Abstract
The prevalence of inflammatory bowel disease (IBD), including Crohn's disease (CD) and ulcerative colitis (UC), is increasing markedly in China. The present study performed pedigree analysis of 4 families with a history of IBD and investigated the association of genetic and environmental factors with susceptibility to IBD. A total of 10 IBD patients (8 CD patients and 2 UC patients) and 90 family members were included in the present study. The clinical characteristics of familial subjects were compared with those of patients with sporadic IBD. Previously reported mutations, namely interleukin-10 receptor (IL10R)-A Thr84Ile, IL10RA Gly141Arg, IL10RB Trp159X, X-linked inhibitor of apoptosis (XIAP) Cys203Tyr, nucleotide-binding oligomerization domain-containing protein 2 (NOD2) Arg702Trp, NOD2 Gly908Arg and NOD2 Leu1007fsinsC, were screened in the patients with IBD, and selected demographic factors were compared between the patients and their unaffected family members. It was observed that single-gene and multi-gene inheritance patterns contributed to IBD in Chinese families. Based on data from the registry system, the ratio of patients with a family history of IBD was 1.25%, which was lower than that in the Western population. First-degree relatives were found to be more susceptible to IBD, and siblings were affected more frequently. Furthermore, the median age of diagnosis was younger in familial patients than in sporadic patients (29.0 vs. 36.0 for CD; 35.5 vs. 41.0 for UC). However, none of the 7 susceptibility loci were present in any of the familial patients. Immigration was a significant risk factor of IBD (odds ratio: 4.667; 95% confidence interval: 1.165–18.690; P=0.021). In conclusion, genetic heterogeneity exits between Chinese families with IBD and the Western population. The present findings suggest that genetic background and environmental factors serve a role in the pathogenesis of IBD.
Keywords: pedigree analysis, inflammatory bowel disease, Crohn's disease, ulcerative colitis, genetic susceptibility, genetic heterogeneity
Introduction
The exact cause of inflammatory bowel disease (IBD) remains unclear. Cronus's disease (CD) and ulcerative colitis (UC) are the two main types of IBD (1). Previous studies have indicated that a genetic predisposition to IBD may exist (2,3). First-degree relatives of patients with IBD were more susceptible to IBD, and sibling patients were identified to have a high concordance of disease type and location (4). In addition, a substantially higher disease concordance has been observed for CD and UC in monozygotic twins compared with that in dizygotic twins (5). It has also been reported that the pair concordance rate in monozygotic pairs is ~50% (6).
Three independent mutations in nucleotide-binding oligomerization domain-containing protein 2 (NOD2), namely Arg702Trp, Gly908Arg and Leu1007fsinsC, are the first verified genetic variants to be associated with CD in the Caucasian population (7,8). Mutation of Cys203Tyr in the X-linked inhibitor of apoptosis (XIAP) gene has also been indicated to cause immunodeficiency in a child with CD (9). In addition, mutations in the interleukin-10 receptor (IL10R) subunit proteins IL10RA (Thr84Ile and Gly141Arg) and IL10RB (Trp159X) have been associated with early-onset enterocolitis in two unrelated families (10).
The incidence and prevalence of IBD used to be high in the western population (11), and is now increasing markedly in China (12,13). Controversies have been observed regarding genetic variants in populations in Japan and Hong Kong (14,15). Therefore, reassessment of the role of previously reported single nucleotide polymorphisms in specific populations is required. Few studies have reported familial aggregations in China, and to the best of our knowledge, there is no report of concordant monozygotic twin pairs with CD in China. In the present study, samples were collected from 4 Han Chinese families with a history of IBD, including 8 CD patients and 2 UC sisters, and the distributions of 7 reported genetic variants associated with IBD were investigated. Associations between specific environmental factors and IBD were also analyzed.
Materials and methods
Subjects
A total of 100 patients from 4 families were studied, including 54 males and 46 females. Patients were recruited between March 2010 and March 2015. The mean age was 30.3 years old. Patients with enterophthisis suffintestinal tumor were excluded. The familial patients included in the present study were recruited from the Department of Gastroenterology at the Zhongnan Hospital of Wuhan University (Wuhan, China) and the Hubei Clinical Center and Key Laboratory of Intestinal and Colorectal Diseases (Wuhan, China). The registered IBD database of the Hubei Clinical Center and Key Laboratory of Intestinal and Colorectal Diseases (Wuhan, China) was used to compare familial and sporadic patients. Written informed consent was obtained from all patients in the registered IBD database and the parents of all enrolled children. The present study was approved by the Ethics Committee of the Zhongnan Hospital of Wuhan University as part of the human subjects' protocol to study the pathogenesis of IBD.
