With no capital costs, inexpensive Oxford Nanopore sequencing can be applied to novel ∼1-Gb plant genomes.
Abstract
Updates in nanopore technology have made it possible to obtain gigabases of sequence data. Prior to this, nanopore sequencing technology was mainly used to analyze microbial samples. Here, we describe the generation of a comprehensive nanopore sequencing data set with a median read length of 11,979 bp for a self-compatible accession of the wild tomato species Solanum pennellii. We describe the assembly of its genome to a contig N50 of 2.5 MB. The assembly pipeline comprised initial read correction with Canu and assembly with SMARTdenovo. The resulting raw nanopore-based de novo genome is structurally highly similar to that of the reference S. pennellii LA716 accession but has a high error rate and was rich in homopolymer deletions. After polishing the assembly with Illumina reads, we obtained an error rate of <0.02% when assessed versus the same Illumina data. We obtained a gene completeness of 96.53%, slightly surpassing that of the reference S. pennellii. Taken together, our data indicate that such long read sequencing data can be used to affordably sequence and assemble gigabase-sized plant genomes.
INTRODUCTION
The last few years have seen tremendous developments in sequencing technologies, which have in turn led to substantial advances in plant genomics. To date, the genomes of ∼200 plant species have been published (www.plabipd.de) (Bolger et al., 2017), yet sequencing plant genomes remains comparatively difficult due to their large sizes and high repeat content (Jiao and Schneeberger, 2017). Long-range data are extremely valuable for resolving repetitive genomic regions and several new technologies have made substantial advances in this area. Some of these technologies track the larger, many kilobase DNA fragments from which shorter Illumina reads were derived, facilitating assembly. In the plant genomics field, one such method, synthetic long reads, has been included to help sequence a new maize (Zea mays) cultivar (Hirsch et al., 2016). By contrast, other new technologies are PCR-free and either directly sequence or produce a sequence barcode from single molecules. For instance, long PacBio reads have been tested successfully in assembling the genome of the model plant Arabidopsis thaliana (Berlin et al., 2015), and optical mapping (restriction barcoding) has been used to improve contiguity in the latest 3.0 release of the cultivated tomato genome (www.solgenomics.net). Another example driving genome technology is the use of dovetail Hi-C proximity ligation. This has been used to vastly improve the lettuce (Lactuca sativa) genome (Reyes-Chin-Wo et al., 2017), and it offers the future possibility of improving fragmented plant genome assemblies to chromosome scale. Combinations of new long-range sequencing technologies are also powerful and have been used to sequence, e.g., a desiccation tolerant grass (VanBuren et al., 2015) and quinoa (Jarvis et al., 2017). However, these long-range sequencing technologies rely on previous extraction of high quality, high molecular weight DNA, which can be an additional challenge in many plants due to both cell walls and secondary metabolite content.
Technological improvements have reduced sequencing costs and increased accessibility; however, large challenges remain for individual labs attempting a genome project. Many of the above-mentioned methods rely on expensive, specialized machinery. State-of-the-art sequencing equipment, however, requires high capital investments and quickly depreciates in value due to new technological developments in the genomics field (compared with Glenn [2011] and companion online updates for recent developments). Thus, it is often financially advantageous for a standard lab to outsource some of the sequencing. Outsourcing, in turn, substantially slows down any necessary iteration in the sequencing project, be it to optimize the DNA quality or library preparation or simply to progressively add to total data.
Recently, Oxford nanopore has emerged as a competitor for long-read sequencing. Notably, Oxford nanopore produces a mini-sequencer, the MinION, requiring only a start-up fee of $1000, which includes two flow cells and a library preparation kit (https://store.nanoporetech.com/minion/sets/?___SID=U). Furthermore, recent updates in nanopore sequencing technology that became commercially available in late 2016 made it possible to obtain gigabases of sequence data from a single flowcell. Prior to this, due to relatively low output, the nanopore sequencing technology was mainly used to analyze and assemble microbial samples (Loman et al., 2015; Quick et al., 2015; Jain et al., 2016; Kranz et al., 2017). Notably, early reports of Oxford nanopore reads indicate that they are exceptionally long (Weirather et al., 2017) but have a high (Judge et al., 2015), and nonrandom, error rate (Deschamps et al., 2016).
A new Solanum pennellii accession has been identified with traits that make it an interesting target for de novo sequencing. S. pennellii is a wild, green-fruited tomato species native to Peru that exhibits beneficial traits such as abiotic stress resistances (Lippman et al., 2007; Koenig et al., 2013). The previously sequenced accession LA716 (Bolger et al., 2014a) has been used to generate a panel of introgression (Eshed and Zamir, 1995) and backcrossed introgression (Ofner et al., 2016) lines that have been used to identify many interesting quantitative trait loci (Alseekh et al., 2015; Fernandez-Moreno et al., 2017), thus complementing large-scale genomic panel studies for tomato (Lin et al., 2014; Tieman et al., 2017). However, the accession LA716 does not perform well in the field and carries the NECROTIC DWARF gene on chromosome 6, which reduces plant vigor when introduced into a Solanum lycopersicum background (Ranjan et al., 2016). A novel divergent accession LYC1722 was identified in a large panel of tomato accessions obtained from the IPK gene bank in Germany as a self-compatible, phenotypically uniform biotype of S. pennellii that does not exhibit these negative traits of LA716. We chose to sequence and assemble the LYC1722 accession de novo using Oxford nanopore technology. The availability of a reference quality genome for the LA716 S. pennellii accession also made it an excellent genome with which to evaluate not just the practicality, but also the resulting quality of Oxford nanopore sequencing for assembling a gigabase-sized plant genome.
Here, we report the de novo sequencing and assembly of S. pennellii LYC1722 using Oxford nanopore long reads, complemented with Illumina short reads for polishing. Genome contiguity, genic completeness, and other quality measures showed the resulting assembly was of comparable or better quality than the Illumina-based LA716 assembly. The genome was already of sufficient quality for comparing gene content within and between species and Oxford nanopore data allowed novel analyses like direct methylation measurement.
RESULTS
Initial Characterization of the S. pennellii LYC1722 Accession
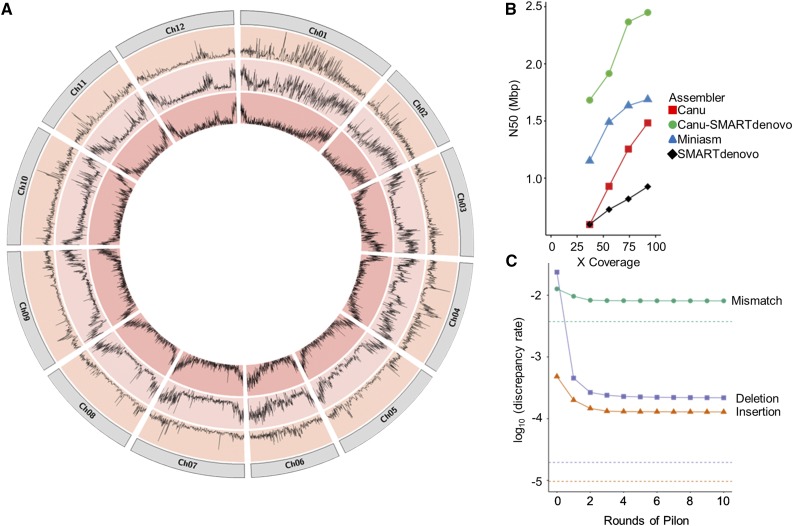
To obtain first insights into the genome of the new S. pennellii accession LYC1722, we generated ∼39 Gb of 2x 300-bp Illumina reads. A kmer analysis of this data set indicated that this accession of S. pennellii has a genome size between 1 and 1.2 Gb (Supplemental Figure 1), similar to the estimate for the reference S. pennellii LA716. Furthermore, the target LYC1722 accession is relatively homozygous (Supplemental Figure 1) in line with its self-compatibility, a trait found in some southern S. pennellii populations, including LA716 and LA2963, and which contrasts with the strict self-incompatibility and high heterozygosity typical of this species as a whole (Rick and Tanksley, 1981). Using the short-read sequencing data to identify variants such as single nucleotide polymorphisms (SNPs) and small insertions and deletions (InDels) versus the S. pennellii LA716 reference revealed 6.2 million predicted variants where the highest variant rate was found on chromosomes 1 and 4 (Figure 1A; Supplemental Table 1). For comparison, in a large panel of cultivated tomatoes (S. lycopersicum) there were only a few cases where more than 2 million variants were found (Aflitos et al., 2014). In addition, the metabolite content of LYC1722 differed from that of LA716 (Supplemental Figure 2). Taken together, these characteristics highlight the high level of diversity within S. pennellii (Rick and Tanksley, 1981) and show that LA716 and LYC1722 are relatively diverged accessions, which as such might provide different beneficial traits or alleles (Aflitos et al., 2014).
