ZYGO1, a novel F-box protein that interacts with OSK1, mediates bouquet formation to promote pairing, synapsis, and CO formation in rice meiosis.
Abstract
Telomere bouquet formation, a highly conserved meiotic event, plays an important role in homologous pairing and therefore progression of meiosis; however, the underlying molecular mechanism remains largely unknown. Here, we identified ZYGOTENE1 (ZYGO1), a novel F-box protein in rice (Oryza sativa), and verified its essential role in bouquet formation during early meiosis. In zygo1 mutants, zygotene chromosome aggregation and telomere clustering failed to occur. The suppressed telomere clustering in homologous pairing aberration in rice meiosis1 (pair1) zygo1 and rice completion of meiotic recombination (Oscom1) zygo1 double mutants, together with the altered localization of OsSAD1 (a SUN protein associated with the nuclear envelope) in zygo1, showed that ZYGO1 has a significant function in bouquet formation. In addition, the interaction between ZYGO1 and rice SKP1-like protein 1 suggested that ZYGO1 might modulate bouquet formation as a component of the SKP1-Cullin1-F-box complex. Although double-strand break formation and early recombination element installation occurred normally, zygo1 mutants showed defects in full-length pairing and synaptonemal complex assembly. Furthermore, crossover (CO) formation was disturbed, and foci of Human enhancer of invasion 10 were restricted to the partially synapsed chromosome regions, indicating that CO reduction might be caused by the failure of full-length chromosome alignment in zygo1. Therefore, we propose that ZYGO1 mediates bouquet formation to efficiently promote homolog pairing, synapsis, and CO formation in rice meiosis.
INTRODUCTION
To achieve faithful segregation of homologous chromosomes, several critical events take place during prophase I of meiosis, including homology searching, recombination, pairing, and synaptonemal complex (SC) assembly. This series of highly coordinated events causes observable changes in the structure of chromosomes. Accordingly, prophase I has been divided into five cytological substages: leptotene, zygotene, pachytene, diplotene, and diakinesis. Zygotene is a complicated and dynamic stage, during which homologous recombination is ongoing, and pairing as well as synapsis initiate (Zickler and Kleckner, 2015). Meanwhile, a highly polarized nuclear reorganization occurs at this stage, involving an aggregation of chromosomes and/or the formation of a “bouquet” in which telomeres are spatially tethered to a limited area of the inner nuclear membrane (INM) (Zickler and Kleckner, 1998).
Nuclear reorganization is a widely conserved event that occurs in most species. In Caenorhabditis elegans, chromosomes cluster to one side of the nucleus and form a crescent-shaped configuration during the zygotene stage (MacQueen and Villeneuve, 2001). In plants, this polarized configuration was also observed at early zygotene stage (Dawe et al., 1994; Ross et al., 1996). Importantly, mutation of genes required for nuclear reorganization causes severe meiotic defects, indicating the essential role of nuclear reorganization during meiosis. For example, the C. elegans checkpoint kinase2 (chk-2) and the Arabidopsis thaliana skp1-like1 (ask1) mutants lack the major spatial reorganization and were also found to be defective in homologous pairing, synapsis, crossover (CO) formation, and chromosome separation (Yang et al., 1999; MacQueen and Villeneuve, 2001; Zhao et al., 2006). Moreover, the correlation between nuclear reorganization and pairing in chk-2 mutants suggests that zygotene nuclear reorganization might be required for homologous pairing (MacQueen and Villeneuve, 2001). Nevertheless, the mechanism of chromosome reorganization is poorly understood compared with that of telomere clustering.
Along with chromosomal movement, telomere bouquets form in many species; these structures are widely thought to efficiently facilitate homology searching and pairing by bringing distant chromosomes into a more confined area (Trelles-Sticken et al., 2000; Scherthan, 2001; Harper et al., 2004). Factors affecting telomere bouquet formation mainly participate in two aspects of telomere behavior: telomere attachment to the INM and subsequent telomere movement along the INM. Many proteins have been identified that connect telomere proteins with INM proteins, such as Bouquet 1 (Bqt1), Bqt2, Bqt3, and Bqt4 in fission yeast (Schizosaccharomyces pombe) (Chikashige et al., 2006, 2009), Nondisjunction 1 (Ndj1) in budding yeast (Saccharomyces cerevisiae) (Conrad et al., 1997; Trelles-Sticken et al., 2000), as well as Telomere repeat binding bouquet formation protein 1 (TERB1), TERB2, and membrane-anchored junction protein (MAJIN) in mouse (Mus musculus) (Shibuya et al., 2014, 2015). These proteins tether telomeres to the INM to ensure bouquet formation. Then, at the INM, telomeres are associated with the transmembrane linker of nucleoskeleton and cytoskeleton (LINC) complex that is composed of SUN and KASH domain proteins (Ding et al., 2007; Yoshida et al., 2013). This transmembrane complex transduces cytoskeletal forces to telomeres and drives telomere clustering along the INM and, therefore, chromosome movements (Starr and Fridolfsson, 2010). However, so far, the molecular mechanisms underlying telomere bouquet formation in plant species are largely unknown. In Arabidopsis, two SUN-domain proteins (AtSUN1 and AtSUN2) were identified to have overlapping function in telomere dynamics and homologous pairing during meiosis (Varas et al., 2015). In addition, the plural abnormalities of meiosis1 (pam1) and the desynaptic mutants were reported to be defective in telomere clustering in maize (Zea mays) (Golubovskaya et al., 2002; Murphy and Bass, 2012), but the candidate genes of these two mutants are still unknown.
The ubiquitin-proteasome system functions through ubiquitylation and subsequent regulation or degradation of target proteins. In the ubiquitin-proteasome system, the SKP1-Cullin1-F-box (SCF) E3 ligase complex has roles in multiple cellular processes (Hua and Vierstra, 2011). The F-box protein determines the substrate specificity of the SCF complex, and SKP1 is the subunit that bridges Cullin1 and the F-box protein. F-box proteins are involved in the mitotic cell cycle by governing the ubiquitination and subsequent degradation of short-lived proteins, and dysregulation of the function of these F-box proteins results in an aberrant mitotic cell cycle and finally tumorigenesis (Wang et al., 2014). Additionally, some components of the SCF complex also participate in the meiotic cell cycle. For example, the C. elegans F-box protein PROM-1 and the Arabidopsis SKP1 protein ASK1 have been shown to be required for meiotic progression (Yang et al., 1999; Jantsch et al., 2007). Moreover, the MEIOTIC F-BOX (MOF) protein in rice (Oryza sativa) was reported to be necessary for male meiotic double-strand break (DSB) repair (He et al., 2016). However, the regulatory function of F-box proteins in zygotene chromosome reorganization and bouquet formation remains enigmatic.
In this study, we identified ZYGOTENE1 (ZYGO1), a novel F-box protein in rice, and verified its function using a combination of genetic and cytological methods. Based on our data, ZYGO1 is essential for telomere bouquet formation during early prophase I. Furthermore, loss of ZYGO1 also caused failures in full-length pairing and synapsis in addition to defects in CO formation. Together, our results uncovered the essential role of this component in SCF complex during rice meiosis.
