CAMTA3-mediated repression of SA pathway genes in nonstressed plants involves action of a repression module that acts independently of calmodulin binding, a finding that challenges current models.
Abstract
Arabidopsis thaliana calmodulin binding transcription activator (CAMTA) factors repress the expression of genes involved in salicylic acid (SA) biosynthesis and SA-mediated immunity in healthy plants grown at warm temperature (22°C). This repression is overcome in plants exposed to low temperature (4°C) for more than a week and in plants infected by biotrophic and hemibiotrophic pathogens. Here, we present evidence that CAMTA3-mediated repression of SA pathway genes in nonstressed plants involves the action of an N-terminal repression module (NRM) that acts independently of calmodulin (CaM) binding to the IQ and CaM binding (CaMB) domains, a finding that is contrary to current thinking that CAMTA3 repression activity requires binding of CaM to the CaMB domain. Induction of SA pathway genes in response to low temperature did not occur in plants expressing only the CAMTA3-NRM region of the protein. Mutational analysis provided evidence that the repression activity of the NRM was suppressed by action of the IQ and CaMB domains responding to signals generated in response to low temperature. Plants expressing the CAMTA3-NRM region were also impaired in defense against the bacterial hemibiotrophic pathogen Pseudomonas syringae pv tomato DC3000. Our results indicate that the regulation of CAMTA3 repression activity by low temperature and pathogen infection involves related mechanisms, but with distinct differences.
INTRODUCTION
The calmodulin (CaM) binding transcription activator (CAMTA) transcription factors (also known as Signal Response [SR] proteins) are highly conserved among plants and other multicellular eukaryotes (Finkler et al., 2007). Arabidopsis thaliana has six CAMTA proteins that regulate the expression of genes that impart tolerance to abiotic stresses and defense against bacterial pathogens (Shen et al., 2015). The six CAMTA proteins can be divided into two classes based on the presence or absence of a TIG (transcription-associated immunoglobulin) domain (Rahman et al., 2016), which functions in nonspecific DNA binding: whereas CAMTA4, CAMTA5, and CAMTA6 have a TIG domain, CAMTA1, CAMTA2, and CAMTA3 do not. Other than this difference, each Arabidopsis CAMTA protein has four functional domains positioned in the same relative order from the N- to C-terminal end (Rahman et al., 2016): the CG-1 DNA binding domain, an ankyrin (ANK) repeat domain, an IQ domain composed of two IQ motifs, and a CaM binding (CaMB) domain. Both the IQ and CaMB domains bind CaM in a calcium-dependent manner (Bouché et al., 2002; Choi et al., 2005; Du et al., 2009; Nie et al., 2012).
In healthy plants grown at warm temperatures (∼22°C), CAMTA1, CAMTA2, and CAMTA3 (also known as SR1) function in a largely additive manner to repress the expression of salicylic acid (SA) immunity pathway genes (Du et al., 2009; Galon et al., 2008; Kim et al., 2013; Kidokoro et al., 2017). These genes include ISOCHORISMATE SYNTHASE1 (ICS1), which encodes isochorismic acid synthase, the primary rate-limiting enzymatic step in SA biosynthesis in Arabidopsis (Dempsey et al., 2011; Wildermuth et al., 2001); CALMODULIN BINDING PROTEIN 60-LIKE.g (CBP60g) and SAR DEFICIENT1 (SARD1), which encode transcription factors that bind to the promoter of ICS1 and stimulate its transcription (Truman et al., 2013; Zhang et al., 2010); and ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and PHYTOALEXIN DEFICIENT4 (PAD4), which encode putative lipase-like proteins that promote SA accumulation by a poorly understood mechanism (Feys et al., 2001; Venugopal et al., 2009). In camta3 and camta1 camta2 camta3 triple mutant plants, the transcript levels for ICS1, CBP60g, SARD1, EDS1, and PAD4 are much higher than they are in wild-type plants, resulting in the biosynthesis of high levels of SA and the induction of PATHOGENESIS RELATED1 (PR1) and other SA-regulated defense genes (Du et al., 2009; Kim et al., 2013). More broadly, the transcript levels for more than 1000 genes are greater in camta1 camta2 camta3 mutant plants than in wild-type plants and are highly enriched for the GO terms “defense response,” “innate immune response,” and “response to SA stimulus” (Kim et al., 2013), consistent with CAMTA1, CAMTA2, and CAMTA3 having an important role in repressing expression of the SA immunity defense pathway in nonstressed plants (Du et al., 2009; Galon et al., 2008; Kim et al., 2013; Kidokoro et al., 2017).
How do the CAMTA proteins repress expression of SA pathway genes in nonstressed plants? At present, little is known, though there is evidence that it involves action of both the CaMB and IQ domains. Du et al. (2009) identified a mutation within the CAMTA3 CaMB domain that impaired binding of CaM to CAMTA3 and found that the variant protein was defective in repressing expression of ICS1, PR1, and other SA pathway genes. Thus, the investigators concluded that the ability of CAMTA3 to repress gene expression required binding of CaM to the CaMB domain. As for the IQ domain, Nie et al. (2012) and Jing et al. (2011) independently identified mutations, designated sr1-4D and sard3, respectively, that caused plants to be more susceptible to infection by biotrophic bacterial and fungal pathogens. The sr1-4D and sard3 mutations proved to be identical, causing an amino acid change within the first IQ motif of CAMTA3 that resulted in a gain-of-function phenotype. In particular, introduction of the sr1-4D mutation into the enhanced disease resistance2 (edr2) background (the edr2 mutation causes constitutive high-level expression of PR1 and enhanced resistance to powdery mildew) resulted in dramatic downregulation of PR1 and greatly reduced resistance of the plants to infection by the powdery mildew pathogen (Nie et al., 2012). Also, SA levels and transcript levels for ICS1, PAD4, and EDS1 were lower in the sr1-4D mutant than they were in wild-type plants. The mechanism whereby the sr1-4D and sard3 IQ mutations impart gain-of-function repression of SA-mediated immunity is not known but does not appear to result from an altered interaction of CaM with the IQ domain, as both CAMTA3 and the IQ mutant protein were found to bind CaM in a calcium-dependent manner (Nie et al., 2012).
The ability of the CAMTA proteins to repress SA immunity pathway genes is overcome in response to infection by biotrophic and hemibiotrophic pathogens (Poovaiah et al., 2013; Fromm and Finkler, 2015). It is generally thought that this regulation involves changes in the interaction of CAMTA3 with CaM caused by fluctuations in the levels of intracellular calcium that are known to occur in response to pathogen attack (Reddy et al., 2011). In addition, Zhang et al. (2014) presented evidence that CAMTA3 is degraded in response to pathogen attack. This degradation was shown to involve action of SR1IP1, a protein proposed to act as an adaptor recruiting CAMTA3 to a cullin3 E3 ligase, resulting in the ubiquitination of CAMTA3 and degradation by the 26S proteasome.
Repression of the SA immunity pathway by CAMTA proteins is also overcome in plants exposed to low temperature for a prolonged period. Scott et al. (2004) first reported that SA levels increase in Arabidopsis plants at chilling temperatures (5°C), but not until plants were exposed to this temperature for more than a week. Kim et al. (2013) confirmed this finding and showed that SA levels did not increase in cold-treated plants carrying the sid2-1 mutation, a null allele of ICS1 (Wildermuth et al., 2001), indicating that SA biosynthesis at low temperature proceeds through the isochorismic acid synthase pathway. In plants exposed to low temperature for more than a week, the transcript levels for SA pathway genes were found to be high, including those for ICS1, CBP60g, SARD1, and PR1 (Kim et al., 2013).
The primary goal of this study was to better understand how CAMTA3 represses the expression of SA pathway genes in healthy nonstressed plants and how this repression is overcome in response to low temperature. Our results led us to propose a model in which the role of CaM binding to the CAMTA3 CaMB domain is reversed from current thinking: Instead of CaM binding to the CaMB domain being required for CAMTA3 repression activity in nonstressed plants, we propose that it is required to downregulate CAMTA3 repression activity in response to signals generated by stress. In addition, our results provide evidence that the regulation of CAMTA3 repression activity in response to low temperature and pathogen infection involves related mechanisms but that there are also significant differences.
RESULTS
The N-Terminal End of CAMTA3 Functions as a Repression Module
As a step toward understanding how CAMTA3 represses the expression of SA pathway genes in healthy nonstressed plants and how this repression is overcome in response to low temperature, we made CAMTA3-GFP variants with modifications in known or potential functional domains and determined whether the proteins were able to repress the expression of SA pathway genes at warm (22°C) and cold (4°C) temperatures. All of the CAMTA3-GFP variant proteins were placed under control of the endogenous CAMTA3 promoter and transformed into camta2 camta3 double mutant plants. We chose to transform the construct into the camta2 camta3 mutant, as opposed to the camta3 single mutant, because SA biosynthesis and expression of SA pathway genes are greater in the camta2 camta3 mutant than they are in the camta3 mutant (Kim et al., 2013). In addition, we chose the camta2 camta3 mutant over the camta1 camta2 camta3 triple mutant, as the triple mutant is tiny in size and difficult to work with.
