Abstract
Prostate cancer (PC) is the most prevalent urological cancer in men. T cells serve a central role in the cancer's immunological microenvironment. In the present study, we applied multiplex PCR and Illumina next-generation sequencing to study the clonal diversity of the T-cell receptor (TCR) repertoire in cancer tissues and paracancer tissues from patients with PC. It was found that the TCR repertoire in the PC samples had a notably more skewed clonotype composition, with a greater number of highly expanded clones (HECs) compared with the prostate paracancer samples. The amino acid sequences ATSRVAGETQY (1.008 vs. 0.002%), ATSRTGRWETQY (3.985 vs. 0.007%), ATSDSSDYEQY (12.464 vs. 0.027%), ATSDFRGQPQETQY (2.205 vs. 0.06%), ASSQQDEAF (1.109 vs. 0.002%) and ARPTRTEETQY (1.263 vs. 0.002%) were found to vary markedly between cancer and paracancer tissues, respectively. In conclusion, the present study identified PC-specific HECs, which are critical to improving understanding of the TCR repertoire in PC. This may accelerate the screening process for potential new autoantigens and provide information for generating more effective T cell-targeted diagnostic and therapeutic strategies.
Keywords: prostate cancer, T-cell receptor, next-generation sequencing, immune repertoire
Introduction
Prostate cancer (PC) is the most prevalent urological cancer and has become the leading cause of cancer-related mortality among males in many countries. Patients with PC are often treated via surgery, radiotherapy and androgen deprivation therapy (1). Manipulation of the immune system represents a promising approach to controlling advanced and metastatic PC; examples of immunotherapeutic and immunomodulatory agents include sipuleucel-T, ipilimumab, tasquinimod, Prostvac®-VF, and GVAX (2,3). However, despite these clinical advances, the underlying mechanisms are complex and remain elusive, and therapies that provide long-term survival are still required. Therefore, further studies are necessary to increase understanding of the characteristics of the immunological microenvironment. The identities, traits, functional status, distribution, kinetics, and interactions of inflammatory cells in the prostate and in PC must be fully characterized in order to improve and combine the current promising immunotherapies (4).
T cells serve a central role in the immunological microenvironment. The T-cell receptor (TCR), a molecule found on the surface of T cells, is responsible for recognizing fragments of antigens in the form of peptides bound to major histocompatibility complex (MHC) molecules (5). The human adaptive immune system harbors a vast array of TCRs that target a wide variety of pathogens, collectively referred to as the TCR repertoire. Generally, each T cell encodes a single unique TCR (5,6). TCRs consist of two chains, most often an α and a β chain, and each chain comprises a variable region (V region), consistent region (C region), transmembrane domain and cytoplasmic domain. The V region of the α and β chains contains three hypervariable complementarity-determining regions (CDRs), namely CDR1, CDR2 and CDR3, of which CDR3 is the most variable and directly determines the antigen-binding specificity of the TCR. The structural diversity of TCRs and their chains is generated by somatic random recombination of V, diversity (D) and joining (J) gene segments, including the contribution of non-germline, non-templated (N) nucleotides, which are added in a manner independent of the template at the V-(D)-J joints (7).
The number of distinct TCRs has been estimated to be ~2.5×107 in a normal adult, making the repertoire particularly difficult to analyze. Profiling the TCR repertoire by next-generation sequencing (NGS) is a relatively new method of analyzing the adaptive immune system response by elucidating the relative abundance and diversity of T-cell clones, which makes possible the study of TCR repertoire diversity and selection mechanisms at a greater depth than in the past (8–10). More recent study has focused on clone tracking, repertoire properties, and the identification of public clones using TCR sequencing technology (11,12). Common TCR ‘signatures’ raised against specific antigens could provide important diagnostic biomarkers (13).
In the present study, NGS was used to characterize the complex alterations in the TCR β-chain repertoire between paired sequential samples from cancer tissues and paracancer tissues. The CDRs of TCRs were subjected to amplification by multiplex-PCR followed by NGS to explore the different TCR repertoires between the cancer and paracancer tissues, with millions of TCR reads obtained for each sample. Such large-scale sequencing of the TCR repertoire in PC may reform our perception of the immune system. Furthermore, a deeper understanding of the TCR repertoire in PC may improve our knowledge of the characteristics of the immunological microenvironment, which may potentially lead to the development of more effective targeted therapies in order to better control the disease course and minimize treatment-related adverse events.
Materials and methods
Clinical samples
Cancer and paracancer tissues from five patients with PC were collected at the Second Clinical Medical College of Jinan University (Shenzhen People's Hospital, Guangdong, China). The patients enrolled in the group were required to have a confirmed pathological diagnosis of PC without metastasis. The patient cohort had a mean Gleason score of 7.40±1.34 (range, 6–9), a mean age of 68.80±9.23 years (range, 58–78 years) (Table I). All patients provided written informed consent.
Table I.