RESULTS
Identification of ZYGO1
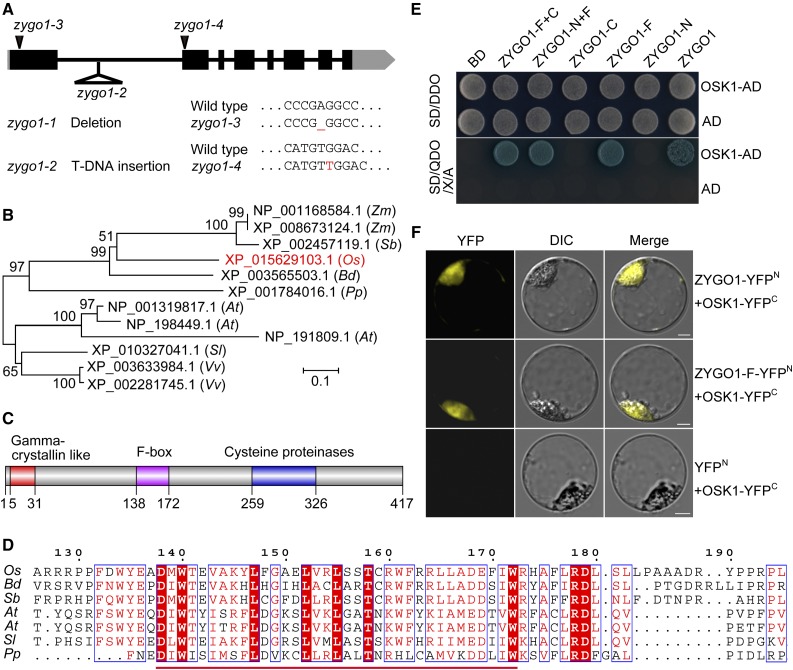
A sterile mutant was obtained from the rice variety Yandao 8 induced by 60Co γ-ray irradiation. The mutant did not exhibit any defects in vegetative growth but was completely sterile (Supplemental Figures 1A and 1B). Observation of the anthers showed that the pollen grains were completely inviable in the mutant (Supplemental Figures 1C and 1D). No seeds were set when the mutant flowers were pollinated with wild-type pollen, suggesting that this mutant is both male and female sterile. Using map-based cloning, the target gene was preliminarily mapped to a 201-kb region on the short arm of chromosome 1. Upon further analysis, we determined that 82,877 bp was deleted in the mutant (Supplemental Figure 2). This deletion mutant was designated zygo1-1. Within the deleted region, we identified a T-DNA insertion mutant line (PFG_2d-00585), which has a T-DNA insertion in the first intron of LOC_Os01g11990 (Figure 1A). Analysis of the chromosome behavior in PFG_2d-00585 during prophase I showed that it has a very similar phenotype to zygo1-1 (Supplemental Figures 3B, 3F, and 3J); therefore, we designated this mutant zygo1-2, and LOC_Os01g11990 was accordingly designated ZYGO1.
Figure 1.
ZYGO1 Encodes a Novel F-Box Protein in Rice.
(A) Gene structure of ZYGO1 and the mutation sites of zygo1 mutants. Black blocks and lines represent exons and introns, respectively. Untranslated regions are shown as gray boxes.
(B) An unrooted tree based on the full-length protein sequences of ZYGO1 and its homologs from seven plant species. ZYGO1 is shown in red. Zm, Zea mays; Sb, Sorghum bicolor; Os, Oryza sativa; Bd, Brachypodium distachyon; Pp, Physcomitrella patens; At, Arabidopsis thaliana; Sl, Solanum lycopersicum; Vv, Vitis vinifera.
(C) ZYGO1 contains a gamma-crystallin-like motif, an F-box domain, and a cysteine proteinase motif.
(D) Multiple sequence alignment of the F-box domain of ZYGO1 with its homologs from five plant species. Amino acid sequence of the F-box domain is underlined and conserved amino acids are highlighted in red.
(E) ZYGO1 interacts with OSK1 in yeast two-hybrid assays. Interactions were verified by the growth and the blue color of yeast cells on SD/QDO/X/A (SD-Ade-His-Leu-Trp + X-α-gal + Aureobasidin) medium. BD, bait vector; AD, prey vector; ZYGO1-N, the N-terminal residues (amino acids 1–137) of ZYGO1; ZYGO1-F, the F-box domain of ZYGO1 (amino acids 138–172); ZYGO1-C, the C-terminal residues (amino acids 173–417) of ZYGO1; ZYGO1-N+F, 1 to 172 amino acids of ZYGO1; ZYGO1-F+C, 138 to 417 amino acids of ZYGO1.
(F) BiFC assays showing the interaction between OSK1 and the full-length protein sequence or the F-box domain of ZYGO1 in rice protoplasts. Bars = 5 µm.
To confirm that the meiotic defects of zygo1-1 indeed result from the mutation of ZYGO1, a complementation test was conducted that succeeded in rescuing the meiotic phenotype of zygo1-1 (Supplemental Figures 3A, 3E, and 3I). To validate the complementation result, point mutations of ZYGO1 were generated using CRISPR-Cas9. Specifically, a 1-bp deletion in the first exon or a 1-bp insertion in the second exon led to the frameshift mutation and finally the formation of a premature stop codon (Figure 1A). These two additional mutants were designated zygo1-3 and zygo1-4, respectively. Further cytological observations revealed that the meiotic defects in zygo1-3 and zygo1-4 were also similar to that of zygo1-1 (Supplemental Figures 3C, 3D, 3G, 3H, 3K, and 3L). Thus, mutations in ZYGO1 were indeed responsible for the meiotic defects observed in these zygo1 mutants. The zygo1-3 mutant allele was used for observation and data generation throughout our research.
ZYGO1 Encodes a Novel F-Box Protein in Rice
According to the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice), ZYGO1 is a conserved gene with unknown function. The cDNA sequence of ZYGO1 was redefined by RT-PCR and RACE PCR (Supplemental Figure 4). The whole length of ZYGO1 is 3265 bp, comprising eight exons and seven introns (Figure 1A). ZYGO1 encodes a 417-amino acid peptide. A neighbor-joining tree was constructed based on the full-length protein sequences of ZYGO1 and its homologs in plants. The result indicated that ZYGO1 is more closely related to its homologs from monocots than those from dicots (Figure 1B). Furthermore, there is only one copy of ZYGO1 in rice, whereas there are several homologs of ZYGO1 in some plant species, for example, in Arabidopsis and maize (Figure 1B).