Patients with IBD were all born from non-consanguineous Chinese parents. All diagnoses were made according to the criteria on the basis of clinical, radiological, endoscopic and histological examinations Clinical blood tests included screening for anemia and infection, and the fecal occult blood test was performed to test for blood in the stool. For radiological diagnosis, an iodinated oil solution (Guerbet, Villepinte, France) was used as a contrast medium reveal the contours of the esophagus, stomach, small bowel or colon. Endoscopies were performed to view the entire colon. The optical instrument is introduced through the mouth and advanced through the esophagus into the stomach and duodenum. During the procedure, small samples of tissue were harvested for laboratory analysis to confirm diagnoses. Histological assessments were performed. The histological features were then used to assess the severity of diagnosis, ranging from definite Crohn's disease to definite ulcerative colitis (16). Demographic data were acquired from reviews of medical records, phone interviews and face-to-face interviews. For each patient, the information collected included: Sex, age, initial diagnosis age, disease extent, disease activity score at diagnosis (using the CD activity index and the UC activity index) (17,18) and treatment history. For family members, the information collected included the following: Sex, age, residence, immigration status, education, occupation, marital status, smoking status, history of other autoimmune diseases, vaccination history and possession of pet animals.
Pedigrees analysis
The familial patients were identified as having a familial history of IBD when first and/or second-degree relatives also had IBD. A total of 4 Han Chinese families, including 10 familial patients with IBD, were all from Hubei province. The pedigrees of the 4 families were acquired on the basis of epidemiological investigation. Detailed information on the whereabouts and cause of mortality were obtained for each patient. The health conditions of each unaffected family member were assessed by a self-report, telephone inquiry and face-to-face interview.
Genotyping
Genomic DNA was isolated from EDTA-anticoagulated peripheral venous blood using a QIAamp DNA Blood Midi kit according to the manufacturer's protocol (Qiagen GmbH, Hilden, Germany). The genotypes of 7 reported IBD mutations (shown in Table I) were screened by Sanger sequencing of polymerase chain reaction (PCR) products. PCR was performed utilizing PCR amplification kit with Taq DNA polymerases according to manufacturer's protocol (New England Biolabs Inc., Ipswich, MA, USA). The PCR was performed as follows: 95°C for 5 min followed by 30 cycles of 45 sec at 95°C, 45 sec at 65°C and 1 min at 72°C. The primers were designed by Primer 5.0 software (Premier Tech Ltd., Rivière-du-Loup, Canada) and synthesized by Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The primers and PCR conditions are specified in Table II. The product length was obtained from gene sequencing. The sequencing reactions were conducted by the Chinese National Human Genome Center (SinoGenoMax Co., Ltd., Beijing, China). DNA sequence analysis and variation identification were performed using DNAssist version 2.0, Chromas version 2.0 (Technelysium Pty., Ltd., South Brisbane, Australia) and the Basic Local Alignment Search Tool database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). To identify whether the twins were monozygotic or dizygotic, short tandem repeat markers were analyzed on an ABI 3730XL DNA Analyzer (Thermo Fisher Scientific, Inc.) at the Gene Diagnosis Center of Zhongnan Hospital (Wuhan University).
Table I.
Mutations screened in the study.
| Gene | Exon | Nucleotide change | Amino acid change |
|---|---|---|---|
| IL10RA | 3 | (C>T) 608 | Thr84Ile |
| IL10RA | 4 | (C>A) 253 | Gly141Arg |
| IL10RB | 4 | (G>A) 421 | Trp159X |
| XIAP | 1 | (G>T) 608 | Cys203Tyr |
| NOD2 | 4 | (C>T) 2104 | Arg702Trp |
| NOD2 | 8 | (G>C) 2722 | Gly908Arg |
| NOD2 | 11 | 3020 insC | Leu1007sinsC |
IL10R, interleukin-10 receptor; XIAP, X-linked inhibitor of apoptosis; NOD2, nucleotide-binding oligomerization domain-containing protein 2; A, adenine, T, thymine; G, guanine; C, cytosine; ins, insertion; X, stop codon.
Table II.
Primers and polymerase chain reaction conditions.
| Wild-type gene (starting position) | Primers, 5′-3′ | Annealing temperature, °C | Product length, bp |
|---|---|---|---|
| IL10RA (608C) | Forward: GGCCTCTTGCGTCTCCC | 56 | 359 |
| Reverse: GCAGACATGGTGAGCTATGG | |||
| IL10RA (253C) | Forward: ATTCTGGAGGCAAAGTCTCG | 56 | 348 |
| Reverse: AGTTCCCAATGGCACACAAG | |||
| IL10RB (421G) | Forward: CTACCCTTCTTAGCCATGTCA | 56 | 777 |
| Reverse: TCCGATCAGATCTTTTGACTC | |||
| XIAP (608G) | Forward: TTGAGAACTGGGCAGGTTGT | 59 | 179 |
| Reverse: CACTGCACTTGGTCACCAAT | |||
| NOD2 (2104C) | Forward: CTCCGCAAGCACTTCCACTC | 59 | 280 |
| Reverse: CCACGCCAATGTCACCCAC | |||
| NOD2 (2722G) | Forward: GAGGAGGACTGTTAGTTCATG | 57 | 290 |
| Reverse: TTCCTTACTCCATTGCCTA | |||
| NOD2 (3020T) | Forward: TGGTACTGAGCCTTTGTT | 57 | 461 |
| Reverse: GAAATCATTGCTCCCTACT |
IL10R, interleukin-10 receptor; XIAP, X-linked inhibitor of apoptosis; NOD2, nucleotide-binding oligomerization domain-containing protein 2; C, cytosine; G, guanine; T, thymine.