Clinical information for the samples.
| Sample | Age (years) | Gleason score | IHC | Diagnosis |
|---|---|---|---|---|
| 1 | 58 | 6 | P504S(+), P63(−), 34βE12(−) | Prostate cancer, no metastasis |
| 2 | 72 | 9 | – | Prostate cancer, no metastasis |
| 3 | 60 | 6 | P504S(+), P63(−), 34βE12(−) | Prostate cancer, no metastasis |
| 4 | 78 | 8 | CK(−), P504S(−), 3βE12(−) | Prostate cancer, no metastasis |
| 5 | 76 | 8 | AR(+), CK(+), CK20(−), 34βE12(−), P504S(+) P63(−), CD56(−), CDX2(−), CgA(−), Syn(−) | Prostate cancer, no metastasis |
DNA extraction and mixing
For extraction of total genomic DNA, 5–10 cancer and paracancer tissue sections were obtained from each patient, and DNA was extracted using standard methods. Briefly, dewaxing was performed with xylene, followed by overnight proteinase K digestion. A QIAamp DNA Mini kit (Qiagen, Hilden, Germany) was further used for DNA extraction following the manufacturer's instructions. DNA quality was evaluated by electrophoresis on 0.8% agarose gels, and the quality of the template DNA was assessed by Qubit™ dsDNA High Sensitivity Assay. DNA samples from the cancer tissues of five patients were mixed together in a 1:1:1:1:1 ratio according to DNA concentration (Qubit value), and this mixture was relabeled as one PC sample. Similarly, DNA from the paracancer tissues of the five patients was mixed together in a 1:1:1:1:1 ratio according to DNA concentration, and the mixture was relabeled as one prostate paracancer sample.
Multiplex-PCR amplification of TCR-β CDR3 regions
The human TCR-β sequences were downloaded from IMGT (http://www.imgt.org/) (14). A relative conserved region in frame region 3, upstream of CDR3, was selected for the putative forward primer region. A cluster of primers corresponding to the majority of the V gene family sequence was selected. Similarly, reverse primers corresponding to the J gene family were designed (14,15). In total, 30 forward primers and 13 reverse primers were used for multiplex PCR to amplify the rearranged TCR-β CDR3 regions. The reaction mixtures (50 µl total) comprised 2 µl pooled TCR-β variable gene (TRBV; 10 µM), 2 µl pooled TCR-β joining gene (TRBJ; 10 µM), 25 µl 2X Qiagen Multiplex PCR Master Mix, 5 µl 5X Q-solution, 500 ng template DNA (10 µl) and 6 µl H2O. The PCR conditions comprised 95°C for 15 min; followed by 25 cycles of 94°C for 15 sec and 60°C for 3 min; followed by a final extension for 10 min at 72°C. The PCR products were purified by AMPure XP beads to remove primer sequences (Beckman Coulter, Inc., Brea, CA, USA). A second round of PCR was performed to add a sequencing index to each sample. In this round, each reaction mixture (50 µl total) consisted of 13.5 µl H2O, 0.5 µl 2X Q5 DNA polymerase, 10 µl 5X Q5 buffer, 1 µl dNTPs (10 mM), 1 µl P1 (10 µM), 23 µl DNA, and 1 µl index (10 µM). The PCR conditions comprised 98°C for 1 min; followed by 25 cycles of 98°C for 20 sec, 65°C for 30 sec and 72°C for 30 sec; and a final extension for 5 min at 72°C. The library was separated on an agarose gel, and the target region was isolated and cleaned by QIAquick Gel Extraction kits (Qiagen).
NGS and data analysis
The library was quantitated using the Agilent 2100 Bioanalyzer instrument (Agilent DNA 1000 reagents) and real-time quantitative PCR (TaqMan probes), and sequenced by Illumina MiSeq. Briefly, the adaptor reads and low quality reads were filtered from the raw data, and the clean data was used in further alignments. Subsequently, the clean data was aligned to the human IGH database and analyzed by the online IMGT/HighV-QUEST tool. The data included V, D and J assignment, CDR3 length distribution, clustering and other analyses (16).
Results
Summary of sequencing
A total of five patients with PC were recruited for the present study, and 5–10 cancer and paracancer pathological sections were obtained from each patient for DNA extraction. The PC and paracancer DNA samples were created by pooling DNA from the PC and paracancer tissues, respectively, of the five patients. We used a combination of multiplex-PCR, Illumina MiSeq sequencing and IMGT (ImMunoGeneTics)/HighV-QUEST to investigate TCR repertoire features in PC and prostate paracancer samples at sequence-level resolution. In total, 641,209 reads were obtained for the pooled cancer sample and 377,706 reads were obtained for the pooled paracancer sample. After filtering, including the removal of contamination as well as adaptor sequences and low-quality reads, the data were aligned to the human Texas Cancer Research Biobank database. The numbers of mapping immune sequence reads were 627,234 and 372,660 for the cancer and paracancer samples, respectively. Among them, the productive sequence reads totaled 446,108 and 301,203, and the in-frame sequence reads totaled 457,951 and 312,270 for the cancer and paracancer samples, respectively. The CDR3 sequences were identified by their conserved motif. The abundance of each CDR3 clone and the number of distinct CDR3 clone species were calculated: There were 435,110 total CDR3 sequences for the cancer sample and 295,793 for the paracancer sample, with unique CDR3 amino acid (aa) sequences totaling 10,433 and 19,948, respectively (Table II).