A SMART search for conserved domains revealed that ZYGO1 contains an N-terminal gamma-crystallin-like motif (amino acids 5–31), an F-box domain (amino acids 138–172), and a cysteine proteinase motif (amino acids 259–326) (Figure 1C). Multiple alignment of the full-length protein sequence of ZYGO1 with its homologs among plants revealed that the F-box domain in ZYGO1 is highly conserved (Figure 1D). Besides, ZYGO1 showed limited sequence similarity to the mouse F-box protein FBL12 (28.6% identity and 60.0% similarity among the F-box domain; 23.5% identity and 37.3% similarity among the cysteine proteinase motif; Supplemental Figure 5).
ZYGO1 Interacts with OSK1 within an SCF Complex
To study the function of ZYGO1, we performed yeast two-hybrid (Y2H) screening to identify ZYGO1-interacting proteins. A cDNA library from rice panicle was used as the prey, and the full-length ZYGO1 was used as the bait. After screening 2 × 106 yeast transformants, 20 positive clones were obtained. Of these, 13 clones were in the right reading frame and were derived from six proteins (Supplemental Table 1). Interestingly, six out of these 13 clones were found to encode the rice SKP1-like protein 1 (OSK1), which belongs to the SKP1 protein family in rice and was previously proved to interact with some known F-box proteins (Kahloul et al., 2013). We therefore focused on the potential interaction between ZYGO1 and OSK1. To verify this interaction, the full-length coding sequence of OSK1 was cloned into pGADT7 and the full-length ZYGO1 was cloned into pGBKT7. Yeast cells cotransformed with these two vectors grew well and turned blue on QDO/X/A medium, indicating that ZYGO1 interacts with OSK1 (Figure 1E). Furthermore, Y2H assays using ZYGO1 fragments and OSK1 revealed that the interaction between ZYGO1 and OSK1 was mediated by the F-box domain of ZYGO1 (Figure 1E). To further validate these interactions, bimolecular fluorescence complementation (BiFC) assays were conducted using rice protoplasts. YFP signals were detected in cells coexpressing ZYGO1-YFPN and OSK1-YFPC (Figure 1F), demonstrating that ZYGO1 interacts with OSK1 in vivo. The interaction between the F-box domain of ZYGO1 and OSK1 was also verified (Figure 1F). Hence, ZYGO1 might be a functional component of a rice SCF complex.
The zygo1 Mutant Shows a Defect in Zygotene Chromosome Organization
To investigate the reason for sterility in the zygo1 mutants, we investigated meiotic progression of both the wild type and zygo1. Two different fixatives were used to accurately identify the stages of meiosis I. Formalin-acetic acid-alcohol (FAA) solution can fix the nuclear membrane well, maintaining the original status of the nucleus. When fixed with this solution, acetocarmine-stained chromosomes were widely dispersed throughout the nucleus at leptotene in the wild type (Supplemental Figure 6A). On entering into zygotene, chromosomes clustered into a mass at one side of the nucleus (Supplemental Figure 6B). At pachytene, the synapsed chromosomes were dispersed throughout the nucleus once again (Supplemental Figure 6C). Then, bivalents were observed at diakinesis (Supplemental Figure 6D). In zygo1, the obvious abnormality was that the huddled chromosome mass was not observed throughout early prophase I (Supplemental Figures 6E to 6G). Typical pachytene cells were also not detected. At diakinesis, a mixture of bivalents and univalents was observed (Supplemental Figure 6H). Therefore, meiocytes in zygo1 during early prophase I were distinguished as being at leptotene or post-leptotene stages according to the stage description of Atsun1-1 Atsun2-2, which also lacks typical zygotene and pachytene stage cells (Varas et al., 2015).
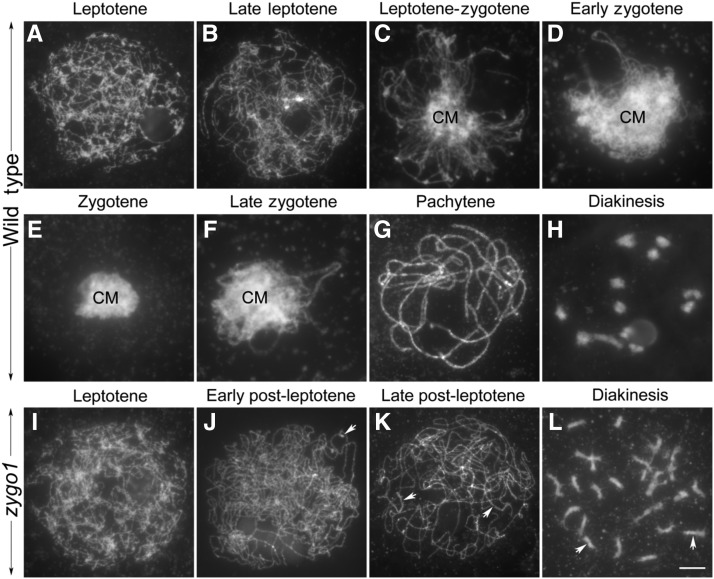
To further confirm that zygotene chromosome organization was absent in zygo1, we checked the chromosome behavior of meiocytes fixed with ethanol-acetic acid (3:1) solution and stained with 4′,6-diamidino-2-phenylindole (DAPI). When meiocytes are fixed with this solution, their cytoplasm and nuclear membrane are not well maintained, and the fixed chromosomes are easily spread. In the wild type, chromosomes at leptotene were observed to be in single threads within loose status (Figures 2A and 2B). From leptotene to zygotene, chromosomes started to aggregate together and a small chromosomal mass, with diffused chromosomes, was present in the central region of the nucleus (Figure 2C). During early zygotene, the mass region increased as chromosomes began pairing together, while unpaired regions of chromosomes outside the mass were still visible as fine threads (Figure 2D). At the typical zygotene stage, all chromosomes were huddled together in a compact mass and had lost their linear characteristics (Figure 2E). Then, the aggregated chromosome mass gradually resolved, and chromosomes outside the mass had distinct synapsed regions typical of late zygotene (Figure 2F). When meiocytes reached pachytene, the mass region was not observed, and all the synapsed chromosomes appeared scattered within the nucleus (Figure 2G). At the end of prophase I (diakinesis), 12 bivalents were observed in the nucleus (Figure 2H).
Figure 2.
The Chromosome Behavior of Meiocytes in the Wild Type and zygo1.
(A) to (H) The detailed chromosome behavior of meiocytes from leptotene to diakinesis in the wild type. Leptotene (A), late leptotene (B), leptotene to zygotene (C), early zygotene (D), zygotene (E), late zygotene (F), pachytene (G), and diakinesis (H). CM, chromosome mass.
(I) to (L) The chromosome behavior of meiocytes from leptotene to diakinesis in zygo1.
(I) and (J) Leptotene (I) and early post-leptotene (J). Chromosomes remained dispersed in the nucleus in zygo1.
(K) Late post-leptotene. Some partially paired regions are indicated with arrows.
(L) Diakinesis. Some univalents are indicated with arrows. Chromosomes were fixated with ethanol-acetic acid (3:1) solution and stained with DAPI. Bar = 5 µm.
