Abstract
MicroRNAs (miRs) are small RNAs that do not code for proteins, but instead decrease the stability and suppress the translation of target mRNAs by binding with complementary sequences in their 3′-untranslated regions (3′-UTRs). In the present study, it is reported that breast cancer tumor tissue, as well as irradiated MCF7 breast cancer cells, exhibit decreased levels of miR-26b expression compared with normal breast tissue and MCF7 cells without exposure to radiation. Additionally, a luciferase reporter assay was used to demonstrate that miR-26b directly targetsDNA damage-regulated autophagy modulator 1 (DRAM1). MCF7 cells that were transfected with an miR-26b mimicexhibited the downregulated expression of DRAM1 protein and a reduced level of irradiation-induced autophagy. Inhibiting miR-26b resulted in the upregulation of DRAM1 and increased levels of irradiation-induced autophagy in MCF7 cells. These results suggest that therapeutic strategies to target miR-26b may increase the efficacy of certain types of cancer therapy.
Keywords: microRNA-26b, DNA damage-regulated autophagy modulator 1, autophagy, breast cancer
Introduction
MicroRNAs (miRNAs/miRs) typically contain 20–24 nucleotides and do not code for proteins, but instead modulate the expression of specific target genes. miRNAs bind to 3′-untranslated regions of target mRNA molecules to repress translation or enhance degradation (1). As each miRNA has multiple targets, a single microRNA may produce diverse effects by functioning as a tumor initiator or suppressor (2). miRNAs may regulate various critical signaling networks to modulate carcinogenesis (3–6). The differential expression of miR-26b has been demonstrated in several types of tumor, including lung (7), cervical (8), tongue (9) and breast cancer (10), suggesting that it has specific roles in tumor pathology. Previous studies conducted with clinical samples of breast tumor tissue have indicated that miR-26b may inhibit tumor formation, and that its overexpression induces apoptosis and inhibits proliferation (10–12).
DNA damage-regulated autophagy modulator 1 (DRAM1) is a lysosomal protein upregulated by p53 that induces the macro-autophagy activity associated with cell death; DRAM is critical for the induction of programmed cell death, contributing to the tumor-suppressive effects of p53 (13–15).
Despite the previous studies on miR-26b, the functions of miR-26b and its target genes remain to be fully characterized, including in human breast cancer. In the present study, the function of miR-26b in breast cancer was determined using the human breast cancer cell line MCF7. The results of the present study demonstrated low-expression of miR-26b in breast cancer tumor tissue. In addition, it was identified that miR-26b downregulated DRAM1 by targeting the 3′-UTR directly. Inhibiting miR-26b upregulated DRAM1 and increased irradiation-induced autophagy. Therefore, the results of the present studymay providea novel insight for targeting therapy of cancer.
Materials and methods
Tissue samples
Three paired samples of breast cancer tissue and adjacent noncancerous tissue were provided by the Breast and Thyroid Surgery Department of the China-Japan Union Hospital (Changchun, China) between June 2013 and December 2013, with a mean age of 41 years (range, 37–49 years). All samples of breast cancer tissue had been pathologically confirmed as invasive ductal carcinoma. None of the patients had been treated with chemotherapy or radiation therapy prior to surgery. The study was approved by the China-Japan Union Hospital of Jilin University and informed, written consent was provided by all patients.
Cell culture and transfection
Human MCF7 breast cancer cells were purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China) and cultured at 37°C in Dulbecco's Modified Eagle's Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing fetal bovine serum (10%; HyClone; GE Healthcare, Logan, UT, USA), penicillin (100 units/ml), and streptomycin (100 units/ml), in a humidity-controlled incubator with an atmosphere containing 5% CO2. Following culture, the cells were transfected with 100 nmol of an miR-26b mimic or inhibitor, or a negative control miRNA (NC; GenePharma, Shanghai, China) using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol; they were then exposed to ionizing radiation (IR) as subsequently described. The NC was designed not to target any human gene. The sequence for the miR-26b mimic was sense, 5′-UCAAGUAAUUCAGGAUAGGU-3′, antisense, 5′-CUAUCCUGAAUUACUUGAAUU-3′; the sequence for the miR-26b inhibitor was sense, 5′-ACCUAUCCUGAAUUACUUGAA-3′, antisense, 5′-CAGUACUUUUGUGUAGUACAA-3′; the sequence for the NC was sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. Specific siRNAs for human DRAM (catalog no. sc-96209) and control siRNA (catalog no. sc-44230) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Irradiation
Cells were given for IR 24 h after transfection and harvested by centrifugation at 13,201 × g for 3 min at room temperature for analysis. IR was delivered at a rate of 0.38 Gy/min (180 kV; 18 mA) using an XSZ-Z20/20 X-ray generator (Kang Jia Instrument Equipment Co., Ltd., Dandong, China).