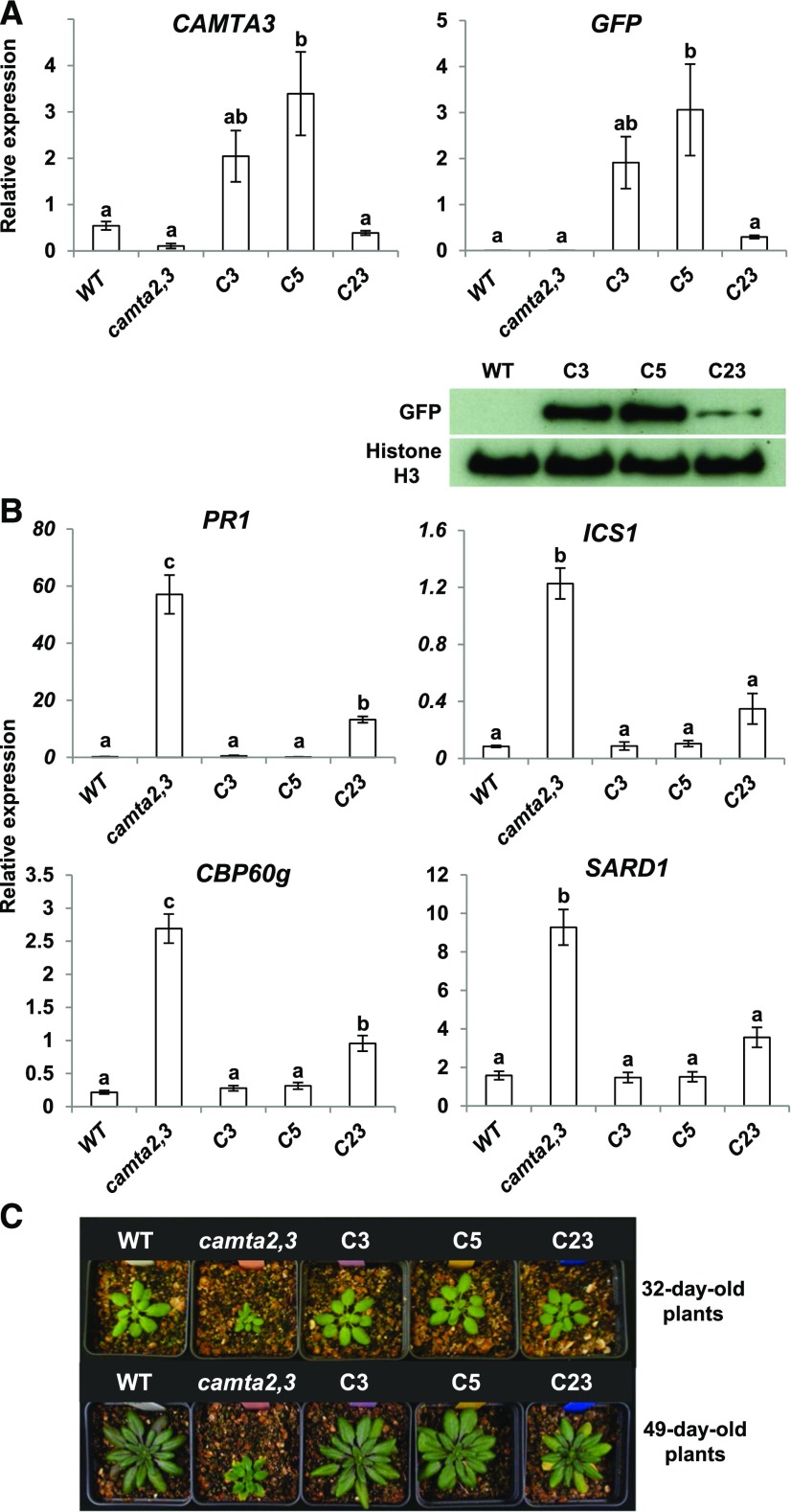
In our initial experiments, we asked whether the CAMTA3-GFP protein fusion was functional. The transcript levels for the CAMTA3-GFP transgene in three transgenic lines ranged from ∼50% less (line C23) to about 6-fold more (line C5) than that for the endogenous CAMTA3 gene in wild-type plants (Figure 1A). The level of the CAMTA3-GFP protein in each line was consistent with the transcript levels for the transgene (Figure 1A). As shown previously (Kim et al., 2013), the transcript levels for the SA pathway genes PR1, ICS1, CBP60g, and SARD1 and were much higher in camta2 camta3 plants than in wild-type plants (Figure 1B), and the camta2 camta3 plants were considerably smaller in size than were wild-type plants (Figure 1C). Expression of the CAMTA3-GFP transgene repressed expression of the SA pathway genes (Figure 1B) and suppressed the small stature phenotype of the camta2 camta3 plants (Figure 1C). These results indicated that the GFP-tagged CAMTA3 protein was active.
Figure 1.
CAMTA3-GFP Represses Expression of SA Pathway Genes in camta2 camta3 Plants.
(A) The CAMTA3p:CAMTA3-GFP construct was transformed into camta2 camta3 mutant plants and three transgenic lines—C3, C5, and C23—were characterized. Plants were grown for 20 d at 22°C under a 12-h photoperiod and relative transcript levels were determined by RT-qPCR. In the immunoblot analysis, anti-GFP antibody was used to detect the CAMTA3-GFP protein and antihistone H3 antibody was used to detect histone H3, which served as the loading control. Genes used for normalization for RT-qPCR are indicated in Methods, and data were subjected to ANOVA as detailed in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05)
(B) Transcript levels of PR1, ICS1, CBP60g, and SARD1 were determined in the transgenic lines C3, C5, and C23 grown under the same conditions as in (A). Data were subjected to ANOVA as detailed in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05)
(C) Photographs of wild-type, camta2 camta3, and transgenic plants after growth for 32 and 49 d at 22°C under a 12-h photoperiod.
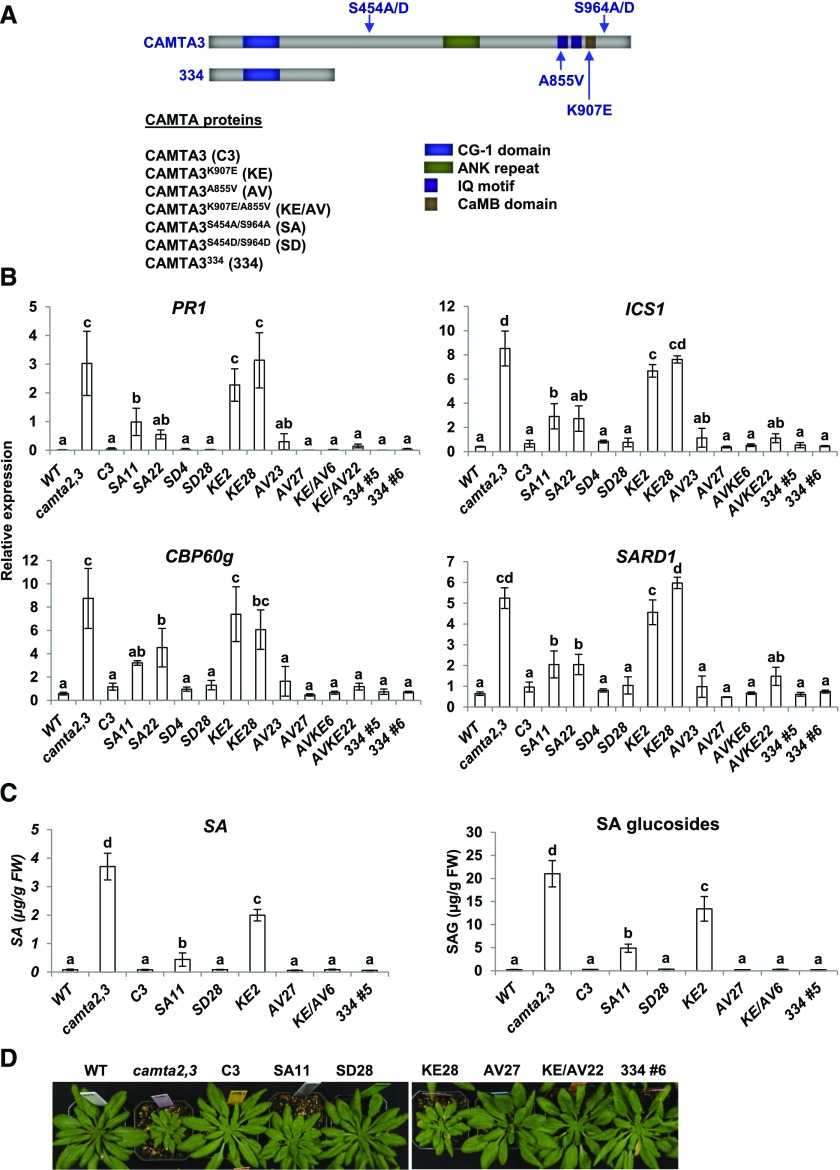
We next made six CAMTA3-GFP protein variants (for simplicity, henceforth referred as CAMTA3 variants) (Figure 2A): CAMTA3K907E, CAMTA3A855V, CAMTA3K907E/A855V, CAMTA3S454A/S964A, CAMTA3S454D/S964D, and CAMTA3334. Two lines for each protein variant were used in our analyses, most of which expressed the transgene at the transcript (Supplemental Figure 1) and protein (Figure 3A) level close to those of CAMTA3 in the CAMTA3 C3 transgenic plants (the exceptions were lines SA22, KE2, and KE28, which expressed the transgenes at 4- to 6-fold higher levels than CAMTA3 in the C3 transgenic line).
