Genome-wide association mapping and protein characterization reveal that the small heat shock-like protein SLI1 restricts phloem feeding by aphids and improves seed set during heat stress.
Abstract
The role of phloem proteins in plant resistance to aphids is still largely elusive. By genome-wide association mapping of aphid behavior on 350 natural Arabidopsis thaliana accessions, we identified the small heat shock-like SIEVE ELEMENT-LINING CHAPERONE1 (SLI1). Detailed behavioral studies on near-isogenic and knockout lines showed that SLI1 impairs phloem feeding. Depending on the haplotype, aphids displayed a different duration of salivation in the phloem. On sli1 mutants, aphids prolonged their feeding sessions and ingested phloem at a higher rate than on wild-type plants. The largest phenotypic effects were observed at 26°C, when SLI1 expression is upregulated. At this moderately high temperature, sli1 mutants suffered from retarded elongation of the inflorescence and impaired silique development. Fluorescent reporter fusions showed that SLI1 is confined to the margins of sieve elements where it lines the parietal layer and colocalizes in spherical bodies around mitochondria. This localization pattern is reminiscent of the clamp-like structures observed in previous ultrastructural studies of the phloem and shows that the parietal phloem layer plays an important role in plant resistance to aphids and heat stress.
INTRODUCTION
Aphids are phloem-feeding insects that impose substantial economic damage to a wide variety of crops. In temperate regions, Aphididae constitute 25% of the major pest insects on food crops, such as maize (Zea mays), wheat (Triticum aestivum), potatoes (Solanum tuberosum), and tomatoes (Solanum lycopersicum) (Hill, 1987; Dedryver et al., 2010). By maneuvering their needle-like mandibles, called stylets, through the cell wall matrix, aphids reach phloem sieve elements to ingest phloem sap (Dixon, 1998). Several plant resistance strategies against aphids have been described in the last few decades (Züst and Agrawal, 2016). One of the most effective mechanisms has been found in lettuce (Lactuca sativa), where black currant-lettuce aphids (Nasonovia ribisnigri) biotype Nr:0 could successfully reach the sieve elements but were virtually unable to ingest any phloem sap (ten Broeke et al., 2013). The absence of (sustained) phloem feeding has been thought to be caused by the occlusion of the aphid’s food canal (Tjallingii, 2006) or the sieve tube (Dreyer and Campbell, 1987; Will and van Bel, 2006). Transmission electron microscopy of stylets in sieve elements showed that fibrous and amorphous, electron-dense material accumulated in the food canal of N. ribisnigri and Aphis fabae (Tjallingii and Hogen Esch, 1993). As aphids are passive feeders and rely on the turgor pressure in sieve elements, occlusion of the sieve tube could also restrict phloem sap ingestion (Will and van Bel, 2006). Callose deposition around the sieve plate pores can plug the sieve plate and reduce the phloem flow rate (Mullendore et al., 2010). In addition, phloem proteins can obstruct sap translocation (Knoblauch and Van Bel, 1998). “Phloem protein” is a collective term for structural proteins with a wide variety of shapes (e.g., amorphous, crystalline, and filamentous) that have been observed in the sieve element lumen and at the sieve element margins (Ehlers et al., 2000; Knoblauch and Mullendore, 2013). Upon injury or temperature fluctuations, Ca2+ influx in the sieve tube can cause phloem protein dispersal into the lumen where they form coagulations that press against the sieve plate within seconds (Knoblauch and Van Bel, 1998; Knoblauch et al., 2001; Furch et al., 2007). Aphids may partially counteract phloem protein aggregations by secreting watery saliva with Ca2+-scavenging proteins into the phloem (Tjallingii, 2006; Will et al., 2007). Recently, it has been shown that the Ca2+-induced expansion of forisomes, which are spindle-shaped crystalline phloem protein bodies in fabaceous plants, reduced phloem feeding by the generalist aphid Myzus persicae (Medina-Ortega and Walker, 2013, 2015). In Brassicaceae, a phloem protein, the lectin AtPP2-A1 has been shown to decrease aphid feeding as well (Zhang et al., 2011), possibly by hampering digestion in the insect midgut (Kehr, 2006). However, the role of the vast majority of phloem proteins remains unknown.
In this study, we performed an unbiased screening of 350 natural Arabidopsis thaliana accessions for resistance to aphids. Feeding behavior of M. persicae aphids was quantified on these plant lines with an automated video-tracking platform (Kloth et al., 2015). Genome-wide association (GWA) mapping revealed an association between aphid feeding behavior and polymorphisms in the small heat shock-like gene SLI1, with hitherto unknown function. We used a near-isogenic line (NIL) and T-DNA insertion lines to show that SLI1 restricts the duration of feeding and the rate of sap ingestion. In addition, SLI1 increased the inflorescence growth rate and seed production at high ambient temperature. In transgenic lines, fluorescently labeled SLI1 proteins, expressed under the native promoter, lined virtually the complete sieve element margin and formed spherical bodies around mitochondria. Based on its protein structure and localization, we discuss the possible role of SLI1 in phloem-based resistance to aphids and heat stress.
RESULTS
Identification of SLI1
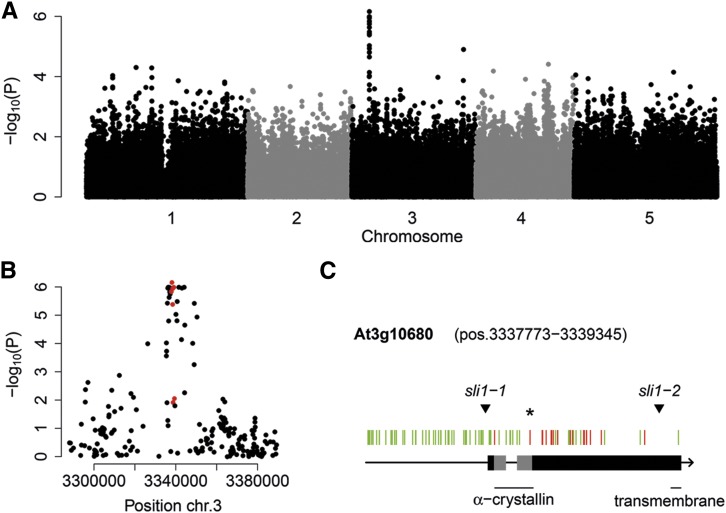
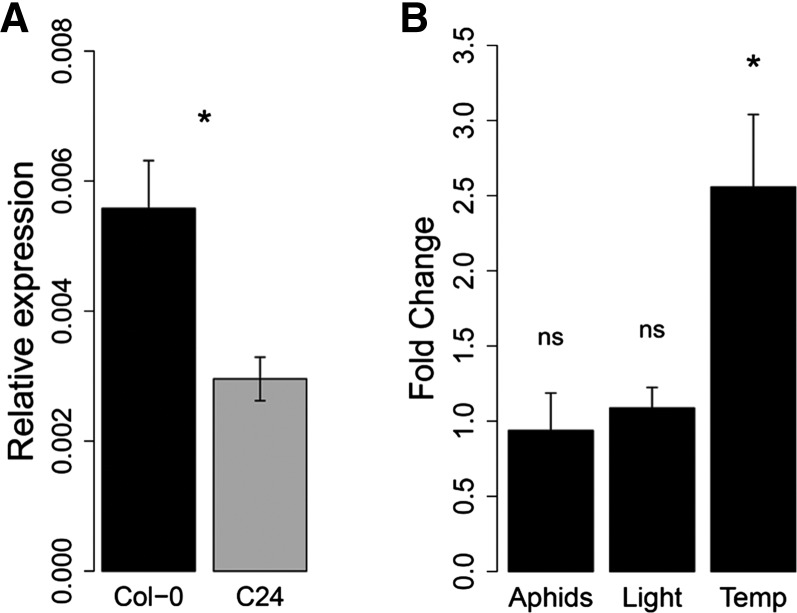
To explore natural variation in Arabidopsis resistance to aphids, we assessed M. persicae feeding behavior on 350 natural Arabidopsis accessions, which had been selected and genotyped in previous studies (Atwell et al., 2010; Li et al., 2010; Platt et al., 2010; Horton et al., 2012). Aphid probing behavior was quantified using a high-throughput video-tracking platform (Kloth et al., 2015, 2016). This platform consisted of a 96-well plate, with a leaf disc and an adult aphid in each enclosed well (Figure 1). With a top-view camera and video-tracking software, aphid body movements were recorded to estimate the number and duration of probes. GWA mapping with 214 k single nucleotide polymorphisms (SNPs) revealed an association between short probes made by the aphids and polymorphisms in the hsp20-like gene SLI1 (At3g10680), with unknown function (Figures 2A and 2B; Supplemental Data Set 1 and Supplemental Table 1). The most significant SNP [–log10(P) = 6.2] caused an amino acid change in the alpha-crystallin domain of SLI1. Other SNPs in linkage were mainly located in the single intron and promoter region (Figure 2C). To test if the polymorphisms in the intron resulted in differential splicing, we compared cDNA fragment lengths of the Col-0 and the C24 allele, conferring a resistant and susceptible phenotype, respectively. No splice variants were observed (Supplemental Figure 1), but RT-qPCR showed that the Col-0 allele had a higher transcript abundance than the C24 allele (Figure 3A; Supplemental Data Set 2). We assessed whether aphid infestation induced SLI1 expression but did not observe any differences in transcript abundance with or without aphids (Figure 3B; Supplemental Data Set 2). Considering that SLI1 is a small heat shock-like gene, we also tested its expression under different temperature and light conditions and found that SLI1 expression was 2-fold higher when plants were grown at 26°C—the temperature at which the 350 Arabidopsis accessions had been grown—than at 20°C (Figure 3B; Supplemental Data Set 2). Light intensity did not affect SLI1 transcription.
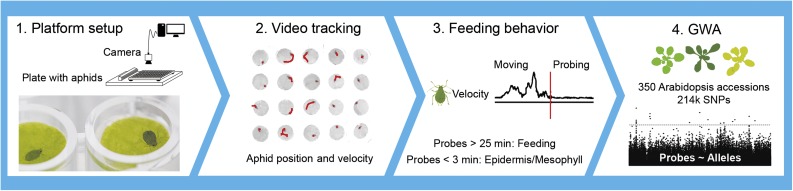
Figure 1.
Phenotyping Pipeline for GWA Mapping.
Aphid feeding behavior was quantified on 350 natural Arabidopsis accessions with a video-tracking platform. Each aphid was released in an enclosed well of a 96-well plate, containing a leaf disc embedded on agar. With a top-view camera and video-tracking software, aphid position (on leaf disc or wall) and velocity were recorded and used for the estimation of the number and duration of probes (Kloth et al., 2015). Probes were classified into short probes (<3 min) that were most likely associated with superficial cell layers, and long probes (>25 min) that were potentially related to phloem feeding. Aphid probing parameters were mapped to 214 k polymorphisms on the Arabidopsis genome to identify genes involved in plant resistance to aphids.