Table II.
TCRB sequence statistics.
| Data type | Cancer sample | Paracancer sample |
|---|---|---|
| Total reads number | 641,209 | 377,706 |
| Immune sequences number | 627,234 | 372,660 |
| Unknown sequences number | 13,975 | 5,046 |
| Productive sequences number | 446,108 | 301,203 |
| Non-productive sequences number | 181,126 | 71,457 |
| In-frame sequences number | 457,951 | 312,270 |
| Out-of-frame sequences number | 164,464 | 58,365 |
| Total CDR3 sequences number | 435,110 | 295,793 |
| Unique CDR3 nt sequences number | 13,214 | 23,093 |
| Unique CDR3 aa sequences number | 10,433 | 19,948 |
Highly expanded clones (HECs) and TCR repertoire diversity
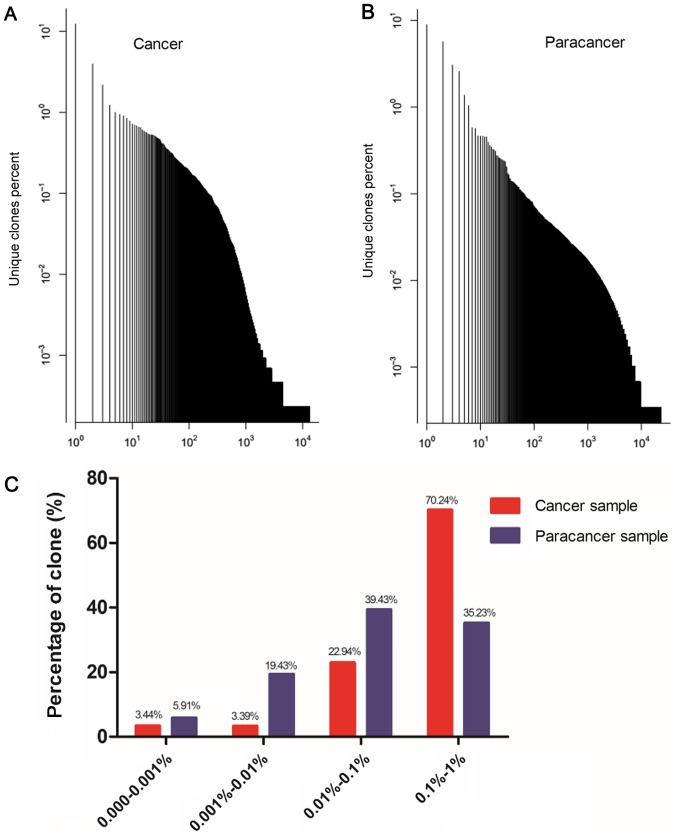
In order to obtain an accurate and comprehensive description of TCR repertoire diversity in the PC and paracancer samples, the expression level of each clone was calculated according to the identity of each sequence after alignment, and the degree of expansion of each clone was based on the frequency of each unique CDR3 sequence. TCR clones with a frequency >0.5% of the total reads in a sample were defined as HECs. In the cancer sample, 26 clones were HECs, with a HEC ratio of 0.34. Comparatively, in the paracancer sample, only 8 clones were defined as HECs, with a HEC ratio of 0.24 (Table III). A plot of the frequency distribution of the TCR repertoire in cancer and paracancer samples is shown in Fig. 1A and B. A majority of the TCR clones were present at low frequencies, with fewer sequences occurring at much higher frequencies in the cancer and paracancer samples. The percentages of clones with different degrees of clone expression (based on the number of reads for that clone out of the total number of reads in the sample) were further calculated. In the cancer sample, clones with a degree of expression of 0.1–1% accounted for 70.24% of the T cell sequences present, while clones with a degree of expression of 0.01–0.1% accounted for 22.94%. In the paracancer sample, clones with an expression degree of 0.01–0.1% accounted for the highest percentage (39.43%), while clones with an expression degree of 0.1–1% accounted for 35.23% (Fig. 1C). Therefore, the TCR repertoire in the PC sample had more HECs (expression degree >0.5%), a higher HEC ratio, and greater percentage of higher degree (>0.1%) clones compared with the prostate paracancer sample.
Table III.
HEC and diversity in cancer and paracancer sample.
| Data type | Cancer sample | Paracancer sample |
|---|---|---|
| HEC number | 26 | 8 |
| HEC ratio | 0.34 | 0.24 |
| Shannon entropy | 0.46 | 0.59 |
HEC, highly expanded clone.
Figure 1.