Figure 1.
Characteristics of the S. pennellii Genome and Its Assembly.
(A) Circos visualization of variant distribution between S. pennellii LYC1722 and S. pennellii LA716. Distribution of single nucleotide polymorphisms (outer layer) and InDels (middle layer) is compared with the gene density (inner layer) for each chromosome of S. pennellii LA716 based on generated Illumina data for S. pennellii LYC1722.
(B) The effect of randomly downsampling pass reads on the N50 produced by different assemblers.
(C) Discrepancies between the assembly and the Illumina data over several rounds of Pilon correction. Dotted lines approximate expected discrepancy rates if Illumina data were mapped to a perfect reference.
Oxford Nanopore Sequence Statistics and Metrics for S. pennellii Reads
Having established substantial differences between the accessions and a within-range genome size, we continued with Oxford nanopore sequencing. Using Oxford nanopore sequencing served both to allow full de novo assembly and avoid reference-based bias and to test the performance of Oxford nanopore sequencing in the plant field. To obtain high coverage of long reads for the gigabase-sized genome in an economic fashion, the majority of the libraries were prepared with an optimized protocol. This protocol included gel-based size selection allowing for a less extreme trade-off between length and yield than the official protocols from early 2017. We thus sequenced the genome of this new self-compatible S. pennellii accession with Oxford nanopore reads. Thirty-one flowcells yielded 134.8 Gb of data in total, of which 110.96 Gb (representing ∼100-fold coverage) were classified as “passed filter,” by the Oxford nanopore Metrichor 1.121 basecaller, representing data of somewhat higher quality. As shown in Supplemental Table 2, total yield per flowcell varied between 1.1 and 7.3 Gb before and 0.96 and 6.02 Gb after filtering. Most data were obtained within the first 24 h of sequencing (Supplemental Figure 3). The average quality Q-score was around 6.88 before and 7.44 after filtering (Supplemental Figure 4), indicating a read error rate of 20 and 18%, respectively. Realigning the reads against the finalized genome assembly (see below) revealed a typical read identity value of aligned bases of 80.97% (Supplemental Figure 5), in line with the estimated quality values (Supplemental Figure 6). However, these values are lower than those observed in microbial data (Ip et al., 2015; Loman et al., 2015), which may be explained by the fact that the basecaller was not trained for unamplified plant DNA.
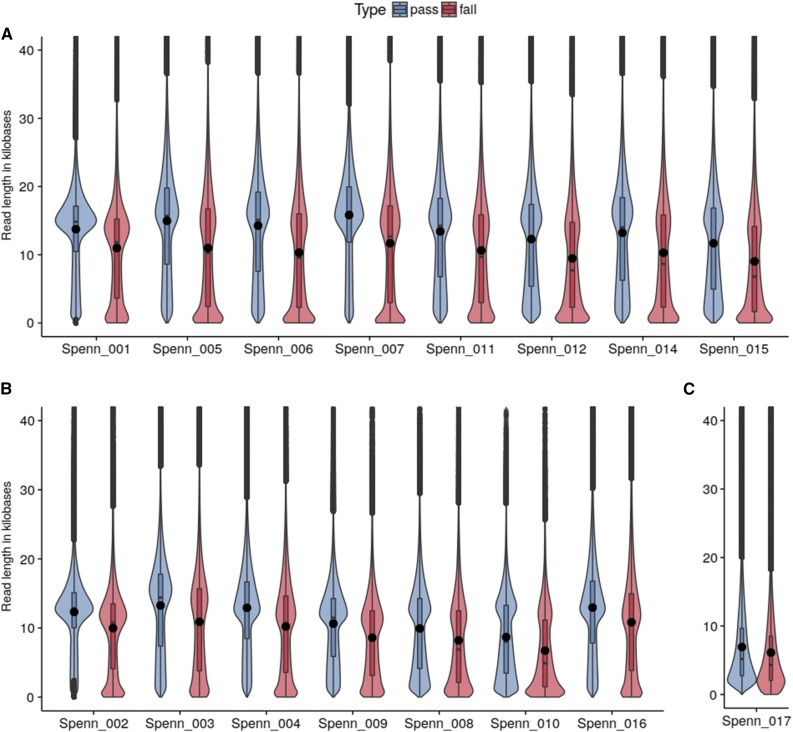
The average read length for the libraries varied between 6760 (6925) and 14,807 (15,822) with library preparation optimizations before (and after) quality filtering (Figure 2; Supplemental Table 3). Notably, it was possible to routinely achieve libraries with an average read length of 12.7 kb when gel-based size selection was employed. The longest read that passed quality filter was 153,099 bases long with an alignment length of 132,365 bases.
Figure 2.
Violin Plots of Read Length per Library for Three Different Size-Selection Protocols.
Read length distribution is shown for all 16 S. pennellii MinION libraries and the corresponding pass (blue) and failed (red) classified reads. Libraries are grouped by size selection protocol: (A) 15-kb cutoff, (B) 12 kb cutoff, and (C) 0.4x bead size selection. Filled dots indicate mean read length.
Genome Assembly Strategies and Metrics
Several assembly options were compared, as it was unclear which would perform best for Oxford nanopore reads from a highly repetitive plant genome (Jiao and Schneeberger, 2017). The data were assembled using Canu (Koren et al., 2017) and SMARTdenovo, which represent state-of-the-art assemblers known to support Oxford nanopore sequencing technology (Istace et al., 2017). Furthermore, data were assembled with miniasm (Li, 2016), which is a fast assembler without a consensus step, thus necessitating a postassembly polishing and/or consensus step. In addition, we used Canu to precorrect the original reads and assembled the resulting data using SMARTdenovo (hereafter Canu-SMARTdenovo) as described in the Supplemental Methods. Assemblies of the genome with the hybrid assembler dbg2olc (Ye et al., 2016) and an early version of the wtdbg assembler had subpar N50 values and were thus not analyzed further (Supplemental Data Sets 1A and 1B).
Statistically, the most contiguous assembly was the one obtained by Canu-SMARTdenovo, with an N50 value of 2.45 Mb and just 899 total contigs. The largest contig in this assembly was 12.32 Mb. Of the single-assembler options, Miniasm had the highest N50 of 1.69 Mb, versus 1.48 Mb for Canu and 1.03 Mb for SMARTdenovo (Table 1) after parameter tuning (Supplemental Data Sets 1C and 1D). Computational requirements varied greatly, with Canu 1.4-0c206c9 needing almost two orders of magnitude more CPU hours than Miniasm or SMARTdenovo (Table 1). However, a newer version of Canu significantly lowered the consumed CPU hours from ∼80k to 14.36k CPU hours, closing the speed gap to the other assemblers.
Table 1. Assembly Statistics and Run-Time Statistics by Assembly and Postprocessing.
| Assembler | k CPU Hours | Memory (GB) | N50 | L50 | Total Size | Largest Contig | Total Contigs | Illumina Mapping Rate (%) | Qualimap Discrepancy Rate | % Complete BUSCO |
|---|---|---|---|---|---|---|---|---|---|---|
| Raw | ||||||||||
| Canu | 80.42 | 199.87 | 1.48 | 169 | 922.94 | 9.63 | 2010 | 98.52 | 3.74 | 26.46 |
| SMARTdenovo | 0.72 | 55.60 | 1.03 | 271 | 929.99 | 5.68 | 1901 | 98.65 | 4.22 | 26.74 |
| Miniasm | 1.86 | 51.93 | 1.69 | 158 | 956.29 | 9.28 | 2704 | 95.53 | 9.11 | 0.21 |
| Canu-SMARTdenovo | 10.68 | 131.32 | 2.45 | 106 | 889.92 | 12.32 | 899 | 98.73 | 3.68 | 29.1 |
| Pilon Polished 5x | ||||||||||
| Canu | – | – | 1.55 | 169 | 961.83 | 10.01 | 2010 | 98.95 | 0.82 | 96.46 |
| SMARTdenovo | – | – | 1.06 | 270 | 955.31 | 5.84 | 1901 | 98.99 | 0.91 | 96.11 |
| Miniasm | – | – | 1.75 | 156 | 977.78 | 9.49 | 2704 | 98.24 | 2.48 | 85.69 |
| Canu-SMARTdenovo | – | – | 2.52 | 106 | 915.60 | 12.72 | 899 | 98.98 | 0.85 | 96.46 |
All sequence lengths are in megabases. –, CPU and memory resources were not tracked for polishing. See Supplemental Data Set 1 for additional polishing data.