Figure 2.
Repression of SA Pathway Genes, SA Biosynthesis, and Small Stature Phenotype of camta2 camta3 Plants Expressing CAMTA3 Protein Variants.
(A) Diagram of CAMTA3 protein variants.
(B) Transcript levels of SA pathway genes in transgenic lines expressing CAMTA3 variant proteins in camta2 camta3 plants. Plants were grown at 22°C for 21 d under a 12-h photoperiod and harvested at the end of the light period (ZT12). Transcript levels were determined by RT-qPCR. Genes used for normalization are indicated in Methods. Data were subjected to ANOVA as detailed in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05)
(C) Levels of SA and SA glucosides in transgenic lines expressing CAMTA3 variants in camta2 camta3 plants. Plants were grown for 28 d at 22°C under a 12-h photoperiod and harvested 3 to 4 h after the start of the light period. Two biological replicates were tested and the values for one experiment are shown. Data were square root transformed to reduce heteroscedasticity and analyzed by ANOVA. Error bars indicate sd (n = 4 technical replicates). Bars marked with different letters are significantly different (LSD, P < 0.05). Values shown in the figure are untransformed for clarity.
(D) Photographs of wild-type and camta2 camta3 plants expressing the CAMTA3 variants. Plants were grown for 42 d under a 12-h photoperiod at 22°C.
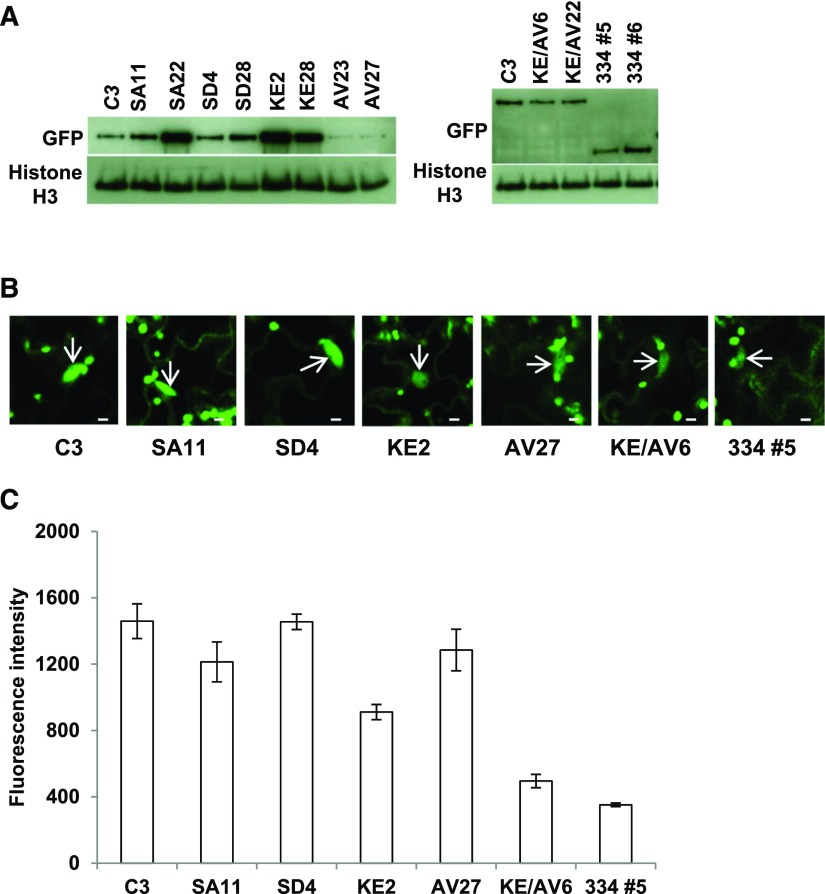
Figure 3.
CAMTA3 Variant Proteins Are Present in the Nucleus.
(A) Protein levels for CAMTA3 protein variants. Plants were grown under a 12-h photoperiod at 22°C for 21 d, and protein levels for the CAMTA3 variant proteins were determined by immunoblot analysis using anti-GFP antibody. Histone H3 served as the loading control.
(B) Confocal optical sections of leaves from camta2 camta3 plants expressing variant CAMTA3 proteins. Plants were grown in soil at 22°C under a 12-h photoperiod for 21 d. The arrows indicate nuclei. Bars = 5 µm.
(C) Quantification of GFP fluorescence in individual nuclei from separate cells. The fluorescence intensity was measured in the nuclei using a fixed region of interest (ROI) as described in Methods. Error bars indicate se; n = 20 to 73 nuclei per sample.
Given the results of Du et al. (2009) indicating that the ability of CAMTA3 to repress the expression of SA pathway genes required CaM binding to the CaMB domain, we anticipated that CAMTA3334, which does not include the CaMB domain, would not be able to repress expression of SA pathway genes. Instead, we found that CAMTA3334 was highly effective in repressing the expression of PR1, ICS1, CBP60g, and SARD1 (Figure 2B). In addition, CAMTA3334 suppressed SA biosynthesis (Figure 2C) and the small stature phenotype (Figure 2D) of camta2 camta3 plants. The ability of CAMTA3334 to repress gene expression appeared to be somewhat greater than that of CAMTA3 as the fluorescence intensity of the CAMTA3334 in the nucleus was less than that of the CAMTA3 protein (Figures 3B and 3C). Taken together, these results indicated that the N-terminal region of CAMTA3 (residues 1–344) functions as an N-terminal repression module (NRM) that is sufficient to inhibit expression of SA pathway genes in nonstressed plants.
NRM Repression Activity Is Regulated by the CaMB and IQ Domains
Du et al. (2009) showed that CAMTA3K907E, which has an amino acid substitution within the CaMB domain, is unable to bind CaM and does not repress the biosynthesis of SA or expression of SA pathway genes. Consistent with these results, we found that CAMTA3K907E did not repress the high level expression of PR1, ICS1, CBP60g, or SARD1 in the camta2 camta3 plants (Figure 2B) and was only marginally effective in repressing SA biosynthesis (Figure 2C) and suppressing the small stature phenotype of the camta2 camta3 plants (Figure 2D). The inability of CAMTA3K907E to repress expression of the SA immunity pathway was not due to degradation of the protein as the CAMTA3K907E protein levels were actually greater than those of CAMTA3 (Figure 3A). The fluorescence intensity of the CAMTA3K907E protein in the nucleus was reduced somewhat compared with the CAMTA3 protein, indicating that import of the CAMTA3K907E into the nucleus might have been impaired somewhat, but the fluorescence intensity of CAMTA3K907E was greater than that of CAMTA3334, indicating that the CAMTA3K907E protein was much less active than CAMTA3334 (Figures 3B and 3C). In sum, these results indicated that a mutation within the CaMB domain that prevents CaM binding suppresses the NRM repression activity.
Nie et al. (2012) showed that CAMTA3A855V, which has an amino acid substitution within the IQ domain, has a gain-of-function phenotype inhibiting the induction of SA pathway genes in plants that have a constitutively active R gene. Similarly, we found that CAMTA3A855V repressed the expression of SA pathway genes (Figure 2B) and SA biosynthesis (Figure 2C) in the camta2 camta3 plants and effectively suppressed the small stature phenotype of the camta2 camta3 plants (Figure 2D). Moreover, we found that the A855V mutation was “dominant” over the K907E mutation: Whereas CAMTA3K907E did not repress the high level expression of SA pathway genes in the camta2 camta3 plants (Figure 2B), CAMTA3K907E/A855V repressed the expression of SA pathway genes (Figure 2B) and SA biosynthesis (Figure 2C) in the camta2 camta3 plants and effectively suppressed the small stature phenotype of the camta2 camta3 plants (Figure 2D). These results provided evidence that the IQ and CaMB domains interact to regulate the NRM repression activity.