Statistical analysis
SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. Data were analyzed with a χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
Pedigree analysis
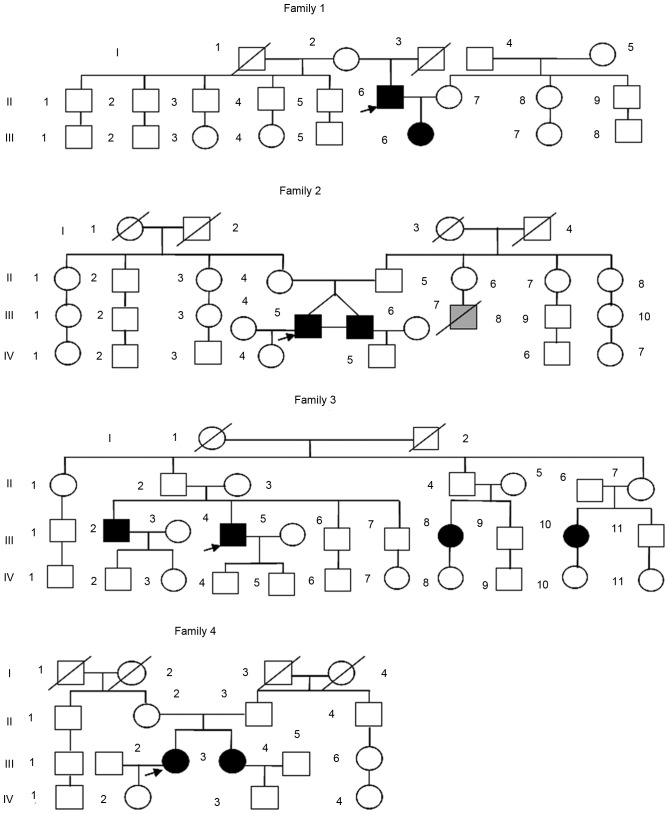
The 4 families identified as having familial IBD in the present study had a total of 100 members, including 10 patients with IBD (8 patients with CD and 2 with UC). A total of 78 subjects (10 patients + 68 unaffected relatives) were interviewed directly, and the health histories of the remaining 22 unaffected relatives were acquired by telephone inquiry or from statements made by relatives. The pedigrees are illustrated in Fig. 1. Family 1 had 2 patients with CD (a father and his daughter). Family 2 had 2 monozygotic twin brothers with CD. Family 3 had 4 cousins with CD (2 females and 2 males, two of whom were siblings). Family 4 had 2 sisters with UC. This pedigree analysis indicated that an autosomal recessive inheritance may have accounted for the disease in family 1; however, the other 3 families appeared to follow a pattern of polygenetic or multifactorial inheritance rather than single-gene inheritance.
Figure 1.
Pedigrees of 4 Chinese families with inflammatory bowel disease. In family 1, a father and his daughter had CD. Family 2 had 2 monozygotic twin brothers with CD. Family 3 had 4 cousins with CD (2 females and 2 males, two of whom were siblings). Family 4 had 2 sisters with ulcerative colitis. Squares, male family members; circles, female family members; black, affected patients; gray, suspected patients; slashes, deceased family members; arrow, the first diagnosed patient of the family. CD, Crohn's disease.
Clinical characteristics
The clinical characteristics of the 10 patients with familial IBD are summarized in Tables III and IV. The registered database at the Hubei Clinical Center and Key Laboratory of Intestinal and Colorectal Diseases included 571 CD patients and 227 UC patients who were admitted between January 2002 and December 2013, with a 1.25% ratio of patients with a family history of IBD. Consistent with reports in the Western population, first-degree relatives were more susceptible to IBD and siblings were affected more frequently (10-fold increase) compared with the general population (19). Among the 4 families, the onset of IBD in the daughter of family 1 was 16 years earlier than her father's, and her infection was more severe and extensive, as CDAI scores were 11 and 4 for the daughter and father, respectively. In family 3, siblings were identified to have a high concordance of clinical type and location according to the location of the lesion, behavior and treatment. In all 4 families, the susceptibility was the same between males and females (5 male and 5 female). Among the couples investigated, there were no instances of both partners being affected by IBD.
Table III.