Clone frequency distribution in the Texas Cancer Research Biobank sequences of prostate cancer sample and paracancer sample. (A) Plot of the unique clones' percent in cancer sample; (B) plot of the unique clones' percent in paracancer sample; (C) percentage of unique clones in different degree of expansion in cancer sample and paracancer sample.
In addition, we applied a normalized Shannon entropy index to quantify the TCR diversity of the cancer and paracancer samples. The normalized Shannon entropy index ranges from 0 to 1, in which 1 indicates the highest degree of diversity and 0 indicates no diversity. The normalized entropy of the cancer sample was found to be less than that of the paracancer sample (0.46 vs. 0.59). These results suggest that the TCR repertoire of cancer tissue has a much more skewed clonotype composition than that of paracancer tissue.
Shared TCR clones in cancer and paracancer samples
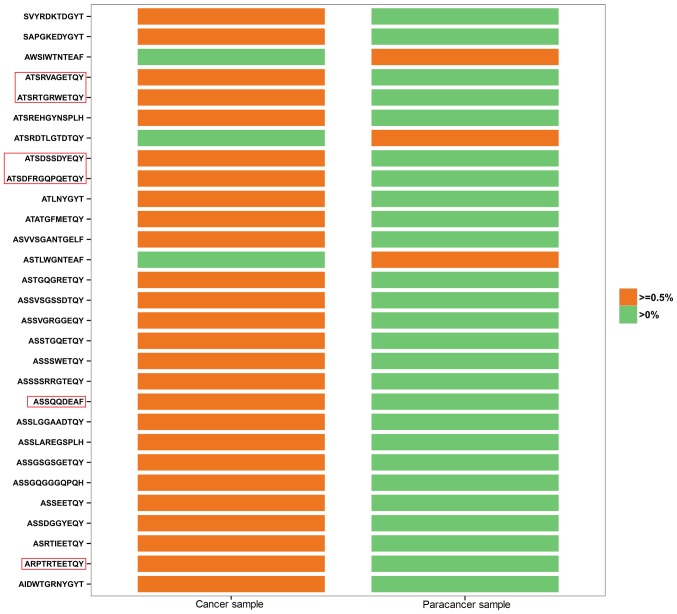
The shared (or public) T cells among healthy individuals and those with disease are of great interest to researchers. We further investigated the clones common to both the PC and the paracancer samples, in which HEC clones were analyzed in depth. All 26 HECs in the cancer sample had low expression (<0.5%) in the paracancer sample (Fig. 2). By contrast, 3 of the 8 HECs in the paracancer sample were expressed at a low level (<0.5%) in the cancer sample, whereas the other 5 HECs were not expressed at all (Fig. 2). In Fig. 2, HEC aa sequences accounting for >1% of the cancer sample are labeled with a red frame. ATSRVAGETQY (1.008 vs. 0.002%), ATSRTGRWETQY (3.985 vs. 0.007%), ATSDSSDYEQY (12.464 vs. 0.027%), ATSDFRGQPQETQY (2.205 vs. 0.06%), ASSQQDEAF (1.109 vs. 0.002%), and ARPTRTEETQY (1.263 vs. 0.002%) were found to vary markedly between the two samples in terms of their degree of expression. Identification of prostate-specific HECs may accelerate the screening process for possible new autoantigens, which is critical to increase understanding of the TCR repertoire in PC tissues.
Figure 2.
Shared HECs in prostate cancer and paracancer sample. Orange represents HEC (>0.5% of total reads), green represents sequences with low abundance (<0.5%). HEC sequences (aa) ~1% in cancer sample were labeled with red frame. HECs, highly expanded clones.