To test the structural correctness of the genome, we aligned the “best” assemblies from Canu-SMARTdenovo, Canu, SMARTdenovo, and Miniasm against the LA716 reference genome. We argued that despite differences in the two accessions on the small-scale level, general structure should be conserved. Indeed, we observed that all four assemblies were comparable with the reference (Supplemental Figure 7), although Miniasm had a perceptibly lower overall alignment rate, as expected due to its lack of a consensus step.
Effects of Read Coverage and Length on Genome Assembly Statistics
To assist in future experimental design, we evaluated the effects of coverage and read length on assembly contiguity. Considering the larger (>1 Gb) genome and the less-established technology, this project aimed for twice the coverage (100x) as the 50x of PacBio reads, which had produced a highly contiguous assembly of the model plant Arabidopsis featuring a genome N50 of 5 Mb (Koren et al., 2017). To assess whether 100x was saturating, or if lower coverage would be sufficient, we subsampled the “passed filter” data set to 40, 60, and 80%, and assembled these with each pipeline. As can be seen in Figure 1B, the N50 was still rising with the inclusion of the full data set, although the increase in N50 from one assembler, Miniasm, was starting to taper. When we reanalyzed the data not versus assembly size but with a maximal genome size estimation of 1.2 Gb (NG50), while the order of the assemblies stayed the same, many assembly NG50 values started to taper (Supplemental Figure 8).
Given the good results for Arabidopsis with PacBio data (Koren et al., 2017), we determined the effect of read length at medium coverage. We produced several subsamples of the data set representing 30x coverage but with different average read lengths, and we assessed the continuity of assembling these subsets with SMARTdenovo. This analysis showed a positive correlation between the resulting N50 value and the average read length at constant 30x coverage. The highest N50 of over 1 Mb was slightly higher than the N50 of the whole data set assembled with SMARTdenovo (Table 1). This was achieved when the average read length surpassed 20 kb. On the other hand, an N50 of only ∼0.2 Mb was produced when the average read length was less than 13 kb (Supplemental Figure 9). This drop in contiguity was more dramatic than that caused by randomly subsampling the data to 40%, where all assemblers produced an N50 value above 0.5 Mb (Figure 1B). Notably, libraries with the higher target for gel-based size selection produced an average of 48.1k reads per flowcell over 20 kb (15%), while the overall higher-yielding standard library produced just 34.1k reads per flowcell over 20 kb (3%). These data indicate that the protocol optimization provided both absolute and relative gains in some of the most valuable reads for assembly.
Prior to Polishing, Genome Error Rate Is Substantial
Assembly quality is dependent on more than simple contiguity, so we checked other important quality measures such as base accuracy and gene content. To estimate base error rate, the nanopore assemblies were compared with the same Illumina data that were used above to predict genome size and small variants versus the reference LA716. To put an upper bound on error rate, we used Qualimap (Okonechnikov et al., 2016), which totals all the discrepancies between the individual raw read data and the reference. To put a lower bound on error rate, we used samtools (Li et al., 2009) to call variants that have consistent support of Illumina reads. For simplicity, the qualimap-based upper bound will be referred to as discrepancy rate, and the variant calling-based lower bound as error rate.
While some raw assemblies performed better than others, all showed high error and discrepancy rates. The error rate was estimated at 2.66, 1.54, 1.2, and 1.1% for the raw assemblies from miniasm, SMARTdenovo, Canu-SMARTdenovo, and Canu, respectively (Supplemental Data Set 1E). For the same raw assemblies, the total discrepancy rate was much higher at 9.11, 4.22, 3.68, and 3.74% for Miniasm, SMARTdenovo, Canu-SMARTdenovo, and Canu, respectively (Supplemental Data Set 1F). Deletions in the assembly were the most common discrepancy, with insertions being an order of magnitude less common (Figure 1C; Supplemental Data Set 1F). The substantial differences between error and discrepancy rate may be attributable to true errors being large enough to disrupt alignment and therefore downstream error and discrepancy rate calculations as well as errors in the short-read data and remaining heterozygosity, which cannot be resolved in qualimap.
As expected from the many base errors, the raw assemblies showed a low genic completeness. BUSCO (Simão et al., 2015) was used to identify and count orthologs from orthologous groups generally conserved in plants. BUSCO estimated the genic completeness at 0.21, 26.46, 26.74, and 29.1% for the assemblies from miniasm, Canu, SMARTdenovo, and Canu-SMARTdenovo, respectively (Table 1; Supplemental Data Set 1G). This pattern suggests that, as anticipated, all four assemblies—while being structurally mostly correct—can be considered only predrafts and should not be regarded as useful for gene definition.
To determine if the high error rates and low genic completeness may be addressable with nanopore data alone, a consensus nanopore data polishing tool, Racon (Vaser et al., 2017), was used for the miniasm, Canu, and Canu-SMARTdenovo assemblies. Racon reduced the discrepancy rates by ∼0.5% and led to a decreased imbalance in the discrepancy rate for insertions and deletions. More dramatically, however, Racon improved the genic completeness by over 15%, giving final scores of 43.19 and 44.58% for Canu and Canu-SMARTdenovo, respectively (Supplemental Data Set 1G). To assess whether Racon would be an adequate post assembly step for the miniasm assembler, the miniasm assembly was polished five times using Racon, which consumed between 712 and 835 CPU hours for each iteration. Indeed, after one polishing step, the discrepancy rate fell from 9.11 to 3.47%, the latter value being comparable to the other raw assemblies. After three and four additional polishing rounds, the error rate fell to 2.93 and 2.92%, respectively (Supplemental Data Set 1F). Similarly, one round of Racon polishing increased the genic completeness of the miniasm assembly to 40.97%, whereas five total polishing rounds yielded a completeness score of 47.78% (Supplemental Data Set 1G). Taken together, these data indicate that four to five total rounds of Racon polishing can be beneficial for miniasm assemblies.
As the execution speed of the tool nanopolish (Loman et al., 2015) was adapted for larger genomes in mid 2017, we assessed the effect this tool had on the nanopore assembly using the most contiguous Canu-SMARTdenovo assembly after the application of Racon. This approach proved beneficial as the discrepancy rate decreased to 2.15% and the genic completeness rose to 84.1% (Supplemental Data Sets 1F and 1G); however, polishing took ∼37.5k CPU hours.
Notably, each self-consensus or correction step with nanopore reads generally improved base error and genic completeness metrics, yet not enough to reach the standards expected of a high-quality genome assembly.
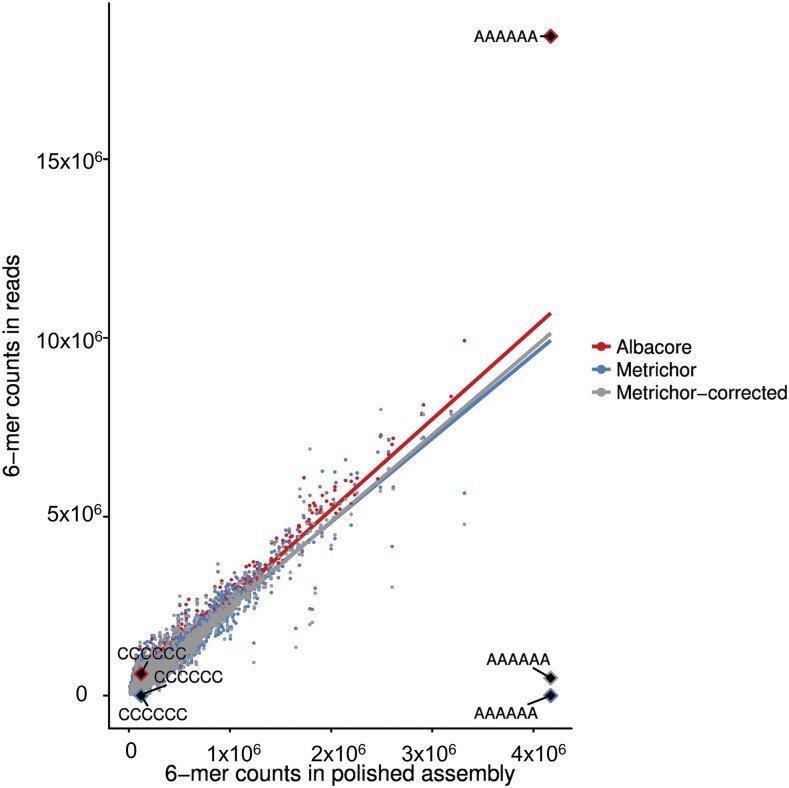
The kmer bias in raw and Canu-corrected reads was analyzed to evaluate whether nonrandom errors may have contributed to the limited efficacy of nanopore self-correction. As expected based on the basecaller used (Lu et al., 2016), the nanopore reads contained (almost) no homohexamers and were depleted in shorter homopolymers compared with the final (see below) Illumina polished assembly (Figure 3). Unsurprisingly, only slight reductions in this bias could be seen in Canu-corrected reads, indicating limitations on self-correction of basecalled reads. By contrast, a rerun of a subset of the basecalling with Oxford Nanopore’s new basecaller, Albacore, showed less depletion or even enrichment of homopolymers relative to the Illumina polished assembly (Figure 3).