Figure 2.
GWA Mapping of M. persicae Aphid Feeding Behavior.
(A) Genome-wide associations of the total time aphids spent on short probes (<3 min, n = 3–6, 350 accessions, 214 k SNPs).
(B) The 80-kb region around the most significant SNP in SLI1 [chromosome 3, position 3338114, At3g10680, –log10(P) = 6.2]. The SNPs in SLI1 are annotated in red.
(C) SLI1 and its 1000-bp promoter region (green, silent SNPs and SNPs in promoter or intron; red, nonsynonymous SNPs; *, position 3338114; arrowhead, T-DNA insertion).
Figure 3.
SLI1 Expression.
(A) SLI1 expression in Col-0 and C24 leaves (Mann-Whitney U test, P = 0.03, n = 4, plants were grown at 26°C).
(B) Expression in Col-0 rosette leaves under different light intensity, temperature, and aphid treatments (measured by RT-qPCR): clean plants versus plants 6 h postinfestation (P = 0.57, n = 4); light intensity 70 versus 120 μmol m−2 s−1 (P = 0.74, n = 3); 26°C versus 20°C (P = 0.042, n = 8), one-way ANOVA; data represent mean ± se.
SLI1 Haplotype and Loss-of-Function Effects on Aphids
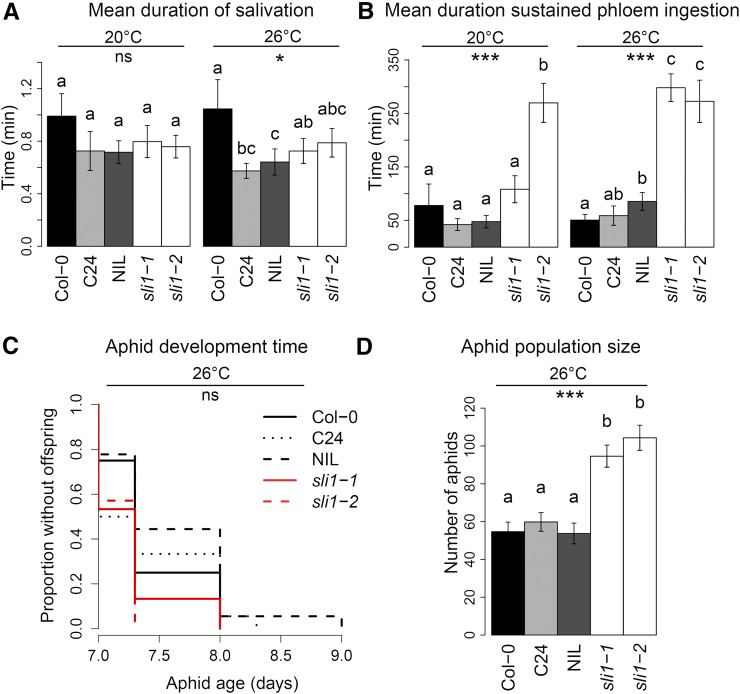
To test whether SLI1 plays a role in resistance to aphids, aphid feeding behavior was characterized by electrical penetration graph (EPG) recording (McLean and Kinsey, 1964; Tjallingii, 1988). With this dedicated technique, we assessed which tissue the aphids penetrated (epidermis/mesophyll or vascular bundle) and what stylet activities they performed (e.g., salivation, phloem ingestion, and xylem ingestion). On accession C24, aphids displayed a shorter salivation phase before feeding (Figure 4A) and more sustained feeding events (Table 1) than on Col-0, when plants were grown at 26°C. On a NIL with Col-0 background and a 3000-kb C24 introgression in the genomic region around SLI1, aphids also spent less time salivating (Figure 4A) and showed an increased mean duration of sustained phloem ingestion (Figure 4B) compared with Col-0 when grown at 26°C. To test whether these effects were caused by SLI1, EPG recording was performed on two T-DNA lines with a homozygous insertion in SLI1 (sli1-1 and sli1-2; Figure 2C). Aphids showed more sustained phloem feeding on both mutants compared with the wild type (Table 1). At 26°C the largest effects were observed: Aphids spent less time on penetration of the epidermis and mesophyll, had a lower contribution of salivation to phloem-feeding events, and showed a 2- to 3-fold increase in the mean duration of sustained phloem ingestion on both sli1 mutants (Figures 4A and 4B). In the long term, aphids did not develop significantly faster from neonate to adult on the mutants (Figure 4C), but produced almost twice as many offspring on sli1-1 and sli1-2 compared with the wild type (Figure 4D). The C24 accession and NIL did not reveal a haplotype effect on aphid population size.
Figure 4.
Performance of M. persicae Aphids on the Arabidopsis Accessions Col-0 and C24, a NIL with Col-0 Background, and a C24 Insertion in the SLI1 QTL Region, and the T-DNA Mutants sli1-1 and sli1-2.
(A) and (B) Mean duration of salivation in the phloem (A) and mean duration of sustained (>10 min) phloem ingestion (B) (8-h electrical penetration graph recordings; 20°C: Col-0 n = 13, C24 n = 12, NIL n = 12, sli1-1 n = 16, sli1-2 n = 13; 26°C: Col-0 n = 20, C24 n = 14, NIL n = 14, sli1-1 n = 20, sli1-2 n = 16).
(C) and (D) Aphid age at start of reproduction (Kaplan-Meier curve of Cox proportional hazards model) (C) and aphid population size (D), 2 weeks after the infestation with one neonate aphid per plant (Col-0 n = 16, C24 n = 18, NIL n = 18, sli1-1 n = 15, sli1-2 n = 15). Bars represent mean ± se; ns, nonsignificant; *P < 0.05, **P < 0.01, and ***P < 0.001 (Kruskal-Wallis group-wise comparisons within temperature treatment), different letters refer to significant differences (P < 0.05, Mann-Whitney U pairwise tests)).
Table 1. Aphid Feeding Behavior, Measured by 8-h Electrical Penetration Graph Recordings on the Natural Arabidopsis Accessions Col-0 and C24, a NIL of Col-0 with a C24 Introgression Covering the Region around SLI1, and the T-DNA Lines sli1-1 and sli1-2.
| Variable |
20°C |
26°C |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Col-0 | C24 | NIL | sli1-1 | sli1-2 | Col-0 | C24 | NIL | sli1-1 | sli1-2 | |
| Total duration nonprobing | 196 ± 30 | 118 ± 18* | 169 ± 27ns | 164 ± 15ns | 107 ± 22* | 141 ± 14 | 98.3 ± 13* | 129 ± 18ns | 55.2 ± 12*** | 119 ± 24ns |
| Total duration epidermis/mesophyll | 150 ± 21 | 169 ± 24ns | 154 ± 16ns | 121 ± 15ns | 91.5 ± 25ns | 181 ± 13 | 176 ± 16ns | 152 ± 19ns | 69.7 ± 9*** | 93.9 ± 16** |
| Number of probes <3 min | 26.8 ± 4.4 | 23.6 ± 3.5ns | 29.8 ± 5.2ns | 28.5 ± 4.2ns | 26.2 ± 9.2ns | 27.4 ± 3.9 | 29.9 ± 5ns | 27.1 ± 4.2ns | 8.4 ± 2.2*** | 21.5 ± 5.6ns |
| Total duration difficulties | 0 ± 0 | 0 ± 0ns | 0 ± 0ns | 18.3 ± 8.5* | 0 ± 0ns | 9.5 ± 6.9 | 2.6 ± 2ns | 3.2 ± 3.2ns | 1.4 ± 1.4ns | 11.8 ± 5.7ns |
| Number of difficulties | 0 ± 0 | 0 ± 0ns | 0 ± 0ns | 0.8 ± 0.3* | 0 ± 0ns | 0.2 ± 0.1 | 0.1 ± 0.1ns | 0.1 ± 0.1ns | 0 ± 0ns | 0.8 ± 0.4ns |
| Latency to 1st salivation | 156 ± 27 | 128 ± 17ns | 122 ± 18ns | 166 ± 26ns | 128 ± 22ns | 116 ± 22 | 83.4 ± 14ns | 99 ± 23ns | 102 ± 17ns | 179 ± 36ns |
| Total duration salivation | 6.4 ± 1.6 | 5.6 ± 0.8ns | 6.1 ± 1.2ns | 3.9 ± 0.7ns | 1.4 ± 0.3* | 9.4 ± 2.3 | 6.1 ± 0.9ns | 4.5 ± 0.9* | 1.8 ± 0.5*** | 1.3 ± 0.3*** |
| Number of salivations | 6.5 ± 1.2 | 8.8 ± 1.3ns | 8.6 ± 1.4ns | 5.4 ± 1ns | 1.8 ± 0.3* | 9.2 ± 0.9 | 11.1 ± 1.4* | 7.1 ± 0.9ns | 2.2 ± 0.3*** | 1.7 ± 0.4*** |
| Number of single salivations | 0.9 ± 0.4 | 0.2 ± 0.1ns | 0.5 ± 0.2ns | 0.2 ± 0.1ns | 0.2 ± 0.1ns | 0.6 ± 0.3 | 0.2 ± 0.2ns | 0.5 ± 0.2ns | 0.3 ± 0.1ns | 0.1 ± 0.1* |
| Mean duration salivation | 1 ± 0.2 | 0.7 ± 0.1ns | 0.7 ± 0.1ns | 0.8 ± 0.1ns | 0.8 ± 0.1ns | 1 ± 0.2 | 0.6 ± 0.1* | 0.6 ± 0.1* | 0.7 ± 0.1ns | 0.8 ± 0.1ns |
| % Salivation in phloem phase | 11.9 ± 4.9 | 4.3 ± 0.8ns | 8.2 ± 2.4ns | 3.1 ± 0.8ns | 2 ± 1.5** | 8.5 ± 1.5 | 3.6 ± 0.6* | 5.8 ± 2ns | 0.6 ± 0.2*** | 4.2 ± 2.4** |
| Latency to 1st phloem feeding | 161 ± 27 | 138 ± 19ns | 123 ± 18ns | 167 ± 26ns | 138 ± 28ns | 129 ± 23 | 83.9 ± 14ns | 114 ± 24ns | 110 ± 19ns | 186 ± 39ns |
| Total duration phloem feeding | 118 ± 34 | 187 ± 34ns | 151 ± 36ns | 173 ± 20ns | 280 ± 41* | 137 ± 20 | 198 ± 23* | 192 ± 35ns | 345 ± 18*** | 251.1 ± 41* |
| Total duration sustained feeding | 107 ± 34 | 169 ± 35ns | 132 ± 37ns | 166 ± 21* | 279 ± 41* | 119 ± 21 | 170 ± 25ns | 174 ± 36ns | 344 ± 18*** | 249 ± 42* |
| Number of phloem feedings | 5.2 ± 1 | 8.2 ± 1.2ns | 7.3 ± 1.3ns | 4.9 ± 0.9ns | 1.7 ± 0.3* | 7 ± 0.9 | 10 ± 1.2ns | 6.3 ± 1ns | 1.8 ± 0.2*** | 1.5 ± 0.3*** |
| Number of sustained feedings | 2.2 ± 2.2 | 4.4 ± 4.4** | 2.7 ± 2.7ns | 2.8 ± 2.8ns | 1.2 ± 1.2ns | 2.8 ± 2.8 | 4.3 ± 4.3* | 2.3 ± 2.3ns | 1.4 ± 1.4*** | 0.9 ± 0.9*** |
| Mean duration phloem feeding | 54.7 ± 38 | 33.8 ± 12ns | 30 ± 11ns | 62.6 ± 13ns | 232 ± 44** | 26 ± 5.7 | 39.5 ± 17ns | 45.2 ± 16ns | 254 ± 31*** | 222 ± 42** |
| Mean duration sustained feeding | 78 ± 40 | 42.2 ± 11ns | 47.8 ± 12ns | 108 ± 25ns | 270 ± 36*** | 50.6 ± 10 | 59 ± 18ns | 85.5 ± 17* | 298 ± 26*** | 273 ± 39*** |
| % Aphids phloem feeding | 84.6 | 100 ± 0ns | 100 ± 0ns | 100 | 92.3 | 100 | 100 | 100 | 100 | 93.8 |
| Total duration xylem ingestion | 9.8 ± 5.8 | 0 ± 0* | 0 ± 0* | 0 ± 0* | 0.5 ± 0.5ns | 1.9 ± 1.7 | 0 ± 0ns | 0 ± 0ns | 6.6 ± 3.1ns | 2.8 ± 2.8ns |
| Number of xylem ingestions | 0.5 ± 0.2 | 0 ± 0* | 0 ± 0* | 0 ± 0* | 0.1 ± 0.1ns | 0.1 ± 0.1 | 0 ± 0ns | 0 ± 0ns | 0.2 ± 0.1ns | 0.1 ± 0.1ns |
Time is shown in minutes. Mean ± se. Mann-Whitney U pairwise comparisons were performed between Col-0 and the plant lines C24, NIL, sli1-1, and sli1-2 for temperature treatments separately (*P < 0.05, **P < 0.01, and ***P < 0.001). Significant differences between 20 and 26°C within plant lines are shown in bold (Mann-Whitney U pairwise comparisons, P < 0.05). “Difficulties” involve penetration difficulties. “Sustained” involves events > 10 min. Latency was measured from the start of the recording. Single salivations were phloem salivations that were not directly followed or preceded by phloem ingestion. Between 12 and 20 biological replicates were measured (20°C: Col-0 n = 13, C24 n = 12, NIL n = 12, sli1-1 n = 16, sli1-2 n = 13; 26°C: Col-0 n = 20, C24 n = 14, NIL n = 14, sli1-1 n = 20, sli1-2 n = 16).