Comparison of TRBV, TRBD and TRBJ repertoires
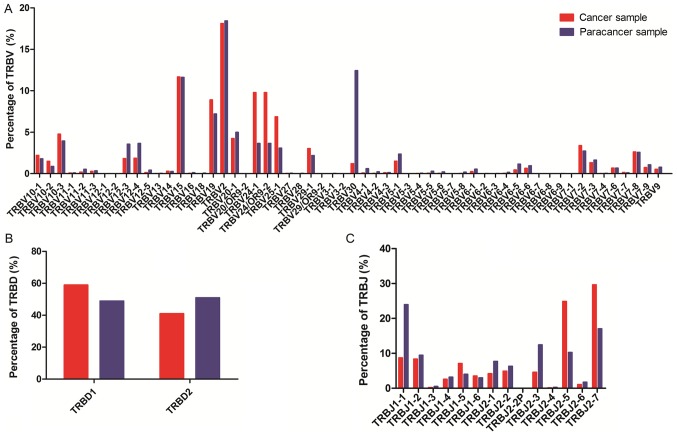
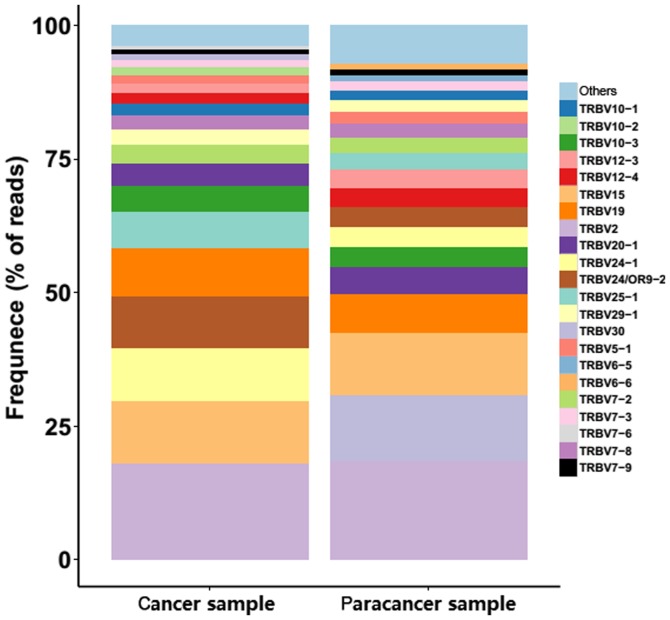
To determine whether any disease-specific differences exist in the TRBV, TRBD and TRBJ repertoires, the expression levels of the respective TRBV, TRBD and TRBJ repertoires in cancer and paracancer samples were compared. In total, 58 TRBV segments, 2 TRBD segments and 14 TRBJ segments were included. For the 58 TRBV segments, the gene expression level of TRBV2 was highest in both the cancer and paracancer samples. The usage ratios of TRBV2 (18.12 vs. 18.43%), TRBV15 (11.68 vs. 11.63%), and TRBV7-8 (2.64 vs. 2.58%) in the TCRs were similar (±10%) between the cancer and paracancer samples (Fig. 3). For the other distinct TRBV sequences, the usage ratios of each TRBV gene exhibited extreme variation. The frequencies of the top 20 V gene family in the PC and paracancer samples are shown in Fig. 4. The majority of the top 20 V gene family in the cancer sample could also be found in the paracancer sample, with the exception of TRBV10-2 (1.5%) and TRBV7-6 (0.68%), whereas TRBV6-5 (1.16%) and TRBV6-6 (0.98%) were specific to the paracancer sample (Fig. 4). As for the 2 TRBD segments, TRBD1 seemed to be more frequent in the cancer sample compared with the paracancer sample. Furthermore, the usage ratio of each TRBJ gene exhibited extreme variation in the cancer and paracancer samples. The usage ratios of this set of TRBJ genes ranged from 0.13% (for TRBJ2-4) to 29.65% (for TRBJ2-7) in the cancer sample, and from 0.27% (for TRBJ2-4) to 23.95% (for TRBJ1-1) in the paracancer sample. These data suggest that there may be clone expansion in specific V and J genes in the PC sample.
Figure 3.
Relative usage percentages (%) of each TRBV, TRBD, TRBJ gene from prostate cancer sample (red) and paracancer sample (blue). (A) Usage percentage of TRBV gene; (B) usage percentage of TRBD gene; (C) usage percentage of TRBJ gene. TRBV, T cell receptor-β variable gene; TRBJ, T cell receptor-β joining gene.
Figure 4.
V gene usage analysis in prostate cancer and paracancer sample. Stacked bar chart for the frequency of 20 top V gene family usages in prostate cancer and paracancer sample. The V gene families were shown in color bar. TRBV, T cell receptor-β variable gene.
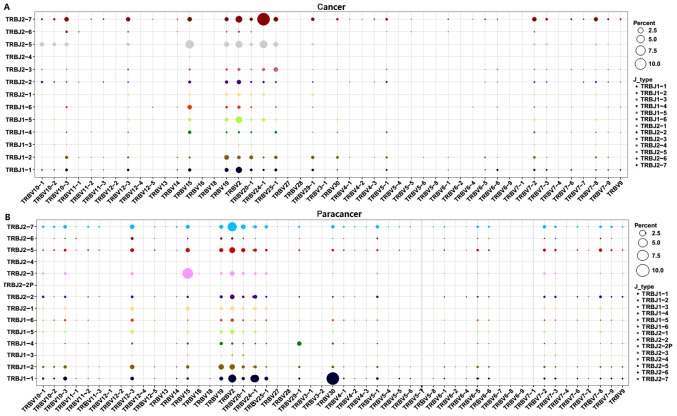
Subsequently, V-J combinations frequencies for each sample were obtained (Fig. 5). In total, 9 V-J gene pairings [TRBV11-1-TRBJ1-3 (0.02%), TRBV13-TRBJ1-3 (0.02%), TRBV13-TRBJ1-6 (0.07%), TRBV6-9-TRBJ2-3 (0.07%), TRBV6-9-TRBJ2-6 (0.02%), TRBV7-4-TRBJ2-4 (0.02%), TRBV7-7-TRBJ1-3 (0.02%), TRBV7-7-TRBJ1-4 (0.02%), and TRBV7-7-TRBJ2-6 (0.05%)] were present specifically in the cancer sample at a low frequency (Fig. 5). The other high expression V-J gene pairings (expression degree >1%) are listed in Table IV. The TRBV15-TRBJ2-5, TRBV2-TRBJ2-5, TRBV2-TRBJ2-7, TRBV2-TRBJ1-1, TRBV19-TRBJ1-2, TRBV2-TRBJ2-2, and TRBV12-3-TRBJ2-7 V-J gene pairings (>1%) were overlapping in the PC and paracancer samples. On the contrary, other V-J gene pairings exhibited marked variation between the PC and paracancer samples. For example, the TRBV24-1-TRBJ2-7 pairing was highest in the cancer sample (12.43%), with low expression in the paracancer sample (<1%); TRBV30-TRBJ1-1 was highest in the paracancer sample (10.05%), with low expression in the cancer sample (<1%). These data indicated that particular V-J gene pairings were dominant and specific to the PC tissues.