We also found that in the context of the complete CAMTA3 protein, phosphorylation at S454 or S964 or both was required for the protein to be fully active in repressing the SA pathway genes. CAMTA3S454A/S964A and CAMTA3S454D/S964D have amino acid substitutions at S454 and S964, which are phosphorylated in nonstressed plants (Jones et al., 2009; Wang et al., 2013). CAMTA3S454A/S964A, which cannot be phosphorylated at S454 and S964, was partially impaired in repressing the expression of SA pathway genes (Figure 2B) and SA biosynthesis (Figure 2C) in camta2 camta3 plants and the plants expressing CAMTA3S454A/S964A were not as big as wild-type plants (Figure 2D). The impaired function of CAMTA3S454A/S964A was not due to degradation of the protein as its level was the same or greater than that of CAMTA3 (Figure 3A). In addition, CAMTA3S454A/S964A accumulated in the nucleus to nearly the same level as did CAMTA3 (Figures 3B and 3C).
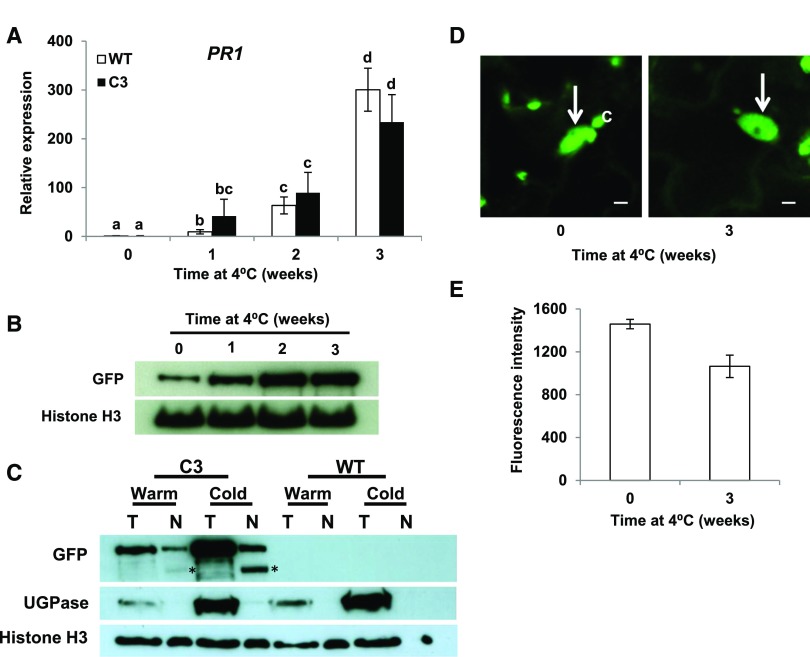
Induction of SA Pathway Genes in Response to Low Temperature Does Not Result from Degradation of CAMTA3 or Exclusion of CAMTA3 from the Nucleus
The ability of CAMTA1, CAMTA2, and CAMTA3 to repress expression of SA pathway genes is overcome in plants exposed to low temperature for more than a week (Kim et al., 2013). Consistent with these results, we found that PR1 was induced in wild-type and CAMTA3 C3 transgenic plants that were exposed to low temperature for more than a week (Figure 4A). The loss of CAMTA3 repression activity did not result from decreased levels of the CAMTA3 protein; indeed, CAMTA3 protein levels actually increased somewhat in response to low temperature (Figure 4B). Fluorescence intensity measurements indicated that the loss of CAMTA3 repression activity in response to low temperature did not result from exclusion of the protein from the nucleus (Figures 4C and 4D). In addition, full-length CAMTA3 protein was detected in nuclei isolated from cold-treated plants and was at greater levels in these plants than it was in plants grown at warm temperature (Figure 4E; a second band, marked with asterisk, was detected in the nuclear preparations, but not in the total protein preparations, indicating that it resulted from proteolytic cleavage of CAMTA3 during isolation of the nuclei). The small apparent discrepancy between the relative amounts of CAMTA3 protein in the nuclei of warm- and cold-treated plants observed in the fluorescence and immunoblotting assays was likely due to the interfering effects that SA has been reported to have on the fluorescence of GFP protein fusions in vivo (de Jonge et al., 2017). Taken together, our results indicated that the induction of SA pathway genes in cold-treated plants involved changes in the ability of the CAMTA3 protein to repress gene expression.
Figure 4.
Induction of PR1 in Plants Exposed to Low Temperature Does Not Involve Degradation of CAMTA3 or Exclusion of CAMTA3 from the Nucleus.
(A) Expression of PR1 in plants exposed to low temperature. wild-type and C3 transgenic plants were grown at 22°C under a 12-h photoperiod and transferred to 4°C for the indicated times. Relative transcript levels of PR1 were determined by RT-qPCR. Data were subjected to ANOVA as detailed in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05).
(B) CAMTA3-GFP levels in the C3 transgenic line. Protein levels were determined by immunoblot analysis using anti-GFP antibody. Histone H3 served as the loading control. Plants were grown as in (A).
(C) CAMTA3-GFP levels in total (T) and nuclear (N) protein preparations in C3 and wild-type plants grown at warm (warm) temperature or grown at warm temperature and cold-treated (cold) for 3 weeks at 4°C. Approximately equal amounts of nuclear protein were run in each lane, indicated by amounts of histone H3. UGPase antibody was used to detect the cytoplasmic protein maker, UDP-glucose pyrophosphorylase. The bands marked with an asterisk are degradation product of CAMTA3-GFP (see text).
(D) Confocal optical sections from leaves of the C3 transgenic line showing CAMTA3-GFP present in the nucleus (arrow). Chloroplasts (C) also appear green due to chlorophyll autofluorescence. Plants were grown as in (A). Bars = 5 µm.
(E) Quantification of CAMTA3-GFP fluorescence intensity of nuclei. Fluorescence intensity of individual nuclei from separate cells was measured using a fixed ROI as described in Methods. Error bars indicate se: n = 36 (0 weeks) and 51 (3 weeks) nuclei.
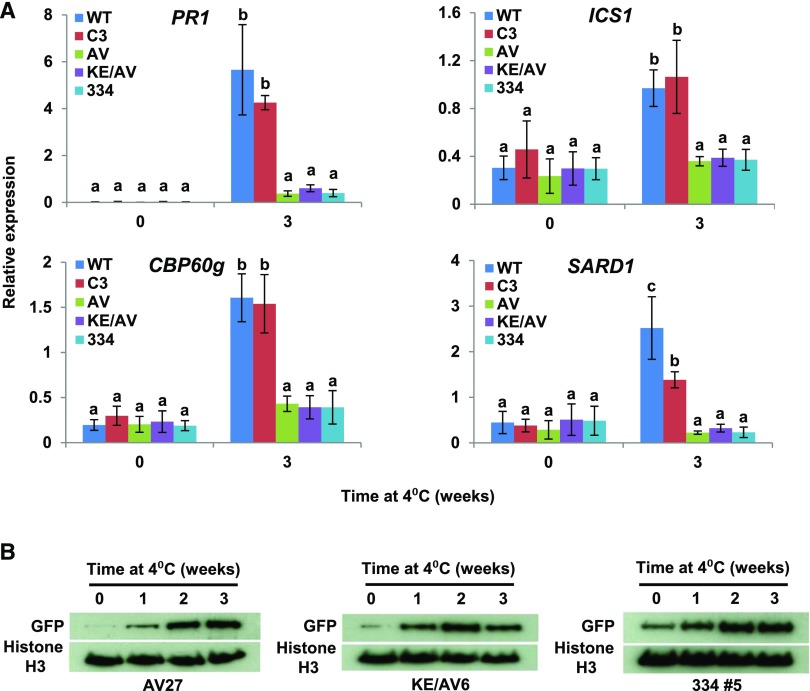
Downregulation of NRM Repression Activity by Low Temperature Requires Action of the C-Terminal Region of CAMTA3
The results presented above indicated that CAMTA3334 was able to repress the expression of SA pathway genes in nonstressed camta2 camta3 mutant plants (Figure 2B). Furthermore, we found that CAMTA3334, unlike CAMTA3, repressed the expression of SA pathway genes in plants exposed to low temperature for 3 weeks (Figure 5A). These results indicated that the C-terminal end of CAMTA3 was required to downregulate the ability of the NRM to repress SA pathway gene expression in response to low temperature. Downregulation of NRM repression activity was also impaired in plants expressing CAMTA3A855V or CAMTA3K907E/A855V: Like CAMTA3334, these proteins repressed the expression of SA pathway genes in plants exposed to low temperature for 3 weeks (Figure 5A). These results provided evidence that the IQ domain has a role in downregulating NRM repression activity in response to low temperature. The protein levels of CAMTA3334, CAMTA3A855V, and CAMTA3K907E/A855V (Figure 5B), like those of CAMTA3 (Figure 4B), increased somewhat in response to low temperature, indicating that the differences in the activities of these proteins did not involve major differences in protein stability at low temperature.