Figure 3.
The 6-mer Counts in the Polished Assembly versus Those in the Raw Reads.
The 6-mers were counted both in the polished assembly and in the raw reads. Each 6-mer represents counts to both itself and to its reverse complement, i.e., AAAAAA represents both AAAAAA and TTTTTT. Red indicates the new Albacore basecaller, whereas blue and gray dots represent the raw and Canu-corrected Metrichor data. In each case, a trend line is added.
After Polishing, Genome Quality Is Competitive with That of the Reference Genome
As Illumina data are known to have a lower overall error rate, with most errors being mismatches and not InDels, further polishing was performed with the Illumina data using Pilon (Walker et al., 2014). A single round of Pilon already brought the genic completeness up to 95.76% and peaked after four rounds at 96.53% for the Canu-SMARTdenovo assembly (Supplemental Data Set 1G). This was slightly (three orthogroups) better than the reference LA716 genome at 96.32%. Iteration of Pilon applications almost continuously decreased discrepancy and error rate, although diminishing returns were apparent after approximately five rounds (Figure 1C; Supplemental Data Sets 1E and 1F). Ultimately, the overall discrepancy rate fell to the same order of magnitude as was expected for errors in the Illumina data (0.84% in Canu-SMARTdenovo versus the expected 0.37%), and the remaining discrepancies were dominated by mismatches. More conservatively, the variant calling-based error rate fell to or below 0.02% for all assemblies, with more insertion or deletion variants remaining than mismatches (Supplemental Data Set 1E). The lowest error rate was found for the polished Canu assembly, with fewer than 90,000 homozygous variants in the 840 Mb of genomic regions covered by at least five reads, representing an error rate approaching 0.01%. The structurally most contiguous assembly, Canu-SMARTdenovo, reached an error rate of 0.016% after polishing. Using an independent Illumina data set featuring a different base detection method (NextSeq) largely confirmed this error rate with a value of 0.025% (Supplemental Data Set 1E).
Notably, over 10 rounds of Pilon polishing could not bring the Miniasm assembly up to a comparable quality as the others, with the discrepancy rate leveling around 2.46% and BUSCO’s genic completeness score leveling just above 85%. However, when first polishing the Miniasm assembly five times with Racon, a single round of Pilon polishing was enough to yield a genic completeness of 94.86% and a discrepancy rate of 0.78%.
Final Genome Quality Sufficient for Intergenic Comparisons
Considering the differences in phenotype of the target LYC1722 and the reference LA716 accessions and the different results of introgressing these lines with S. lycopersicum, we wondered whether there would be apparent differences in gene presence/absence between the accessions and species. For a more detailed perspective on this than could be obtained by, e.g., BUSCO, we called de novo gene models for the Canu-SMARTdenovo, 4x Pilon assembly with Augustus (Stanke and Waack, 2003; Stanke et al., 2008), and created orthogroups between Arabidopsis, S. lycopersicum, S. pennellii LA716, and the new S. pennellii LYC1722 with OrthoFinder (Emms and Kelly, 2015). Orthogroups lacking a representative from a single species were carefully checked against genome, other proteins, and where possible, accession-specific RNA to confirm whether a gene was missing (see Methods for details). We could identify only two candidate genes that were found in LYC1722, S. lycopersicum and Arabidopsis but not in the LA716 genome. The two genes were IRX9 (Solyc09g007420) and the ribosomal gene RPS17 (Solyc03g120630). However, as irx9 mutants have a strong cell wall and morphological phenotype in Arabidopsis (Peña et al., 2007), we suspected an assembly problem in LA716. Indeed, we were able to find RNA-seq data from the accession LA716 mapping well to the LYC1722 IRX9 and RPS17 loci, confirming the presence of these genes in the LA716 reference accession. Furthermore, aligning the regions of LA716 that contained the IRX9 locus in LYC1722 against LYC1722 and S. lycopersicum, a gap in the LA716 genome of ∼6 kb relative to LYC1722 (or 4 kb relative to S. lycopersicum) was identified which was marked by a single “N” base indicating incomplete gap filling (Supplemental Figure 10). Similarly for RPS17, we found parts of the region surrounding RPS17 in the S. lycopersicum genome on the S. pennellii LA716 chromosome 00, meaning the immediate region was not well assembled and placed. In both cases, fragments of the genes were found on very small scaffolds that had ultimately been filtered from the final assembly of LA716.
By contrast, we were not able to identify any gene that could be found in S. lycopersicum, LA716, and Arabidopsis but not in the LYC1722 genome. Furthermore, an analysis of lineage-specific tandem duplications and overall number of tandems and strong ortholog candidates supported by syntenic blocks revealed a high completeness of the LYC1722 genome (Supplemental Data Sets 1I and 1J), which is in line with BUSCO results.
The same strategy to find potentially missing genes using orthogroups and detailed analyses was used to identify five genes present in both S. pennellii genomes and the Arabidopsis genome but not the S. lycopersicum cultivated tomato genome. These were a maternally expressed gene (Sopen08g025790.1), an unassigned gene (Sopen11g020600.1), a strictosidine synthase-like gene (Sopen11g013260), a lactamase family gene (Sopen02g001700), and an ATSNM1 homolog (Sopen02g039260). None of the above could be identified in RNA-seq data from a S. lycopersicum expression atlas (Tomato Genome Consortium, 2012). Four of the above genes occurred in larger regions of S. pennellii that appeared to be absent in S. lycopersicum, while interestingly, the region around the maternally expressed gene was well conserved, but specifically the exons of this gene were missing in S. lycopersicum (Supplemental Figure 11).
While the strictosidine synthase-like gene was phylogenetically distant from its characterized namesake and unlikely to synthesize strictosidine, it might be involved in stress responses (Sohani et al., 2009). The ATSNM1 homolog is likely involved in DNA repair after oxidative damage (Molinier et al., 2004).
DISCUSSION
Many crop and other plant genomes have been sequenced in the last years and one can observe a general trend toward better genomes driven by third-generation sequencing technologies and novel techniques such as Hi-C or optical mapping to gain long-range contiguity information, achieving similar contig N50s as the S. pennellii genome assembled here (VanBuren et al., 2015, Wang et al., 2017). Despite this general trend toward long read incorporation, the application of Oxford nanopore sequencing to plants remains in its infancy. This was mainly due to the large size of crop plant genomes, which made Oxford nanopore technology economically unfeasible. The dramatically higher yield of the nanopores using the 9.4 chemistry has largely resolved this issue
Previous plant-related projects employing Oxford nanopore sequencing were therefore focused on plant pathogens such as Rhizoctonia solani (Datema et al., 2016) or Agrobacterium tumefaciens (Deschamps et al., 2016), or on algal species with comparatively small genomes (Davis et al., 2016). Recently, an Arabidopsis accession was sequenced using Oxford nanopores (Michael et al., 2017).