Similarly, no difference was observed between the wild type and zygo1 at leptotene (Figure 2I). However, the aggregated chromosome mass and fully synapsed chromosomes had not been observed throughout early prophase I in the 245 meiocytes examined (Figures 2I to 2K). Although some homologous chromosomes were partially paired (arrows), most chromosomes remained as single threads in zygo1 (Figures 2J and 2K). Besides, many univalents were observed at diakinesis (Figure 2L, arrows). Finally, unequal separation of homologous chromosomes during meiosis resulted in sterile gametes in zygo1. Taken together, zygo1 showed complicated meiotic defects, the most striking of which is the absence of zygotene chromosome organization.
Loss of ZYGO1 Causes Failure of Telomere Bouquet Formation
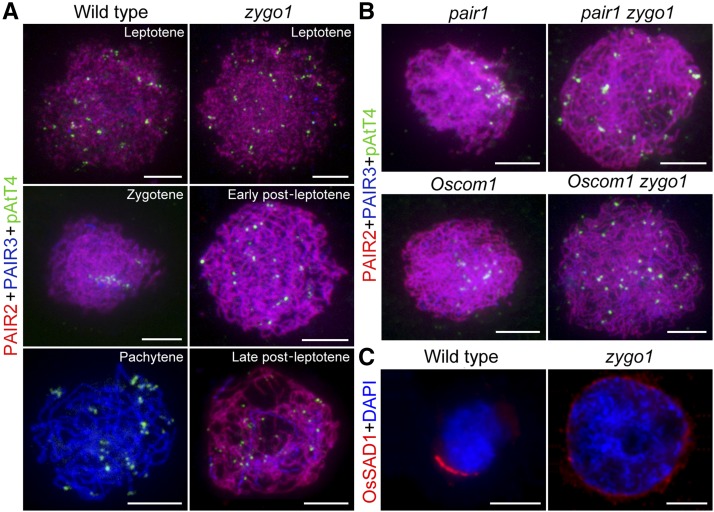
During early zygotene, telomere bouquets form in most organisms, making homologous chromosomes meet and pair more efficiently (Scherthan, 2001; Harper et al., 2004). The absence of typical zygotene chromosome organization in zygo1 triggers our suspicion about the impacts of ZYGO1 deficiency on telomere bouquet formation. To verify this, we performed immunostaining-fluorescence in situ hybridization (immuno-FISH) analysis using antibodies against PAIR2 and PAIR3 as well as the telomere probe pAtT4. In rice, PAIR3 is associated with both unsynapsed axial elements and synapsed lateral elements of the SC during prophase I, whereas PAIR2 is only associated with unsynapsed axial elements at leptotene and zygotene (Wang et al., 2011). So, the loading pattern of PAIR2 and PAIR3 precisely indicates the stage of meiocytes. In the wild type, the discrete foci of PAIR2 and PAIR3 were colocalized and telomere foci were widely scattered in the nucleus at leptotene (Figure 3A, top panel). At early zygotene, most linear foci of PAIR2 colocalized well with PAIR3 along the entire length of chromosomes and a few PAIR2 proteins released from the partially synapsed chromosomes (Figure 3A, middle panel). We investigated the telomere foci of 69 meiocytes with this loading pattern of PAIR2 and PAIR3 in the wild type, and nearly 72% of these meiocytes showed typical telomere clustering on the chromosome mass. At pachytene, nearly no PAIR2 foci were observed, but the linear foci of PAIR3 were still distributed along the full-length chromosomes (Figure 3A, third panel). Meanwhile, the telomere foci dispersed again and localized at the end of chromosomes.
Figure 3.
ZYGO1 Is Essential for Bouquet Formation.
(A) Telomere behavior in the wild type and the zygo1 mutant during early prophase I. Immuno-FISH analysis was performed using antibodies against PAIR2 (red) and PAIR3 (blue) as well as the telomere probe pAtT4 (green). The loading pattern of PAIR2 and PAIR3 indicates the stage of meiocytes in the wild type and zygo1. Telomere bouquet was observed in wild-type meiocytes at early zygotene, whereas it was not observed in zygo1 throughout the post-leptotene stage.
(B) Distribution of telomere foci in pair1, Oscom1, pair1 zygo1, and Oscom1 zygo1 at early zygotene or post-leptotene stage. Telomere clustering in pair1 and Oscom1 was suppressed in pair1 zygo1 and Oscom1 zygo1 double mutants.
(C) Immunofluorescent investigation of OsSAD1 signal (red) in the wild type and zygo1. Chromosomes were stained with DAPI (blue). The polarized enrichment of OsSAD1 foci was not observed in zygo1 throughout the post-leptotene stage. Bars = 5 µm.
In zygo1, no obvious defect was observed at leptotene (Figure 3A, top panel). When entering into post-leptotene stage, although PAIR2 and PAIR3 were observed as linear foci along the entire length of chromosomes, they were looser than those in the wild type. We surveyed the telomere foci of meiocytes that showed similar loading pattern of PAIR2 and PAIR3 to that of wild-type meiocytes at bouquet stage. In zygo1, 212 meiocytes with this loading pattern were surveyed; however, their telomeres did not congregate to a certain region but were still scattered over the dispersed chromosomes (Figure 3A, middle panel). At the later stage of post-leptotene, some telomeres were eventually paired, along with the progression of partial synapsis in zygo1. There was a significant reduction (P < 0.0001) in the number of pAtT4 foci in meiocytes with partial synapsis (36.2 ± 0.6, n = 34) compared with that in meiocytes at the early stage of post-leptotene (45.5 ± 0.4, n = 37) (Supplemental Figure 7). However, most telomeres remained scattered and unpaired in zygo1 (Figure 3A, third panel). Together, these results suggested that ZYGO1 deficiency disrupts telomere clustering, resulting in failure of bouquet formation.
ZYGO1 Is Required for Telomere Clustering in pair1 and Oscom1
To further confirm the essential role of ZYGO1 in bouquet formation, we constructed pair1 zygo1 and Oscom1 zygo1 double mutants. PAIR1 is the rice homolog of AtPRD3, which is required for DSB formation in Arabidopsis (De Muyt et al., 2007, 2009). OsCOM1 is essential for DSB repair in rice (Ji et al., 2012). Although mutations in PAIR1 and OsCOM1 lead to severe defects in homologous recombination, telomere bouquets were still present in both pair1 and Oscom1 single mutants at early zygotene (Figure 3B, left panels). Among these meiocytes with the loading pattern of PAIR2 and PAIR3 at early zygotene, ∼75% (n = 60) and 80% (n = 76) of meiocytes were observed to have telomere clustering in pair1 and Oscom1, respectively. These results indicated that telomere clustering is independent of PAIR1-mediated DSB formation or OsCOM1-mediated DSB repair pathway. However, telomeres in pair1 zygo1 and Oscom1 zygo1 double mutants showed a scattered distribution similar to that of zygo1 at post-leptotene stage (Figure 3B, right panels). Among the meiocytes with similar loading pattern of PAIR2 and PAIR3 in pair1 zygo1 (n = 133) and Oscom1 zygo1 (n = 162) double mutants, none was with a typical telomere bouquet. These results provided convincing evidence demonstrating that ZYGO1 is a prerequisite to establish the telomere bouquet in pair1 and Oscom1.