Clinical characteristics of the Crohn's disease patients.
| Case | Year of birth | Sex | Year of onset | Year of diagnosis | Location of lesion | Disease phenotype | Treatment | EIM | Surgery | CDAI |
|---|---|---|---|---|---|---|---|---|---|---|
| F1-II6 | 1968 | M | 2000 | 2002 | Terminal ileum + rectum | Stricturing | Mesalazine + corticosteroids | No | No | 4 |
| F1-III6 | 1996 | F | 2012 | 2012 | Terminal ileum + ileocecus + ascending colon + rectum + upper gastrointestinal tract | Penetrating | Mesalazine + corticosteroids + biological agents + azathioprine | Yes | No | 11 |
| F2-III5 | 1981 | M | 2004 | 2006 | Terminal ileum + ileocecus + rectum | Stricturing | Mesalazine + corticosteroids + azathioprine | No | Yes | 9 |
| F2-III6 | 1981 | M | 2009 | 2009 | Terminal ileum + ileocecus | Stricturing | Mesalazine + corticosteroids + biological agents + azathioprine | No | Yes | 11 |
| F3-III2 | 1973 | M | 2013 | 2013 | Terminal ileum + ileocecus | Non-penetrating + non-stricturing | Mesalazine | No | No | 1 |
| F3-III4 | 1975 | M | 2009 | 2010 | Terminal ileum + ileocecus + rectum | Penetrating | Mesalazine + corticosteroids + azathioprine | No | Yes | 11 |
| F3-III8 | 1987 | F | 2012 | 2012 | Terminal ileum + ileocecus + rectum | Non-penetrating + non-stricturing | Mesalazine | No | No | 6 |
| F3-III10 | 1979 | F | 2013 | 2013 | Terminal ileum + ileocecus | Non-penetrating + non-stricturing | Mesalazine | No | No | 1 |
F1, family 1; F2, family 2; F3, family 3; M, male; F, female; EIM, extraintestinal manifestations; CDAI, Crohn's disease activity index.
Table IV.
Clinical characteristics of the ulcerative colitis patients.
| Case | Year of birth | Sex | Year of onset | Year of diagnosis | Location of lesion | Treatment | UCAI |
|---|---|---|---|---|---|---|---|
| F4-III3 | 1972 | F | 2008 | 2010 | Descending colon + sigmoid colon + rectum | Mesalazine + corticosteroids | 8 |
| F4-III4 | 1974 | F | 2009 | 2010 | Descending colon + sigmoid colon + rectum | Mesalazine + corticosteroids | 8 |
F4, family 4; F, female; UCAI, ulcerative colitis activity index.
The median age of onset was 29.0 years old for the 8 CD patients and 35.5 years old for the 2 UC patients. A previous study by our group observed that the median age of diagnosis was 36 years old for CD patients and 41 years old for UC patients in mainland China (20). In the present study, the median age of diagnosis was younger in familial patients compared with sporadic patients in China (data not shown).
A comparison between the familial and sporadic patients was performed using the data in our registered IBD database. The percentage of extraintestinal manifestations was 12.5% (1/8) of familial patients, which was similar to the previously reported result of 13.1% of sporadic patients (21). Among the 4 families, one CD patient (family 1 member 6, F1-III6) exhibited an extraintestinal manifestation of aphthous stomatitis. Of the 8 CD patients, 3 male CD patients (37.5%; F2-III5, F2-III6 and F3-III4) underwent surgical enterectomy, which was a similar percentage to that observed in a previous study (26.9%) (22). A total of 2 CD patients (F1-III6 and F2-III6) exhibited an acute and fulminant onset and 2 CD patients (F1-III6 and F2-III5) exhibited an anal fistula. A total of 4 CD (F1-II6, F2-III5, F2-III6, F3-II4) patients suffered severe attacks, while the other 4 CD patients (F3-III8, F3-III10, F3-III2, F1-II6) and 2 UC sisters (F4-III3, F4-III4) underwent a mild-moderate progression.
Risk analysis of demographic factors
Selected demographic factors were compared between the patients and their unaffected family members. As depicted in Table V, the stressful life event of immigration to a new living place significantly increased the risk of IBD (odds ratio: 4.667; 95% confidence interval: 1.165–18.690; P=0.021). The confidence intervals of all the demographic factors was 95%. Other demographic factors, namely residence, education, labor intensity, marital status, smoking, history of other autoimmune diseases, vaccinations and possession of a pet animal were also evaluated; however, no significant correlations were observed (P>0.05).
Table V.
Comparison of demographic factors between familial patients and interviewed unaffected relatives.
| Factor | IBD (n=10) | Unaffected (n=68) | Odds ratio | P-value |
|---|---|---|---|---|
| Residence | 0.929 | 0.915 | ||
| City | 6 | 42 | ||
| Town | 4 | 26 | ||
| Immigration before diagnosis | 5 | 12 | 4.667 | 0.021 |
| Education | 1.826 | 0.632 | ||
| High school or below | 6 | 46 | ||
| University or college | 4 | 22 | ||
| Occupation | 0.941 | |||
| Student | 1 | 6 | 1.481 | |
| Manual worker | 4 | 24 | 1.267 | |
| Mental worker | 5 | 38 | ||
| Marital status | 1.222 | 0.772 | ||
| Single | 4 | 24 | ||
| Married | 6 | 44 | ||
| Current smoker | 1 | 18 | 0.309 | 0.257 |
| History of other autoimmune diseases | 0 | 3 | 0 | 0.498 |
| Vaccinations | 5 | 43 | 0.581 | 0.422 |
| Pet animal | 3 | 22 | 0.986 | 0.882 |
IBD, inflammatory bowel disease.