Genome Completeness
One major achievement of the S. pennellii LYC1722 assembly presented here is its high contig contiguity and very good gene representation, as estimated both by BUSCO and a more detailed gene loss analysis between tomato genomes and the genome of Arabidopsis. Although the genome could undoubtedly be improved by large-scale scaffolding relying on e.g., optical mapping and/or Hi-C technologies, the essence of the genome for gene calling and functional studies is already very complete when using Oxford nanopore technology in combination with a small amount of Illumina data for polishing alone. Furthermore, simply by relying on linkage maps, one should be able to place most of the contigs on pseudochromosomes, as we have done earlier for similarly sized Illumina scaffolds of the reference S. pennellii accession LA716 (Bolger et al., 2014a). We have shown that there is a strong dependence of assembly contiguity on read length. Thus, new library preparation methods to produce long reads will potentially allow even better N50 values to be obtained. Also, new preparation techniques are being commercially developed to avoid the need for long molecule purification, which represents an additional cumbersome step.
Error Rates
The base error rates assessed by samtools within our assemblies were lower than 2 bases in 10 kb and ∼2.5 bases in 10 kb using a complementary data set. These rates approach those of Sanger sequence-based assemblies and are only one order of magnitude worse than the reference PacBio and Illumina-based assemblies that are just being released (Jiao et al., 2017). However, it should be noted that even when using an independent Illumina data set to estimate error rates, there could be an ascertainment bias toward a lower error rate as the whole genome might not be covered by Illumina data.
The Illumina-based quality control could also detect a decrease in error rates when the nanopore-based polisher Racon and/or nanopolish were used. However, even when using multiple rounds of Racon and combining with nanopolish, the error rates remained higher, whereas gene completeness remained lower than when polished with Illumina data. This result is in line with recent data from the model plant Arabidopsis, where even after three rounds of Racon polishing, an additional round of Illumina data decreased error rates drastically (Michael et al., 2017). Thus, at the current stage, one would still recommend including Illumina data for polishing errors. Nevertheless, the combination of Racon and nanopolish yielded a genic completeness of almost 85%, bringing it close to that of very early draft genome versions. Our findings indicate that a hybrid strategy should be followed where an optional nanopore data correction step would be followed by Illumina data polishing. This strategy should definitely be chosen if miniasm were to be used, as this assembler lacks a consensus step during the assembly, so raw error rate is expected to be similar to that of the reads.
Short-term developments are expected to improve the overall accuracy of Oxford Nanopore reads. The new basecaller Albacore (Supplemental Figures 9 and 10) already provides slightly improved accuracy using the same data and reduces the homopolymer depletion problems seen in the Metrichor base-called data (Supplemental Figures 12 and 13). Also, this basecaller is currently under active development and even better accuracies have already been achieved. These software improvements are complemented by a new pore allowing so-called 1D2 reads, by basically leveraging a “pull-through” of the second strand leading to a coupled sequencing of forward and reverse strands and, thus, improved accuracy. In addition, the basecaller became more user friendly and needs fewer resources, as intermediate steps have been omitted. Starting with nanopolish 0.8 (Loman et al., 2015; Simpson et al., 2017), this polisher also no longer relies on intermediate data (http://simpsonlab.github.io/2017/09/06/nanopolish-v0.8.0/), making it easier to use.
However, it must be stressed that despite dramatic improvements in assemblers and nanopore technology as a whole, data need to be analyzed and polished carefully to obtain meaningful error rates.
Genome Assemblies Made Cheaper and Easier
In conclusion, we demonstrated that it is possible to obtain functional and highly contiguous genome assemblies covering most of the gene space for gigabase-sized plant genomes using nanopore-based long-read data. Given a bulk discount price of about $500 per flow cell, and a cost of $215 for library preparation, which is sufficient for up to three flow cells, consumable costs for medium-sized plant genomes (<2 Gb) would thus be estimated to be below $25,000. The additional major cost factors are the computational resources, the costs of which are falling, especially with the release of more precise and eukaryote-optimized basecallers and the development of more tailored bioinformatics pipelines. This development is evidenced already in the drastic speed improvement in Canu. In addition, further methodological improvements to obtain even higher average read length (compared with Figure 2) will decrease computational requirements and would also bring the coverage requirement down (Supplemental Figure 9), potentially allowing analysis of a multitude of accessions. Indeed, our data would indicate that both LYC1722 and LA2963 represent the same original accession, i.e., LA2963 (Supplemental Data Set 1K).
As an added benefit, Oxford Nanopore data sets already provide CpG methylation data, (Simpson et al., 2017) and may potentially offer more plant-relevant methylation patterns in the future. As this information would come at no extra cost, it would allow researchers to potentially hone in on epialleles that play a role in tomato, e.g., for vitamin E accumulation and ripening (Zhong et al., 2013; Quadrana et al., 2014).
Overall, we conclude that while Oxford nanopore technology does “democratize” genome sequencing, it is mandatory to check genome quality and gene content and carefully polish the genome. In addition to using established techniques such as BUSCO, comparing the whole plant gene set data against the backdrop of closely related species promises to become a versatile tool (Bolger et al., 2017) and the comparison can be largely automated (Lohse et al., 2014; Lyons and Freeling, 2008).
METHODS
Plant Growth
Solanum pennellii LYC1722 seeds were surface sterilized in a 10% hydrogen peroxide solution for 10 min, rinsed three times with sterile water, and transferred to 0.8% half strength Murashige and Skoog Gelrite plates supplemented with 1% sucrose and 10 µM gibberellic acid. Seeds were incubated for 7 d under constant light at 22°C in a CLF Percival mobile plant chamber at 110 µmol m−2 s−1 light intensity generated using Philips TL-D 18W/840 fluorescent tubes. Seedlings were transferred to soil and further cultivated in a greenhouse supplemented with artificial light to a light intensity of at least 200 µmol m−2 s−1 generated using Phillips hpi-t plus 400w/645 metal-halide lamps for 16 h a day.
S. pennellii LA2963 seeds were obtained from the C.M. Rick Tomato Genetics Resource Center and germinated the same way as S. pennellii LYC1722. Plantlets were transplanted to Rockwool cubes irrigated with Hoagland media solution over a continuous dripping system in a phytochamber with 400 µmol m−2 s−1 light intensity generated with Iwasaki Electric MF400LSH/U and NH360 metal-halide lamps to provide 12 h of light at 18°C and 70% humidity during light cycles and 15°C and 80% humidity during dark cycles.
Long Fragment Enriched 1D R9.4 Library Preparation
To take advantage of the long-read technology, an optimized protocol for enrichment of DNA fragments of 12 to 20 kb was developed based on Oxford Nanopore’s “1D gDNA selection for long reads” protocol. For compatibility with the R9.4 SpotON MIN106 flow cells, the Ligation Sequencing Kit 1D (R9.4) was used (Oxford Nanopore Technologies; SQK-LSK108). For each library, 20 μg of high molecular weight DNA was sheared using a g-Tube (Covaris) in a total volume of 150 μL nuclease free water at 4500 to 6000 rpm depending on the desired fragment size. Enrichment for long fragments was achieved by BluePippin size selection (Sage Science). Approximately 35 μL per lane was run together with an S1 marker reference lane on a 0.75% Agarose Cassette (Biozyme) using the high pass protocol and a collection window of 12 to 80 kb or 15 to 80 kb. Upon completion of the elution, the sample was allowed to settle for at least 45 min to allow the long DNA fragments to dissociate from the elution well membrane. All subsequent bead clean-ups were performed with an equal volume of Agencourt AMPure XP beads (Beckman) with elongated bead binding and elution time of 15 min on a Hula Mixer (Grant) at 1 rpm. Bead binding was performed at room temperature and elution at 37°C. Subsequently, up to 5 μg of DNA was used for NEBNext FFPE DNA Repair (New England Biolabs) in a total volume of 155 μL including 16.3 μL NEBNext FFPE DNA Repair Buffer and 5 μL NEBNext FFPE DNA Repair Mix. The reaction was incubated for 15 min at 20°C. To reduce DNA shearing during the following bead clean-up, the sample was split in two 77.5-μL aliquots that were each eluted in 50.5 μL nuclease free water. For NEBNext Ultra II End Repair/dA-Tailing treatment (New England Biolabs), 100 μL of FFPE repaired DNA, together with 14.0 μL NEBNext Ultra II End Prep Reaction Buffer and 6 μL NEBNext Ultra II End Prep Enzyme Mix, was incubated for 30 min at 20°C followed by 20 min at 65°C and 4°C until further processing. For purification, the sample was split again into two aliquots of 60.0 μL and subjected to a bead clean up. Twenty microliters of Oxford Nanopore 1D Adapter Mix (1D AMX; Oxford Nanopore Technologies; catalog no. SQK-LSK108) was ligated to 30 μL of end repaired and adenylated DNA with 50 μL NEB Blunt/TA Master Mix (New England Biolabs; catalog no. M0367L) for 20 min at 25°C. As the motor protein is already part of the adapter, beads were resuspended twice with Oxford Nanopore Adapter Bead Buffer (Oxford Nanopore Technologies). The final library was eluted in 13 to 37 μL of Oxford Nanopore Elution Buffer (Oxford Nanopore Technologies) depending on how many flow cells were run in parallel. The final sequencing library was kept on ice until sequencing, but time was kept as short as possible. An overview of intermediate DNA quantifications and clean-up recoveries can be found in Supplemental Table 4.