Polarized Localization of OsSAD1 on the Nuclear Envelope Is Dependent on ZYGO1
SUN-domain proteins form a key component of the LINC complex that promotes telomere clustering and chromosomal movement (Starr and Fridolfsson, 2010). SUN proteins, such as SUN-1, AtSUN1, AtSUN2, and ZmSUN2, show polarized localization on one side of the nuclear envelope during early prophase I in C. elegans, Arabidopsis, and maize (Penkner et al., 2009; Murphy et al., 2014; Varas et al., 2015). In rice, there are four SUN-domain-containing proteins, among which OsSAD1 is closely related to AtSUN1 and AtSUN2 (Murphy et al., 2010). To verify whether loss of ZYGO1 affects the localization of SUN protein, an antibody against OsSAD1 was generated to investigate its localization in rice meiocytes. In the wild type, the foci of OsSAD1 were associated with the nuclear envelope and enriched on one side of the nuclear envelope at zygotene, with the polarized chromosome mass toward the same side of the nucleus in the 43 meiocytes examined (Figure 3C, left panel). However, in the zygo1 mutants, OsSAD1 was uniformly located on the nuclear envelope and the polarized enrichment of OsSAD1 foci was not observed throughout the post-leptotene stage in the 61 meiocytes examined (Figure 3C, right panel). These results indicated that the polarized localization of OsSAD1 on the nuclear envelope is altered in zygo1.
ZYGO1 Is Required for Full-Length Homologous Pairing and Synapsis
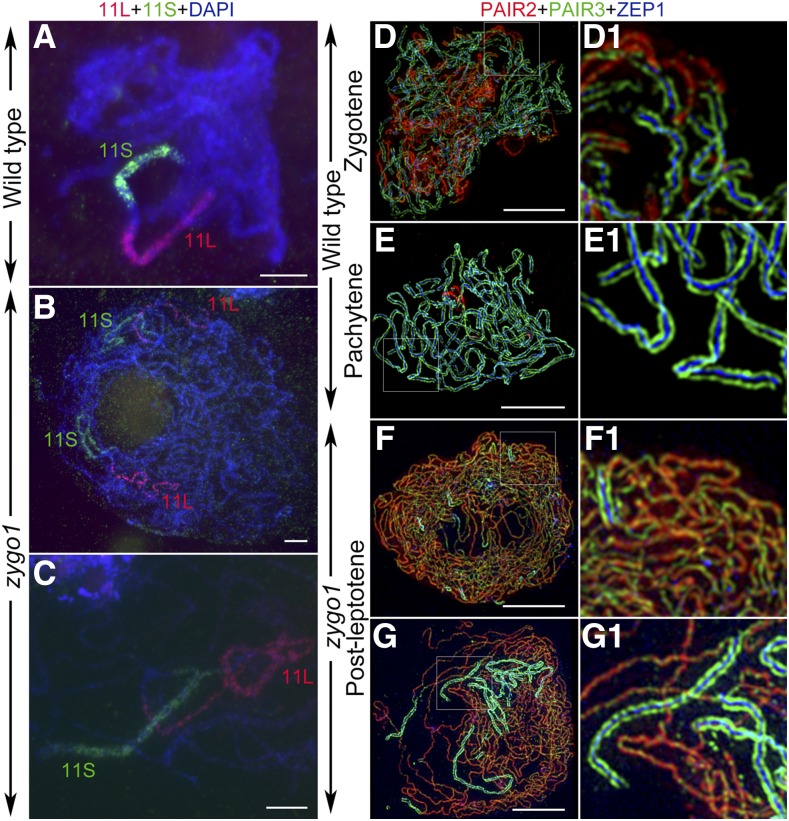
Telomere bouquet efficiently facilitates homologous pairing, so we wondered if the bouquet deficiency in zygo1 affects homologous pairing. To monitor the homologous pairing status, two bulked oligonucleotide probes, specific to the short (11S) and long arms (11L) of chromosome 11, were prepared and used in the FISH assay. In the wild type, the signal arrays of these two probes were well paired at pachytene in the 65 meiocytes examined (Figure 4A). In contrast, separated signals of these two probes were observed in 92.4% of the meiocytes (n = 79) in zygo1 at post-leptotene stage (Figure 4B). In only 7.6% of the meiocytes in zygo1, partially paired signals of either one or two of these probes were observed (Figure 4C). These results suggested that full-length pairing of homologous chromosomes is not achieved in zygo1.
Figure 4.
ZYGO1 Is Necessary for Full-Length Homologous Pairing and Synapsis.
(A) to (C) The chromosome pairing status revealed by FISH assays in the wild type and zygo1. Bulked oligonucleotide probes, 11S (green) and 11L (red), were used to track the short and long arms of chromosome 11, respectively. Chromosomes were stained with DAPI (blue).
(A) Wild-type meiocyte with well-paired probe signals at pachytene.
(B) zygo1 meiocyte with separated signals of these two probes.
(C) zygo1 meiocyte with partially paired signals of these two probes.
(D) to (G) Super-resolution images of the triple immunolocalization of PAIR2 (red), PAIR3 (green), and ZEP1 (blue) in the wild type and zygo1 during early prophase I.
(D) Wild type. Zygotene with partial synapsis.
(E) Wild type. Pachytene with full synapsis.
(F) zygo1. Early post-leptotene with short fragments of ZEP1 signals.
(G) zygo1. Late post-leptotene with more extensive but incomplete ZEP1 signals.
(D1), (E1), (F1), and (G1) show magnified images of the framed parts in (D), (E), (F), and (G), respectively. Bars = 5 µm.
Then, SC assembly was investigated by immunofluorescence, using a super-resolution structured illumination microscope. A similar distribution of PAIR3 was observed between the wild type and zygo1 (Figures 4D to 4G). However, the disassociation of PAIR2 was severely delayed relative to that of the wild type (Figures 4D to 4G), indicating defects in the synapsis completion in zygo1. ZEP1 is the transverse filament protein of the SC in rice (Wang et al., 2010). In the wild type, ZEP1 was detected as discontinuous linear signals at zygotene (Figures 4D and 4D1). When entering pachytene, ZEP1 signals stretched along entire chromosomes and localized in the center between the two parallel PAIR3 signals (Figures 4E and 4E1). However, the short fragments of ZEP1 signals never stretched along the entire length of chromosomes in zygo1 throughout post-leptotene stage (Figures 4F, 4F1, 4G, and 4G1). Measurements of the SC length, based on ZEP1 signals, revealed that the mean SC length in zygo1 (66.2 ± 4.7 µm, n = 44) was ∼21.3% of that in the wild type (310.1 ± 9.1 µm, n = 30) at pachytene (Supplemental Figure 8), indicating that full-length synapsis is also impaired in zygo1.