Figure 5.
V-J combinations percentage for each sample. TRBV segments were arranged on the x-axis, and TRBJ segments were arranged and colored differently on the y-axis. And the different percentages of V-J combinations for each sample were showed by balloon size. The results of the (A) cancer and (B) paracancer samples are shown. TRBV, T cell receptor-β variable gene; TRBJ, T cell receptor-β joining gene.
Table IV.
V-J paring usage (>1%) in prostate cancer and paracancer samples.
| V-J pairing in cancer sample | Usage in cancer sample (%) | V-J pairing in paracancer sample | Usage in paracancer sample (%) | ||
|---|---|---|---|---|---|
| TRBV24-1 | TRBJ2-7 | 12.43 | TRBV30 | TRBJ1-1 | 10.05 |
| TRBV15 | TRBJ2-5 | 5.48 | TRBV15 | TRBJ2-3 | 7.11 |
| TRBV24-1 | TRBJ2-5 | 4.66 | TRBV2 | TRBJ2-7 | 5.22 |
| TRBV2 | TRBJ2-5 | 3.79 | TRBV2 | TRBJ1-1 | 3.95 |
| TRBV2 | TRBJ2-7 | 3.45 | TRBV24-1 | TRBJ1-1 | 3.59 |
| TRBV2 | TRBJ1-5 | 3.33 | TRBV2 | TRBJ2-5 | 2.04 |
| TRBV2 | TRBJ1-1 | 2.86 | TRBV19 | TRBJ1-2 | 1.73 |
| TRBV19 | TRBJ2-5 | 1.90 | TRBV2 | TRBJ1-2 | 1.71 |
| TRBV25-1 | TRBJ2-5 | 1.65 | TRBV20-1 | TRBJ2-7 | 1.27 |
| TRBV19 | TRBJ1-2 | 1.61 | TRBV2 | TRBJ2-3 | 1.22 |
| TRBV25-1 | TRBJ2-7 | 1.52 | TRBV2 | TRBJ2-2 | 1.21 |
| TRBV25-1 | TRBJ2-3 | 1.50 | TRBV12-3 | TRBJ2-7 | 1.20 |
| TRBV10-1 | TRBJ2-5 | 1.45 | TRBV12-3 | TRBJ1-2 | 1.16 |
| TRBV15 | TRBJ2-7 | 1.42 | TRBV29-1 | TRBJ1-4 | 1.13 |
| TRBV10-3 | TRBJ2-7 | 1.42 | TRBV15 | TRBJ2-5 | 1.01 |
| TRBV15 | TRBJ1-6 | 1.39 | TRBV2 | TRBJ2-1 | 1.00 |
| TRBV7-2 | TRBJ2-7 | 1.39 | |||
| TRBV2 | TRBJ2-2 | 1.35 | |||
| TRBV12-3 | TRBJ2-7 | 1.29 | |||
| TRBV7-8 | TRBJ2-7 | 1.17 | |||
| TRBV10-3 | TRBJ2-5 | 1.15 | |||
| TRBV19 | TRBJ2-7 | 1.15 | |||
| TRBV19 | TRBJ1-1 | 1.07 | |||
| TRBV20-1 | TRBJ1-2 | 1.02 | |||
Discussion
The immunological treatment of PC has been proposed due to the association between the immunological microenvironment of the primary tumor and the patient outcome (4). Arap et al found that the intratumoral absence or presence of T cells and regulatory T cells (Tregs) is functionally relevant and may have an impact on the clinical course of PC (17). T cells serve an important role in adaptive immunity, recognizing MHC-bound peptides via TCRs on the T-cell surface. The CDR3 region of TCRs is responsible for the unique molecular structure that allows the diversity of a T-cell population. In the present study, we used a novel NGS protocol to investigate the TCR repertoire in PC and prostate paracancer samples at the sequence level. In total, 641,209 reads for the cancer sample and 377,706 reads for the paracancer sample were obtained, providing extensive information on the TCR repertoire in PC.