Genotypes of selected loci
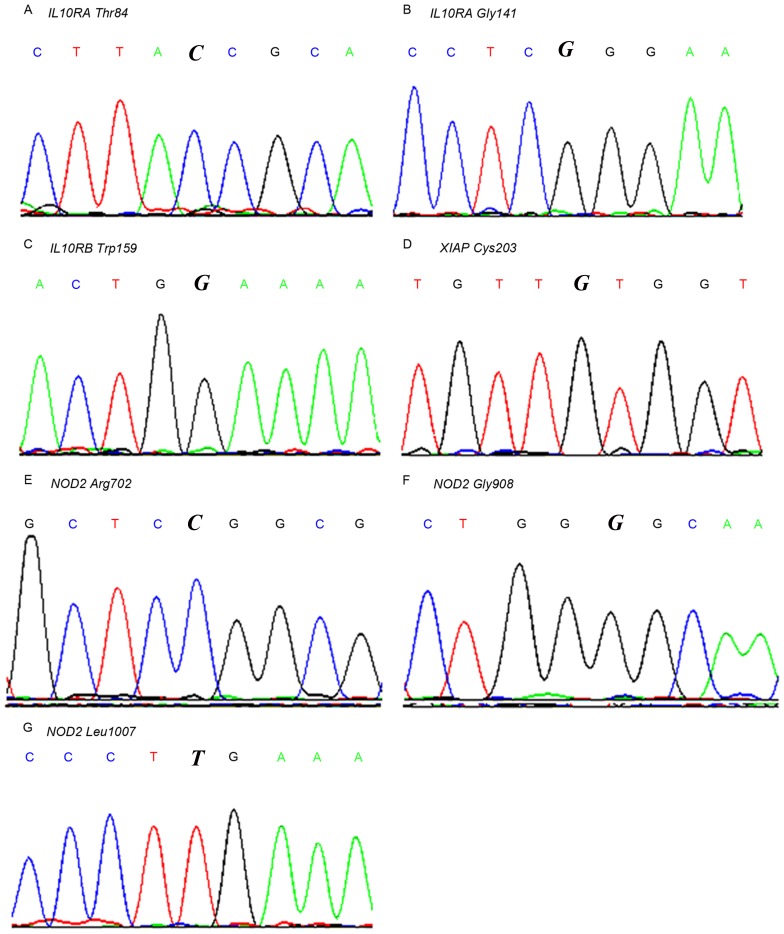
The 7 reported gene mutations associated with IBD (Table I) were screened in the familial patients. No patients carried any of the mutations. Nearby sequences within the same exon of the 7 loci were also evaluated (3rd and 4th exon of IL10RA; 4th exon of IL10RB; 1st exon of XIAP; and 4th, 8th and 11th exon of NOD2). No homozygous or heterozygous mutations were identified. Partial wild-type sequence chromatograms of these gene loci are depicted in Fig. 2. Single-site mutations were observed in 7 genes, including IL10R Thr84, IL10R Gly141, IL10R Trp159, XIAP Cys203, NOD2 Arg702, NOD2 Gly908 and NOD2 Leu1007.
Figure 2.
Partial wild-type sequence chromatograms of the 7 gene loci detected. (A) IL10RA Thr84; (B) IL10RA Gly141; (C) IL10RB Trp159; (D) XIAP Cys203; (E) NOD2 Arg702; (F) NOD2 Gly908; (G) NOD2 Leu1007. The italicized letters indicate the mutation sites gene coding the coresponding proteins. IL10R, interleukin-10 receptor; XIAP, X-linked inhibitor of apoptosis; NOD2, nucleotide-binding oligomerization domain-containing protein 2.
Discussion
A large number of patients with familial IBD have been reported in Western countries (23,24). Halme et al (25) previously documented that the ratio of patients with a family history of IBD varied between 2–14%. Gearry et al (26) reported a family history prevalence rate of 33.2% in IBD. Furthermore, Ng et al (27) reported that the ratios of patients with a family history of IBD were 3 and 17% in Asia and Australia, respectively. In the present study, the ratio of patients with a family history of IBD was 1.25%, which was lower than those reported previously. The phenomenon of less familial clustering in the Chinese population has been discussed in a previous study (28). Other major findings concerning familial clustering in the present study were consistent with previous studies (4,29). The present results indicated that offspring had an earlier onset and more severe infection than the parental generation. In addition, compared with sporadic patients, the median age of diagnosis was younger in familial patients.