ZYGO1 Is Not Required for Early Recombination Processes
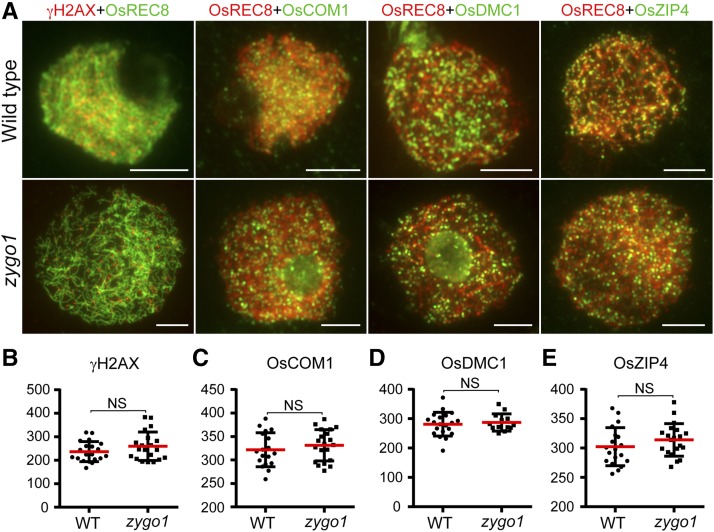
To explore whether homologous recombination is affected in zygo1, first, an antibody against phosphorylated H2AX (γH2AX) was used to monitor DSB formation in both wild-type and zygo1 meiocytes. No significant difference in γH2AX signals was observed between meiocytes of zygo1 and the wild type at early prophase I (Figures 5A and 5B), suggesting that the initiation of homologous recombination was not disturbed in zygo1. We also investigated the localizations of OsCOM1, OsDMC1, and OsZIP4, which are all essential elements involved in rice DSB repair (Ji et al., 2012; Shen et al., 2012; Wang et al., 2016). In the wild type, the signals of OsCOM1, OsDMC1, and OsZIP4 were observed as punctuate foci at zygotene. In zygo1, the same distribution patterns of OsCOM1, OsDMC1, and OsZIP4 were observed at post-leptotene stage (Figure 5A). Statistical analysis also indicated that there were no significant differences in the numbers of these foci between the wild type and zygo1 (Figures 5C to 5E). These results suggested that ZYGO1 is dispensable for early homologous recombination processes.
Figure 5.
ZYGO1 Is Not Required for the Localization of γH2AX, OsCOM1, OsDMC1, and OsZIP4 onto Chromosomes.
(A) Immunolocalization of γH2AX (red), OsCOM1 (green), OsDMC1 (green), and OsZIP4 (green) at zygotene or post-leptotene stage in the wild type and zygo1. Bars = 5 µm.
(B) to (E) Quantification of γH2AX (B), OsCOM1 (C), OsDMC1 (D), and OsZIP4 (E) foci per meiocyte in the wild type and zygo1, respectively. Values are means ± se. Statistical analyses revealed no significant differences (NS) between the wild type (n values are 20, 18, 20, and 19, respectively) and zygo1 (n values are 20, 20, 13, and 20, respectively). The P values are 0.1746, 0.4010, 0.6400, and 0.2404, respectively, from two-tailed Student’s t tests.
CO Formation Is Affected in the zygo1 Mutant
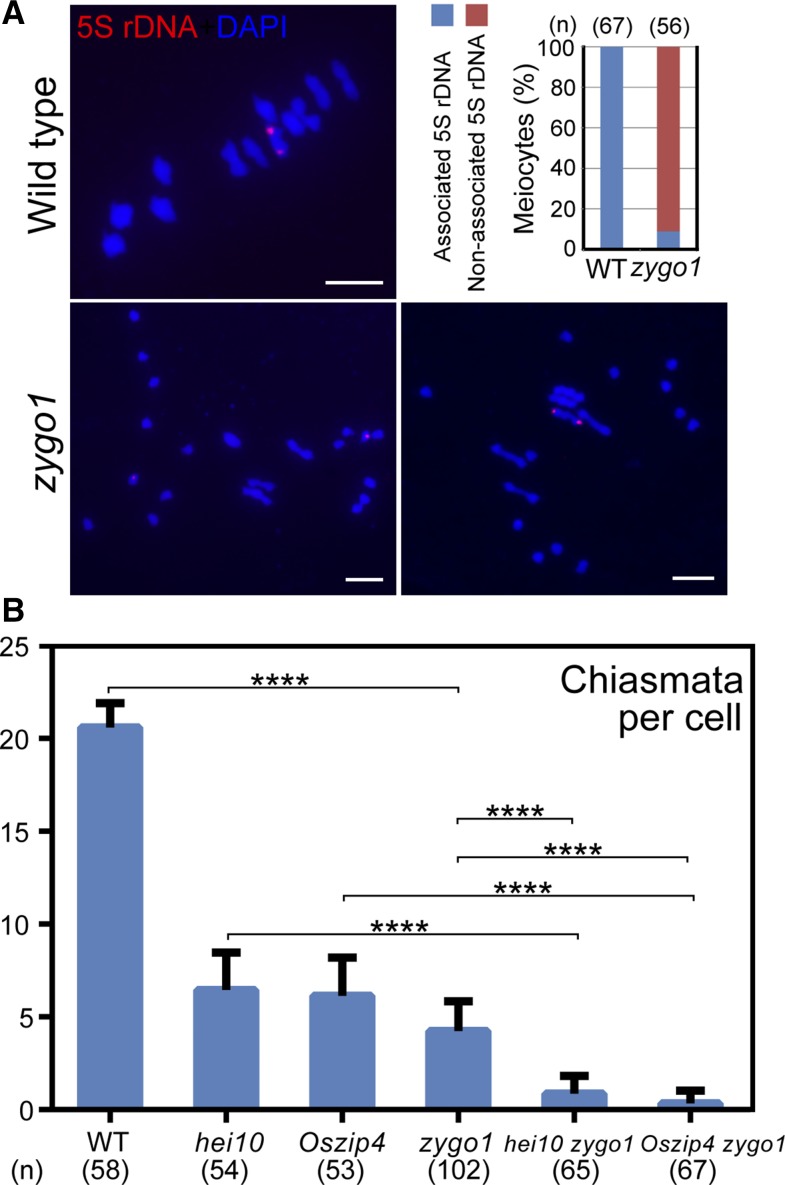
To investigate whether the residual bivalents in zygo1 result from homologous recombination, we performed FISH assays using 5S rDNA, which is located on the short arm that close to the centromere of chromosome 11. The results revealed that two obvious 5S rDNA signals were located on the same bivalent in all of the wild-type meiocytes at metaphase Ι (n = 67; Figure 6A). In zygo1, however, two 5S rDNA foci were observed to be on two separated univalents in most meiocytes (91.1%, n = 56). We only observed these two foci on the same bivalent in 8.9% of the meiocytes. The distribution of 5S rDNA indicated that the residual bivalents in zygo1 are still composed of homologous chromosomes.
Figure 6.
CO Formation Is Affected in zygo1.