Detection of miR-26b by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
At 48 h after transfection, total cellular RNA was extracted using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA samples were then frozen at −80°C until use in further analyses. The expression of miRNA was detected usinga Hairpin-it™ miRNAs RT-PCR Quantization kit, containing miR-26b primers and U6 primers (E22002; GenePharma) and an MX3000p thermocycler (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). Reverse transcription was performed with a 20 ng sample of RNA, which was incubated for 60 min at 42°C followed by 5 min at 95°C. Following synthesis, the cDNA template was diluted 80-fold with nuclease-free H2O. Then qPCR was performed as follows: Denaturation at 95°C for 10 min, followed by 40 amplification cycles of 95°C for 10 sec and 60°C for 1 min (ramp-rate, 1.6°C/sec). All samples were normalized to the U6 expression level using the 2−ΔΔCq method (16).
Plasmid construction
DRAM1 was identified as a potential target of miR-26b with TargetScan (http://www.targetscan.org/), which was used to identify autophagy-associated genes. The wild-type (WT) 3′-untranslated region (3′UTR) of DRAM1, including the putative miR-26b-binding site, was extracted by performing PCR with genomic DNA obtained from the extracted blood of a healthy human donor. DNA was extracted using Rapid DNA extraction and detection kit (Tiangen Biotech Co., Ltd., Beijing, China). The amplified product was then cloned into the HindIII and XhoI sites of the pMIR vector (Promega Corporation, Madison, WI, USA). PCR was also performed to generate a DRAM1 3′UTR with a mutated miR-26b binding site. The primer sequences were WT forwards, 5′-GTCAAGCTTCTGCAGCACATCCAGGACTTGAATTTCATTACGAGTTCCT-3′, WT reverse, 5′-CCACTCGAGACCAGGCGATACAGACTATT-3′; mutant forwards, 5′-GTCAAGCTTCTGCAGCACATCCAGGACAACAATTTCATTACGAGTTCCT-3′, mutant reverse, 5′-CCACTCGAGACCAGGCGATACAGACTATT-3′. PCR was performed as follows: Denaturation at 95°C for 5 min, 40 amplification cycles of 95°C for 10 sec, 55°C for 15 sec and 72°C for 90 sec, followed by 72°C for 5 min (ramp-rate, 1.6°C/sec). PrimeSTAR® Max DNA Polymerase (R045Q; Takara Biotechnology Co., Ltd., Dalian, China) was used for PCR amplification. DNA sequencing was used to verify the identities of the PCR products incorporated into the plasmids.
Luciferase reporter assays
MCF7 cells were cultured in 96-well plates (5×104 cells/well). Lipofectamine® 2000 was used to transfect the cells with 200 ng of pMIR-DRAM1 3′UTR and 5 ng of the pRL-SV40 vector containing the Renilla luciferase gene used to normalize transfection efficiency (Promega Corporation), in addition to 100 nmol of the miR-26b mimic or NC. At 48 h after transfection, the Dual-Luciferase Reporter assay (Promega Corporation) was used to examine firefly and Renilla luciferase activities. Each transfection was repeated 5 times in three independent experiments.