Among the 435,110 total CDR3 sequences acquired for the cancer sample and 295,793 for the paracancer sample, TCR clones with a frequency >0.5% of the total number of reads in a sample were defined as HECs. The TCR repertoire in the PC sample had more HECs (>0.5%), a higher HEC ratio, and a greater percentage of higher degree (>0.1%) clones than the prostate paracancer sample. Furthermore, Shannon entropy was utilized to investigate the immunological diversity in patients with PC; this method was introduced by Claude Shannon as part of information theory, and is the mostly widely used method as it integrates the number of different species as well as the relative proportion of each species (18). Our results suggested that the entire TCR repertoire of the pooled cancer tissue sample had a markedly more skewed clonotype composition than that of the paracancer sample. Numerous previous studies have assessed the association between specific immune cells and PC (3,4), most of which investigated CD3+, CD4+ or CD8+ cells, revealing pro-tumorigenic activities in the prostatectomy or biopsy tissues of patients with PC (4). Research has also focused on the presence of immunosuppressive immune cells, such as Tregs (19–21), myeloid-derived suppressor cells (22,23), and T helper 17 cells (21), in prostate tissue, which were found to be suggestive of more aggressive disease. However, notably few studies were able to equate this with clinical prognosis. In the present study, we concentrated on only the TCR repertoire in paired cancer and paracancer tissues from patients with PC. The amount of different T cell subsets was not well distinguished, and the repertoire of different T cell subsets in PC tissue requires further research.
Prostate-associated antigens, such as prostate-specific antigen, prostate-specific membrane antigen, prostate stem-cell antigen, and prostatic acid phosphatase, which are weak or non-immunogenic self-antigens (3), have been used as a screening approach for PC. In the current study, we further investigated the shared clones in PC and paracancer samples, in which HEC clones were analyzed in depth. ATSRVAGETQY (1.008 vs. 0.002%), ATSRTGRWETQY (3.985 vs. 0.007%), ATSDSSDYEQY (12.464 vs. 0.027%), ATSDFRGQPQETQY (2.205 vs. 0.06%), ASSQQDEAF (1.109 vs. 0.002%), and ARPTRTEETQY (1.263 vs. 0.002%) were found to vary markedly between the two samples. Identification of PC-specific HECs may accelerate the screening process for possible new autoantigens.
While this study have some limitations. In our present study, the peripheral blood of the same patients haven't been collected in time and the data can not be included. For further sample collection, the peripheral blood sample will be included to give us more information. An ideal tumorigenesis model evaluate normal and tumoral tissue, the normal tissues are too hard to be collected for clinical sample collection. We confirmed that the paracancer tissue samples were taken from non-malignant surrounding tissue of prostate tumor of the same patients. And the paracancer tissues were confirmed by pathological detection without cancer cells, which we thought the most close to the normal tissue. The TCR in precancerouse tissue, cancer tissue and metastatic tissue could be better explaining the effect of immunologic system on PC aggression and progression. The mice model will also included in our further study to better understand the role of TCR impact on progression to prostate cancer. This is a descriptive and preliminary study in a very small group of patient, while we used a novel NGS protocol to investigate the TCR repertoire in PC and prostate paracancer samples at the sequence level for the first time. In total, 641,209 reads for the cancer sample and 377,706 reads for the paracancer sample were obtained, providing extensive information on the TCR repertoire in PC. A more comprehensive study is needed to evaluate the clinical impact of the information, where more samples will be included.
In conclusion, the present study demonstrated a successful approach to profiling an entire TCR repertoire at sequence-level resolution, providing knowledge of the characteristics of the immunological microenvironment in PC tissues. We hope that this will lead to the generation of more effective targeted therapies, which could have benefits for controlling the disease course and minimizing treatment-related adverse events in PC.
Acknowledgements
This study was supported by grants from the Science and Technology Plan of Shenzhen (no. JCYJ20140416095712714), Technology Plan of Shenzhen, Guangdong (no. JCYJ20160422150329190), China Postdoctoral Science Foundation Grant (no. 2017M610575), Shenzhen Pingshan new district health research project (no. 201613) and Guangdong-Hong Kong Technology Cooperation Funding Scheme (TCFS) 2015–2016, under the Innovation and Technology Fund (ITF) (no. GHP/003/16GD).