SLI1 Localizes at Sieve Element Margins and around Mitochondria
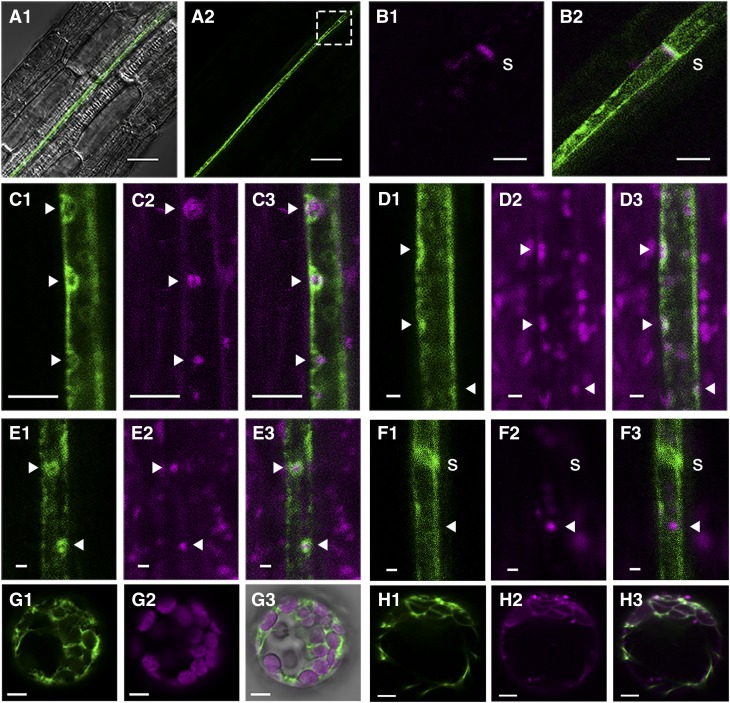
SLI1 is a member of the alpha-crystallin domain (ACD) superfamily, also referred to as the small heat shock-like or HSP20-like chaperone superfamily (superfamily code SSF49764), which includes 47 genes in Arabidopsis (Bondino et al., 2012). SLI1 is a homolog of RESTRICTED TOBACCO ETCH POTYVIRUS MOVEMENT2 (RTM2) (Bondino et al., 2012), which is required for the restriction of tobacco etch potyvirus transport through the sieve tubes via an unknown mechanism (Mahajan et al., 1998; Whitham et al., 1999, 2000; Chisholm et al., 2001; Cosson et al., 2010; Cayla et al., 2015). Both SLI1 and RTM2 have a transmembrane domain, which is rare among ACD proteins. RTM2 proteins localize to sieve tubes, where they line the sieve element margins (Chisholm et al., 2001; Cayla et al., 2015). To retrieve the protein localization of SLI1, fluorescent reporter lines were constructed with protein fusions expressed under the native promoter. Confocal microscopy was performed on seedling roots because of good nonintrusive imaging possibilities of the inner cell layers. SLI1 proteins localized to the vascular bundle (Figure 5A) and colocalized with aniline blue-stained sieve plates (Figure 5B), indicating that they were confined to sieve elements. SLI1 lined virtually the complete margin of mature sieve elements and sometimes aggregated into round or circular shapes, similar to the spherical RTM2 bodies observed by Cayla et al. (2015). To assess whether these spherical structures were associated with organelles, we used FM4-64 as a membrane-selective marker. FM4-64 dye is generally used to track endocytosis and vesicle trafficking, but will label different organelle membranes upon longer exposures (Bolte et al., 2004). After 3 h of staining, we observed small membrane compartments in the sieve tubes, most likely corresponding to vesicles, but no obvious colocalization with SLI1 (Supplemental Figure 2). However, when seedlings were incubated for 30 h, membrane compartments colocalized with the spherical SLI1 bodies (Figures 5C). Staining with MitoTracker Deep Red indicated that at least part of these structures involved mitochondria (Figure 5D), which was confirmed in seedlings with both pSLI1:EYFP:SLI1 and the mCherry construct of the mitochondria transit peptide ScCOX4 of CYTOCHROME C OXIDASE (Nelson et al., 2007) (Figure 5E). Colocalization around plastids was tested with the plastid marker tpCab:mCherry (Kim et al., 2013), containing the transit peptide of CHLOROPHYLL A/B BINDING PROTEIN. We did not observe any overlap between SLI1 and the plastid marker (Figure 5F), indicating that the spherical SLI1 bodies only associate around mitochondria. To see other putative organelle associations, we expressed SLI1 transiently in Arabidopsis protoplasts. Here, SLI1 formed a lose mesh in between chloroplasts (Figure 5G) and sometimes appeared to localize to the nucleus. Staining with ER Tracker Red indicated that SLI1 proteins partially overlapped with the endoplasmic reticulum (ER) (Figure 5H).
Figure 5.
Localization of SLI1 in Arabidopsis.
(A1) to (B2) Confocal images of pSLI1:EYFP:SLI1 (green) in seedling roots ([A1] and [A2]), with details in the boxed region showing aniline blue staining (magenta) of a sieve plate (B1) and merged signals of aniline blue and SLI1 (B2).
(C) FM4-64 staining (30 h incubation; 1, SLI1; 2, FM4-64; 3, merged).
(D) MitoTracker staining (1, SLI1; 2, MitoTracker Deep Red; 3, merged).
(E) Cotransformation with the mitochondria marker ScCOX4 (Nelson et al., 2007) (1, SLI1; 2, ScCOX4:mCherry; 3, merged).
(F) Cotransformation with the plastid marker tpCab (Kim et al., 2013) (1, SLI1; 2, tpCab:mCherry; 3, merged).
(G) Arabidopsis protoplast with transient expression of 35S:EYFP:SLI1 (1, SLI1; 2, chloroplasts; 3, merged).
(H) Protoplast stained with ER Tracker Red (1, SLI1; 2, ER Tracker; 3, merged).
Images represent consistent results in at least two independent transgenic lines. s, sieve plate; solid arrows, phloem organelle location. Bars = 25 µm in (A), 5 µm in (B) and (C), 1 µm in (D) to (F), 5 µm in (G) and (H).