Western blot analysis
MCF7 cells were irradiated for 24 h (a time selected based on a time-course curve) and harvested; 1 ml lysis buffer (50 mM NaCl, 10 mM HEPES, 1 mM EDTA, 1 mM DTT, 1% NP-40, 0.1% SDS and 1 mM PMSF) and 10 µl protease inhibitor cocktail (P8340; Sigma-Aldrich; Merck KGaA) was added to the cell pellets. Subsequently, protein was quantified using BCA Protein Assay kit (P0012S; Beyotime Institute of Biotechnology, Haimen, China). SDS-PAGE was used to separate 60 µg of total cellular protein extract; the separated bands were transferred onto a nitrocellulose membrane for western blotting. Membranes were blocked with 5% skimmed milk in Tris-Buffered Saline Tween-20 for 1 h at room temperature. Subsequently, horseradish peroxidase-conjugated secondary antibodies [HRP-Goat anti-Rabbit IgG (cat no. bs-0295G-HRP) and HRP-Goat anti-Mouse IgG (cat no. bs-0296G-HRP; both 1:2,000); Boster Biological Technology, Pleasanton, CA, USA] was added for 1 h at room temperature. Primasry antibodies were used against DRAM1 (1:300; ab72171; Abcam; Cambridge, UK), light chain 3 (LC3) I and II (1:300; 4108S; Cell Signaling Technology; Danvers, MA, USA), and GAPDH (1:1,000; sc-32233; Santa Cruz Biotechnology, Inc.). Immunoreactive bands were detected by chemiluminescence (sc-2048; Santa Cruz Biotechnology, Inc.). The staining intensities of individual proteins were measured using ImageJ software (version 1.41; National Institutes of Health, Bethesda, MD, USA), and expressed relativeto the intensity of the GAPDH band.
Monodansylcadaverine (MDC) staining assay
MCF7 cells (4×105 cells/well) were seeded in 60-mm cell culture dishes and let sit overnight at room temperature. Each group of cells received the specified dose of IR. After 24 h, they were washed twice in cold PBS solution. The cells were incubated in DMEM containing 50 mM MDC for 1 h at 37°C to label the autophagic vacuoles; they were then washed with PBS and fixed by a 15 min immersion in 4% paraformaldehyde solution at room temperature. The cells were analyzed for fluorescence with a FACSort Flow Cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed using Cell Quest software (version 5.1; BD Biosciences) and FlowJo software (version 7.6.5; Tree Star Inc., Ashland, OR, USA).
Statistical analysis
The Student's t test and the χ2 test were used to analyze experimental data, and data are presented as the mean ± standard deviation using SPSS software (version 12.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-26b expression is reduced in breast cancer tissue and irradiated MCF7 cells
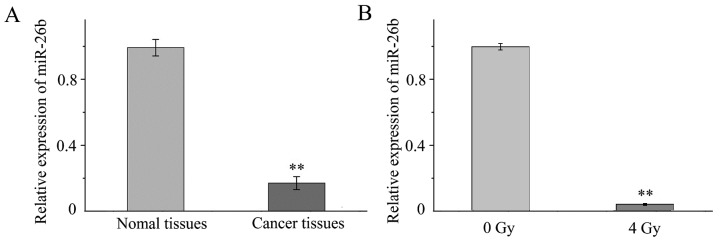
RT-qPCR was performed to investigate the expression of miR-26b in tissue samples and MCF7 cells. It was demonstrated that the expression of miR-26b was significantly lower in the three samples of breast cancer tissue when compared with expression in the noncancerous adjacent tissue (P<0.001; Fig. 1A). In addition, miR-26b expression was significantly downregulated in MCF7 cells exposed to 4-Gy IR compared with MCF7 cells that were not irradiated (P<0.01; Fig. 1B).
Figure 1.
miR-26b relative expression was decreased expression in cancer tissue and irradiated MCF7 cells. Reverse transcription-quantitative polymerase chain reaction was used to quantify the level of miR-26b in tissue samples and cells. (A) Level of miR-26b in breast cancer tissue compared with adjacent noncancerous tissue. (B) Level of miR-26b expression in MCF7 cells exposed to 4 Gy of radiation compared with MCF7 control cells. **P<0.01. miR, microRNA.