Accumulating evidence suggests that genetic factors are involved in the development of IBD (30). A total of 7 mutations within 4 genes were investigated in the present study. Mutations in the NOD2 gene (Arg702Trp, Gly908Arg and Leu1007fsinsC) have been widely studied and are typically associated with IBD in Caucasian individuals (8,31,32). It has been demonstrated that NOD-induced nuclear factor-κB may be dependent on XIAP through an indirect interaction between the baculoviral IAP repeats (BIR2) and receptor-interacting serine/threonine-protein kinase 2 (RIP2) (33–35). Furthermore, a previous study observed that a single mutation in the XIAP gene (Cys203Tyr) caused intractable IBD of a male child, which was cured by a hematopoietic progenitor cell transplant (9). In addition, XIAP has been implicated as a critical factor for toll-like receptor-modulated signaling in response to infection (36). Previous results have suggested that mutations in IL10RA (Thr84Ile and Gly141Arg) and IL10RB (Trp159X) may result in a loss of protein function and have been associated with hyperinflammatory reactions caused by intestinal bacteria (10,37,38). However, none of the 7 susceptibility mutations were detected in the familial patients of the present study. Inoue et al (14) also reported a lack of common NOD2 variants in Japanese patients with CD, and Wang et al (39) reported that the common variants of NOD2 had no correlation with IBD in the Zhejiang province of China (39). One of our previous studies indicated that the NOD2 3020insC frame-shift mutation did not contribute to the susceptibility of CD and UC in sporadic patients in central China (40). These results indicate that inconsistencies exist in different ethnic groups. Furthermore, a recent trans-ancestry association study of IBD demonstrated genetic heterogeneity between divergent populations (41).
In the present study, the stressful life event of immigration was a potential risk factor of IBD. Stress may alter the microenvironment of the intestinal tract (42). A previous study observed that fried food and stress were associated with an increased risk of IBD (39). However, diet information was not recorded in our database; therefore, the association between diet and IBD was not analyzed in the present study. These findings suggest that even for familial patients, environmental factors are involved in the pathogenesis of IBD, in addition to genetic background.
In China, the prevalence of IBD has increased markedly in the past two decades (43,44). Compared with the Western population, less familial clustering is observed in the Chinese population (28). The one-child policy also makes it difficult to conduct family research in the Chinese population. These factors may explain the relatively small number of familial patients enrolled in the present study. However, to the best of our knowledge, the present study observed the first cases of concordant CD to occur in monozygotic twins in China. In addition, the electronic medical database enabled comparisons between familial and sporadic patients.
A number of limitations were included in the present study. Firstly, selection bias may have occurred, as the study was performed using a hospital-based registry instead of a population-based resource. However, universal health systems in China currently contain incomplete data on rural populations, which may result in some families with IBD being omitted from wide-scale population studies. Secondly, only 7 common genetic variants were tested, and no positive associations were identified. Other previously identified or unidentified genes may also contribute to disease progression in familial patients. For instance, the DLG1 gene was recently identified as a potential gene associated with CD in Chinese patients (45). Two genome-wide association studies also identified a number of novel susceptibility loci in Japanese and Korean individuals (46,47). Therefore, further studies with larger sample sizes and extensive coverage are required to map the susceptibility loci of IBD in the Chinese population.
In conclusion, the results of the present study indicated that the percentage of patients with a family history of IBD was 1.25%, which was lower than that reported previously. Furthermore, the present results indicated that offspring had an earlier onset and more severe infection compared with parents. In addition, the median age of diagnosis was younger in familial patients compared with sporadic patients. Genetic factors are involved in the development of IBD and immigration was identified as a potential risk factor for IBD due to alterations to the intestinal tract microenvironment. These findings suggest that the genetic background but and environmental factors are involved in the pathogenesis of IBD, even in familial patients.
Acknowledgements
The authors would like to thank Professor Bing Xia (from the Hubei Clinical Center and Key Laboratory for Intestinal and Colorectal Diseases, Wuhan, China) for his assistance. The present study was supported by the Natural Science Foundation of China (grant no. 81500427).
