 | 24 893 788 | 1 891 927 888 | |
| Malpighi_3 | 76 | 45 213 164 | 3 436 200 464 | |
| Midgut | Midgut_1 | 76 | 23 350 138 | 1 774 610 488 |
| Midgut_2 | 76 | 24 597 972 | 1 869 445 872 | |
| Midgut_3 | 76 | 50 949 986 | 3 872 198 936 | |
| Head | Head_1 | 76 | 26 526 276 | 2 015 996 976 |
| Head_2 | 76 | 26 581 124 | 2 020 165 424 | |
| Head_3 | 76 | 40 900 456 | 3 108 434 656 | |
| Integument | Skin_1 | 76 | 24 592 846 | 1 869 056 296 |
| Skin_2 | 76 | 42 775 430 | 3 250 932 680 | |
| Skin_3 | 76 | 35 043 570 | 2 663 311 320 | |
| Fat body | Fat Body_1 | 76 | 24 637 810 | 1 872 473 560 |
| Fat Body_2 | 76 | 24 037 494 | 1 826 849 544 | |
| Fat Body_3 | 76 | 40 817 582 | 3 102 136 232 | |
| Anterior-middle/silk gland | AM/Silk Gland_1 | 76 | 21 399 638 | 1 626 372 488 |
| AM/Silk Gland_2 | 76 | 24 292 386 | 1 846 221 336 | |
| AM/Silk Gland_3 | 76 | 37 331 530 | 2 837 196 280 | |
| Posterior/silk gland | P/Silk Gland_1 | 76 | 27 359 580 | 2 079 328 080 |
| P/Silk Gland_2 | 76 | 23 300 962 | 1 770 873 112 | |
| P/Silk Gland_3 | 76 | 39 421 430 | 2 996 028 680 | |
| Testis | Testis_1 | 76 | 40 890 404 | 3 107 670 704 |
| Testis_2 | 76 | 45 733 846 | 3 475 772 296 | |
| Testis_3 | 76 | 44 985 224 | 3 418 877 024 | |
| Ovary | Ovary_1 | 76 | 40 797 628 | 3 100 619 728 |
| Ovary_2 | 76 | 40 409 752 | 3 071 141 152 | |
| Ovary_3 | 76 | 42 417 892 | 3 223 759 792 |
Table 2:
Summary statistics of generated Illumina synthetic long read (Moleculo) library
| 500–1499 bp | ≥1500 bp | |
|---|---|---|
| No. of assembled reads | 302 132 | 342 738 |
| No. of bases in assembled read | 268 853 717 | 1 205 349 082 |
| N50 length of assembled read | 960 | 4031 |
Genome assembly and evaluation
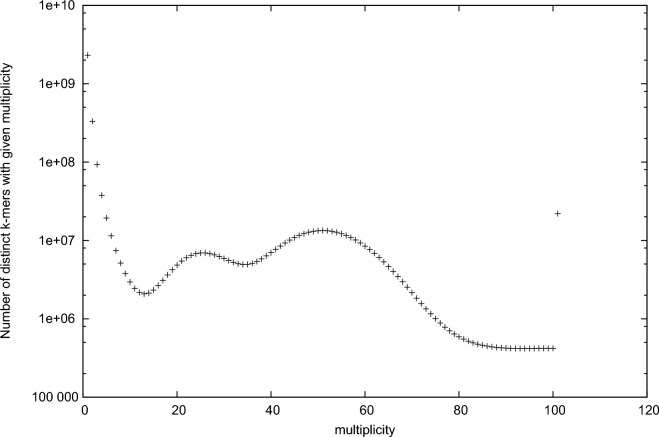
Before conducting genome assembly, we conducted k-mer distribution analysis using a 350-bp paired-end library in order to estimate the size and characteristics of the A. yamamai genome. The quality of our generated raw data was checked using FASTQC (FastQC, RRID:SCR_014583) [16]. Sequencing artifacts such as adapter sequences and low-quality bases were removed using Trimmomatic (Trimmomatic, RRID:SCR_011848) [17]. Jellyfish [18] was used to count the k-mer frequency for estimation of the genome size of A. yamamai. Figure 2 shows the 19-mer distribution of the A. yamamai genome using a 350-bp paired-end library. In the 19-mer distribution, the second peak, at approximately half the coverage value (x-axis) of the main peak, indicates heterozygosity. Although the inbred line used in this study was the single pair sib-mating maintained for more than 10 generations, high heterozygosity still remains. This phenomenon has been observed in a previous genomic study of the Diamondback moth (Plutella xylostella), and sustained heterozygosity as an important genomic characteristic was hypothesized to be a result of environmental adaption [19]. The underlying mechanism of the sustained heterozygosity is unclear, but associative overdominance can be one of the candidate explanations of this phenomenon [20, 21]. Based on the result of 19-mer distribution analysis, the genome size of A. yamamai was estimated to be 709 Mb. However, this size might be larger than the real genome size of A. yamamai because high heterozygosity could affect the estimation of genome size based on the K-mer distribution. Next, we conducted error correction