(A) The distribution of 5S rDNA foci (red) revealed by FISH assays in the wild type and zygo1. Chromosomes at metaphase I were stained with DAPI (blue). Frequencies of associated or nonassociated 5S rDNA signals in the wild type and zygo1 are displayed. Bars = 5 µm.
(B) Chiasmata per cell in the wild type, hei10, Oszip4, zygo1, hei10 zygo1, and Oszip4 zygo1. Values are means ± se. ****P < 0.0001. The n values are 58, 54, 53, 102, 65, and 67, respectively, from two-tailed Student’s t tests.
However, CO formation was severely affected in zygo1. Statistical analysis showed that the chiasmata frequency in zygo1 was 4.2 ± 0.2 (n = 102), which is significantly (P < 0.0001) less than that in the wild type (20.6 ± 0.2, n = 58; Figure 6B). HEI10 and OsZIP4 are both ZMM proteins and have been proved to be essential for class I CO formation in rice (Shen et al., 2012; Wang et al., 2012). The chiasmata frequency in hei10 and Oszip4 mutants was 6.4 ± 0.3 (n = 54) and 6.1 ± 0.3 (n = 53), respectively (Figure 6B; Supplemental Figure 9). Strikingly, the chiasmata frequency of hei10 zygo1 and Oszip4 zygo1 double mutants was reduced to 0.8 ± 0.1 (n = 65) and 0.3 ± 0.1 (n = 67), respectively. Considering that nearly 80% of COs were lost in zygo1 and CO numbers were further reduced in hei10 zygo1 and Oszip4 zygo1 double mutants, we reasoned that ZYGO1 deficiency impacts both Class I and Class II CO formation in rice.
ZYGO1 Promotes Homologous Pairing and Synapsis Independent of ZMM Proteins
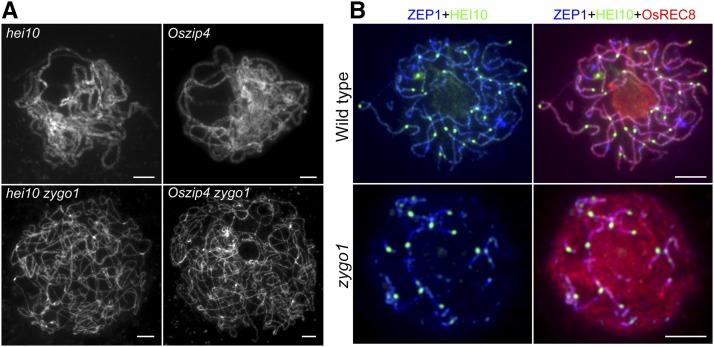
Compared with ZYGO1, HEI10 and OsZIP4 have nearly no effects on homologous pairing and SC assembly in rice (Shen et al., 2012; Wang et al., 2012). The hei10 and Oszip4 single mutants underwent complete synapsis at pachytene. Nevertheless, hei10 zygo1 and Oszip4 zygo1 double mutants showed severe synapsis defects at late post-leptotene stage (Figure 7A), mimicking the phenotype of the zygo1 single mutant. Therefore, the function of ZYGO1 in promoting homologous chromosome alignment is independent of ZMM proteins.
Figure 7.
ZYGO1 Promotes Homologous Pairing Independent of HEI10 and OsZIP4, but Affects HEI10 Distribution.
(A) Chromosome behavior of meiocytes in hei10, Oszip4, hei10 zygo1, and Oszip4 zygo1 at pachytene or late post-leptotene stage. Full-length chromosome pairing in hei10 and Oszip4 is suppressed in hei10 zygo1 and Oszip4 zygo1 double mutants. Chromosomes were stained with DAPI.
(B) Triple immunolocalization of OsREC8 (red), HEI10 (green), and ZEP1 (blue) in the wild type and zygo1 at late pachytene or late post-leptotene stage. The bright punctuated signals of HEI10 are restricted to the short ZEP1 stretches in zygo1. Bars = 5 µm.
The Distribution of HEI10 Is Limited to the Partial Synapsis Region in zygo1
To further explore the cause of chiasmata frequency decrease in zygo1, immunofluorescent examinations were performed using antibodies against ZEP1 and HEI10. In the wild type, linear signals of HEI10 colocalized well with ZEP1 from zygotene to pachytene, and the bright dot-like foci of HEI10 appeared at the faint linear signals at late pachytene (Figure 7B), which are indicative of class I COs (Chelysheva et al., 2012; Wang et al., 2012). However, the bright dot-like signals of HEI10 were only associated with the short fragments of ZEP1 signals in zygo1 (Figure 7B). The restricted distribution of HEI10 foci implied that HEI10 might be able to promote CO formation on the partially synapsed chromosomes, but might lose its function in the unsynapsed region. As normal CO maturation depends on normal chromosome alignment (Lambing et al., 2015; Zickler and Kleckner, 2015), the defects in CO formation might be caused by the failure in full-length chromosome alignment in zygo1.