Figure 5.
CAMTA3334, CAMTA3A855V, and CAMTA3K907E/A855V Repress Expression of SA Pathway Genes in camta2 camta3 Plants Exposed to Low Temperature for a Prolonged Period.
(A) Expression of SA pathway genes in camta2 camta3 plants expressing CAMTA3 variant proteins. Plants were grown at 22°C under a 12-h photoperiod followed by exposure to 4°C for the indicated times. Transcript levels were determined by RT-qPCR. Genes used for normalization are indicated in the Methods. Data were subjected to ANOVA as detailed in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05). Transgenic lines used were AV27, KE/AV6, and 334 #5.
(B) Protein levels of CAMTA3A855V, CAMTA3K907E/A855V, and CAMTA3334 in camta2 camta3 plants exposed to low temperature (4°C) for the indicated times. Protein levels were detected by immunoblot analysis using anti-GFP antibody. Histone H3 was used as a loading control.
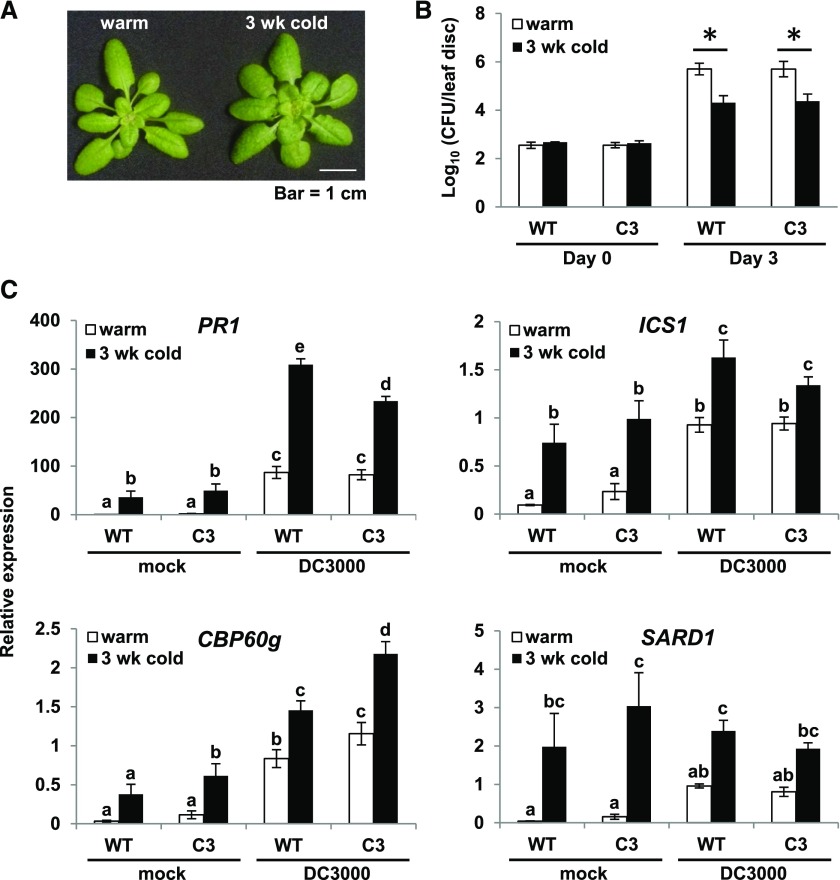
Prolonged Exposure to Low Temperature Increases Immunity against the Bacterial Plant Pathogen Pseudomonas syringae pv tomato DC3000
We previously speculated (Kim et al., 2013) that the increase in SA pathway gene expression that occurs in plants exposed to low temperature for a prolonged period might result in an increase in plant immunity. To test this possibility, wild-type plants and camta2 camta3 plants expressing CAMTA3 that had been grown at warm temperature (22°C) or grown at warm temperature and then cold-treated (4°C) for 3 weeks (Figure 6A) were inoculated at warm temperature with the virulent bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000) (Xin and He, 2013) and bacterial numbers were determined immediately or after 3 d of incubation at warm temperature (22°C). The results indicated that the plants that had been cold-treated were more resistant to infection than were the plants grown at warm temperature (Figure 6B). Consistent with these results was our finding that the expression levels of PR1, ICS1, CBP60g, and SARD1 were greater in the pathogen-infected plants that had been exposed to low temperature for 3 weeks than they were in the plants that had been grown at warm temperature (Figure 6C).
Figure 6.
Prolonged Exposure to Low Temperature Results in an Increase in Immunity against Pst DC3000.
(A) Photograph of wild-type plants grown at either 22°C for 4 weeks (warm) or grown at 22°C for 4 weeks followed by 3 weeks at 4°C (3 wk cold).
(B) Wild-type and C3 transgenic plants that had been grown at 22°C for 4 weeks (warm) or grown at 22°C for 4 weeks followed by 3 weeks at 4°C (3 wk cold) were inoculated with Pst DC3000 (OD600 = 0.0001). Bacterial growth was measured at 0 and 3 d postinoculation as described in Methods. Three biological replicates were performed; the figure shows a representative experiment. Error bars indicate sd (n = 4 [day 0] and n = 8 [day 3] technical replicates). An asterisk indicates a difference between warm and cold-treated plants (P < 0.05, Student’s t test).
(C) Wild-type and C3 transgenic plants that had been grown at 22°C for 4 weeks (warm) or at 22°C for 4 weeks followed by 3 weeks at 4°C (3 wk cold) were inoculated with Pst DC3000 (DC3000) (OD600 = 0. 001) or treated with 10 mM MgCl2 (mock). Leaf tissue was collected at 24 h postinoculation. Transcript levels were determined by RT-qPCR as detailed in Methods. Data were subjected to ANOVA as described in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05). Transgenic lines AV27, KE/AV6, and 334 #5 were used in the experiments.
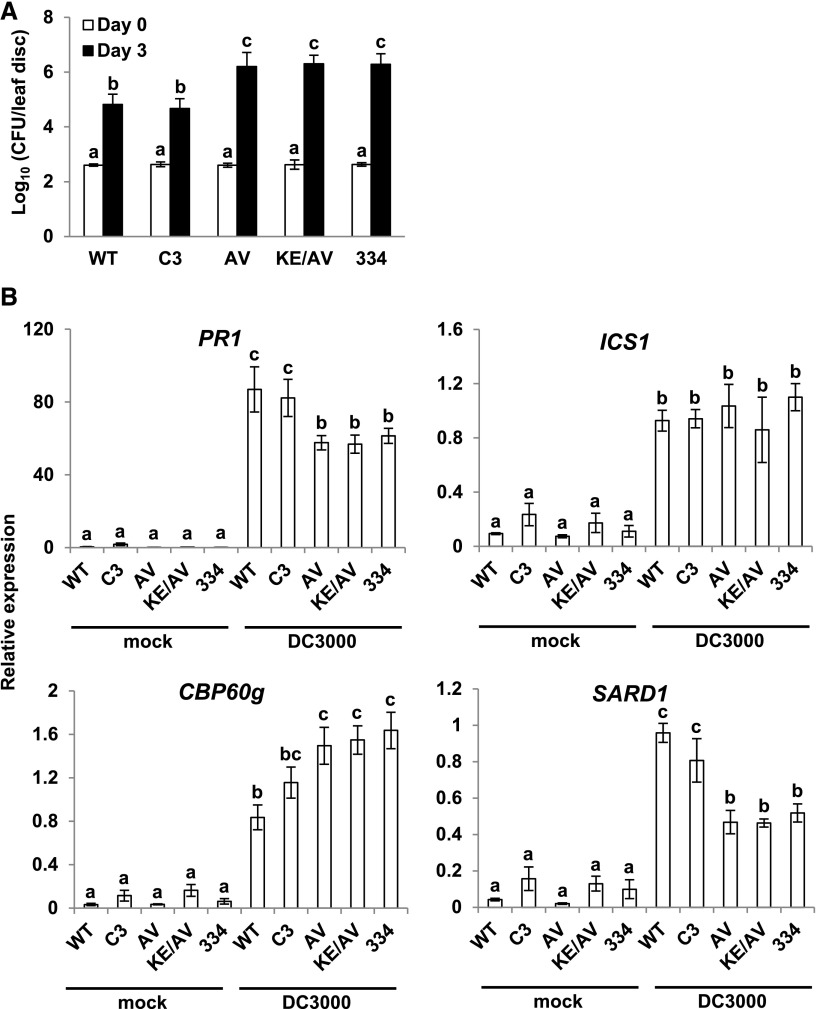
Immunity against Pst DC3000 Is Compromised in Plants Expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V
Our finding that cold induction of SA immunity genes was severely impaired in camta2 camta3 plants expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V prompted us to determine whether the immunity of these plants was also compromised. To test this, wild-type plants and camta2 camta3 plants expressing either CAMTA3 or the CAMTA3 variants were grown at warm temperature (22°C) and inoculated with Pst DC3000, and bacterial numbers were determined immediately or after 3 d of incubation at warm temperature. The results indicated that the camta2 camta3 plants expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V were indeed more susceptible to infection by Pst DC3000 than were wild-type plants or camta2 camta3 plants expressing CAMTA3 (Figure 7A). Therefore, we anticipated that pathogen-induced expression of SA pathway genes would be impaired in plants expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V. This speculation proved to be only partially true (Figure 7B): The induction of PR1 and SARD1 was impaired in plants expressing these CAMTA3 variants, but induction of ICS1 was not affected and the induction of CBP60g was actually greater in these plants. It should also be noted that whereas pathogen-induced expression of PR1 and SARD1 was reduced in plants expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V (Figure 7B), cold-induced expression of these genes was essentially eliminated in plants expressing these CAMTA3 variants (Figure 5B).