on Illumina paired-end libraries using the error correction module of Allpaths-LG [22] before the initial contig assembly process (ALLPATHS-LG, RRID:SCR_010742). After error correction, initial contig assembly with 350-bp and 700-bp libraries was conducted using SOAP denovo2 [23] with the parameter option set at K = 19; this approach showed the best assembly statistics compared with other assemblers and parameters (SOAPdenovo2, RRID:SCR_014986). Quality control processing for mate-pair libraries and scaffolding was conducted using Nxtrim [24] and SSPACE (SSPACE, RRID:SCR_011848) [25], respectively. At each scaffolding step, SOAP Gapcloser [23] with -l 155 and -p 31 parameters was repeatedly used to close the gaps within each scaffold. In order to obtain a higher-quality genome assembly of A. yamamai, we employed several long read scaffolding strategies using SSPACE-LongRead [26]. First, we used an Illumina synthetic long read sequencing platform called Moleculo, which has been proven valuable for the study of highly heterozygous genomes in previous studies [27, 28]. After scaffolding was performed using SSPACE-LongRead with Illumina synthetic long read data, the total number of assembled scaffolds was effectively reduced from 398 446 to 24 558. The average scaffold length was also extended from 1.7 Kb to 24.8 Kb. However, there was no impressive improvement in N50 length (approximately 91 Kb to 112 Kb) of assembled scaffolds. Therefore, we employed another type of long read data generated from 10 cells of Pacbio RS II system with P6-C4 chemistry. After final scaffolding processing using Pacbio long reads, the number of scaffolds was reduced to 3675, and N50 length was effectively extended from 112 Kb to 739 Kb. Summary statistics of the assembled A. yamamai genome are provided in Table 5. Final assembly of the A. yamamai genome was 656 Mb (>2 kb) long with 3675 scaffolds, and the N50 length of assembly was 739 Kb with a 34.07% GC ratio. To evaluate the quality of the assembled genome, we conducted Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis [29] using BUSCO v2.0 with insecta_odb9 including 1658 BUSCOs from 42 species (BUSCO, RRID:SCR_015008). From BUSCO analysis, 96.7% of BUSCOs were completely detected in the assembled genome (1576: complete and single-copy; 27: complete and duplicated) among 1658 tested BUSCOs. The numbers of fragmented and missing BUSCOs were 21 and 34, respectively. Based on the result of BUSCO analysis, the genome of A. yamamai presented here was considered properly constructed for downstream analysis.
Figure 2:
19-mer distribution of A. yamamai genome using jellyfish with 350-bp paired-end whole genome sequencing data.
Table 5:
Summary statistics of the A. yamamai genome (>2 kb)
| Assembled genome | |
|---|---|
| Size, 1n | 656 Mb |
| GC level | 34.07 |
| No. of scaffolds | 3675 |
| N50 of scaffolds, bp | 739 388 |
| No. of bases in scaffolds, % | 19 257 439 (2.93) |
| Longest (shortest) scaffolds, bp | 3 156 949 (2003) |
| Average scaffold length, bp | 178 657.53 |
Repeat identification and comparative repeat analysis
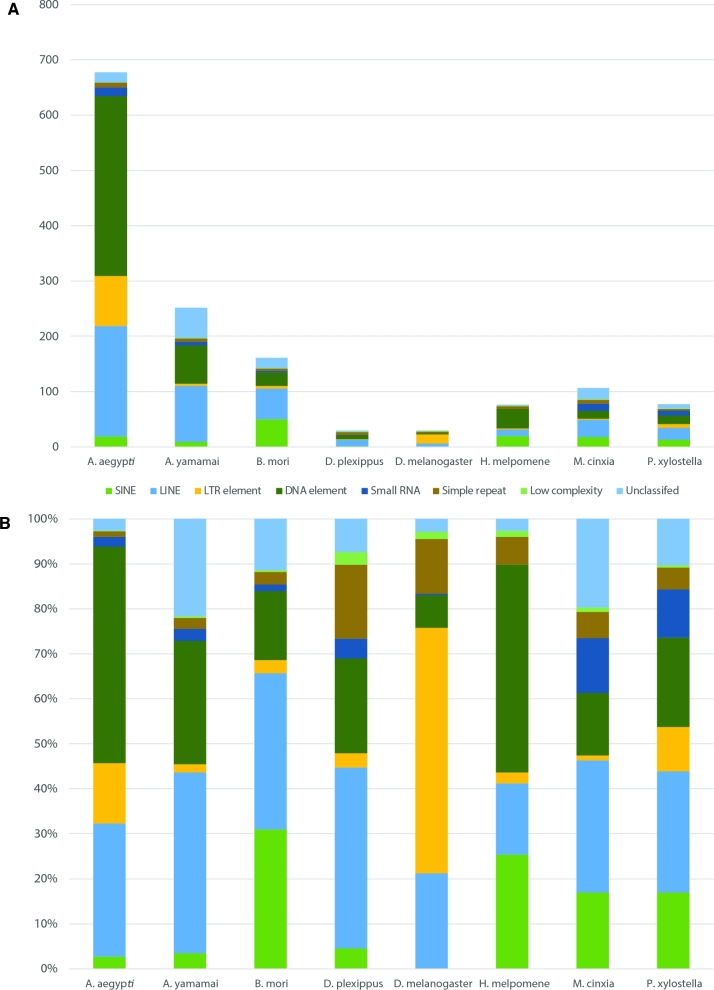