Restricted Phloem Sap Ingestion
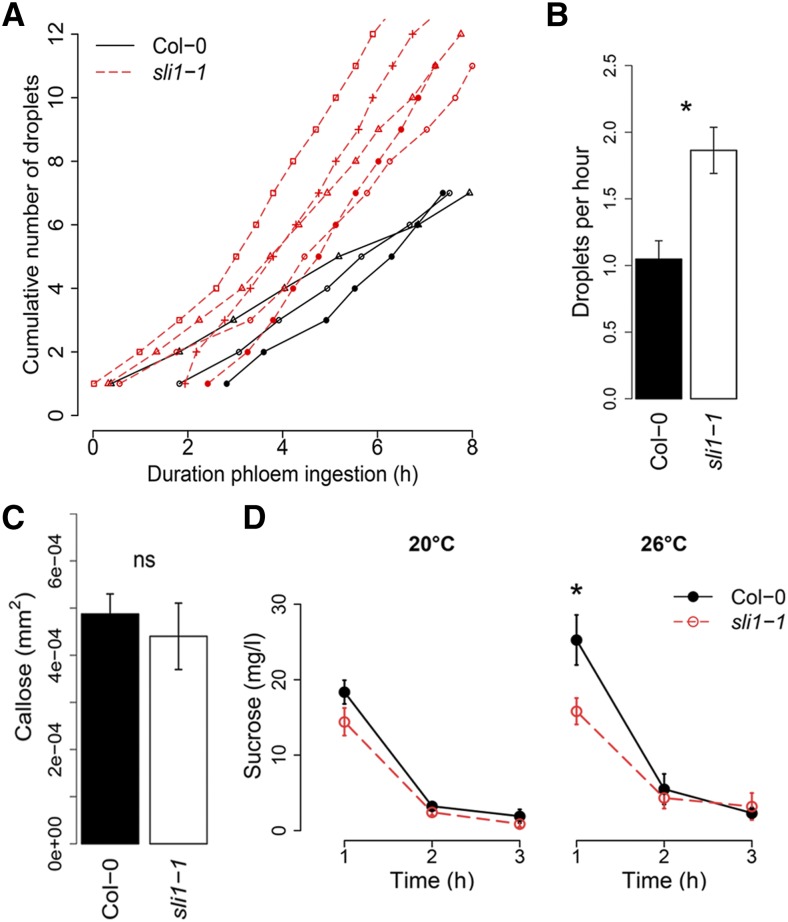
Considering that SLI1 is associated with sieve tube membranes and organelles, we tested its effect on sap ingestion by aphids. The honeydew droplet excretion rate, which is highly correlated with the sap ingestion rate (Tjallingii, 1995), was quantified simultaneously with feeding behavior. During hour-long feeding events, we observed that honeydew droplets were excreted at a significantly lower rate on wild-type plants compared with sli1-1 plants (Figures 6A and 6B; Supplemental Data Set 2). Considering that aphids are passive feeders, this reduced ingestion rate could be caused by the obstruction of sieve tubes. As SLI1 shows coexpression with three genes involved in callose formation on sieve plates, i.e., SUCROSE SYNTHASE5 (SUS5), SUS6, and GLUCAN SYNTHASE-LIKE7 (GSL7) (Barratt et al., 2009, 2011) (Supplemental Table 2), we tested whether SLI1 was involved in callose-mediated plugging of sieve plates. However, aniline blue staining of the main leaf vein did not reveal significant differences in callose accumulation between the wild type and sli1-1 (Figure 6C; Supplemental Data Set 2). In addition, we assessed the phloem exudation rates of excised leaves. Sucrose was used as an estimate of exudate rate, assuming that the sucrose concentration of the sap was similar between plant lines. At 20°C, no differences were observed. At 26°C, wild-type exudates contained higher sucrose levels during the first hour (Figure 6D; Supplemental Data Set 2), indicative of a higher exudation rate, whereas a lower rate was expected if sieve tubes were occluded. Overall, these observations did not support the hypothesis that SLI1-mediated restriction of phloem ingestion was caused by sieve tube obstruction.
Figure 6.
Honeydew Excretion, Callose, and Phloem Sap Exudation.
(A) Honeydew droplet excretion by M. persicae aphids on wild-type and sli1-1 plants. Droplets were collected and counted during events of continuous phloem ingestion. Every line represents an aphid, and each symbol represents a honeydew droplet.
(B) Mean honeydew excretion rate. P = 0.018, one-way ANOVA, plants were grown at 26°C, Col-0 n = 3, sli1-1 n = 5; data represent mean ± se.
(C) Callose content of the leaf mid vein according to quantification with aniline blue. P > 0.05, n = 6, one-way ANOVA; plants were grown at 26°C.
(D) Sucrose content of phloem exudates, collected in a 3-h time course at 20°C (P > 0.05, n = 4, one-way ANOVA) and 26°C (*P < 0.05, n = 4, one-way ANOVA, corrected for leaf fresh weight, data represent mean ± se).
Increased Plant Fitness during Moderate Heat Stress
In view of the temperature-dependent phenotype of SLI1, we quantified several plant fitness-related variables at growth conditions of 20°C and moderate heat stress of 26°C. Relative to the wild type, sli1-1 displayed a stunted growth of the inflorescence in the first phase after bolting and developed fewer and shorter siliques at 26°C (Table 2). Nonstratified sli1-1 seeds that had been collected from plants grown at 26°C displayed a higher germination rate than wild-type seeds (Supplemental Figure 3 and Supplemental Data Set 2), indicative of a reduced dormancy. In addition, the correlation between climate conditions within Europe and natural variation in SLI1 was calculated. When taking into account the population structure, the SLI1 haplotype distribution did not reveal any associations with the 52 climate variables that were tested (Table 3; Supplemental Table 3). However, we found that the Col-0 haplotype that confers enhanced resistance to aphids is biased toward the east of Europe (Supplemental Figure 4). As the Eurasian Arabidopsis population exhibits isolation by distance (Platt et al., 2010) and correction for kinship might eliminate geographical patterns, we also tested the climate variables without correction for population structure and found 12 significant climate associations (Table 3; Supplemental Table 3). The majority involved temperature-related factors and indicated that the resistant SLI1 haplotype occurred more often in regions with large seasonal temperature fluctuations.
Table 2. Rosette, Inflorescence, and Silique Development of Wild-Type and sli1-1 Plants.
| Variable | 20°C |
26°C |
||
|---|---|---|---|---|
| Col-0 | sli1-1 | Col-0 | sli1-1 | |
| Number of leaves (15 d) | 6.2 ± 0.1 | 6.1 ± 0.1ns | 7.8 ± 0.1 | 7.6 ± 0.1ns |
| Number of leaves (35 d) | 21.8 ± 0.8 | 20.9 ± 0.7ns | 30.2 ± 0.6 | 29.4 ± 0.4ns |
| Rosette fresh weight (g) | 0.4 ± 0 | 0.3 ± 0ns | 1.3 ± 0 | 1.2 ± 0.1ns |
| Flowering time (d) | 51.4 ± 0.4 | 51 ± 0.6ns | 52.8 ± 0 | 54 ± 1ns |
| Inflorescence stem height (cm, 58 d) | 5 ± 1.6 | 6.7 ± 2.7ns | 8.9 ± 1.9 | 2.3 ± 0.9* |
| Inflorescence stem height (cm, 60 d) | 10.8 ± 1.8 | 12.1 ± 2.8ns | 16.1 ± 1.5 | 7.6 ± 2.2* |
| Inflorescence stem height (cm, 63 d) | 22 ± 1.5 | 23 ± 2.1ns | 23.3 ± 1.2 | 14.3 ± 3.5* |
| Inflorescence stem height (cm, final) | 32.7 ± 0.8 | 31.5 ± 1.5ns | 41.5 ± 1 | 41.2 ± 1.7ns |
| Number of primary side branches | 6.8 ± 0.5 | 7 ± 0.4ns | 8.5 ± 0.4 | 8.2 ± 0.3ns |
| Number of secondary side branches | 0 ± 0 | 5.5 ± 2.6* | 3.8 ± 0.8 | 13.2 ± 4.2ns |
| Number of siliques main stem | 32.3 ± 1.9 | 33.3 ± 1.4ns | 40.2 ± 1.5 | 42.5 ± 2.1ns |
| Number of siliques side branches | 97.2 ± 8.9 | 81.8 ± 7.6ns | 110.7 ± 4.8 | 64 ± 14.3* |
| Number of siliques (total) | 129.5 ± 8.6 | 115.2 ± 8.9ns | 150.8 ± 5.7 | 106.5 ± 14.4* |
| Silique length (cm) | 1.3 ± 0 | 1.3 ± 0ns | 1 ± 0 | 0.9 ± 0* |
Means ± se; *P < 0.05, **P < 0.01, and ***P < 0.001 (Mann-Whitney U pairwise comparisons of sli1-1 with Col-0 within temperature treatments). Significant differences between 20 and 26°C within the plant lines Col-0 and sli1-1 are shown in bold (Mann-Whitney U pairwise comparisons, P < 0.05). Number of leaves at 15 d, n = 13; number of leaves at 35 d and fresh weight, n = 8; flowering time, inflorescence stem height, number of branches, siliques, and silique length, n = 5 to 6.
Table 3. Climate Variables with a Significant Haplotype Effect for the SNP on Chromosome 3, Position 3338114, in the ACD of SLI1.
| Climate Variable | Source | Col-0 Allele (59%) | Rare Allele (41%) | Mixed Model | M-W |
|---|---|---|---|---|---|
| Temp. seasonality (sd) (°C) | worldclim.org | 6.1 ± 0.1 | 5.5 ± 0.2 | ns | *** |
| Temp. annual range (°C) | worldclim.org | 24.5 ± 0.4 | 22.6 ± 0.6 | ns | *** |
| Isothermality (diurnal/annual range) | worldclim.org | 3.0 ± 0 | 3.2 ± 0.1 | ns | *** |
| Mean temp. wettest quarter (°C) | worldclim.org | 13.5 ± 0.4 | 10.8 ± 0.5 | ns | *** |
| Mean temp. coldest quarter (°C) | worldclim.org | 0.6 ± 0.2 | 2.3 ± 0.3 | * | *** |
| Mean temp. driest quarter (°C) | worldclim.org | 3.6 ± 0.4 | 7.3 ± 0.6 | ** | *** |
| Min temp. coldest month (°C) | worldclim.org | −2.7 ± 0.3 | −1 ± 0.3 | ns | *** |
| Mean monthly temp. Nov.–Feb. (°C) | nelson.wisc.edu/sage | 1.4 ± 0.2 | 3 ± 0.3 | * | *** |
| Number of frost days 1980–2010 | cgiar-csi.org | 2409 ± 46 | 2113 ± 72 | ns | *** |
| Precipitation warmest quarter (mm) | worldclim.org | 209.9 ± 5 | 183.6 ± 6 | * | *** |
| Relative humidity in May (%) | cru.uea.ac.uk/cru/data | 72.6 ± 0.3 | 74.6 ± 0.4 | ns | *** |
| Vegetation index MOD13C2 in March | modis.gsfc.nasa.gov | 7.0e7 ± 2e6 | 8.0e7 ± 2e6 | ns | *** |
In total, 52 climate variables were tested (Supplemental Table 3); the 12 variables with P < 0.001 (Mann-Whitney U test) are shown. Means ± se. M-W, Mann-Whitney U test; Mixed Model, mixed model with correction for kinship [*P < 0.05, ** P < 0.01, and ***P < 0.001; below the Bonferroni threshold (0.05/52 = 0.001)].