miR-26b may regulate IR-induced autophagy in MCF7 cells
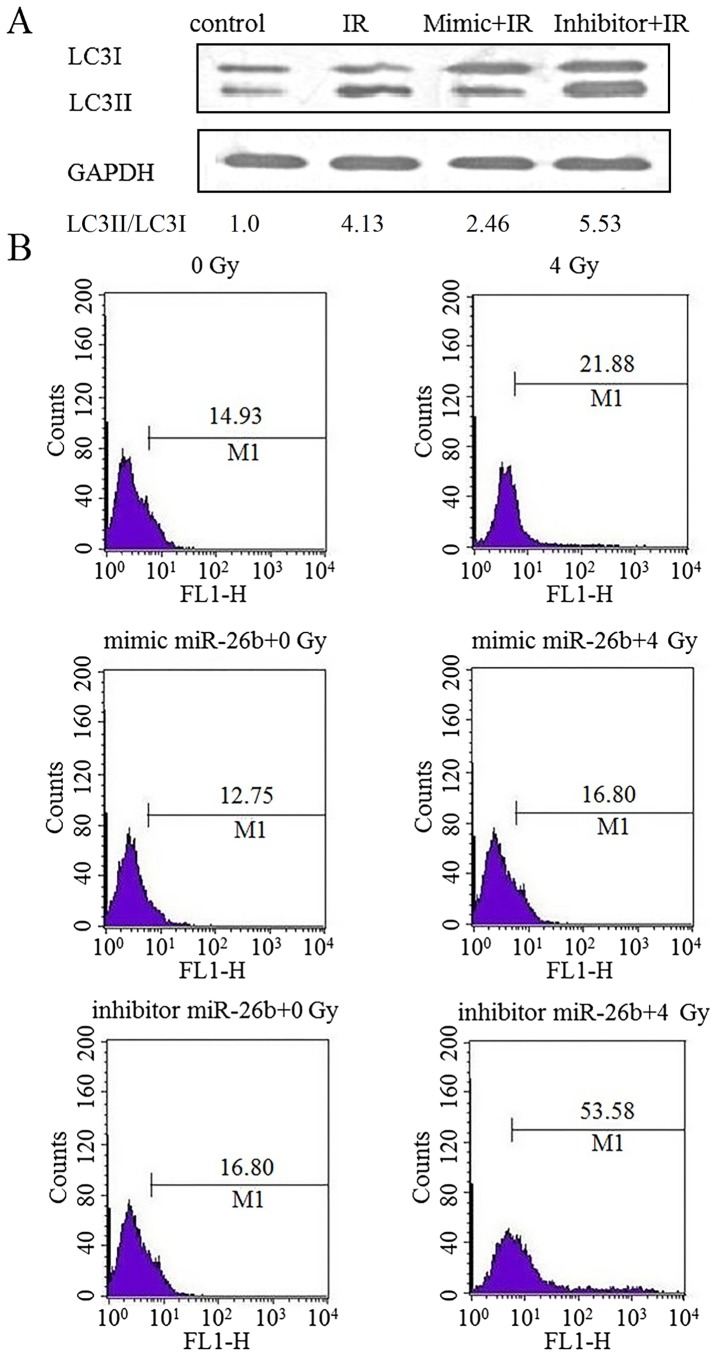
The effect of miR-26b on autophagy in MCF7 cells was examined by performing a western blot analysis for LC3II protein expression. Cells exposed to IR exhibited an increased level of LC3II protein expression, which was suppressed following transfection with an miR-26b mimic. Furthermore, LC3II protein was overexpressed in cells treated with the miR-26b inhibitor (P<0.01; Fig. 2A). MDC staining demonstrated that exposure to IR increased the rate of autophagy by 21.88% in the control cells, by 16.80% in the miR-26b mimic-treated cells and by 53.58% in cells treated with the miR-26b inhibitor, suggesting that miR-26b can inhibit IR-induced autophagy in MCF7 cells (P<0.001; Fig. 2B).
Figure 2.
miR-26b regulated autophagy in MCF7 cells exposed to IR. MCF7 cells that were transfected with NC, miR-26b mimic or the miR-26 inhibitor were subsequently exposed to IR. (A) miR-26b promoted the LC3II protein expression induced by IR. (B) MCF7 cells were exposed to 4 Gy of IR. Staining with monodansylcadaverine was detected by flow cytometry at 24 h. miR, microRNA; IR, ionizing radiation; IR, ionizing radiation; NC, negative control; LC3II, light chain 3-II.