To identify repeat elements of the A. yamamai genome, a custom repeat library was constructed using RepeatModeler with RECON [30], RepeatScout [31], and TRF [32]. The resulting constructed custom repeat library for A. yamamai was further curated using the CENSOR [33] search, and the curated library was employed in RepeatMasker [34] with Repbase [35]. RepeatMasker (RepeatMasker, RRID:SCR_012954) was conducted with RMBlast and the “no_is” option for skipping bacterial insertion element check. Table 6 summarizes the proportion of identified mobile elements in the A. yamamai genome. The most prevalent repeat elements in the A. yamamai genome were LINE elements (101 Mb, 15.31% of total genome), and total repeat elements accounted for 37.33% of the total genome. In order to compare the repeat elements of A. yamamai with those of other genomes, we conducted the same process for 7 public genomes that are close neighbors of A. yamamai—Aedes aegypti [36], Bombyx mori [37], Danaus plexippus [38], Drosophila melanogaster [39], Heliconius Melpomene [40], Melitaea cinxia [41], and Plutella xylostella [19]. Figure 3 displays the amount and proportion of identified repeat elements from the 8 species. Despite the small genome size of B. mori, the total amount of identified SINE elements in the B. mori genome was 5.77 times larger than that of A. yamamai. The top 5 expanded repeat elements in the A. yamamai genome were DNA/RC, LINE/L2, LINE/RTE-BovB, DNA/TcMar-Mariner, and LINE/CR1. Among these, DNA/TcMar-Mariner was the specifically expanded repeat element in A. yamamai among the 8 species. In B. mori, SINE/tRNA-CR1, LINE/Jockey, DNA/RC, LINE/CR1-Zenon, and LINE/RTE-BovB were the top 5 expanded repeat elements. When comparing the repeat elements of A. yamamai with those of B. mori, which are both producers of the same type of silk, repeat elements showed family and species-specific patterns in the 2 silk moth linages. Particularly, we found that the mariner repeat element, which was found specifically expanded in the A. yamamai genome, was also included in the fibroin gene. A previous sequencing study also showed that the mariner repeat element was inserted in the 5’-end of the fibroin gene of A. yamamai [42]. Fibroin is the core component of the silk protein found in the silk moth, and the physical characteristics of silk mainly depend on the types and unique repeat motifs of the fibroin [43]. This gene is known to have hundreds of tandem repeat motifs, and these kinds of tandem repeats can be derived through transposable elements. This indicates that the mariner repeat element, specifically expanded in the A. yamamai genome, may play an important role in the development of the unique silk of A. yamamai, and the lineage-specific repeat elements may be one of the candidate evolution forces related to host-specific phenotype during genome evolution.
Table 6:
Summary of identified repeat elements in the A. yamamai genome
| Repeat element | No. of elements | Length, % |
|---|---|---|
| SINE | 59 968 | 8 615 338 (1.30) |
| LINE | 426 522 | 101 251 176 (15.31) |
| LTR element | 53 977 | 4 552 386 (0.69) |
| DNA element | 512 760 | 69 071 227 (10.44) |
| Small RNA | 43 645 | 6 691 619 (1.01) |
| Simple repeat | 135 989 | 6 256 839 (0.95) |
| Low complexity | 19 937 | 932 829 (0.14) |
| Unclassified | 294 190 | 54 552 009 (8.25) |
Figure 3:
Amount and proportion of identified repeat element from 8 species including A. yamamai. A) Absolute amount of repeat element classified into 8 different categories. B) Proportion of each repeat element in identified total repeat element.