DISCUSSION
From Aphid Behavior to a Novel Quantitative Resistance Gene
This study identifies and validates a novel, quantitative aphid resistance gene by GWA mapping. Phenotyping is often a major bottleneck for elucidating the quantitative genetics of plant resistance to herbivores because of time-consuming measurements and multigenic mechanisms, involving numerous small-effect genes (Kloth et al., 2012; Kliebenstein, 2014). In this study, a high-throughput video-tracking platform was instrumental in acquiring replicated data of 350 plant lines with reduced trait complexity. The compromises we made in phenotypic accuracy (automated annotation of video footage instead of high-precision EPG recording) and representativeness of the natural system (leaf discs instead of whole plants) did not hinder the identification of this causal gene. GWA mapping of the time that aphids spent on short probes revealed a strong association with polymorphisms in the small heat shock-like gene SLI1. Surprisingly, the SNPs of this phloem protein were associated with short probes, which are located in the epidermis and mesophyll, and not long probes, which are associated with phloem feeding. However, this discrepancy can be explained by the fact that a phloem-based mechanism indirectly affects the epidermis/mesophyll phase. Aphids that fail in sustained phloem feeding can spend more time on probing activities in other tissues. The 2-fold higher SLI1 expression in Col-0 in comparison to C24 might indicate that SNPs in the promoter region are causal for the increased salivation on the Col-0 haplotype, although effects of other polymorphisms cannot be excluded.
SLI1 Restricts Phloem Feeding but Does Not Occlude Sieve Tubes
The experiments with near-isogenic and mutant lines showed that SLI1 confers partial resistance to aphids. All the experiments were performed on plants in the vegetative state, which were unaffected in their development and growth (Table 2). The haplotype effect on feeding behavior did not result in differences in aphid population size, possibly because of a marginal effect of size of the SNPs and adaptation of the aphids. However, the impact on loss-of-function mutants was substantial. Aphids produced almost twice as many offspring on sli1-1 and sli1-2 than on wild-type plants. SLI1 expression was not affected by aphids, but was temperature-dependent and increased 2-fold at 26°C compared with 20°C. The effects of sli1-1 on aphid feeding behavior were largest at 26°C. The sli1-2 mutant also showed relatively large effects at 20°C for an unknown reason. SLI1 had a particularly striking effect on the phloem phase. Feeding events lasted on average 40 to 50 min on wild-type plants, compared with 4 h on loss-of-function mutants (Figure 4B). The sap ingestion rate was almost 2-fold lower on wild-type plants, and before sap ingestion could start, aphids needed to salivate longer (Table 1; proportion of salivation during phloem phase). The watery saliva that aphids secrete in the sieve tube contains many calcium binding components, which could counteract occlusion mechanisms (Tjallingii and Hogen Esch, 1993; Will et al., 2007). All three elements, i.e., lower ingestion rates, loss of sustained feeding, and longer salivation, point toward an occlusion of either the sieve tube or the aphid food canal. However, we observed neither callose accumulation, nor reduced phloem exudation, which are both markers of sieve tube occlusion. As those traits were measured on clean plants, the possibility that they differentiate during or after aphid infestation cannot be excluded. Yet, we consider SLI1-mediated callose deposition not very likely, since it is not supported by a sieve plate- or plasmodesmata-specific localization. SLI1 was found along the whole sieve element margin and around mitochondria. In addition, our phenotypic data did not support SLI1 being involved in sieve tube occlusion. On the contrary, plants with functional SLI1 proteins displayed higher exudation rates than mutants (Figure 6D), faster elongation of the inflorescence stem, and more seed set during heat stress (Table 2).
Is SLI1 a Clamp Protein?
Fluorescent reporter lines revealed that SLI1 proteins are confined to sieve element margins (Figures 5A and 5B). Sieve elements are highly specialized cells without a nucleus and largely empty for the purpose of phloem sap transport under high turgor pressure (Windt et al., 2006; Knoblauch and Oparka, 2012). SLI1 is localized in the region where there is a small, peripheral strip of cytoplasm, which is enclosed by the plasma membrane and the sieve element reticulum, consisting of stacked or fenestrated ER (Sjölund and Shih, 1983). This parietal layer contains other elements, such as filamentous protein structures, amorphous ground matrices, phloem plastids, and individual or clustered mitochondria, which are often partially covered by ER (White, 2013). SLI1 forms both straight and spherical bodies along the parietal layer. With fluorescent dyes and genetic markers, we could show that the spherical SLI1 bodies localize around the periphery of mitochondria (Figures 5C to 5E). SLI1 might require one or more phloem-specific factors for its association with mitochondria, possibly via a comparable mechanism as its homolog, RTM2. Sofer et al. (2017) showed in a transient expression assay that RTM2 requires BRANCHED-CHAIN AMINOTRANSFERASE4 (BCAT4) for its recruitment to the chloroplast periphery. Without BCAT4, RTM2 formed small aggregates in the cytoplasm of Nicotiana benthamiana epidermal cells. The SLI1 protein structure (as well as the RTM2 structure) contains an ACD chaperone domain on the N terminus and a transmembrane domain on the C terminus (Figure 2C). Possibly, SLI1 attaches to proteins, such as mitochondrial envelope proteins, with its chaperone domain and to a membrane, such as the plasma membrane and/or ER, with its transmembrane domain. Remarkably, the observed SLI1 localization along the sieve tube margin and around mitochondria is strongly reminiscent of the clamp-like structures, which are described in transmission electron microscopy studies. In Arabidopsis a “halo” of 34.5 nm is seen around mitochondria to which often protein filaments and membranes attach (Froelich et al., 2011). Arabidopsis phloem plastids do generally not have a halo of clamp-like structures, but only a stable attachment to the margin (Knoblauch and Van Bel, 1998), which is consistent with our observation that the spherical SLI1 bodies are not associated with plastids (Figure 5F). In tomato and fava bean (Vicia faba), similar clamp-like structures have been observed between ER membranes, between ER and plastids, and between ER and mitochondria and are thought to anchor organelles to the sieve element margin (Ehlers et al., 2000). Further ultrastructural studies are necessary to elucidate the membrane specificity of SLI1 and its putative involvement with these clamp-like structures.
The Role of the Parietal Layer in Restricted Sap Uptake
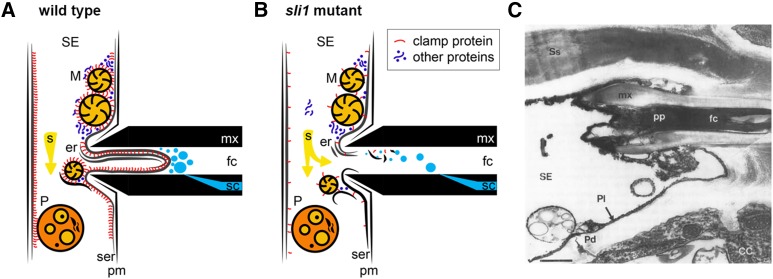
Coagulation of dispersed proteins is considered a major cause for occlusion of the aphid food canal (Tjallingii, 2006; Will et al., 2007; Will and Vilcinskas, 2015). Surprisingly, the role of nondispersed factors in the parietal sieve element layer has hardly received any attention in this context. The sieve element plasma membrane is covered by a thin layer of cytoplasm with sieve element reticulum membranes, organelles, and protein layers (Sjölund et al., 1983; White, 2013). After puncturing the plasma membrane, aphids need to breach this layer to arrive at the sieve element lumen for passive uptake of phloem sap. Aphids salivate for ∼30 to 90 s before phloem feeding starts, and their watery saliva is rich in proteases and Ca2+ binding proteins. These proteins are considered to absorb the Ca2+ influx after penetration of the plasma membrane and prevent the expansion of forisomes and possibly the coagulation of other phloem proteins (Will et al., 2007; van Bel and Will, 2016). We would expect these effectors to play an important role in the degradation of the parietal layer too, particularly in places where it contains organelles and a thick protein matrix. These regions are both membrane-dense and protein- and calcium-rich, since many proteins are synthesized and/or stored in the ER, plastids, and mitochondria, and since mitochondria generally play a role in calcium regulation (Behnke, 1981; Sparkes et al., 2009; Kühlbrandt, 2015). One hypothesis is that SLI1 is a membrane-bound scaffold for other proteins, including mitochondrial envelope proteins. It would then be involved in anchoring organelles and phloem proteins and would increase the firmness and thickness of the parietal layer and decrease the accessible area for aphid effectors. Consequently, aphids would have to salivate longer in order to breach the parietal layer and reach the sieve element lumen for sap ingestion. This would explain the prolonged salivation period in the Col-0 haplotype (Figure 4A). The high number of prematurely interrupted feeding events and lower ingestion rate in the wild type compared with sli1 mutants are indicative of an even greater, unsolvable problem for aphids (Figure 4B). Sieve tubes are under high turgor pressure (Windt et al., 2006; Knoblauch and Oparka, 2012), and when the parietal layer is not degraded well or fast enough, it may be pushed into the food canal and thereby reduce the phloem ingestion rate or force the aphid to retract their stylets from the sieve element (Figures 7A and 7B). Such an occlusion would resemble the plugged A. fabae stylets in V. faba sieve elements as observed by Tjallingii and Hogen Esch (1993) (Figure 7C), although the possibility cannot be excluded that these images reflect artifacts of the stylectomy and fixation procedure.
Figure 7.
Working Model of SLI1-Mediated Resistance to Aphids.
(A) In wild-type plants, the plasma membrane (pm) is mechanically punctured by the stylets (mx), but the parietal layer remains intact due to SLI1 clamp proteins, which anchor organelles and proteins to a parietal membrane. The high turgor pressure in the sieve element pushes the parietal layer in the aphid food canal (fc) where it occludes the entrance.
(B) The sli1 mutant only has a few other clamp proteins. Consequently, organelles and proteins are loosely attached to the sieve element reticulum (ser). After plasma membrane puncture, effectors in the aphid saliva successfully disassemble the parietal layer and break down the fragmentized membrane, organelle, and protein components. The aphid food canal will not be occluded and passive uptake of phloem sap can take place.
(C) Transmission electron micrograph of an A. fabae stylet in a V. faba sieve element (Tjallingii and Hogen Esch, 1993; reproduced with the authors’ permission). The aphid food canal is filled with electron-dense material, presumed to be coagulating proteins, resembling the membrane structures in (A).
Bar = 0.5 µm. er, stacked endoplasmic reticulum cisternae; M, mitochondria; P, plastid; mx, maxillary stylets; pm/Pl, plasma membrane; sc, salivary canal; s, phloem sap; Pd, plasmodesma; cc, companion cell; Ss, salivary sheet material; pp, presumed phloem proteins.
The Role of SLI1 during Heat Stress
Temperature played an important role in the functionality of SLI1. At high temperature, SLI1 expression was induced (Figure 3) and phenotypic effects on aphids were substantially larger (Figure 4, Table 1). In addition, the SLI1 natural haplotype distribution is correlated with temperature-related climate factors. Several plant fitness parameters confirmed that SLI1 increases resistance to heat stress. The reduced seed dormancy in loss-of-function mutants (Supplemental Figure 3) suggests that sli1 mutants are more sensitive to high temperatures than Col-0 (He et al., 2014). Vegetative growth did not show any irregularities, but sli1-1 displayed a stunted growth of the inflorescence stem in the first weeks after bolting at 26°C and suffered from reduced seed set (Table 2). Carbohydrate allocation is crucial during this stage, and compromised allocation could be a reason for the retarded development of inflorescences and siliques. Whether or not protein bodies can obstruct the sap translocation rate is still debatable (Furch et al., 2010; Knoblauch et al., 2014). However, if SLI1 is involved in anchoring organelles and proteins to the sieve tube margin, it would be worth studying its effects on the volumetric flow rate in sieve tubes during developmental stages with high sink demands, particularly in water-deficient conditions, such as heat or drought stress.