References
- 1.Zatorski H, Sałaga M, Zielińska M, Fichna J. Genetic factors in pathogenesis, course and treatment of inflammatory bowel diseases. Postepy Hig Med Dosw (Online) 2015;69:335–344. doi: 10.5604/17322693.1145172. (In Polish) [DOI] [PubMed] [Google Scholar]
- 2.Huang C, Haritunians T, Okou DT, Cutler DJ, Zwick ME, Taylor KD, Datta LW, Maranville JC, Liu Z, Ellis S, et al. Characterization of genetic loci that affect susceptibility to inflammatory bowel diseases in African Americans. Gastroenterology. 2015;149:1575–1586. doi: 10.1053/j.gastro.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lappalainen M, Halme L, Turunen U, Saavalainen P, Einarsdottir E, Färkkilä M, Kontula K, Paavola-Sakki P. Association of IL23R, TNFRSF1A and HLA-DRB1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm Bowel Dis. 2008;14:1118–1124. doi: 10.1002/ibd.20431. [DOI] [PubMed] [Google Scholar]
- 4.Colombel JF, Grandbastien B, Gower-Rousseau C, Plegat S, Evrard JP, Dupas JL, Gendre JP, Modigliani R, Bélaïche J, Hostein J, et al. Clinical characteristics of Crohn's disease in 72 families. Gastroenterology. 1996;111:604–607. doi: 10.1053/gast.1996.v111.pm8780563. [DOI] [PubMed] [Google Scholar]
- 5.Halfvarson J, Jess T, Bodin L, Järnerot G, Munkholm P, Binder V, Tysk C. Longitudinal concordance for clinical characteristics in a Swedish-Danish twin population with inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1536–1544. doi: 10.1002/ibd.20242. [DOI] [PubMed] [Google Scholar]
- 6.Halfvarson J, Bodin L, Tysk C, Lindberg E, Jäenerot G. Inflammatory bowel disease in a Swedish twin cohort: A long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–1773. doi: 10.1016/S0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 8.van der Linde K, Boor PP, Houwing-Duistermaat JJ, Crusius BJ, Wilson PJ, Kuipers EJ, de Rooij FW. CARD15 mutations in Dutch familial and sporadic inflammatory bowel disease and an overview of European studies. Eur J Gastroenterol Hepatol. 2007;19:449–459. doi: 10.1097/01.meg.0000236887.44214.6a. [DOI] [PubMed] [Google Scholar]
- 9.Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, et al. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 10.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. New Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogel O, Richard-Miceli C, Tost J. Epigenetic changes in chronic inflammatory diseases. Adv Protein Chem Struct Biol. 2017;106:139–189. doi: 10.1016/bs.apcsb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Cao Q, Si JM, Gao M, Zhou G, Hu WL, Li JH. Clinical presentation of inflammatory bowel disease: A hospital based retrospective study of 379 patients in eastern China. Chinese Med J (Engl) 2005;118:747–752. [PubMed] [Google Scholar]
- 13.Wang YF, Ouyang Q, Hu RW. Progression of inflammatory bowel disease in China. J Dig Dis. 2010;11:76–82. doi: 10.1111/j.1751-2980.2010.00421.x. [DOI] [PubMed] [Google Scholar]
- 14.Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Núñez G, Kishi Y, Koike Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 15.Leong RW, Armuzzi A, Ahmad T, Wong ML, Tse P, Jewell DP, Sung JJ. NOD2/CARD15 gene polymorphisms and Crohn's disease in the Chinese population. Aliment Pharmacol Ther. 2003;17:1465–1470. doi: 10.1046/j.1365-2036.2003.01607.x. [DOI] [PubMed] [Google Scholar]
- 16.Tertychnyi AS, Akhrieva KM, Maev IV, Zayratyants OV, Selivanova LS. Diagnostic problems of histological remission in patients with inflammatory bowel disease. Arkh Patol. 2017;79:3–9. doi: 10.17116/patol20177933-9. [DOI] [PubMed] [Google Scholar]
- 17.Lahat A, Kopylov U, Neuman S, Levhar N, Yablecovitch D, Avidan B, Weiss B, Ben-Horin S, Eliakim R, on behalf of the Israeli IBD research Network (IIRN) Helicobacter pylori prevalence and clinical significance in patients with quiescent Crohn's disease. BMC Gastroenterol. 2017;17:27. doi: 10.1186/s12876-017-0588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamani H, Barzin G, Yousefinia M, Mohammadkhani A, Ostovaneh MR, Sharifi AH, Tayebi S, Malekzadeh R, Ansari R. Diagnostic value of fecal calprotectin in patient with ulcerative colitis. Middle East J Dig Dis. 2013;5:76–80. [PMC free article] [PubMed] [Google Scholar]
- 19.Orholm M, Munkholm P, Langholz E, Nielsen OH, Sørensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Ng SC, Lei Y, Yi F, Li J, Yu L, Zou K, Dan Z, Dai M, Ding Y, et al. First prospective, population-based inflammatory bowel disease incidence study in mainland of China: The emergence of ‘western’ disease. Inflamm Bowel Dis. 2013;19:1839–1845. doi: 10.1097/MIB.0b013e31828a6551. [DOI] [PubMed] [Google Scholar]
- 21.Yi F, Chen M, Huang M, Li J, Zhao J, Li L, Xia B. The trend in newly diagnosed Crohn's disease and extraintestinal manifestations of Crohn's disease in central China: A retrospective study of a single center. Eur J Gastroenterol Hepatol. 2012;24:1424–1429. doi: 10.1097/MEG.0b013e3283583e5c. [DOI] [PubMed] [Google Scholar]
- 22.Jiang L, Xia B, Li J, Ye M, Yan W, Deng C, Ding Y, Luo H, Hou W, Zhao Q, et al. Retrospective survey of 452 patients with inflammatory bowel disease in Wuhan city, central China. Inflamm Bowel Dis. 2006;12:212–217. doi: 10.1097/01.MIB.0000201098.26450.ae. [DOI] [PubMed] [Google Scholar]