Figure 7.
Plants Expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V Are Impaired in Immunity against Pst DC3000.
(A) Plants that had been grown at 22°C for 4 weeks were inoculated with Pst DC3000 (OD600 = 0.0001) and bacterial growth was measured at 0 and 3 d postinoculation as described in Methods. Transgenic lines AV27, KE/AV6, and 334 #5 were used in the experiments. Three biological replicates were performed yielding similar results; the results from a representative experiment are shown. Error bars indicate sd (n = 4 [day 0] and n = 8 [day 3] technical replicates). Statistical significance of pathogen growth among the different genotypes was determined using one-way ANOVA and Tukey HSD test when statistical significance was found. Means with the same letter were not significantly different, LSD P < 0.01.
(B) Wild-type and camta2 camta3 mutant plants expressing the indicated CAMTA3 variants that had been grown at 22°C for 4 weeks were inoculated with Pst DC3000 (DC3000) (OD600 = 0. 001). Leaf tissue was collected at 24 h postinoculation. Transcript levels of the indicated genes were determined by RT-qPCR. Data were subjected to ANOVA as described in Methods. Error bars indicate se (n = 3 biological replicates). Bars marked with different letters are significantly different (LSD, P < 0.05). Transgenic lines AV27, KE/AV6, and 334 #5 were used in the experiments.
DISCUSSION
In healthy plants grown at moderate temperatures, CAMTA1, CAMTA2, and CAMTA3 act in an additive manner to repress the expression of SA immunity pathway genes (Du et al., 2009; Galon et al., 2008; Kim et al., 2013; Kidokoro et al., 2017), thus preventing the allocation of valuable resources away from growth and development toward unneeded defense. However, upon prolonged exposure to low temperature or infection by biotrophic and hemibiotrophic pathogens, CAMTA-mediated repression of the SA pathway is alleviated and plant defense genes are expressed (Poovaiah et al., 2013; Fromm and Finkler, 2015). Understanding how the CAMTA proteins repress the expression of SA pathway genes in nonstressed plants and how this repression is overcome in response to abiotic and biotic stresses is integral to an overall understanding of the plant immune response.
Here, we present results that force a rethinking of how CAMTA3 functions to repress the expression of SA pathway genes in nonstressed plants. Du et al. (2009) found that the K907E substitution within the CaMB domain of CAMTA3 impaired binding of CaM to the CaMB domain and greatly reduced the ability of CAMTA3 to repress expression of SA pathway genes in nonstressed plants. Based on these results, the investigators reasonably proposed that the ability of CAMTA3 to repress expression of the SA pathway requires binding of CaM to the CaMB domain. However, as noted by Fromm and Finkler (2015), there is a tension between this model and two other considerations: i.e., that binding of CaM to the CAMTA3 CaMB domain is calcium-dependent and that basal calcium levels in the nucleus are expected to be low (Pauly et al., 2001). Thus, in nonstressed plants, CAMTA3 would not be predicted to effectively bind CaM and therefore would not effectively repress SA pathway genes, a prediction that is opposite to what is observed.
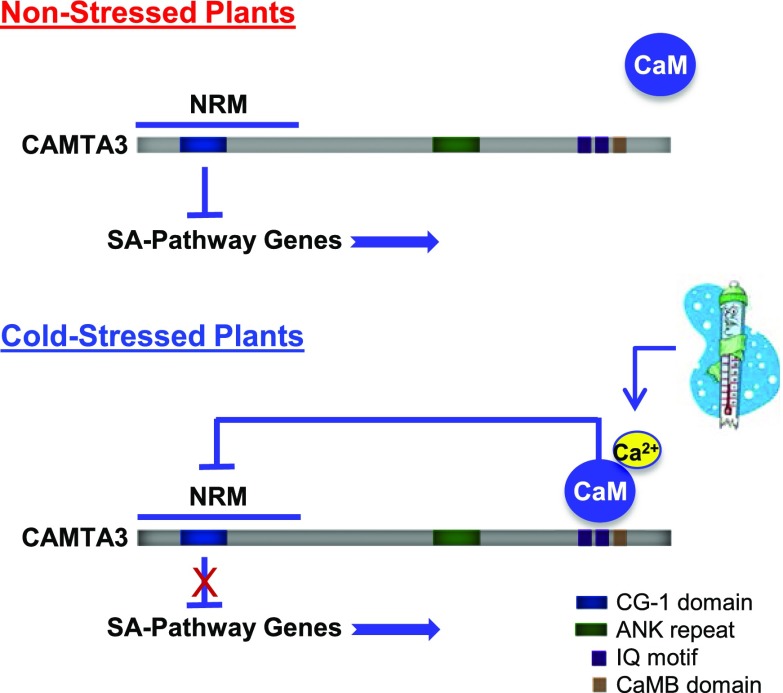
Our results support an alternative model that eliminates this apparent contradiction (Figure 8). In particular, we show that CAMTA3334, a severely truncated variant of CAMTA3 that contains the CG-1 DNA binding domain but lacks the IQ and CaMB domains, is highly effective in repressing the expression of SA pathway genes in nonstressed plants (Figure 2B). This result provides direct evidence that the N-terminal region of CAMTA3 comprises a repression module—the NRM—that can function autonomously from the CaM binding domains to repress gene expression in nonstressed plants. In addition, our results indicate that cold induction of SA pathway genes is severely impaired in plants expressing CAMTA3334 (Figure 5A). A straightforward interpretation of this finding is that the C-terminal end of CAMTA3 is required to downregulate the ability of the NRM to repress SA gene expression in response to low temperature. Finally, our finding that cold induction of SA pathway genes is severely impaired in plants expressing CAMTA3A855V (Figure 5A) suggests that the CaM binding IQ domain has a role in downregulating the repression activity of the NRM in cold-stressed plants. Thus, in our model, the role of CaM binding to CAMTA3 is reversed from current thinking: Instead of enabling CAMTA3 to repress SA pathway genes in nonstressed plants, we propose that CaM binding to CAMTA3 has a role in nullifying the ability of the CAMTA3 NRM to repress gene expression in cold-stressed plants. We realize that this model requires a radical change in current thinking about the role of CaM binding in regulating CAMTA3 repression activity and that its validation will require corroboration through further testing that includes complementary experimental approaches.
Figure 8.
Model for CAMTA3 Regulation of SA Pathway Genes in Nonstressed and Cold-Stressed Plants.
In nonstressed plants, CAMTA3 binds to the promoters of certain SA pathway genes and represses their transcription through action of the NRM region of the protein. When plants are exposed to low temperature for more than a week, a calcium signature is generated that promotes binding of CaM to the IQ and/or CaMB domain. The binding of CaM causes a change in protein conformation or charge that blocks the repression activity of the NRM through intraprotein interactions. The loss of NRM repression activity allows the target genes to be transcribed resulting in activation of the SA pathway. See Discussion for details.
How might low temperature lead to downregulation of NRM repression activity? At present, we can only speculate. One simple scenario would be that low temperature generates a signal that brings about an increase in calcium resulting in CaM binding to the IQ and/or CaMB domains causing downregulation of NRM repression activity. Loss of CAMTA3 repression would then lead to induction of positive regulators of ICS1, including CBP60g, SARD1, and EDS1, resulting in increased levels of SA and the induction of SA-regulated defense genes (Du et al., 2009; Kim et al., 2013). However, while this overall scheme may be broadly correct, it does not fully explain all available data. Specifically, the results of Kim et al. (2013) indicate that exposure of wild-type plants to low temperature for 3 weeks results in the induction of CBP60g, SARD1, EDS1, and ICS1 but that these genes are not induced in sid2 mutant plants exposed to low temperature for 3 weeks. Thus, it would seem that normal low basal levels of SA might have an integral role in the induction of SA pathway genes in response to low temperature.