METHODS
Plant Material
A collection of 350 natural accessions of Arabidopsis thaliana was obtained from the ABRC Stock Center (Baxter et al., 2010). This set was selected in a previous study to represent most intraspecific genetic variation and minimal redundancy (Platt et al., 2010) and was genotyped for ∼214,000 SNPs with Atsnptile1 arrays (Atwell et al., 2010; Li et al., 2010; Horton et al., 2012). The T-DNA lines SALK_027475 (sli1-1) and SAIL_1269_C01 (sli1-2), both in the Col-0 background, were obtained from the European Arabidopsis Stock Centre (NASC). Homozygous T-DNA plants were selected by PCR (Supplemental Table 4) and harvested for seeds for subsequent experiments. The location of the T-DNA inserts was validated via sequencing and At3g10670 and At3g10680 expression was measured by RT-qPCR (Supplemental Figure 5). The natural accession C24 and the near-isogenic Col-0 line with a C24 introgression on chromosome 3 between positions 276917 and 3463232 (line 94/7) were obtained from the Leibniz Institute of Plant Genetics and Crop Research (IPK), Gatersleben, Germany (Törjék et al., 2008).
Plant Growth and Insect Rearing
Arabidopsis seeds were cold stratified for 72 h at 4°C before they were sown into pots (5 cm diameter) with pasteurized (4 h at 80°C) Arabidopsis potting soil (Lentse Potgrond) in a climate room at 26 ± 1°C, 50 to 70% relative humidity, an 8-h:16-h light (L):dark (D) photoperiod, and a light intensity of 200 μmol m−2 s−1 (Philips TL5 HO 54W/840). To assess temperature effects, plants were also grown in a climate cabinet at 20 ± 1°C, 60 to 70% relative humidity, an 8-h:16-h L:D photoperiod, and light intensity of 120 μmol m−2 s−1 (Philips TL5 HO F54T5/841). For assessment of light intensity effects on gene expression, light intensity in the climate cabinet was changed to 70 μmol m−2 s−1 (Philips TL5 HO F54T5/841). Green peach aphids, Myzus persicae, were reared on radish (Raphanus sativus) at 19°C, 50 to 70% relative humidity, and a 8-h:16-h L:D photoperiod. Only wingless aphids have been used in experiments.
Automated Video Tracking
Aphid behavior was tracked on 350 natural accessions of Arabidopsis according to the methodology of Kloth et al. (2015). One biological replicate consisted of a leaf disc of a unique plant. An aphid was introduced into a well of a 96-well plate containing a leaf disc of 6-mm diameter, abaxial side up, on 1% agar substrate. Wells were covered with cling film, to avoid aphid escape, and 20 aphids were recorded on 20 different accessions simultaneously with a camera mounted above the plate, in a climate-controlled room at 22 ± 1°C. EthoVision XT 8.5 video-tracking and analysis software (Noldus Information Technology) was used for automated acquisition of aphid position and velocity. The number and duration of probes were subsequently calculated with the statistical computing program R (R Core Team, 2013). Leaf discs (one disc per plant) were punched from 4- to 5-week-old Arabidopsis plants, just below the tip of leaf 8–14 (Mousavi et al., 2013). Aphid behavior was recorded for 85 min, starting immediately after inoculation with the aphids. The video-tracking assay was performed in an incomplete block design with each complete replicate consisting of 18 blocks of 20 accessions. Sixty plants were screened each day across three blocks, and one replicate of the complete Hapmap collection was acquired in 6 d. An alpha design was generated with Gendex (http://designcomputing.net/gendex/) to assign accessions to each block.
GWA Mapping
GWA mapping was performed on the total time spent on short probes (<3 min, data arcsine transformed) with scan_GLS (Kruijer et al., 2015). Asreml (VSN International) was used to apply a mixed model to correct for population structure, which was estimated with a kinship matrix including all SNPs. SNPs with a minor allele frequency of below 0.05 were excluded from analysis. Blocks and replicates were included in the model as covariates. Generalized heritability was calculated according to Oakey et al. (2006).
LD, Protein Structure, and Coexpression
Linkage disequilibrium (LD) between At3g10680 and other genes was taken into account for polymorphisms in a 150-kb window around the highest SNP, with an LD of 0.3 as the lower threshold. LD was calculated using the resequenced data of more than 500 Arabidopsis accessions from the 1001 Genomes Project (www.1001genomes.org; Cao et al., 2011) using the SNP LD tool as described by Bac-Molenaar et al. (2015). In total, 597 coexpressed genes of At3g10680 were identified from three independent coexpression databases: (1) top 300 genes of Atted-II version c4.1 (Obayashi et al., 2014), (2) top 100 genes of BAR Expression Angler Abiotic Stress, Pathogen, and Extended Tissue Compendium (Toufighi et al., 2005), and (3) the top 100 genes of Genevestigator’s Pertubations data set (Zimmermann et al., 2004).
RT-qPCR
Two leaves, with leaf numbers varying between 8 and 14 (Mousavi et al., 2013), per 4- to 5-week-old Arabidopsis plant were harvested between 12 and 3 pm. Samples were immediately frozen in liquid nitrogen and stored at –80°C until processing. RNA was isolated from homogenized leaf material with an InviTrap Spin Plant RNA kit and treated with Ambion TURBO DNA-free according to the manufacturer’s instructions. RNA was quantified with a NanoDrop ND-1000 spectrophotometer, and integrity was assessed with electrophoresis in a 1% (w/v) agarose gel stained with ethidium bromide. DNA-free RNA was converted into cDNA using the Bio-Rad iScript cDNA synthesis kit. RT-qPCR was performed on a Bio-Rad IQ 5 system using SYBR Green. For each primer combination (Supplemental Table 4), RT-qPCR products were sequenced to validate the region of amplification. For determination of splice variants, cDNA bands were isolated from the gel, purified with a Bioké PCR clean-up kit, and sequenced. To test for natural variation in SLI1 expression, leaves were sampled of Col-0 and C24 plants grown at 26°C. To test the effects of light intensity, samples were collected at 70 μmol m−2 s−1 (20°C; Philips TL5 HO F54T5/841 lamps) and 120 μmol m−2 s−1 (20°C; Philips TL5 HO F54T5/841 lamps). For temperature effects, samples were collected from plants grown at 20°C and 26°C. To test induction by aphids, samples with and without aphids were collected at 26°C. Fifteen adult M. persicae aphids were placed on each leaf, and the leaf was enclosed in a Petri dish with an indentation for the petiole, which did not allow aphids to escape. As a control, leaves without aphids were placed in a similar Petri dish. Samples were harvested 6 h after infestation. In all RT-qPCR assays, each biological replicate consisted of leaf material from one unique plant.
Plasmid Construction and Transformations
A 1063-bp promoter region of SLI1 and the coding sequence including the intron (1573 bp) were amplified (see primers in Supplemental Table 4). After SacI and SpeI digestion, the promoter was cloned into the SacI and SpeI-digested vector pK7WGY2 (https://gateway.psb.ugent.be) (Karimi et al., 2005). The coding sequence with intron was cloned into pENTR/D-TOPO (https://www.thermofisher.com/order/catalog/product/K240020), resulting in the plasmid pENTR/D-TOPO:35S:gSLI1. Via LR clonase recombination (https://www.thermofisher.com/order/catalog/product/11791100?ICID=search-product), these two vectors were recombined, resulting in the plasmid pK7WGY2:pSLI1:EYFP:gSLI1. The Agrobacterium tumefaciens strain Agl0 was transformed with pK7WGY2:pSLI1:EYFP:gSLI1 (kanamycin resistance) and used to transform Col-0 by floral dip (Clough and Bent, 1998). Protoplasts were made from Col-0 leaves according to Yoo et al. (2007). The pENTR/D-TOPO:35S:gSLI1 construct was used for transformation of protoplasts with PEG solution according to the methodology described by Yoo et al. (2007). The mitochondria mCherry marker mt-rb CD3-992 (Nelson et al., 2007), containing the first 29 amino acids of yeast (Saccharomyces cerevisiae) cytochrome c oxidase IV (ScCOX4), was obtained from the ABRC. The plastid construct tpCab:mCherry (transit peptide of chlorophyll a/b binding protein) was kindly provided by Inhwan Wang (Pohang University of Science and Technology, Korea; Kim et al., 2013). pSLI1:EYFP:SLI1 lines were transformed with organelle markers by floral dip (Clough and Bent, 1998).
Microscopy
Fluorescence signals were inspected in 7-d-old seedlings with a Zeiss LSM 780 spectral confocal laser scanning microscope. Seedlings were grown on agar plates (1% agar and 0.5× Murashige Skoog [MS] medium) at 22°C and 8-h:16-h L:D photoperiod. In case of staining, seedlings were incubated at room temperature in either 0.1% aniline blue (VWR Chemicals) sodium phosphate solution (70 mM, pH 9) for 1 h, 50 µM FM4-64 (Thermo Fisher Scientific) 0.5× MS solution for 30 h, or 1 µM Mitotracker Deep Red (Thermo Fisher Scientific) 0.5× MS solution for 24 h. Protoplasts were stained in 1 µM ER Tracker Red (Thermo Fisher Scientific) and incubated for 15 min at room temperature. For confocal microscopy, seedlings were mounted in 0.5× MS liquid medium using a spacer of double-sided tape between the slide and cover slip. EYFP was detected using a 514-nm laser and 517- to 598-nm emission filter; aniline blue with a 405-nm laser and 420- to 500-nm emission filter; FM4-64 with a 514-nm laser and 620- to 710-nm emission filter; ER Tracker Red and mCherry with a 561-nm laser and 590- to 640-nm emission filter; and Mitotracker Deep Red with a 633-nm laser and 650- to 760-nm emission filter. Callose depositions in the leaf veins were quantified with a fluorescence microscope (Nikon Instruments Europe; 100×, DM400 filter). One excised leaf of intermediate age per 4.5-week-old plant was placed in 96% ethanol overnight, incubated in sodium phosphate buffer (70 mM, pH 9) for 1 h, and immersed in 0.05% aniline blue (VWR Chemicals) solution of sodium phosphate buffer (70 mM, pH 9) for 24 h. One biological sample consisted of a leaf of a unique plant, for which the complete mid-vein from leaf base to top was captured in ∼30 images. Fluorescent areas were assessed with ImageJ (Schneider et al., 2012), counting all particles of more than five pixels using the threshold adjustment method “Triangle.”