Gene prediction and annotation
Three different algorithms were used for gene prediction of the A. yamamai genome: ab initio, RNA-seq transcript-based, and protein homology-based approaches. For ab initio gene prediction, Augustus (Augustus: Gene Prediction, RRID:SCR_008417) [44], Geneid [45], and GeneMarks-ET [46] were employed. Augustus was trained using known genes of A. yamamai in the NCBI database, and mapping information of RNA-seq data obtained from Tophat (TopHat, RRID:SCR_013035) [47] was also utilized for gene prediction. Geneid was used with predefined parameters for Drosophila melanogaster. GeneMarks-ET was employed using junction information of genes from transcriptome data alignment. For RNA-seq transcript-based prediction, generated transcriptome data from 10 organ tissues of A. yamamai were aligned to the assembled genome and gene information was predicted using Cufflinks (Cufflinks, RRID:SCR_014597) [48]. The longest CDS sequences were identified from Cufflinks results using Transdecoder. For the homology-based approach, all known genes of the order Lepidoptera in the NCBI database were aligned using PASA [49]. Table 7 shows the gene prediction results from each method. Gene prediction results from different prediction algorithms were combined using Evidence Modeler [50], and a consensus gene set of the A. yamamai genome was created. Manual curation was performed based on the 5 types of evidence (3 in silico, known protein, and RNA-seq) using IGV [51] and BLASTP (BLASTP, RRID:SCR_001010). Using IGV with each gene evidence and comparing results with known genes via BLASTP, we mainly focused on removing false positively predicted genes that don’t have enough evidence. Merged and spliced gene structured were corrected by comparing the gene structure with known exon structure in the NCBI NR database. In addition, fibroin and sericin genes, which couldn’t be properly predicted because of their high repeat motif, were also manually identified with previously known sequences [42, 52] with RNA-seq data. The final gene set of the A. yamamai genome contains 15 481 genes. Summary statistics for the consensus gene set are provided in Table 8. The average gene length was 11 016.34 bp with a 34.38% GC ratio, and the number of exons per gene was 5.64. In order to identify the function of predicted genes in A yamamai, 3 nonredundant sequence databases (Swiss-Prot [53], Uniref100 [53], and NCBI NR [54]) as well as the gene information of 2 species (B. mori and D. melanogaster) were used for target databases using BLASTP. Additionally, protein domain searches were conducted on the consensus gene set using InterproScan5 (InterProScan, RRID:SCR_005829) [55]. Figure S1 shows the top 20 identified terms from 7 different InterproScan5 analyses. Among the various analyses conducted using InterproScan5, gene ontology analysis with the Pfam database showed that a large proportion of genes in the A. yamamai genome were related with the function of molecular binding, catalytic activity, internal component of membrane, metabolic process, oxidation-reduction process, and transmembrane transport.
Table 7:
Summary statistics of ab initio, RNA-seq-based, and homology-based gene prediction results
| Evidence type | Programs | Element | Total count | Exon/gene | Total length, bp | Mean length, bp |
|---|---|---|---|---|---|---|
| Gene | 14 576 | 142 415 318 | 9770.53 | |||
| Augustus | 4.85 | |||||
| Exon | 70 733 | 14 736 668 | 208.34 | |||
| Gene | 10 946 | 46 119 402 | 4213.35 | |||
| ab_initio | Geneid | 2.25 | ||||
| Exon | 24 686 | 3 925 563 | 159.01 | |||
| Gene | 27 754 | 273 745 951 | 9863.29 | |||
| GeneMarks-ET | 5.50 | |||||
| Exon | 152 660 | 30 847 503 | 202.06 | |||
| Gene | 36 213 | 840 429 061 | 23 207.94 | |||
| RNA-seq | Cufflinks Transdecoder | 7.03 | ||||
| Exon | 254 770 | 201 721 675 | 791.77 | |||
| Known gene (NCBI lepidoptera) | PASA (gmap) | 44 561 | 22 484 151 | 504.57 |
Table 8:
Summary statistics for the consensus gene set of the A. yamamai genome
| Element | No. of elements | Exon/gene | Avg. length | Total length | Genome coverage, % |
|---|---|---|---|---|---|
| Gene | 15 481 | 11 016.34 | 170 543 958 | 25.78 | |
| 5.64 | |||||
| Exon | 87 346 | 1346.23 | 20 840 925 | 3.31 |
Comparative genome analysis
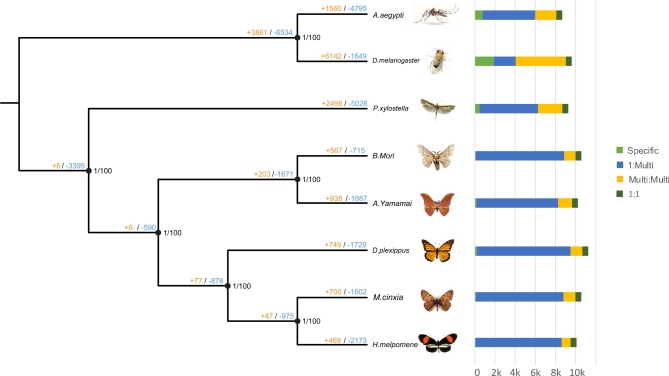
We used OrthoMCL [56] and Reciprocal Best Hit within BLASTP for identification of gene family clusters and 1:1 orthologous gene sets. Gene information of 7 taxa (A. aegypti, B. mori, D. plexippus, D. melanogaster, H. melpomene, M. cinxia, and P. xylostella), the same taxa used in repeat analysis, was employed for OrthoMCL with A. yamamai. A total of 17 406 gene family clusters were constructed, and 3586 1:1 orthologous genes were identified. Before conducting comparative genome analysis, we constructed phylogenetic trees for the 8 species. In order to build the phylogenetic tree, multiple sequence alignment for the 1:1 orthologous genes of all 8 species was conducted using PRANK [57], and Gblocks [58] was used to obtain conserved blocks for the phylogenetic tree. Conserved