- 23.Freeman HJ. Familial Crohn's disease in single or multiple first-degree relatives. J Clin Gastroenterol. 2002;35:9–13. doi: 10.1097/00004836-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 25.Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006;12:3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gearry RB, Richardson AK, Frampton CM, Dodgshun AJ, Barclay ML. Population-based cases control study of inflammatory bowel disease risk factors. J Gastroenterol Hepatol. 2010;25:325–333. doi: 10.1111/j.1440-1746.2009.06140.x. [DOI] [PubMed] [Google Scholar]
- 27.Ng SC, Tang W, Ching JY, Wong M, Chow CM, Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, et al. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-pacific Crohn's and colitis epidemiology study. Gastroenterology. 2013;145:158–165.e2. doi: 10.1053/j.gastro.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Ng SC. Epidemiology of inflammatory bowel disease: Focus on Asia. Best Pract Res Clin Gastroenterol. 2014;28:363–372. doi: 10.1016/j.bpg.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang YF, Zhang H, Ouyang Q. Clinical manifestations of inflammatory bowel disease: East and West differences. J Dig Dis. 2007;8:121–127. doi: 10.1111/j.1443-9573.2007.00296.x. [DOI] [PubMed] [Google Scholar]
- 30.Yuan F, Zhang YH, Kong XY, Cai YD. Identification of candidate genes related to inflammatory bowel disease using minimum redundancy maximum relevance, incremental feature selection, and the shortest-path approach. Biomed Res Int. 2017;2017:5741948. doi: 10.1155/2017/5741948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abreu MT, Taylor KD, Lin YC, Hang T, Gaiennie J, Landers CJ, Vasiliauskas EA, Kam LY, Rojany M, Papadakis KA, et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123:679–688. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 32.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural basis of caspase inhibition by XIAP: Differential roles of the linker versus the BIR domain. Cell. 2001;104:781–790. doi: 10.1016/S0092-8674(01)00273-2. [DOI] [PubMed] [Google Scholar]
- 34.Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Knoefel WT, Reed JC. XIAP mediates NOD signaling via interaction with RIP2; Proc Natl Acad Sci USA; 2009; pp. 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigaud S, Fondanèche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 36.Richardson WM, Sodhi CP, Russo A, Siggers RH, Afrazi A, Gribar SC, Neal MD, Dai S, Prindle T, Jr, Branca M, et al. Nucleotide-binding oligomerization domain-2 inhibits toll-like receptor-4 signaling in the intestinal epithelium. Gastroenterology. 2010;139:904–917. doi: 10.1053/j.gastro.2010.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kühn R, Löhler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
- 38.Paul G, Khare V, Gasche C. Inflamed gut mucosa: Downstream of interleukin-10. Eur J Clin Invest. 2012;42:95–109. doi: 10.1111/j.1365-2362.2011.02552.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZW, Ji F, Teng WJ, Yuan XG, Ye XM. Risk factors and gene polymorphisms of inflammatory bowel disease in population of Zhejiang, China. World J Gastroenterol. 2011;17:118–122. doi: 10.3748/wjg.v17.i1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo QS, Xia B, Jiang Y, Qu Y, Li J. NOD2 3020insC frameshift mutation is not associated with inflammatory bowel disease in Chinese patients of Han nationality. World J Gastroenterol. 2004;10:1069–1071. doi: 10.3748/wjg.v10.i7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alverdy J, Holbrook C, Rocha F, Seiden L, Wu RL, Musch M, Chang E, Ohman D, Suh S. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: Evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann Surg. 2000;232:480–489. doi: 10.1097/00000658-200010000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouyang Q, Tandon R, Goh KL, Ooi CJ, Ogata H, Fiocchi C. The emergence of inflammatory bowel disease in the Asian Pacific region. Curr Opin Gastroenterol. 2005;21:408–413. [PubMed] [Google Scholar]
- 44.Zheng JJ, Zhu XS, Huangfu Z, Gao ZX, Guo ZR, Wang Z. Crohn's disease in mainland China: A systematic analysis of 50 years of research. Chin J Dig Dis. 2005;6:175–181. doi: 10.1111/j.1443-9573.2005.00227.x. [DOI] [PubMed] [Google Scholar]
- 45.Xu S, Zhou F, Tao J, Song L, Ng SC, Wang X, Chen L, Yi F, Ran Z, Zhou R, Xia B. Exome sequencing identifies DLG1 as a novel gene for potential susceptibility to Crohn's disease in a Chinese family study. PLos One. 2014;9:e99807. doi: 10.1371/journal.pone.0099807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamazaki K, Umeno J, Takahashi A, Hirano A, Johnson TA, Kumasaka N, Morizono T, Hosono N, Kawaguchi T, Takazoe M, et al. A genome-wide association study identifies 2 susceptibility Loci for Crohn's disease in a Japanese population. Gastroenterology. 2013;144:781–788. doi: 10.1053/j.gastro.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 47.Yang SK, Hong M, Zhao W, Jung Y, Baek J, Tayebi N, Kim KM, Ye BD, Kim KJ, Park SH, et al. Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63:80–87. doi: 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]