Non-Size-Selected Library Preparation
A total amount of 10 μg high molecular weight DNA in 150 μL was used for g-Tube (Covaris) sheared at 4500 rpm. Directly after shearing, 0.4 volumes of Agencourt Ampure XP beads (Beckman) was added to the sample to deplete small fragments while following the bead clean-up protocol with elongated bead binding and elution as described above. The bead size-selected DNA was eluted in 133.7 μL nuclease-free water. Based on Qubit dsDNA BR quantification, 5 μg of DNA was subjected to the protocol described for long fragment-enriched libraries from NEBNext FFPE DNA Repair to the adapter ligation. The ratio of Agencourt AMPure XP beads (Beckman) for the final bead clean-up of the ligation reaction was adjusted to 0.4x of the sample volume for repeated depletion of small fragments. The library was eluted in 25 μL for Qubit dsDNA BR (ThermoFisher Scientific) quantification and loading of two flow cells.
MinION Sequencing
All sequencing runs were performed on MinION SpotON Flow Cells MK I (R9.4) (Oxford Nanopore Technologies; catalog no. FLO-SPOTR9). Immediately before start of sequencing run, a Platform QC was performed to determine the number of active pores (Supplemental Table 2). Priming of the flow cell was performed by applying 800 μL priming buffer (500 μL Oxford Nanopore Running Buffer RBF and 500 μL nuclease free water) through the sample port. After 5 min incubation at room temperature, 200 μL of priming buffer was loaded through the sample port with opened SpotON port. In parallel, 12 μL of final library was mixed with 25.5 μL Library Loading Beads (Oxford Nanopore Technologies LLB) and 37.5 Running Buffer 1 (Oxford Nanopore Technologies RBF1). Directly after priming, 75 μL of the prepared library was loaded through the SpotON port. Loading amounts of libraries quantified via Qubit dsDNA BR assay are given in Supplemental Table 2. The sequencing script “NC_48Hr_Sequencing_Run_FLO-MIN106_SQK-LSK108” was used. Basecalling was performed upon completion of the sequencing run with Metrichor and the “1D Basecalling for FLO-MIN106 450 bps” workflow (v1.121).
Illumina Sequencing
High molecular weight DNA from one 2-month-old plant of S. pennellii LYC1722 and four individual LA2963 plants was extracted as described earlier (Bolger et al., 2014a).
For S. pennellii LYC1722, 2 µg of this DNA were sheared using a Diagenode Bioruptor Pico Sonicator using five cycles of 5-s sonication interchanging with 60-s breaks to yield fragmented DNA with a target insert size of 550 bp. The fragmented DNA was then used to create an Illumina TruSeq PCR-free library according to the manufacturer’s instructions.
The sequencing library was quantified using the Perfecta NGS Quantification qPCR kit from Quanta Biosciences and sequenced four times on an Illumina MiSeq-Sequencer using three 600 cycle V3 and one 150 cycle V2 Sequencing Kits.
For S. pennellii LA2963, 5 µg of high molecular weight DNA was sheared using a Diagenode Bioruptor using eight cycles of 5-s sonication interchanging with 60-s breaks to yield fragmented DNA with a target insert size of 350 bp. The fragmented DNA was then size selected from 200 to 500 bp using a Blue Pippin with Dye free 1.5% Agarose cartridges and Marker R2.
Size-selected DNA was then purified using Beckman and Coulter Ampure XP beads in a sample to beads ratio of 1:1.6. To repair possible single-strand nicks, DNA was then treated with the New England Biolabs FFPE-repair-mix according to the manufacturer’s instructions followed by another Ampure XP bead clean-up. DNA was then end-prepped and adenylated using the NEBNext Ultra II DNA Library Prep Kit according to the manufacturer’s instructions. For ligation of sequencing adapters, 2.5 μL adapter from the Illumina TruSeq PCR-free kit was used together with the 30 μL of the NEBNext Ultra II Ligation Master Mix, 1 μL NEBNext Ligation Enhancer, and 60 μL of the End Prep Reaction Mixture. These components were mixed and incubated at 20°C for 15 min before adding 3 μL nuclease-free water and incubating at 37°C for 15 min. Afterwards, adapter-ligated DNA was cleaned up with two consecutive bead clean-ups with a 1:1 ratio of sample and beads.
The resulting library was quantified using the NEBNext Library Quant Kit for Illumina and sequenced on an Illumina MiSeq-Sequencer using a 150 cycle V3 Sequencing Kit.
A library for providing additional Illumina data for independent error rate estimation was prepared the same way as the LA2963 libraries using the Illumina LT Index Adapter AD001. This library was then sequenced on a NextSeq500 (Illumina) using a NextSeq 500/550 High Output 150 cycles v2 kit set to 2x 75 cycles for forward and reverse read sequencing.
Assembly
Reads flagged as “passing” were assembled with a variety of different tools to determine whether coverage was saturating. Then parameters and tool combinations were further refined to obtain a handful of “top” assemblies, which were then thoroughly quality controlled. All assemblies were performed with the relevant genome size parameter set to, or coverage calculation based on, a 1.2-Gb genome size.
For coverage curves, pass reads were subset randomly to yield 40, 60, 80, and 100% of reads in each library. Canu version 1.3 + (commit: 37b9b80) was used for initial read correction with the parameters corOutCoverage = 500, corMinCoverage = 2, and minReadLength = 2000 (later used as input for SMARTdenovo). Final Canu assemblies were performed with updated Canu version 1.4 + (commit: 0c206c9) and default parameters. Minimap (Li, 2016) (version 0.2-r124-dirty) was used to find overlaps with -L 1000 -m0 -Sw5, and miniasm (version 0.2-r137-dirty) was used to complete the assembly. For selected top assemblies, miniasm and Canu were run as above.
We tested several data sets as input to SMARTdenovo 61cf13d to compare the contiguity metrics of the resulting assemblies (Supplemental Data Set 1D). The random subsets of reads (Subset040, Subset060, Subset080, and Subset100) were used but we also selected 30X of the longest raw reads and Canu-corrected reads (Supplemental Data Sets 1D and 1I), as it was previously demonstrated (Istace et al., 2017) that using only a subset of the longest reads to SMARTdenovo could be beneficial to the assembly results. The assembler parameters were ‘-c 1’ to run the consensus step and ‘-k 17’, as a larger kmer size than 16 is advised on large genomes. Wtdbg version 3155039 was run with S = 1.02, k = 17. SMARTdenovo was run on 30x coverage of the longest pass reads with k = 17. The 30x coverage of the longest corrected reads was then assembled with SMARTdenovo using k = 17.
Finally, parameters for additional miniasm and SMARTdenovo assemblies are detailed in Supplemental Data Sets 1C and 1D, respectively.
BUSCO
Quality of genomes for gene detection was assessed with BUSCO (version 2.0) (Simão et al., 2015) against the embryophyta_odb9 lineage. BUSCO in turn used Augustus (version 3.2.1) (Stanke and Waack, 2003), NCBI’s BLAST (version 2.2.31+) (Camacho et al., 2009), and HMMER (version 3.1b2) (Eddy, 2011).
De Novo Gene Models and Missing Gene Analysis
Gene calling was performed with Augustus with external homology evidence from Arabidopsis thaliana, Solanum lycopersicum, and S. pennellii LA716 and with external RNaseq evidence from public S. pennellii samples in SRP068871 (Pease et al., 2016b), ERP005244 (Bolger et al., 2014a), and SRP067562 (Pease et al., 2016a). Putative missing genes were identified as orthogroups produced by OrthoFinder that had zero members in just one species. They were then further filtered to remove any gene that had a best BLASTN hit back to a genomic region and sanity checked with accession-specific RNA-seq evidence (where possible) and for a very closely related second orthogroup.