block sequences were sequentially concatenated to obtain 1 consensus sequence for each species. MEGA [59] was used for constructing Neighbor-Joining Trees (bootstrap 1000, maximum composite likelihood, transitions + transversions, and gamma distributed option), and MrBayes (MrBayes, RRID:SCR_012067) [60] was employed for the construction of Bayesian inference trees. To select the best evolution model for our data, Modeltest [61] was conducted and the GTR-based invariant model was chosen based on the AIC value of Modeltest. Figure 4 shows the constructed phylogenetic tree of the 8 species using 3586 orthologous genes. The bootstrap value and Bayesian poster probability value of all nodes were 100 and 1, respectively. The closest neighbor of A. yamamai was B. mori, which is included in Bombycidae family; this result is consistent with that of previous studies. Three butterfly species (D. plexippus, M. cinxia, and H. meplmene) included in Nymphalidae family were also shown to share a common ancestor with the families Saturniidae and Bombycidae.
Figure 4:
Constructed phylogenetic tree and comparative gene family analysis. Node values indicate Bayesian posterior probability, bootstrap and gene expansion, and contraction value. Orange and blue colors indicate expansion and contraction, respectively. Bar chart indicates the number of genes cauterized into 4 groups (Specific, 1:Multi, Multi:Multi, and 1:1) using OrthoMCL.
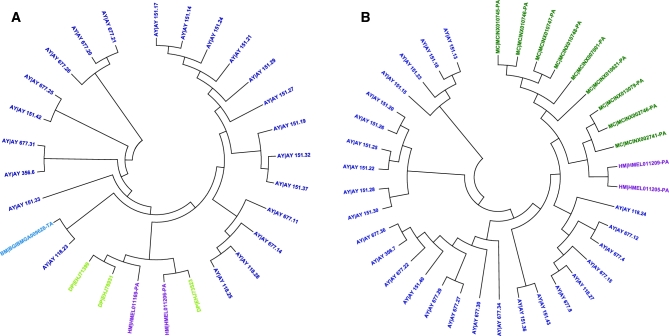
Based on the constructed phylogenetic tree, gene family expansion and contraction analysis were conducted using a 2-parameter model in CAFÉ [62] and the gene tree was constructed using protein sequence via MEGA [59]. Figure 4 shows the result of gene family expansion and contraction analysis of 8 species; 938 and 1987 gene families of A. yamamai and 567 and 715 gene families of B. mori were estimated to be expanded and contracted from the common ancestors, respectively. Among these, 15 gene families in A. yamamai were estimated to be under rapid expansion during the evolution process. Functions of genes in the rapidly expanded gene families of A. yamamai were transposase, fatty acid synthase, zinc finger protein, chorion (eggshell protein), reverse transcriptase, prostaglandin dehydrogenase, RNA-directed DNA polymerase, gag like protein, juvenile hormone acid methyltransferase, facilitated trehalose transporter, and glucose dehydrogenase. Figure 5 shows the gene tree of 2 chorion gene (chorion class A and B) family clusters rapidly expanded in the A. yamamai genome. Chorion, called eggshell protein, composes the surface of the egg and protects the embryo from environmental threats such as desiccation, flooding, freezing, infection of microorganisms, and physical destruction. It also provides channels, such as aeropyle, that enable gas exchange and maintain proper conditions for diapause of the egg [63]. These diverse functions of eggshell are implemented by the specific eggshell structure, and the surface structure of eggshell varies between species for the adaptation in a different environment. The ancestor of Antherea has the unique aeropyle structure called “aerophyle crown” on the eggshell surface [64]. This unique structure is formed by the circular vertical projection of lamellar chorion from the follicle cell, and it surrounds the aeropyles near the end of oogenesis [65]. Acquiring this kind of de novo complex structure requires numerous genetic changes, and a previous study about Antheraea polyphemus has shown that more than 100 chorion-specific polypeptides were involved in this unique ultra-structure [65]. Therefore, the specific rapid expansion of the chorion class A and B gene family in the A. yamamai genome might be one of the convincing molecular explanations for acquiring this unique ultrastructure in the eggshell surface of Antheraea genus. However, this unique ultrastructure tends to be reduced during the current evolution process of the Antheraea genus. Types of eggshell structures in the Antheraea genus can be categorized into multiple classes based on the morphology and regional distribution of the aeropyle [64]. The shape of the aeropyle in the A. yamamai egg is known to be converted to a mound shape from the crown shape, and these aeropyle mounds only exist in the narrow band surrounding the micropyle region [64]. Only a very few, small aeropyle crowns remain, and it is entirely different from the ancestral form of eggshell surface mostly covered by aeropyle crowns. These regional differences were known to be adjusted by regional differences of filler genes during choriogenesis [66], and the additional regulations of related genes for choriogenesis have to be considered. This indicates that the specifically expanded chorion gene families of A. yamamai may be one of the remaining evolutionary tracks in the genome of the Antheraea genus. However, further functional studies must be conducted to resolve the limited understanding about the relationship between these expanded chorion gene families and the current eggshell surface formation of A. yamamai.