Another question fundamental to our proposed model is how might CaM binding to CAMTA3 negate NRM activity. Again, at present, we can only speculate. It would appear that the CaMB domain can negate the activity of the NRM without binding CaM as the K907E mutation in the CaMB domain eliminates both NRM repression activity and CaM binding to the CaMB domain (Du et al., 2009). These results suggest that a change in charge or secondary structure of the CaMB domain caused by the K907E mutation blocks the repressive activity of the NRM, presumably through altered intraprotein interactions. Perhaps CaM binding to the CaMB domain of CAMTA3 brings about a functionally analogous intraprotein interaction that suppresses NRM activity.
As mentioned above, our results indicate that the IQ domain has a role in regulating NRM repression activity in response to low temperature. This is evidenced by the fact that repression of SA pathway genes by CAMTA3A855V is not overcome in plants exposed to low temperature for a prolonged period (Figure 5A). Moreover, the A855V substitution within the IQ domain is dominant over the K907E substitution within the CaMB domain: Whereas CAMTA3K907E did not repress expression of SA immunity genes in nonstressed plants, CAMTA3K907E/A855V did (Figure 2B). These results point to interactions between the IQ domain and the CaMB domain in regulating the repression activity of the NRM. Additional studies will be required to determine the nature of this apparent interaction.
Plants expressing CAMTA3334, CAMTA3A855V, and CAMTA3K907E/A855V were not only impaired in activation of SA pathway genes in response to low temperature (Figure 5A), but were also impaired in their immunity against pathogen attack (Figure 7A). These findings—which are consistent with those of Nie et al. (2012) and Jing et al. (2011), who previously reported that plants expressing CAMTA3A855V were impaired in immunity—suggest that regulation of CAMTA3 NRM activity by low temperature and pathogen attack involve related mechanisms. However, it is also clear that there are significant differences in this regulation. Whereas our results indicate that CAMTA3 protein levels increase somewhat in plants exposed to low temperature (Figure 4B), Zhang et al. (2014) reported that CAMTA3 was degraded in response to pathogen attack. Moreover, whereas induction of ICS1, SARD1, CBP60g, and PR1 in response to low temperature was essentially eliminated in plants expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855 (Figure 5A), their induction in response to infection by Pst DC3000 was variable: Induction of SARD1 and PR1 was reduced ∼30 to 40%, induction of ICS1 was not affected, and induction of CPB60g was actually higher in the plants expressing CAMTA3334, CAMTA3A855V, or CAMTA3K907E/A855V (Figure 7B).
We previously reported that the biosynthesis of SA that occurs in plants exposed to low temperature does not contribute to freezing tolerance and speculated that it might contribute to enhanced immunity (Kim et al., 2013). Here, we show that this is the case: Cold-acclimated Arabidopsis plants were more resistant to infection by Pst DC3000 than were plants that had been grown at moderate temperature (Figure 6B). These results are consistent with previous findings indicating that cold-acclimated plants exhibit enhanced disease resistance (Kuwabara and Imai, 2009; Pagter and Arora, 2013). Thus, a question raised is, what might be the advantage for plants to induce expression of the SA immunity pathway in the absence of pathogens? As we previously speculated (Kim et al., 2013), one possibility is that it might be a preemptive defense strategy. Cold-acclimated plants are better able to survive freezing, but they still suffer freezing injury. Such injury would presumably enable pathogens to gain access to intercellular spaces more easily and result in the plants becoming more susceptible to infection. In addition, wounding would be expected to activate JA signaling which is antagonistic to SA signaling (Robert-Seilaniantz et al., 2011; Thaler et al., 2012). Thus, “waiting” for a pathogen attack to activate expression of the SA pathway might be a “too little, too late” strategy. Indeed, Todesco et al. (2010) identified Arabidopsis accessions that carry alleles of the ACCELERATED CELL DEATH6 (ACD6) gene that result in constitutively elevated levels of SA and enhanced disease resistance. An analysis of worldwide Arabidopsis populations led these investigators to conclude that these alleles of ACD6 are likely to provide substantial fitness benefits despite the decrease in growth rate and biomass that is associated with SA signaling.
The CAMTA proteins not only have a critical role in regulating plant defense genes, but also have an important role in regulating the expression of genes that impart freezing tolerance (Doherty et al., 2009; Kim et al., 2013; Kidokoro et al., 2017). However, in the case of freezing tolerance, the role of the CAMTA proteins is reversed: Whereas the CAMTA transcription factors repress the expression of SA immunity genes, they induce the expression of freezing tolerance genes. Within minutes of exposing plants to low temperature, CAMTA1, CAMTA2, CAMTA3, and CAMTA5 contribute to the induction of three C-repeat binding factor (CBF) genes—CBF1, CBF2, and CBF3 (also known as DREB1b, DREB1c, and DREB1a, respectively)—which encode closely related members of the AP2/ERF family of transcription factors (Doherty et al., 2009; Kim et al., 2013; Kidokoro et al., 2017). Expression of the CBF proteins leads to the induction of more than 100 genes, the CBF regulon; this induction contributes to the increase in freezing tolerance that occurs in response to low temperature (Thomashow, 2010). Determining how the CAMTA proteins respond rapidly to low temperature to induce the expression of freezing tolerance genes and respond slowly to low temperature to relieve repression of SA pathway genes is fundamental to an understanding of low temperature signaling and the regulation of freezing tolerance and plant immunity by the CAMTA proteins. Our results indicate that the IQ and CaMB domains, and thus presumably calcium signaling, have a role in CAMTA regulation of SA pathway genes in response to low temperature. The fact that calcium levels spike rapidly upon transferring Arabidopsis from moderate to low temperature (Knight et al., 1991) and that calcium antagonists impair the induction of some cold-regulated genes (Knight et al., 1996) suggests that calcium signaling also has a role in CAMTA regulation of freezing tolerance genes. However, if true, it would seem that the rapid and delayed cold-induced calcium signatures must be different given the different effects that rapid and delayed low-temperature signaling has on the regulatory activities of the CAMTA proteins.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana plants used in this study were in the Col-0 background. The camta2 camta3 mutant has been described previously (Kim et al., 2013). All plants were grown in pots on soil for 3 to 4 weeks in a growth chamber set at 22°C in a 12-h photoperiod with a light intensity of ∼120 µmol m−2 s−1. Cold treatment was at 4°C in a 12-h photoperiod with a light intensity of ∼35 µmol m−2 s−1. Cool white fluorescent bulbs (T8 type) were used for both plant growth and cold treatment.
Gene Constructs and Plant Transformation
Site-directed mutagenesis was performed on the CAMTA3 coding sequence in a Gateway entry vector (Life Technologies) as described (Ho et al., 1989). All mutations were verified by DNA sequencing. The oligonucleotide primers used in the PCR reactions can be found in Supplemental Table 1. Mutated CAMTA3 sequences were recombined into vector pEGC3PGFP, a binary vector based on pEarleyGate100 (Earley et al., 2006), but containing the CAMTA3 endogenous promoter (2 kb) and an in-frame terminal GFP (derived from pEarleyGate103). Constructs were transformed into camta2 camta3 mutant plants using the floral dip method (Clough and Bent, 1998). Homozygous plants (T3 or higher) were used for all experiments.
Quantification of Transcript Levels
Total RNA was extracted from leaf tissue using an RNeasy plant mini kit (Qiagen) with on-column DNase treatment. Synthesis of cDNA was performed on 100 ng (Figures 6C and 7B) and 250 ng (all other figures) total RNA and random primers using a reverse transcription system (Promega) in a 20-μL volume as detailed by the manufacturer. Real-time RT-qPCR was performed using SYBR Green in a 10-μL volume as described previously (Kim et al., 2013). cDNA was diluted either 10- or 20-fold and 2 μL was used in the quantitative PCR reaction. Either UBQ10 (all genes in Figures 6C and 7B), PP2A (ICS1 and CBP60g), YLS8 (PR1 and SARD1), or IPP2 (CAMTA3 and GFP) were used for normalization. Primer sequences are given in Supplemental Table 2. Three biological replicates were performed for each experiment from plants grown at different times under the same conditions. Leaf tissue was pooled from separate plants for each sample. Relative expression values were calculated using the ddCt method using an inter-run calibrator that was set to one for each plate. In Figures 1B, 4A, 6C, and 7B, dCt was used instead of ddCt. For Figures 1, 2, and 4 to 7, RT-qPCR data were analyzed by ANOVA (Supplemental Table 3). Mean values were separated by LSD at P < 0.05. Data for PR1 were square root transformed to reduce heteroscedasticity of treatment variance prior to ANOVA. Values shown in the figures are untransformed for clarity.