DRAM1 expression in MCF7 cells is suppressed by the overexpression of miR-26b
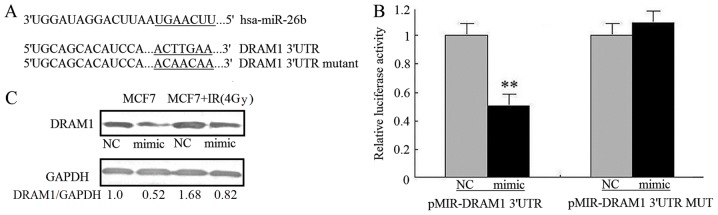
The mechanism by which miR-26b inhibits autophagy was investigated by using TargetScanto search for autophagy-associated potential target genes. The DRAM1 3′UTR contained a nucleotide sequence that matched the seed sequence of miR-26b (Fig. 3A). To obtain experimental evidence that miR-26b targets DRAM1, the 3′-UTR of DRAM1, including the potential miR-26b-binding sequence, was cloned into a firefly luciferase reporter vector; an miR-26b mimic was used to examine how miR-26b affected luciferase activity. Noluciferase activity inhibition was observed in MCF7 cells transfected with mutant 3′UTR vectors, where as the miR-26b mimic significantly inhibited the luciferase activity of cells transfected with WT DRAM1 3′UTR (P<0.01). This result demonstrated the specificity of miR-26b for targeting the DRAM1 3′UTR (Fig. 3B). It was also investigated how miR-26b affected the levels of endogenous DRAM1 protein in MCF7 cells. Transfection with the miR-26b mimic was associated with a reduction in the amount of DRAM1 protein in MCF7 cells compared with the NC. MCF7 cells exposed to IR exhibited upregulated DRAM1 protein expression; miR-26b overexpression attenuated the stimulatory effect of IR on DRAM1 expression (P<0.01; Fig. 3C). This result suggests that miR-26b may inhibit IR-induced autophagy in MCF7 cells by directly targeting DRAM1.
Figure 3.
DRAM1 expression in MCF7 cells was downregulated by miR-26B. (A) Alignment of miR-26b and DRAM1 mRNA 3′UTR sequences. The seed match region is indicated by a solid line. (B) MCF7 cells co-transfected with miR-26b mimic were analyzed for luciferase activity with wild type and mutant DRAM1 3′UTRs. **P<0.01. (C) Western blot images of the expression of DRAM1 in MCF7 cells transfected with miR-26 mimic or exposed to IR. DRAM1, DNA damage-regulated autophagy modulator 1; miR, microRNA; 3′UTR, 3′-untranslated region; IR, ionizing radiation; NC, negative control.
Loss of DRAM1 inhibited IR-induced autophagy
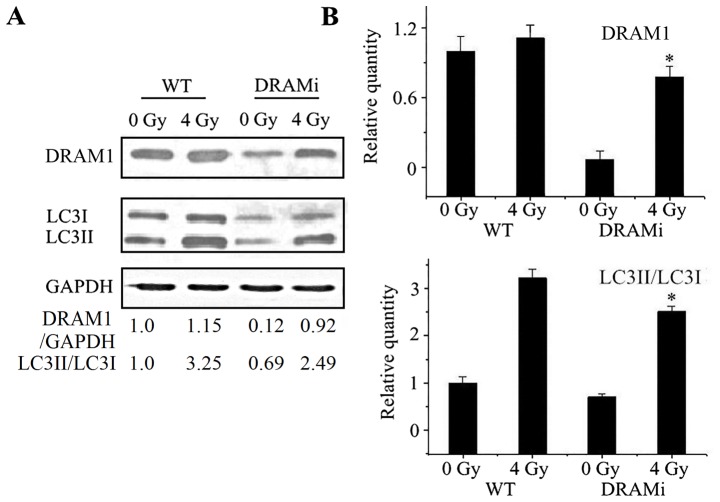
Autophagy was then analyzed in MCF7 cells that were transfected with siRNA against DRAM1. Compared with negative controls, the LC3-II to LC3-I ratio in cells transfected with siRNA against DRAM1 increased following irradiation, whereasthe ratio in MCF7 cells transfected with DRAM1 remained unchanged. Following a 4-Gy IR exposure, the LC3-II to LC3-I ratio in cells transfected with DRAM1 was significantly lower than the control cells (P<0.05; Fig. 4A and B), suggesting that DRAM1 may inhibit IR-induced autophagy.
Figure 4.
Knockdown of DRAM1 suppressed autophagy in MCF7 cells exposed to IR. (A) Knockdown of DRAM1 in MCF7 cells was confirmed by western blotting. Cells treated with small interfering RNA against DRAM1 exhibited markedly suppressed autophagy following exposure to IR. (B) Quantitated western blot analysis of DRAM1 expression and the LC3II/I ratio.*P<0.05 vs. 0 Gy. DRAM1, DNA damage-regulated autophagy modulator 1; IR, ionizing radiation; LC3, light chain 3-II; WT, wild-type; DRAMi, cells with DRAM1 inhibition.