Figure 5:
Expansion of chorion gene in the A. yamamai genome. A, B) The gene trees of chorion A and B in the rapid expanded gene family cluster, respectively. Color of terminal node indicates each taxon identified in the gene family cluster.
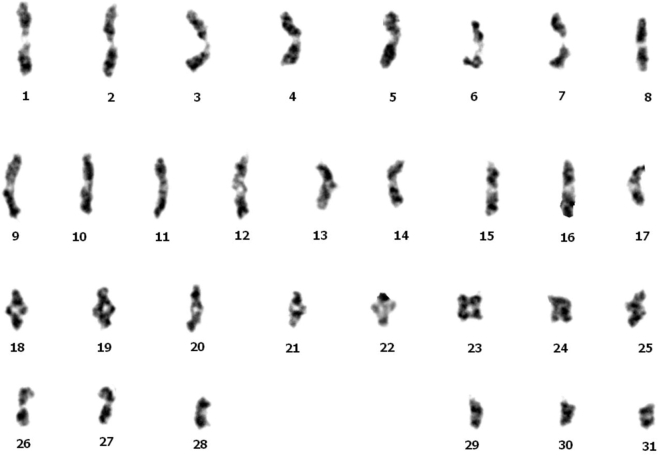
The constructed genome of A. yamamai presented here is the first announced genome in the family Saturniidae, and the karyotyping analysis using gamete in the metaphase showed that the genome of A. yamamai consists of 31 chromosomes (Fig. 6). This constructed genome information provides more insight into the genome evolution and phylogeny of the family Saturniidae, which contains the largest number of species in Lepidoptera. For example, although 2 silk moths, A. yamamai and B. mori, appear similar, comparative genome analysis showed the significant differences in the genome size and the specific expansion of repeat elements and gene families between the families Saturniidae and Bombycidae. In case of molecular phylogeny, most previous phylogenetic studies were limited to few genes due to the lack of genomic information on the family Saturniidae. We expect that our study and resulting constructed genome will resolve some limitations of molecular phylogenetic and ecological research on the Saturniidae species. Additionally, constructed genome information will help researchers better understand the molecular background of wild silk and its production. Silk produced by A. yamamai, referred to as tensan silk, shows unique characteristics that have made it valuable in various fields. However, A. yamamai has not been completely domesticated compared with B. mori, making mass production of tensan silk infeasible. Understanding of the molecular mechanisms behind the tensan silk production process is essential for mass production using biotechnology, and this genome sequence with manually curated gene information is a fundamental resource for related research and industrial improvement. Additionally, the transcriptome data of 10 different organ tissues with the 3 biological replications presented here may be also useful resources for uncovering the molecular mechanisms related to specific phenotypes of A. yamamai and the family Saturniidae.
Figure 6:
Karyotype of A. yamamai using a gamete of testis in metaphase.
Availability of supporting data
The generated genome sequence and gene information of A. yamamai are available in GigaDB [67], and generated raw data are available under project accession PRJNA383008 and PRJNA383025 of the NCBI database.
Additional file
Additional file 1: Figure S1. Top 20 terms in each 7 Interproscan5 analysis (CDD, Pfam, PRINTS, ProSitePatterns, ProSiteProfiles, TIGRF, Gene Ontology).
Additional file 1: Supplementrary_information2.xlsx.
Competing interests
All authors report no competing interests.