EPG Recording
Feeding behavior of M. persicae aphids was measured with EPG recording on 4- to 5-week-old plants, using direct currents according to the methodology of ten Broeke et al. (2013). To adjust the radish-reared aphids to Arabidopsis, aphids were transferred to Col-0 Arabidopsis plants 24 h before the experiments. EPG recording was performed at 22 ± 2°C and a light intensity of 120 μmol m−2 s−1 (Philips TL5 HO 39W/840). For each biological replicate, one unique plant and aphid were used. An electrode was inserted in the potting soil and a thin gold wire of 1.5 cm was gently attached to the dorsum of an adult, wingless aphid with silver glue. The electrical circuit was completed when the aphid’s piercing-sucking stylet mouthparts penetrated the plant cuticle. Electrical signals associated with stylet activities were recorded and annotated with EPG Stylet+ software and further processed in R (Tjallingii, 1988; R Core Team, 2013) (http://www.epgsystems.eu).
Aphid Population Development
To assess aphid developmental rate and population size, each 3.5-week-old Arabidopsis plant was infested with one M. persicae neonate of 0 to 24 h old and placed in a climate room at 26 ± 1°C, 50 to 70% relative humidity, an 8-h:16-h L:D photoperiod, and 200 μmol m−2 s−1 light intensity (Philips TL5 HO 54W/840). A soap-diluted water barrier (2 ± 1 mL l−1 of Piek dish detergent; Van Dam Bodegraven) prevented aphids from moving between plants. None of the aphids developed wings. From day 7 onwards, occurrence of the first offspring was checked twice per day using 5× magnification glasses. The number of aphids per plant was counted 14 d after infestation.
Honeydew Droplet Collection
Aphid excretions were collected with honeydew clocks according to Tjallingii (1995). Honeydew clocks consisted of a 12-h quartz clockwork (Hechinger) with a rotating plastic disc (20-cm diameter) attached. Honeydew droplets were collected on a 2-cm-wide thin-layer chromatography (TLC) paper strip that completely covered the rear of the rotating disc. To visualize the droplets via amino acid staining, TLC paper was treated with ninhydrin (Acros organics ninhydrin spray, 0.5% 1-butanol solvent) and dried before the experiment. To adjust the radish-reared aphids to Arabidopsis, aphids were transferred to Col-0 Arabidopsis plants 24 h before the experiments. Adult, wingless aphids were positioned on the abaxial side of the leaves of clean plants. Each biological replicate consisted of one unique plant and aphid. Honeydew clocks were placed 1 ± 0.5 cm below the aphids. Honeydew collection and EPG recording were performed simultaneously over a 12-h period at 22 ± 2°C and light intensity of 120 μmol m−2 s−1 (Philips TL5 HO 39W/840). Honeydew excretion rates were calculated over 8 h of continuous phloem ingestion.
Phloem Exudation
Phloem exudates were collected according to Tetyuk et al. (2013). For each sample, 15 leaves were pooled from three 5.5-week-old plants grown at 20°C or 26°C. To chelate free calcium ions and prevent the sealing of the phloem, the petioles of the leaves were submerged in one reaction tube containing 1.4 mL 20 mM K2-EDTA solution. The tubes were placed at 20 ± 2°C or 26 ± 2°C in a closed, transparent container, which was lined with wet filter paper to reduce transpiration. After 1 h of incubation in the EDTA, the petioles were washed with distilled, deionized Millipore water to remove the EDTA. Immediately, the 15 leaves were transferred into a tube with the petioles submerged in 1.4 mL autoclaved distilled, deionized Millipore water to collect phloem exudates. The tubes were returned to the closed, transparent container at 20 ± 2°C or 26 ± 2°C. Exudates were collected for the first, second, and third hour separately, by transferring the 15 leaves each hour into a fresh tube of water. Samples were immediately frozen in liquid nitrogen, and stored at –80°C. Sucrose, glucose, and fructose content were measured by analyzing 200 μL exudate using HPLC (Dionex ICS5000; Thermo Scientific), with a Carbopac PA1-column and column temperature of 30°C, and a 20 to 150 mM NaOH eluent gradient over 30 min and a 150 mM NaOH eluent for 10 min. The column was rinsed between runs for 5 min with 500 mM NaAc, followed by 10 min equilibration with 20 mM NaOH. Carbohydrate content was quantified with Chromeleon 7.
Plant Performance
Plants were grown at either 20°C or 26°C, according to the settings described in the Methods section “Plant Growth and Insect Rearing.” For final inflorescence stem height, the number of branches and silique number and length were measured after the last flowers and siliques had been developed. Leaves were counted when they were >0.1 cm in length, and siliques were counted when they were >0.5 cm in length. Silique length was recorded for the six lowest and six highest siliques (>0.5 cm) of the main branch and youngest primary side branch of each plant. For all the above plant performance parameters, one biological replicate consisted of a unique plant. Seed germination values were measured with the Germinator package (Joosen et al., 2010). Seeds were sown in trays containing two layers of blue germination paper (Anchor Paper) with 50 mL of deionized water, and depending on the treatment, were cold stratified at 4°C for 72 h prior to incubation or incubated immediately. For germination, the trays were placed under constant light at either 22°C ± 0.5 or 25°C ± 0.5. Three biological replicates were measured, each consisting of ∼50 seeds collected from a different mother plant. Plants were all grown simultaneously at 26 ± 1°C, 50 to 70% relative humidity, for 4 weeks under 8-h:16-h L:D and an additional 6 weeks under 16-h:8-h L:D photoperiod, with a light intensity of 200 μmol m−2 s−1 (Philips TL5 HO 54W/840). At the start of the experiments, seeds had experienced 2.5 weeks of dry storage at room temperature.
Statistics
Data involving gene expression, honeydew droplet excretion, callose content, sucrose content, and seed germination were tested for normality using the Shapiro test and analyzed using a one-way ANOVA (Supplemental Data Set 2). Data involving EPG variables, aphid population, plant performance, and haplotype distribution were assessed with the Mann-Whitney U test (two groups) and the Kruskal-Wallis test (more than two groups). Aphid development was analyzed with a Cox proportional hazards model. Honeydew droplet excretion rate was calculated as the slope of the linear model between the cumulative number of droplets and time. Climate data were used to test for a haplotype effect using two approaches: (1) a Mann-Whitney U test and (2) a mixed model correcting for population structure. For the latter, haplotype was defined as a fixed effect and the first eight principal components of the marker-based kinship matrix were included as random effects. The Bonferroni threshold of 0.001 was calculated by dividing alpha (0.05) by the number of climate variables that were tested (n = 52). Only European Arabidopsis accessions were tested for geographical distribution patterns and climate variables (latitude: 36°00'N to 63°00'N, longitude: 26°00'E to 40°00'W) due to the dense sampling compared with other regions. Statistical tests and graphs were constructed with R, using the packages “survival” and “asreml” (Butler et al., 2009; R Core Team, 2013; Therneau, 2015).
Accession Numbers
The following Arabidopsis Genome Initiative locus identifiers have been reported: SLI1 (At3g10680), RTM2 (At5g04890), SUS5 (At5g37180), SUS6 (At1g73370), and GSL7 (At1g06490).
Supplemental Data
Supplemental Figure 1. Splicing of SLI1.
Supplemental Figure 2. FM4-64 staining of sieve elements after 3 h of incubation.
Supplemental Figure 3. Germination of wild-type and sli1-1 seeds.
Supplemental Figure 4. SLI1 haplotype distribution within Europe.
Supplemental Figure 5. Gene expression of sli1-1 and sli1-2.
Supplemental Table 1. Genes in linkage with SNP chr.3, position 3338114.
Supplemental Table 2. Coexpression of SLI1.
Supplemental Table 3. All climate variables tested for the SLI1 haplotype distribution.
Supplemental Table 4. Primers used for DNA and cDNA amplification.
Supplemental Data Set 1. The total time M. persicae aphids spent on short probes (<3 min) on 350 natural Arabidopsis accessions.
Supplemental Data Set 2. ANOVA tables.
Acknowledgments
This work was supported by the Netherlands Organization for Scientific Research (NWO) through the Technology Foundation Perspective Programme “Learning from Nature” (STW10989), by the Swedish Foundation ‘Carl Tryggers Stiftelse för Vetenskaplig Forskning’ (51616), and by the Kungliga Skogs- och Lantbruksakademien (GFS2016-0019). We thank Freddy Tjallingii for advice regarding the honeydew clock experiment, Diaan Jamar for the HPLC measurements, Joost van Heerwaarden for compiling the climate data set, Ioanna Antoniadi for kindly providing several fluorescent dyes, and Daniela Liebsch for the helpful instructions regarding transient expression in protoplasts.
AUTHOR CONTRIBUTIONS
K.J.K., H.J.B., M.D., and M.A.J. designed the research. W.K. designed the GWA mapping script. R.C.M. constructed the NILs. K.J.K., J.B.-L., G.L.W., and G.B. performed the experiments. K.J.K. performed the analyses. K.J.K., B.R.A., H.J.B., M.D., and M.A.J. wrote the manuscript.