Illumina Read Trimming
Illumina reads were trimmed for low quality bases and TruSeq-3 adapter sequences using Trimmomatic 0.35 (Bolger et al., 2014b) with a sliding window of four bases and average quality score threshold of 15. Reads below a minimal length of 36 base pairs after trimming were dropped.
kmer Analysis
A total of 25 billion 17-mers were generated from the adapter trimmed Illumina paired-end data using Jellyfish (v2.2.4) (Marçais and Kingsford, 2011). 17-mers with a depth of below 8 were considered error-prone and dropped for further analysis. The remaining 24 billion 17-mers indicated a peak depth of 22 resulting in a genome size estimate of 1.12 Gb.
Polishing
Racon
Racon (Vaser et al., 2017) was used in version 0.5.0 based on overlaps created with the included minimap release. Both tools were used with standard settings except switching off the read quality filtering option in racon (–bq -1). The Racon iterations for the minimap assembly were generated based on overlaps created with minimap2 version 2.0-r296-dirty using the settings recommended for Oxford nanopore sequencing data (-x map-ont).
Nanopolish
Nanopolish v0.7.1 was used for polishing of 50 kb segments with the ‘–faster’ option invoked based on bwa mem v0.7.15-r1140 aligned nanopore reads (Loman et al., 2015). Contigs utg875 and utg4130 were excluded from the polishing step due to a noncorrectable error in the nanopolish_makerange step.
Pilon
Iterative polishing by Pilon (v1.20) (Walker et al., 2014) was achieved by aligning adapter-trimmed paired-end Illumina reads to the corresponding assembly or polished consensus sequence from the previous iteration using bwa mem (v0.7.15-r1140) (Li, 2013). The resulting sorted alignment file (samtools v1.3) (Li et al., 2009) was subjected to Pilon (Walker et al., 2014) (v1.20) together with the corresponding assembly for generation of a new consensus sequence. Pilon was run at default settings to fix bases, fill gaps, and correct local misassemblies.
Qualimap
Illumina reads were mapped to the assemblies with bwa mem, secondary alignments were removed with samtools, and discrepancies were quantified with Qualimap (v.2.2.1).
Read Quality
Expected error rate was quantified across reads and pass/fail subsets of libraries according to the Phred scores in FASTQ files by calculating the sum of 10phred/ −10 at each base position, divided by the number of bases. Empirical read quality was gathered by aligning nanopore reads back to the 4-times Pilon polished Canu assembly using bwa mem -x ont2d (v0.7.15-r1140) and calculating read identity including InDels per mapped bases.
Determination of Summary Statistics
Assembly statistics were computed using quast (Gurevich et al., 2013) (v4.3) for eukaryotes (-e). Oxford Nanopore metadata and fastq sequences were extracted from base called fast5 files using in-house scripts.
Dot Plots
Dot plots were generated using the MUMMER package (Delcher et al., 2003) (v.3.23). The unpolished assemblies were aligned to the reference genome of S. pennellii LA716 (Bolger et al., 2014a) using nucmer. The resulting alignment was filtered for a minimal alignment length of 20 kb (-l 20,000) and 1-to-1 global alignments (-g) and subsequently partitioned based on chromosome. Plotting was performed using mummerplot.
Colinear Block Identification
MCScanX (Wang et al., 2012) was used to count the number of collinear genes between LYC1722 and LA716. Similarly, MCScanX was used to identify collinear regions both between and within both S. pennellii accessions and S. lycopersicum, as well as to identify tandem duplicates. MCScanX was run with default parameters except setting the e-value threshold to 10−10 on the same BLAST results as used for Orthofinder.
To further identify tandem duplicates that occurred after the divergence of the S. pennellii species, a series of filters were applied to the tandem clusters from MCScanX to avoid more complicated homologous relationships, clusters near assembly weak points, and tandem:one relationships caused by misannotations. BLASTN refers to querying the collinear ortholog of a tandem cluster against its own genome (e-value < 10−10). “Neighboring” is used here to mean within 2x the range (maximum coordinate − minimum coordinate) of genes in the tandem cluster. Filters were applied in the following order. Putative predivergence duplications, for which multiple genes in the tandem cluster had collinear orthologs, were excluded. Clusters with ambiguity in collinear match, that is all clusters that didn’t have a 1:1 collinear relationship with the other S. pennellii accession, were excluded. Clusters (or singleton orthologs) in nonscaffolded regions (chr 00) as well was matching singleton orthologs neighboring the sequence end were excluded. Clusters with orthologs with possible missed annotations having a non-self BLASTN hits back to neighboring regions were excluded, as well as those lacking any BLASTN hit. Finally, clusters with a collinear ortholog which nevertheless appeared to generally have promiscuous paralogs (over 50 BLASTN hits) were excluded.
Gas Chromatography-Mass Spectrometry
Extraction and analysis by gas chromatography-mass spectrometry was performed using the same equipment set up and protocol as described by Lisec et al. (2006). Briefly, frozen ground material was homogenized in 700 μL methanol at 70°C for 15 min and 375 μL chloroform followed by 750 μL water were added. The polar fraction was dried under vacuum, and the residue was derivatized for 120 min at 37°C (in 60 μL of 30 mg mL−1 methoxyamine hydrochloride in pyridine) followed by a 30-min treatment at 37°C with 120 μL MSTFA. An autosampler Gerstel Multi Purpose system was used to inject the samples to a chromatograph coupled to a time-of-flight mass spectrometer system (Leco Pegasus HT TOF-MS). Helium was used as carrier gas at a constant flow rate of 2 mL/s, and gas chromatography was performed on a 30 m DB-35 column. The injection temperature was 230°C and the transfer line and ion source were set to 250°C. The initial temperature of the oven (85°C) increased at a rate of 15°C/min up to a final temperature of 360°C. After a solvent delay of 180 s, mass spectra were recorded at 20 scans s−1 with m/z 70 to 600 scanning range. Chromatograms and mass spectra were evaluated using Chroma TOF 4.5 (Leco) and TagFinder 4.2 software.
Accession Numbers
Sequence data from this article are available at http://www.plabipd.de/portal/solanum-pennellii. This will also include additional and updated protocols in the future. In addition, data have been deposited at the EBI under accession PRJEB19787.
Supplemental Data
Supplemental Figure 1. 17-mer distribution for Illumina data.
Supplemental Figure 2. Metabolite profile for leaves from the three S. pennellii accessions LA2963, LA716, and LYC1722.
Supplemental Figure 3. Yield-time plot for all S. pennellii MinION sequencing runs.
Supplemental Figure 4. Q-score distribution per library for S. pennellii nanopore reads.
Supplemental Figure 5. Nanopore read identity for R9.4 chemistry.
Supplemental Figure 6. Comparison of theoretical and empirical error rate for S. pennellii nanopore reads.
Supplemental Figure 7. Dot plot comparison of assemblies against S. pennellii LA716.
Supplemental Figure 8. Assembly and coverage graphs using NG50 instead of N50.
Supplemental Figure 9. SMARTdenovo N50 as a function of average read length.
Supplemental Figure 10. Genetic region comparison of the S. pennellii LA716 region and the corresponding S. lycopersicum genome.
Supplemental Figure 11. Genetic region comparison of the Solanum region encoding genes present in S. pennellii and Arabidopsis but not in the S. lycopersicum genome assembly.
Supplemental Figure 12. Nanopore read identity comparison.
Supplemental Figure 13. Comparison of Albacore and Metrichor basecalled reads.
Supplemental Table 1. Predicted single nucleotide polymorphism and Indel distribution across the chromosomes of S. pennellii LA716.
Supplemental Table 2. Overview of 31 Solanum pennellii MinION runs.
Supplemental Table 3. Read length overview for S. pennellii sequencing libraries.
Supplemental Table 4. Library preparation overview for all prepared libraries for S pennellii.
Supplemental Methods. Script for the hybrid assembly.
Supplemental Data Set 1. Assembly statistics.