Protein Extraction and Immunoblots
To prepare total proteins, leaf samples were ground, heated in SDS loading buffer for 10 min at 95°C, then centrifuged. Nuclear protein was prepared as described previously (Dong et al., 2011). Leaf samples were ground, immersed in chilled extraction buffer 1, and centrifuged to recover the pellets, which were subsequently resuspended in extraction buffer 2 and centrifuged. Remaining pellets were added with extraction buffer 3 and layered over another extraction buffer 3 for centrifugation. Finally, nuclear pellets were recovered and resuspended in nuclear lysis buffer. Protein samples were analyzed on a 4 to 12% BT NuPAGE gel (Life Technologies) and proteins were transferred to a Hybond ECL nitrocellulose membrane (Life Technologies). The membrane was blocked with 5% skim milk in 1× TBST for 2 h at room temperature followed by incubation with the primary antibody (rabbit anti-GFP [Life Technologies], mouse anti-Histone H3 [Millipore], or rabbit anti-UGPase [Agrisera]) in 1% skim milk at 4°C overnight. The secondary antibody was horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Thermo Fisher). SuperSignal West Femto Chemiluminescent Substrate (Thermo Fisher) was used for visualization.
Quantification of SA and SA Glucoside Levels
SA and SA glucosides were determined as described by Kim et al. (2013).
Determination of Pathogen Resistance
Inoculation of plants (grown at 22°C or exposed to 4°C for 3 weeks) with Pseudomonas syringae pv tomato DC3000 was performed by pressure infiltration with a 1-mL needleless syringe as described previously (Yao et al., 2013). After inoculation, four infected leaves were collected from each genotype or treatment to determine the initial pathogen number. Eight representative leaves were collected 3 d after inoculation to examine pathogen growth. The syringe infiltration inoculation method was also used for pathogen-induced gene expression analysis. Leaves infiltrated with either bacteria or 10 mM MgCl2 (mock) were collected 24 h after inoculation for RNA extraction and RT-qPCR gene expression analysis.
Confocal Microscopy
Plants were grown for 3 weeks at 22°C or for 3 weeks at 22°C followed by exposure to 4°C for 3 weeks. Leaves were analyzed using an inverted Laser Scanning Nikon A1Rsi confocal microscope with a PlanFluor 40× oil DIC H N2 objective. Each image was captured in the medial region of the cell with a 512 × 512 pixel resolution with pinhole and laser power kept at the same values for all samples to allow comparison of the fluorescence intensities across the different lines. The GFP signal was detected using 488-nm excitation and 505- to 555-nm emission wavelengths. NIS-Elements Advanced Research software (Nikon) was used for image handling and fluorescence quantification. The fluorescence intensity of the tagged protein was measured in the nucleus using a fixed ROI of 8.1 µm2.
Accession Numbers
Sequence data for genes and proteins presented in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: CAMTA1 (AT5G09410), CAMTA2 (AT5G64220), CAMTA3 (AT2G22300), CBP60g (AT5G26920), ICS1 (AT1G74710), PR1 (AT2G14610), and SARD1 (AT1G73105).
Supplemental Data
Supplemental Figure 1. Transcript Levels for CAMTA3 Variants in Transgenic Lines.
Supplemental Table 1. Primers Used for Site-directed Mutagenesis of CAMTA3.
Supplemental Table 2. Primers Used for Quantitative RT-PCR.
Supplemental Table 3. ANOVA Tables.
Acknowledgments
We thank Sheng Yang He for his critical reading of the manuscript and Lena Sanfilippo and Cynthia Collings for excellent technical assistance in conducting the experiments. We thank Lijun Chen at the Mass Spectrometry Facility at Michigan State University (Department of Biochemistry and Molecular Biology) for help with the SA and SA glucoside measurements. The work to construct the CAMTA3 protein variants was supported by funds to M.F.T. from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy (DE-FG02-91ER20021). The experiments testing the effects of the CAMTA variants on gene expression and plant immunity were supported by funds to M.F.T. from the Michigan AgBioResearch program (MICL02415). L.W. was supported by a scholarship from the China Scholarship Council (No.201406850041). L.R. was supported by the U.S. Department of Energy, Office of Basic Energy Sciences (award number DE-FG02-91ER20021).
AUTHOR CONTRIBUTIONS
Y.S.K., C.A., S.P., S.J.G., and L.W. conducted all of the experiments and analyzed the results with the exception of the confocal microscopy, which was performed and analyzed by L.R. and F.B., and the statistical analysis of results, which was performed by R.G. M.F.T. helped analyze the results, conceptualized the study with contributions from Y.S.K., C.A., S.P., and S.J.G., and wrote the manuscript with input from the other authors.
Footnotes
Articles can be viewed without a subscription.
References
- Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277: 21851–21861. [DOI] [PubMed] [Google Scholar]
- Choi M.S., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280: 40820–40831. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Jonge J., Hofius D., Hennig L. (2017). Salicylic acid interferes with GFP fluorescence in vivo. J. Exp. Bot. 68: 1689–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey D.A., Vlot A.C., Wildermuth M.C., Klessig D.F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulated expression of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ali G.S., Simons K.A., Hou J., Yang T., Reddy A.S.N., Poovaiah B.W. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.A., Parker J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkler A., Ashery-Padan R., Fromm H. (2007). CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 581: 3893–3898. [DOI] [PubMed] [Google Scholar]
- Fromm H., Finkler A. (2015). Repression and de-repression of gene expression in the plant immune response: the complexity of modulation by Ca2+ and calmodulin. Mol. Plant 8: 671–673. [DOI] [PubMed] [Google Scholar]
- Galon Y., Nave R., Boyce J.M., Nachmias D., Knight M.R., Fromm H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582: 943–948. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Jing B., Xu S., Xu M., Li Y., Li S., Ding J., Zhang Y. (2011). Brush and spray: a high-throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiol. 157: 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M.E., MacLean D., Studholme D.J., Serna-Sanz A., Andreasson E., Rathjen J.P., Peck S.C. (2009). Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J. Proteomics 72: 439–451. [DOI] [PubMed] [Google Scholar]
- Kidokoro S., Yoneda K., Takasaki H., Takahashi F., Shinozaki K., Yamaguchi-Shinozaki K. (2017). Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29: 760–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Park S., Gilmour S.J., Thomashow M.F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75: 364–376. [DOI] [PubMed] [Google Scholar]
- Knight H., Trewavas A.J., Knight M.R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M.R., Campbell A.K., Smith S.M., Trewavas A.J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526. [DOI] [PubMed] [Google Scholar]
- Kuwabara C., Imai R. (2009). Molecular basis of disease resistance acquired through cold acclimation in overwintering plants. J. Plant Biol. 52: 19–26. [Google Scholar]
- Nie H., Zhao C., Wu G., Wu Y., Chen Y., Tang D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158: 1847–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagter M., Arora R. (2013). Winter survival and deacclimation of perennials under warming climate: physiological perspectives. Physiol. Plant. 147: 75–87. [DOI] [PubMed] [Google Scholar]
- Pauly N., Knight M.R., Thuleau P., Graziana A., Muto S., Ranjeva R., Mazars C. (2001). The nucleus together with the cytosol generates patterns of specific cellular calcium signatures in tobacco suspension culture cells. Cell Calcium 30: 413–421. [DOI] [PubMed] [Google Scholar]
- Poovaiah B.W., Du L., Wang H., Yang T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 163: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H., Yang J., Xu Y.P., Munyampundu J.P., Cai X.Z. (2016). Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 7: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S.D., Ali G.S., Celesnik H., Day I.S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E., Mur L.A.J. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Yang Y., Du L., Wang H. (2015). Calmodulin-binding transcription activators and perspectives for applications in biotechnology. Appl. Microbiol. Biotechnol. 99: 10379–10385. [DOI] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T., Whiteman N.K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17: 260–270. [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., et al. (2010). Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W., Sreekanta S., Lu Y., Bethke G., Tsuda K., Katagiri F., Glazebrook J. (2013). The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 163: 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal S.C., Jeong R.D., Mandal M.K., Zhu S., Chandra-Shekara A.C., Xia Y., Hersh M., Stromberg A.J., Navarre D., Kachroo A., Kachroo P. (2009). Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 5: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bian Y., Cheng K., Gu L.F., Ye M., Zou H., Sun S.S.M., He J.X. (2013). A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteomics 78: 486–498. [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Xin X.F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Yao J., Withers J., He S.Y. (2013). Pseudomonas syringae infection assays in Arabidopsis. Methods Mol. Biol. 1011: 63–81. [DOI] [PubMed] [Google Scholar]
- Zhang L., Du L., Shen C., Yang Y., Poovaiah B.W. (2014). Regulation of plant immunity through ubiquitin-mediated modulation of Ca2+-calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78: 269–281. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Ding P., Wang D., Cheng Y.T., He J., Gao M., Xu F., Li Y., Zhu Z., Li X., Zhang Y. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 107: 18220–18225. [DOI] [PMC free article] [PubMed] [Google Scholar]