Discussion
miRNAs are differentially expressed in normal tissue compared with cancer tissue; there may be a critical role for dysregulated miRNA expression in tumorigenesis (17–20). However, the mechanisms by which miRNAs regulate autophagy are only starting to be explored. A number of miRNAs have been demonstrated to modulate autophagy in human cancer cells by targeting autophagy-associated genes (21–23). These studies have enriched the understanding of the autophagy signaling process and provided novel therapeutic perspectives. A range of previous studies have investigated the role that microRNAs serve in the cellular response to radiation therapy by examining the process of autophagy (16,24–29).
The downregulation of miR-26b expression in breast cancer tissue compared with noncancerous tissue was previously identified, as well as in breast cancer cells compared with normal breast cells (10). In the present study, upregulated miR-26b expression levels were associated with a reduction in the levels of IR-induced autophagy in MCF7 cells. Additionally, DRAM1 mRNA was identifiedas a potential target for miR-26b. Previous studies have demonstrated that the DRAM1 gene codes for a number of splice variant proteins that are induced by p53, and that these proteins localize to autophagosomes and peroxisomes, where they may be required for the process of p53-induced autophagy (30,31). The luciferase and western blot assays of the present study identified DRAM1 as a target of miR-26b. MCF7 cells transfected with an miR-26b mimic exhibited the reduced expression of DRAM1, which may be due to the 3′UTR of the DRAM1 mRNA being targeted. Additionally, transfection with the miR-26b mimic suppressed the expression of DRAM1 protein in MCF7 cells, regardless of whether they had been exposed to IR. When taken together, the data suggest that miR-26b, at least partially, inhibits IR-induced autophagy in MCF7 cancer cells by inhibiting DRAM1. Overall, the results of the present study may improve the understanding of the various mechanisms by which miRNAs influence cells
It was also demonstrated that IR downregulated miR-26b expression in MCF7 cells. MCF7 cells transfected with an miR-26b inhibitor exhibited reduced miR-26b expression prior to IR and enhanced miR-21b expression following IR; however, the underlying mechanism for this response remains unclear. Previous studies have demonstrated that miRNA modulation may allow an improvement of the clinical effect of radio- or chemotherapy (32–34). However, the mechanisms by which miRNAs produce their effects are not fully understood. Additional studies are required to identify the miRNAs that modulate the effects of chemo- or radiotherapy, as well as their effect on cellular metabolic processes. It will also be necessary to confirm that patients can be safely and effectively treated by a clinical regimen that targets miRNAs.
In summary, it was demonstrated that miR-26b may regulate the basal level of autophagy in MCF7 cells, as well as autophagy resulting from IR exposure. DRAM1 was identified as a target gene through which miR-26b may modulate autophagy. The present study may provide evidence of a specific role for miR-26b in carcinogenesis and cancer therapy effectiveness. This result may enhancethe understanding of the miRNA-modulated networks associated with autophagy, ultimately facilitating improvement of the methods for treating cancer.
Acknowledgements
The present study was supported by NSFC grants (grant nos. 31500682 and 81570897).