References
- Atwell S., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar J.A., Fradin E.F., Becker F.F.M., Rienstra J.A., van der Schoot J., Vreugdenhil D., Keurentjes J.J.B. (2015). Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 27: 1857–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt D.H.P., Kölling K., Graf A., Pike M., Calder G., Findlay K., Zeeman S.C., Smith A.M. (2011). Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol. 155: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt D.H.P., Derbyshire P., Findlay K., Pike M., Wellner N., Lunn J., Feil R., Simpson C., Maule A.J., Smith A.M. (2009). Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 106: 13124–13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I., Brazelton J.N., Yu D., Huang Y.S., Lahner B., Yakubova E., Li Y., Bergelson J., Borevitz J.O., Nordborg M., Vitek O., Salt D.E. (2010). A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke H.D. (1981). Sieve-element characters. Nord. J. Bot. 1: 381–400. [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173. [DOI] [PubMed] [Google Scholar]
- Bondino H.G., Valle E.M., Ten Have A. (2012). Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 235: 1299–1313. [DOI] [PubMed] [Google Scholar]
- Butler D.G., Cullis B.R., Gilmour A.R., Gogel B.J. (2009). ASReml-R Reference Manual. (Brisbane, Australia: Department of Primary Industries and Fisheries; ). [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Cayla T., Batailler B., Le Hir R., Revers F., Anstead J.A., Thompson G.A., Grandjean O., Dinant S. (2015). Live imaging of companion cells and sieve elements in Arabidopsis leaves. PLoS One 10: e0118122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Parra M.A., Anderberg R.J., Carrington J.C. (2001). Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 127: 1667–1675. [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cosson P., Sofer L., Le Q.H., Léger V., Schurdi-Levraud V., Whitham S.A., Yamamoto M.L., Gopalan S., Le Gall O., Candresse T., Carrington J.C., Revers F. (2010). RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain-containing protein. Plant Physiol. 154: 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedryver C.A., Le Ralec A., Fabre F. (2010). The conflicting relationships between aphids and men: a review of aphid damage and control strategies. C. R. Biol. 333: 539–553. [DOI] [PubMed] [Google Scholar]
- Dixon A.F.G. (1998). Aphid Ecology. (London: Chapman and Hall; ). [Google Scholar]
- Dreyer D.L., Campbell B.C. (1987). Chemical basis of host plant resistance to aphids. Plant Cell Environ. 10: 553–561. [Google Scholar]
- Ehlers K., Knoblauch M., van Bel A.J.E. (2000). Ultrastructural features of well-preserved and injured sieve elements: minute clamps keep the phloem transport conduits free for mass flow. Protoplasma 214: 80–92. [Google Scholar]
- Froelich D.R., Mullendore D.L., Jensen K.H., Ross-Elliott T.J., Anstead J.A., Thompson G.A., Pélissier H.C., Knoblauch M. (2011). Phloem ultrastructure and pressure flow: Sieve-Element-Occlusion-Related agglomerations do not affect translocation. Plant Cell 23: 4428–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furch A.C.U., Hafke J.B., Schulz A., van Bel A.J.E. (2007). Ca2+-mediated remote control of reversible sieve tube occlusion in Vicia faba. J. Exp. Bot. 58: 2827–2838. [DOI] [PubMed] [Google Scholar]
- Furch A.C.U., Zimmermann M.R., Will T., Hafke J.B., van Bel A.J.E. (2010). Remote-controlled stop of phloem mass flow by biphasic occlusion in Cucurbita maxima. J. Exp. Bot. 61: 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., de Souza Vidigal D., Snoek L.B., Schnabel S., Nijveen H., Hilhorst H., Bentsink L. (2014). Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J. Exp. Bot. 65: 6603–6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.S. (1987). Agricultural Insect Pests of Temperate Regions and Their Control. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Horton M.W., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen R.V., Kodde J., Willems L.A.J., Ligterink W., van der Plas L.H.W., Hilhorst H.W.M. (2010). GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J. 62: 148–159. [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Kehr J. (2006). Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 57: 767–774. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Xu Z.-Y., Hwang I. (2013). Generation of transgenic Arabidopsis plants expressing mcherry-fused organelle marker proteins. J. Plant Biol. 56: 399–406. [Google Scholar]
- Kliebenstein D.J. (2014). Quantitative genetics and genomics of plant resistance to insects. In Annual Plant Reviews: Insect-Plant Interactions, Vol. 47, C. Voeckel and G. Jander, eds (Wiley-Blackwell), pp. 235–262. [Google Scholar]
- Kloth K.J., Thoen M.P.M., Bouwmeester H.J., Jongsma M.A., Dicke M. (2012). Association mapping of plant resistance to insects. Trends Plant Sci. 17: 311–319. [DOI] [PubMed] [Google Scholar]
- Kloth K.J., Wiegers G.L., Busscher-Lange J., van Haarst J.C., Kruijer W., Bouwmeester H.J., Dicke M., Jongsma M.A. (2016). AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 67: 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloth K.J., Ten Broeke C.J.M., Thoen M.P.M., Hanhart-van den Brink M., Wiegers G.L., Krips O.E., Noldus L.P.J.J., Dicke M., Jongsma M.A. (2015). High-throughput phenotyping of plant resistance to aphids by automated video tracking. Plant Methods 11: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M., Van Bel A.J.E. (1998). Sieve tubes in action. Plant Cell 10: 35–50. [Google Scholar]
- Knoblauch M., Oparka K. (2012). The structure of the phloem--still more questions than answers. Plant J. 70: 147–156. [DOI] [PubMed] [Google Scholar]
- Knoblauch M., Mullendore D.L. (2013). Sieve element occlusion. In Phloem Molecular Cell Biology, Systemic Communication, Biotic Interactions, Thompson G.A., van Bel A.J.E., eds (Oxford, UK: Wiley-Blackwell; ), pp. 141–153. [Google Scholar]
- Knoblauch M., Peters W.S., Ehlers K., van Bel A.J.E. (2001). Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell 13: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M., Froelich D.R., Pickard W.F., Peters W.S. (2014). SEORious business: structural proteins in sieve tubes and their involvement in sieve element occlusion. J. Exp. Bot. 65: 1879–1893. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Boer M.P., Malosetti M., Flood P.J., Engel B., Kooke R., Keurentjes J.J.B., van Eeuwijk F.A. (2015). Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. (2015). Structure and function of mitochondrial membrane protein complexes. BMC Biol. 13: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang Y., Bergelson J., Nordborg M., Borevitz J.O. (2010). Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S.K., Chisholm S.T., Whitham S.A., Carrington J.C. (1998). Identification and characterization of a locus (RTM1) that restricts long-distance movement of tobacco etch virus in Arabidopsis thaliana. Plant J. 14: 177–186. [DOI] [PubMed] [Google Scholar]
- McLean D.L., Kinsey M.G. (1964). A technique for electronically recording aphid feeding and salivation. Nature 202: 1358–1359. [Google Scholar]
- Medina-Ortega K.J., Walker G.P. (2013). Does aphid salivation affect phloem sieve element occlusion in vivo? J. Exp. Bot. 64: 5525–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ortega K.J., Walker G.P. (2015). Faba bean forisomes can function in defence against generalist aphids. Plant Cell Environ. 38: 1167–1177. [DOI] [PubMed] [Google Scholar]
- Mousavi S.A.R., Chauvin A., Pascaud F., Kellenberger S., Farmer E.E. (2013). GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426. [DOI] [PubMed] [Google Scholar]
- Mullendore D.L., Windt C.W., Van As H., Knoblauch M. (2010). Sieve tube geometry in relation to phloem flow. Plant Cell 22: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Oakey H., Verbyla A., Pitchford W., Cullis B., Kuchel H. (2006). Joint modeling of additive and non-additive genetic line effects in single field trials. Theor. Appl. Genet. 113: 809–819. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Okamura Y., Ito S., Aoki Y., Shirota M., Kinoshita K. (2014). ATTED-II in 2014: Evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol. 55: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A., et al. (2010). The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund R.D., Shih C.Y. (1983). Freeze-fracture analysis of phloem structure in plant tissue cultures. I. The sieve element reticulum. J. Ultrastruct. Res. 82: 111–121. [DOI] [PubMed] [Google Scholar]
- Sjölund R.D., Shih C.Y., Jensen K.G. (1983). Freeze-fracture analysis of phloem structure in plant tissue cultures. III. P-Protein, sieve area pores, and wounding. J. Ultrastruct. Res. 82: 198–211. [DOI] [PubMed] [Google Scholar]
- Sofer L., Cabanillas D.G., Gayral M., Téplier R., Pouzoulet J., Ducousso M., Dufin L., Bréhélin C., Ziegler-Graff V., Brault V., Revers F. (2017). Identification of host factors potentially involved in RTM-mediated resistance during potyvirus long distance movement. Arch. Virol. 162: 1855–1865. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., Hawes C. (2009). The plant endoplasmic reticulum: a cell-wide web. Biochem. J. 423: 145–155. [DOI] [PubMed] [Google Scholar]
- ten Broeke C.J.M., Dicke M., van Loon J.J.A. (2013). Performance and feeding behaviour of two biotypes of the black currant-lettuce aphid, Nasonovia ribisnigri, on resistant and susceptible Lactuca sativa near-isogenic lines. Bull. Entomol. Res. 103: 511–521. [DOI] [PubMed] [Google Scholar]
- Tetyuk O., Benning U.F., Hoffmann-Benning S. (2013). Collection and analysis of Arabidopsis phloem exudates using the EDTA-facilitated Method. J. Vis. Exp. 80: e51111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T. (2015). A package for survival analysis in S version 2.38. https://cran.r-project.org/web/packages/survival/index.html.
- Tjallingii W.F. (1988). Electrical recording of stylet penetration activities. In Aphids, Their Biology, Natural Enemies and Control, Minks A.K., Harrewijn P., eds (Amsterdam: Elsevier; ), pp. 95–108. [Google Scholar]
- Tjallingii W.F. (1995). Regulation of phloem sap feeding by aphids. In Regulatory Mechanisms in Insect Feeding, Chapman R.F., de Boer G., eds (New York: Chapman & Hall; ), pp. 190–209. [Google Scholar]
- Tjallingii W.F. (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57: 739–745. [DOI] [PubMed] [Google Scholar]
- Tjallingii W.F., Hogen Esch T. (1993). Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol. Entomol. 18: 317–328. [Google Scholar]
- Törjék O., Meyer R.C., Zehnsdorf M., Teltow M., Strompen G., Witucka-Wall H., Blacha A., Altmann T. (2008). Construction and analysis of 2 reciprocal Arabidopsis introgression line populations. J. Hered. 99: 396–406. [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The botany array resource: e-northerns, expression angling, and promoter analyses. Plant J. 43: 153–163. [DOI] [PubMed] [Google Scholar]
- van Bel A.J.E., Will T. (2016). Functional evaluation of proteins in watery and gel saliva of aphids. Front. Plant Sci. 7: 1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.G. (2013). Cell biology of sieve element-companion cell complexes. In Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions, Thompson G.A., Van Bel A.J.E., eds (Oxford, UK: Wiley-Blackwell; ), pp. 8–29. [Google Scholar]
- Whitham S.A., Yamamoto M.L., Carrington J.C. (1999). Selectable viruses and altered susceptibility mutants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S.A., Anderberg R.J., Chisholm S.T., Carrington J.C. (2000). Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12: 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T., van Bel A.J.E. (2006). Physical and chemical interactions between aphids and plants. J. Exp. Bot. 57: 729–737. [DOI] [PubMed] [Google Scholar]
- Will T., Vilcinskas A. (2015). The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 57: 34–40. [DOI] [PubMed] [Google Scholar]
- Will T., Tjallingii W.F., Thönnessen A., van Bel A.J.E. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. USA 104: 10536–10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windt C.W., Vergeldt F.J., de Jager P.A., van As H. (2006). MRI of long-distance water transport: a comparison of the phloem and xylem flow characteristics and dynamics in poplar, castor bean, tomato and tobacco. Plant Cell Environ. 29: 1715–1729. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang C., Shi H., Chen L., Wang X., Lü B., Zhang S., Liang Y., Liu R., Qian J., Sun W., You Z., Dong H. (2011). Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol. 11: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T., Agrawal A.A. (2016). Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2: 15206. [DOI] [PubMed] [Google Scholar]