Abbreviations
BUSCO: Benchmarking Universal Single-Copy Orthologs; MYA: million years ago.
Author contributions
Sampling: Kee-Young Kim, Su-Bae Kim; sequencing: Kwang-Ho Choi, Seong-Wan Kim genome assembly: Seong-Ryul Kim, Woori Kwak, Jae-Sam Hwang, Seung-Won Park repeat element analysis: Seong-Ryul Kim, Woori Kwak, Seung-Won Park gene prediction: Seong-Ryul Kim, Woori Kwak, Hyaekang Kim, Jae-Sam Hwang comparative genome analysis: Seong-Ryul Kim, Woori Kwak, Min-Jae Kim, Kelsey Caetano-Anolles funding and experimental design: Seong-Ryul Kim, Seung-Won Park.
Supplementary Material
09 Jun 2017 Reviewed
17 Jun 2017 Reviewed
18 Jun 2017 Reviewed
Acknowledgements
This work was supported by a grant from the Rural Development Administration, Republic of Korea (grant No. PJ010442).
References
- 1. Regier JC, Grant MC, Mitter C et al. Phylogenetic relationships of wild silkmoths (Lepidoptera: Saturniidae) inferred from four protein-coding nuclear genes. Syst Entomol 2008;33(2):219–28. [Google Scholar]
- 2. Regier JC, Mitter C, Zwick A et al. A large-scale, higher-level, molecular phylogenetic study of the insect order Lepidoptera (moths and butterflies). PLoS One 2013;8(3):e58568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 2006;22(23):2971–2. [DOI] [PubMed] [Google Scholar]
- 4. Kawahara AY, Barber JR. Tempo and mode of antibat ultrasound production and sonar jamming in the diverse hawkmoth radiation. Proc Natl Acad Sci U S A 2015;112(20):6407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peigler RS. Wild Silks of the World. Am Entomol; 1993;39(3):151–62. [Google Scholar]
- 6. Matsumoto Y-I, Saito H. Load-extension characteristics of composite raw silk of Antheraea yamamai and Bombyx mori. J Sericult Sci Japan 1997;66(6):497–501. [Google Scholar]
- 7. Nakamura S, Saegusa Y, Yamaguchi Y et al. Physical properties and structure of silk. XI. Glass transition temperature of wild silk fibroins. J Appl Polym Sci 1986;31(3):955–6. [Google Scholar]
- 8. Kweon H, Park Y. Structural characteristics and physical properties of wild silk fibres; Antheraea pernyi and Antheraea yamamai. Korean J Sericult Sci (Korea Rep) 1994. [Google Scholar]
- 9. Zheng Z, Wei Y, Yan S et al. Preparation of regenerated Antheraea yamamai silk fibroin film and controlled-molecular conformation changes by aqueous ethanol treatment. J Appl Polym Sci 2010;116(1):461–7. [Google Scholar]
- 10. Omenetto F, Kaplan D, Amsden J et al. Silk based biophotonic sensors. 2011. Google Patents. [Google Scholar]
- 11. Takeda S. New field of insect science: research on the use of insect properties. Entomol Sci 2013;16(2):125–35. [Google Scholar]
- 12. Omenetto F, Kaplan DL. Silk-based multifunctional biomedical platform. 2012. Google Patents. [Google Scholar]
- 13. Serban MA. Silk medical devices. 2016. Google Patents. [Google Scholar]
- 14. Jiang G-L, Collette AL, Horan RL et al. Drug delivery platforms comprising silk fibroin hydrogels and uses thereof. 2010. Google Patents. [Google Scholar]
- 15. Kamiya M, Oyauchi K, Sato Y et al. Structure-activity relationship of a novel pentapeptide with cancer cell growth-inhibitory activity. J Pept Sci 2010;16(5):242–8. [DOI] [PubMed] [Google Scholar]
- 16. Bioinformatics B. FastQC A Quality Control Tool for High Throughput Sequence Data. Cambridge, UK: Babraham Institute, 2011. [Google Scholar]
- 17. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014:btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011;27(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. You M, Yue Z, He W et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet 2013;45(2):220–5. [DOI] [PubMed] [Google Scholar]
- 20. Maruyama T, Nei M. Genetic variability maintained by mutation and overdominant selection in finite populations. Genetics 1981;98(2):441–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pamilo P, Pálsson S. Associative overdominance, heterozygosity and fitness. Heredity 1998;81(4):381–9. [DOI] [PubMed] [Google Scholar]
- 22. Gnerre S, MacCallum I, Przybylski D et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A 2011;108(4):1513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo R, Liu B, Xie Y et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2012;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O’Connell J, Schulz-Trieglaff O, Carlson E et al. NxTrim: optimized trimming of Illumina mate pair reads: Table 1. Bioinformatics 2015;31(12):2035–7. [DOI] [PubMed] [Google Scholar]
- 25. Boetzer M, Henkel CV, Jansen HJ et al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011;27(4):578–9. [DOI] [PubMed] [Google Scholar]
- 26. Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 2014;15(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voskoboynik A, Neff NF, Sahoo D et al. The genome sequence of the colonial chordate, Botryllus schlosseri. Elife 2013;2:e00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCoy RC, Taylor RW, Blauwkamp TA et al. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS One 2014;9(9):e106689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simão FA, Waterhouse RM, Ioannidis P et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015:btv351. [DOI] [PubMed] [Google Scholar]