References
- 1.Kgatle MM, Kalla AA, Islam MM, Sathekge M, Moorad R. Prostate cancer: Epigenetic alterations, risk factors and therapy. Prostate Cancer. 2016;2016:5653862. doi: 10.1155/2016/5653862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silvestri I, Cattarino S, Giantulli S, Nazzari C, Collalti G, Sciarra A. A perspective of immunotherapy for prostate cancer. Cancers (Basel) 2016;8:E64. doi: 10.3390/cancers8070064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thakur A, Vaishampayan U, Lum LG. Immunotherapy and immune evasion in prostate cancer. Cancers. 2013;5:569–590. doi: 10.3390/cancers5020569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strasner A, Karin M. Immune infiltration and prostate cancer. Front Oncol. 2015;5:128. doi: 10.3389/fonc.2015.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 6.Dudley DJ. The immune system in health and disease. Baillieres Clin Obstet Gynaecol. 1992;6:393–416. doi: 10.1016/S0950-3552(05)80003-3. [DOI] [PubMed] [Google Scholar]
- 7.Risitano AM, Maciejewski JP, Green S, Plasilova M, Zeng W, Young NS. In-vivo dominant immune responses in aplastic anaemia: Molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet. 2004;364:355–364. doi: 10.1016/S0140-6736(04)16724-X. [DOI] [PubMed] [Google Scholar]
- 8.Calis JJ, Rosenberg BR. Characterizing immune repertoires by high throughput sequencing: Strategies and applications. Trends Immunol. 2014;35:581–590. doi: 10.1016/j.it.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Zhang D, Yan W, Yang D, Shen B. Translational bioinformatics for diagnostic and prognostic prediction of prostate cancer in the next-generation sequencing era. Biomed Res Int. 2013;2013:901578. doi: 10.1155/2013/901578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrati S, De Summa S, Pilato B, Petriella D, Lacalamita R, Tommasi S, Pinto R. Next-generation sequencing: Advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355–7365. doi: 10.2147/OTT.S99807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robins H. Immunosequencing: Applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Hou XL, Wang L, Ding YL, Xie Q, Diao HY. Current status and recent advances of next generation sequencing techniques in immunological repertoire. Genes Immun. 2016;17:153–164. doi: 10.1038/gene.2016.9. [DOI] [PubMed] [Google Scholar]
- 13.Mirshahidi S, Ferris LC, Sadegh-Nasseri S. The magnitude of TCR engagement is a critical predictor of T cell anergy or activation. J Immunol. 2004;172:5346–5355. doi: 10.4049/jimmunol.172.9.5346. [DOI] [PubMed] [Google Scholar]
- 14.Lefranc MP. Immunoglobulin and T cell receptor genes: IMGT (®) and the birth and rise of immunoinformatics. Front Immunol. 2014;5:22. doi: 10.3389/fimmu.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou X, Lu C, Chen S, Xie Q, Cui G, Chen J, Chen Z, Wu Z, Ding Y, Ye P, et al. High throughput sequencing of T cell antigen receptors reveals a conserved TCR repertoire. Medicine (Baltimore) 2016;95:e2839. doi: 10.1097/MD.0000000000002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui W, Hou X, Zou G, Che W, Yang M, Zheng C, Liu F, Chen P, Wei X, Lai L, Dai Y. Composition and variation analysis of the TCR β-chain CDR3 repertoire in systemic lupus erythematosus using high-throughput sequencing. Mol Immunol. 2015;67:455–464. doi: 10.1016/j.molimm.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Arap W, Trepel M, Zetter BR, Pasqualini R. Meeting report: Innovations in prostate cancer research. Cancer Res. 2008;68:635–638. doi: 10.1158/0008-5472.CAN-07-3232. [DOI] [PubMed] [Google Scholar]
- 18.Magurran AE. Ecology: Linking species diversity and genetic diversity. Curr Biol. 2005;15:R597–R599. doi: 10.1016/j.cub.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 19.Ebelt K, Babaryka G, Frankenberger B, Stief CG, Eisenmenger W, Kirchner T, Schendel DJ, Noessner E. Prostate cancer lesions are surrounded by FOXP3+, PD-1+ and B7-H1+ lymphocyte clusters. Eur J Cancer. 2009;45:1664–1672. doi: 10.1016/j.ejca.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Valdman A, Jaraj SJ, Compérat E, Charlotte F, Roupret M, Pisa P, Egevad L. Distribution of Foxp3-, CD4- and CD8-positive lymphocytic cells in benign and malignant prostate tissue. APMIS. 2010;118:360–365. doi: 10.1111/j.1600-0463.2010.02604.x. [DOI] [PubMed] [Google Scholar]
- 21.Derhovanessian E, Adams V, Hähnel K, Groeger A, Pandha H, Ward S, Pawelec G. Pretreatment frequency of circulating IL-17+CD4+ T-cells, but not Tregs, correlates with clinical response to whole-cell vaccination in prostate cancer patients. Int J Cancer. 2009;125:1372–1379. doi: 10.1002/ijc.24497. [DOI] [PubMed] [Google Scholar]
- 22.Garcia AJ, Ruscetti M, Arenzana TL, Tran LM, Bianci-Frias D, Sybert E, Priceman SJ, Wu L, Nelson PS, Smale ST, Wu H. Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Mol Cell Biol. 2014;34:2017–2028. doi: 10.1128/MCB.00886-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Mitri D, Toso A, Chen JJ, Sarti M, Pinton S, Jost TR, D'Antuono R, Montani E, Garcia-Escudero R, Guccini I, et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515:134–137. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]