Acknowledgments
We want to acknowledge partial funding through the Federal Ministry of Education and Research (0315961, 031A053, and 031A536C), the Ministry of Innovation, Science, and Research within the framework of the NRW Strategieprojekt BioSC (313/323-400-002 13), the Deutsche Forschungsgemeinschaft (Grants US98/7-1 and FE552/29-1) within ERACAPS Regulatome, and support for large equipment from Deutsche Forschungsgemeinschaft (Grossgeräte NextSeq LC-MS) and France Génomique (ANR-10-INBS-09). D.Z. was supported by the Horizon-2020 Grant G2P-SOL (677379). S.K. was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
AUTHOR CONTRIBUTIONS
B.U. designed the project. B.U. and A.M.B. managed the project. M.H.-W.S., A.V., and A.W. developed the DNA extraction and sequencing protocol and generated primary sequencing data. J.M., M.E.B., and A.M.B. processed and analyzed primary data. A.V., A.K.D., A.M.B., B.I., H.v.d.G., S.K., J.-M.A., B.U., and A.R.F. conducted secondary data analyses, assemblies, and statistics. S.A., A.R.F., D.Z., M.-H.W., C.P., U.S., and R.C. analyzed plants and provided materials. F.M. provided material. A.K.D., M.H.-W.S., A.V., B.I., J.-M.A., A.M.B., D.Z., U.S., S.A., A.R.F, and R.C. interpreted data. B.U. and A.K.D. wrote the manuscript with help from all authors.
References
- Aflitos S., et al.; 100 Tomato Genome Sequencing Consortium (2014). Exploring genetic variation in the tomato (Solanum section lycopersicon) clade by whole-genome sequencing. Plant J. 80: 136–148. [DOI] [PubMed]
- Alseekh S., et al. (2015). Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C.S., Drake J.P., Landolin J.M., Phillippy A.M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33: 623–630. [DOI] [PubMed] [Google Scholar]
- Bolger A., et al. (2014a). The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46: 1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014b). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger M.E., Arsova B., Usadel B. (2017). Plant genome and transcriptome annotations: from misconceptions to simple solutions. Brief. Bioinform. pii: bbw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datema E., Hulzink R.J.M., Blommers L., Espejo Valle-Inclan J., Van Orsouw N., Wittenberg A.H.J., De Vos M. (2016). The megabase-sized fungal genome of Rhizoctonia solani assembled from nanopore reads only. bioRxiv doi/10.1101/084772. [Google Scholar]
- Davis A.M., et al. (2016). Using MinION nanopore sequencing to generate a de novo eukaryotic draft genome: preliminary physiological and genomic description of the extremophilic red alga Galdieria sulphuraria strain SAG 107.79. bioRxiv doi/10.1101/076208.
- Delcher A.L., Salzberg S.L., Phillippy A.M. (2003). Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics 10: 10.3.1–10.3.18. [DOI] [PubMed] [Google Scholar]
- Deschamps S., Mudge J., Cameron C., Ramaraj T., Anand A., Fengler K., Hayes K., Llaca V., Jones T.J., May G. (2016). Characterization, correction and de novo assembly of an Oxford Nanopore genomic dataset from Agrobacterium tumefaciens. Sci. Rep. 6: 28625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (2011). Accelerated Profile HMM Searches. PLOS Comput. Biol. 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moreno J.P., Levy-Samoha D., Malitsky S., Monforte A.J., Orzaez D., Aharoni A., Granell A. (2017). Uncovering tomato quantitative trait loci and candidate genes for fruit cuticular lipid composition using the Solanum pennellii introgression line population. J. Exp. Bot. 68: 2703–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T.C. (2011). Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 11: 759–769. [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2016). Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip C.L.C., et al. ; MinION Analysis and Reference Consortium (2015). MinION Analysis and Reference Consortium: Phase 1 data release and analysis. F1000 Res. 4: 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istace B., et al. (2017). de novo assembly and population genomic survey of natural yeast isolates with the Oxford Nanopore MinION sequencer. Gigascience 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Olsen H.E., Paten B., Akeson M. (2016). The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis D.E., et al. (2017). The genome of Chenopodium quinoa. Nature 542: 307–312. [DOI] [PubMed] [Google Scholar]
- Jiao W.-B., Schneeberger K. (2017). The impact of third generation genomic technologies on plant genome assembly. Curr. Opin. Plant Biol. 36: 64–70. [DOI] [PubMed] [Google Scholar]
- Jiao W.-B., et al. (2017). Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27: 778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge K., Harris S.R., Reuter S., Parkhill J., Peacock S.J. (2015). Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes. J. Antimicrob. Chemother. 70: 2775–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D., et al. (2013). Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 110: E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A., Vogel A., Degner U., Kiefler I., Bott M., Usadel B., Polen T. (2017). High precision genome sequencing of engineered Gluconobacter oxydans 621H by combining long nanopore and short accurate Illumina reads. J. Biotechnol. 258: 197–205. [DOI] [PubMed] [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv doi/10.1101/1303.3997v2.
- Li H. (2016). Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32: 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46: 1220–1226. [DOI] [PubMed] [Google Scholar]
- Lippman Z.B., Semel Y., Zamir D. (2007). An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Genet. Dev. 17: 545–552. [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396. [DOI] [PubMed] [Google Scholar]
- Lohse M., Nagel A., Herter T., May P., Schroda M., Zrenner R., Tohge T., Fernie A.R., Stitt M., Usadel B. (2014). Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37: 1250–1258. [DOI] [PubMed] [Google Scholar]
- Loman N.J., Quick J., Simpson J.T. (2015). A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12: 733–735. [DOI] [PubMed] [Google Scholar]
- Lu H., Giordano F., Ning Z. (2016). Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics Proteomics Bioinformatics 14: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Freeling M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53: 661–673. [DOI] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. (2011). A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T. P., Jupe, F., Bemm, F., Motley, S.T., Sandoval, J.P, Loudet, O. Weigel, D., Ecker, J.R. (2017). High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. bioRxiv doi/10.1101/149997. [DOI] [PMC free article] [PubMed]
- Molinier J., Stamm M.E., Hohn B. (2004). SNM-dependent recombinational repair of oxidatively induced DNA damage in Arabidopsis thaliana. EMBO Rep. 5: 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofner I., Lashbrooke J., Pleban T., Aharoni A., Zamir D. (2016). Solanum pennellii backcross inbred lines (BILs) link small genomic bins with tomato traits. Plant J. 87: 151–160. [DOI] [PubMed]
- Okonechnikov K., Conesa A., García-Alcalde F. (2016). Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32: 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease J.B., Haak D.C., Hahn M.W., Moyle L.C. (2016a). Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14: e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease J.B., Guerrero R.F., Sherman N.A., Hahn M.W., Moyle L.C. (2016b). Molecular mechanisms of postmating prezygotic reproductive isolation uncovered by transcriptome analysis. Mol. Ecol. 25: 2592–2608. [DOI] [PubMed] [Google Scholar]
- Peña M.J., Zhong R., Zhou G.K., Richardson E.A., O’Neill M.A., Darvill A.G., York W.S., Ye Z.H. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L., et al. (2014). Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 5: 3027. [DOI] [PubMed] [Google Scholar]
- Quick J., et al. (2015). Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 16: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A., Budke J.M., Rowland S.D., Chitwood D.H., Kumar R., Carriedo L., Ichihashi Y., Zumstein K., Maloof J.N., Sinha N.R. (2016). eQTL regulating transcript levels associated with diverse biological processes in tomato. Plant Physiol. 172: 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8: 14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick C.M., Tanksley S.D. (1981). Genetic variation in Solanum pennellii: Comparisons with two other sympatric tomato species. Plant Syst. Evol. 139: 11–45. [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. (2017). Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14: 407–410. [DOI] [PubMed] [Google Scholar]
- Sohani M.M., Schenk P.M., Schultz C.J., Schmidt O. (2009). Phylogenetic and transcriptional analysis of a strictosidine synthase-like gene family in Arabidopsis thaliana reveals involvement in plant defence responses. Plant Biol (Stuttg.) 11: 105–117. [DOI] [PubMed] [Google Scholar]
- Stanke M., Waack S. (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (Suppl. 2): ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. [DOI] [PubMed] [Google Scholar]
- Tieman D., et al. (2017). A chemical genetic roadmap to improved tomato flavor. Science 355: 391–394. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R., et al. (2015). Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527: 508–511. [DOI] [PubMed] [Google Scholar]
- Vaser R., Sović I., Nagarajan N., Šikić M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., Earl A.M. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., Kissinger J.C., Paterson A.H. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2017). Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 49: 765–772. [DOI] [PubMed] [Google Scholar]
- Weirather, J., de Cesare M., Wang Y., Piazza P., Sebastiano V., Wang X., Buck D., Au K. (2017). Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research 6: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Hill C.M., Wu S., Ruan J., Ma Z.S. (2016). DBG2OLC: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 6: 31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y.R., Zheng Y., Huang M., Vrebalov J., McQuinn R., Gapper N., Liu B., Xiang J., Shao Y., Giovannoni J.J. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31: 154–159. [DOI] [PubMed] [Google Scholar]