- 30. Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 2002;12(8):1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics 2005;21(Suppl 1):i351–8. [DOI] [PubMed] [Google Scholar]
- 32. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999;27(2):573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kohany O, Gentles AJ, Hankus L et al. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 2006;7(1):474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2009:4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- 35. Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 2015;6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nene V, Wortman JR, Lawson D et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007;316(5832):1718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia Q, Zhou Z, Lu C et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004;306(5703):1937–40. [DOI] [PubMed] [Google Scholar]
- 38. Zhan S, Merlin C, Boore JL et al. The monarch butterfly genome yields insights into long-distance migration. Cell 2011;147(5):1171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams MD, Celniker SE, Holt RA et al. The genome sequence of Drosophila melanogaster. Science 2000;287(5461):2185–95. [DOI] [PubMed] [Google Scholar]
- 40. Consortium HG. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 2012;487(7405):94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ahola V, Lehtonen R, Somervuo P et al. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hwang J-S, Lee J-S, Goo T-W et al. Cloning of the fibroin gene from the oak silkworm, Antheraea yamamai and its complete sequence. Biotechnol Lett 2001;23(16):1321–6. [Google Scholar]
- 43. Malay AD, Sato R, Yazawa K et al. Relationships between physical properties and sequence in silkworm silks. Sci Rep 2016;6(1):27573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stanke M, Diekhans M, Baertsch R et al. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008;24(5):637–44. [DOI] [PubMed] [Google Scholar]
- 45. Blanco E, Parra G, Guigó R. Using Geneid to identify genes. Curr Protoc Bioinformatics 2007:4.3.1–4.3.28. [DOI] [PubMed] [Google Scholar]
- 46. Lomsadze A, Burns PD, Borodovsky M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res 2014:gku557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25(9):1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trapnell C, Roberts A, Goff L et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7(3):562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Campbell MA, Haas BJ, Hamilton JP et al. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 2006;7(1):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haas BJ, Salzberg SL, Zhu W et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 2008;9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson JT, Thorvaldsdóttir H, Winckler W et al. Integrative genomics viewer. Nat Biotechnol 2011;29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zurovec M, Yonemura N, Kludkiewicz B et al. Sericin Composition in the Silk of Antheraea yamamai. Biomacromolecules 2016;17(5):1776–87. [DOI] [PubMed] [Google Scholar]
- 53. Consortium U. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 2011:gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 2007;35(Database):D61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jones P, Binns D, Chang H-Y et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014;30(9):1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13(9):2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Löytynoja A, Goldman N. From The Cover: An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A 2005;102(30):10557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000;17(4):540–52. [DOI] [PubMed] [Google Scholar]
- 59. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19(12):1572–4. [DOI] [PubMed] [Google Scholar]
- 61. Posada D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinformatics 2003:6.5.1–6.5.14. [DOI] [PubMed] [Google Scholar]
- 62. De Bie T, Cristianini N, Demuth JP et al. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 2006;22(10):1269–71. [DOI] [PubMed] [Google Scholar]
- 63. Chapman RF. The Insects: Structure and Function. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 64. Regier JC, Paukstadt U, Paukstadt LH et al. Phylogenetics of eggshell morphogenesis in Antheraea (Lepidoptera: Saturniidae): unique origin and repeated reduction of the aeropyle crown. Syst Biol 2005;54(2):254–67. [DOI] [PubMed] [Google Scholar]
- 65. Regier JC, Mazur GD, Kafatos FC. The silkmoth chorion: morphological and biochemical characterization of four surface regions. Devel Biol 1980;76(2):286–304. [DOI] [PubMed] [Google Scholar]
- 66. Hatzopoulos AK, Regier JC. Evolutionary changes in the developmental expression of silkmoth chorion genes and their morphological consequences. Proc Natl Acad Sci U S A 1987;84(2):479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim S, Kwak W, Kim K et al. The Japanese silk moth, Antheraea yamamai, draft genome sequence. GigaScience Database 2017. http://dx.doi.org/10.5524/100382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
09 Jun 2017 Reviewed
17 Jun 2017 Reviewed
18 Jun 2017 Reviewed