References
- 1.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 2.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 3.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et al. Micro-RNAs profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol. 2012;33:1933–1942. doi: 10.1007/s13277-012-0454-8. [DOI] [PubMed] [Google Scholar]
- 5.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozubek J, Ma Z, Fleming E, Duggan T, Wu R, Shin DG, Dadras SS. In-depth characterization of microRNA transcriptome in melanoma. PLoS One. 2013;8:e72699. doi: 10.1371/journal.pone.0072699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang LP, Zhu ZT, He CY. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future Oncol. 2016;12:1105–1115. doi: 10.2217/fon.16.21. [DOI] [PubMed] [Google Scholar]
- 8.Luo M, Shen D, Wang W, Xian J. Aberrant expression of microRNA-26b and its prognostic potential in human cervical cancer. Int J Clin Exp Pathol. 2015;8:5542–5548. [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, Guo T, Dong Q, Zhang J, Li Y. miR-26b is downregulated in human tongue squamous cell carcinoma and regulates cell proliferation and metastasis through a COX-2-dependent mechanism. Oncol Rep. 2015;33:974–980. doi: 10.3892/or.2014.3648. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. Correction: MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013;13:17. doi: 10.1186/1475-2867-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX, Cao DX, He M, Chen GQ, He JR, Zhao Q. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 2011;585:1363–1367. doi: 10.1016/j.febslet.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo H, Tang X, Zeng Z, Liu M. MiRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. Int J Biochem Cell Biol. 2012;44:2051–2059. doi: 10.1016/j.biocel.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 14.Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy. 2007;3:72–74. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 15.Criollo A, Dessen P, Kroemer G. DRAM: A phylogenetically ancient regulator of autophagy. Cell Cycle. 2009;8:2319–2320. doi: 10.4161/cc.8.15.9153. [DOI] [PubMed] [Google Scholar]
- 16.Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, Ma S, Liu X. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett. 2013;587:436–443. doi: 10.1016/j.febslet.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Rutnam ZJ, Yang BB. The involvement of microRNAs in malignant transformation. Histol Histopathol. 2012;27:1263–1270. doi: 10.14670/HH-27.1263. [DOI] [PubMed] [Google Scholar]
- 18.Banno K, Yanokura M, Kisu I, Yamagami W, Susumu N, Aoki D. MicroRNAs in endometrial cancer. Int J Clin Oncol. 2013;18:186–192. doi: 10.1007/s10147-013-0526-9. [DOI] [PubMed] [Google Scholar]
- 19.Mayne GC, Hussey DJ, Watson DI. MicroRNAs and esophageal cancer-implications for pathogenesis and therapy. Curr Pharm Des. 2013;19:1211–1226. doi: 10.2174/138161213804805702. [DOI] [PubMed] [Google Scholar]
- 20.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33:2018–2025. doi: 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 22.Wang P, Zhang L, Chen Z, Meng Z. MicroRNA targets autophagy in pancreatic cancer cells during cancer therapy. Autophagy. 2013;9:2171–2172. doi: 10.4161/auto.26463. [DOI] [PubMed] [Google Scholar]
- 23.Fu LL, Wen X, Bao JK, Liu B. MicroRNA-modulated autophagicsignaling networks in cancer. Int J Biochem Cell Biol. 2012;44:733–736. doi: 10.1016/j.biocel.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, Pirtoli L, Miracco C. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickey JS, Zemp FJ, Martin OA, Kovalchuk O. The role of miRNA in the direct and indirect effects of ionizing radiation. Radiat Environ Biophys. 2011;50:491–499. doi: 10.1007/s00411-011-0386-5. [DOI] [PubMed] [Google Scholar]
- 26.Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ, Kim JH, Yin J, Yoo H, Lee SH, Park JB. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7:e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Devel. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qased AB, Yi H, Liang N, Ma S, Qiao S, Liu X. MicroRNA-18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells. Mol Med Rep. 2013;7:559–564. doi: 10.3892/mmr.2012.1214. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, Zhuang L, Luo J, Chen H, Liu L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143.e1112. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 30.Lorin S, Pierron G, Ryan KM, Codogno P, Djavaheri-Mergny M. Evidence for the interplay between JNK and p53-DRAM signalling pathways in the regulation of autophagy. Autophagy. 2010;6:153–154. doi: 10.4161/auto.6.1.10537. [DOI] [PubMed] [Google Scholar]
- 31.Yen ML, Jim OP, Baudot AD, Attje H, Ryan KM. DRAM-1 encodes multiple isoforms that regulate autophagy. Autophagy. 2012;8:18–28. doi: 10.4161/auto.8.1.18077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Q, Li XJ, Cao PG. MicroRNA-26b enhances the radiosensitivity of hepatocellular carcinoma cells by targeting EphA2. Tohoku J Exp Med. 2016;238:143–151. doi: 10.1620/tjem.238.143. [DOI] [PubMed] [Google Scholar]
- 33.Shi L, Yin W, Zhang Z, Shi G. Down-regulation of miR-26b induces cisplatin resistance in nasopharyngeal carcinoma by repressing JAG1. FEBS Open Bio. 2016;6:1211–1219. doi: 10.1002/2211-5463.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arora H, Qureshi R, Park AK, Park WY. Coordinated regulation of ATF2 by miR-26b in γ-irradiated lung cancer cells. PLoS One. 2011;6:e23802. doi: 10.1371/journal.pone.0023802. [DOI] [PMC free article] [PubMed] [Google Scholar]