DISCUSSION
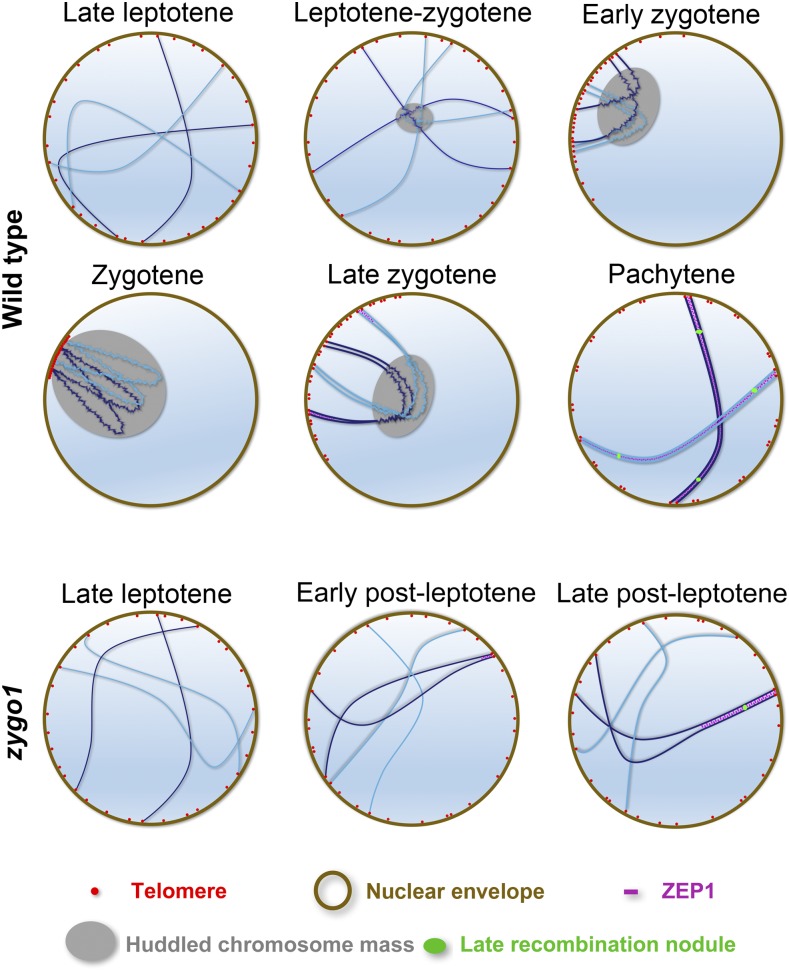
A Functional Model of ZYGO1 in Meiotic Bouquet Formation
During zygotene, accompanying the dramatic nuclear reorganization and dynamic chromosome movement, homologous pairing, synapsis, and recombination take place concurrently at this stage (Zickler and Kleckner, 2015). Therefore, there must be cohesive links between these meiotic events during zygotene. The chromosomal bouquet is an evolutionary conserved meiotic configuration essential for homologous pairing in a wide range of species. However, we lack an understanding of the underlying molecular mechanisms of telomere bouquet arrangement in plants. Here, we characterized the bouquet-deficient mutant zygo1 and identified some key linkages between bouquet formation and other meiotic events. Through a combination of genetic and cytological methods, we validated the essential role of ZYGO1 in telomere bouquet formation. In addition, our results indicate that ZYGO1 deficiency partially suppresses the processes of homologous pairing, SC assembly, and CO formation. Thus, we propose a model for the function of ZYGO1 in rice meiosis (Figure 8). In the wild type, telomeres are gradually clustered into a bouquet from late leptotene to zygotene along with homologous pairing. Then, the telomere bouquet gradually dissolves from zygotene to pachytene during SC assembly and CO maturation. Meanwhile, the chromosome mass gradually evolves and then resolves along with the dynamic movement of telomeres. In zygo1, however, both the telomere bouquet and chromosome mass are destroyed. Full-length pairing and synapsis are not completed, and CO formation is limited to the partially synapsed region of chromosomes. Based on this model, we propose that the absence of telomere bouquet is the primary defect in zygo1. Moreover, the failure to complete pairing/synapsis and recombination suggest that bouquet formation is required for efficient homology searching and homologous recombination. These findings reveal the regulatory role for a novel F-box protein in early meiotic events.
Figure 8.
Proposed Model for ZYGO1 Function during Early Prophase I.
In the wild type, accompanying homologous pairing, telomeres gradually aggregate from late leptotene to zygotene and finally are clustered into a typical bouquet at zygotene. Meanwhile, linear chromosomes gradually aggregate into a fused mass. The telomere bouquet and chromosome mass completely dissolve and homologous chromosomes are fully synapsed at pachytene. In zygo1, however, both the telomere bouquet and chromosome mass do not form. Although some chromosomes initiate pairing and synapsis, full-length chromosome pairing and synapsis are not completed. Finally, crossover formation is restricted to the partially synapsed region of chromosomes in zygo1.
ZYGO1 Efficiently Facilitates Homologous Pairing
Our results reveal that bouquet formation is not disturbed in pair1 and Oscom1 mutants, indicating that homologous recombination is not required for telomere clustering. Moreover, ZYGO1 is not required for DSB formation, DSB end processing, and early recombination element installation. Collectively, these data indicate that bouquet formation and DSB formation are mutually independent. Although early recombination processes are not affected in zygo1, full-length pairing and synapsis were found to be interrupted. In most organisms, CO maturation requires normal SC installation (Zickler and Kleckner, 2015). For example, in the Arabidopsis Atpch2 mutant, recombination initiation and CO designation occur normally, but the maturation of the CO designated intermediates is compromised by the defect in SC formation (Lambing et al., 2015). Similarly, early recombination processes are normal in zygo1, but the bright HEI10 foci are limited to the partial SCs. Thus, ZYGO1 deficiency has an impact on CO formation possibly due to its effects on normal chromosome alignment.
Although homologous recombination is not required for bouquet formation, homologous searching and stable homolog juxtaposition are dependent on meiotic recombination in most organisms (Peoples et al., 2002; Zickler and Kleckner, 2015). In zygo1, the telomere clustering deficiency does not fully suppress homologous pairing and recombination. In other bouquet-deficient mutants, such as in ndj1Δ, pam1, and Atsun1 Atsun2, a level of pairing and recombination are also achieved (Trelles-Sticken et al., 2000; Golubovskaya et al., 2002; Varas et al., 2015). Therefore, telomere bouquet is not absolutely required for homologous pairing, and the partial pairing observed in zygo1 might be promoted by homologous recombination. We propose that ZYGO1 deficiency may disturb homologous pairing to extend to the full-length chromosomes.
ZYGO1 Promotes Bouquet Formation within an SCF Complex
ZYGO1 encodes a novel F-box protein in rice that interacted with OSK1, which was predicted to have similar functions to those of ASK1 in Arabidopsis (Kahloul et al., 2013). ASK1 is involved in many aspects of plant growth and development through the interactions with different F-box proteins (Gray et al., 1999; Zhao et al., 2001; Yao et al., 2016). During male meiosis, ASK1 is also required for chromosome condensation, homologous pairing, synapsis, and chromosome separation (Yang et al., 1999; Zhao et al., 2006). However, the ASK1-interacting F-box proteins and their substrates involved in meiosis remain unknown. MOF, another F-box protein required for meiosis, was previously demonstrated to interact with OSK1 in rice (He et al., 2016). Unlike zygo1, however, the mof mutant is female fertile and its male meiocytes are arrested at prophase I. Therefore, it is clear that MOF and ZYGO1 play distinct roles during meiosis in rice. Interestingly, recent studies reported that chromosomally tethered proteasomes promote pairing and recombination in mouse, budding yeast, and C. elegans, suggesting that the meiotic programmed degradation event is highly conserved among species (Ahuja et al., 2017; Rao et al., 2017). We also found that ZYGO1 shows limited sequence similarity to FBL12 in mouse. SCFFbl12 participates in cellular proliferation and DNA repair through ubiquitylation of some cell cycle inhibitors and KU80 (Kim et al., 2008; Postow and Funabiki, 2013). However, so far, there is no evidence for FBL12 in regulating meiotic progress. Our results suggest that ZYGO1 may function as an essential component of a SCF E3 ligase, which targets a specific protein involved in suppressing telomere clustering and normal meiotic progression for ubiquitin-mediated degradation. However, we cannot exclude the possibility that ZYGO1 may regulate meiotic proteins through the modification pathway. Further investigation toward identification of the target(s) of ZYGO1 will shed light on its specific function during meiosis.
METHODS
Plant Materials
The zygo1-1 mutant was isolated from the japonica rice (Oryza sativa) variety Yandao 8 induced by 60Co γ-ray irradiation. The zygo1-2 mutant was ordered from the Rice T-DNA Insertion Sequence Database (Jeong et al., 2006). The zygo1-3 and zygo1-4 mutants were generated using CRISPR-Cas9 targeting method in the japonica rice variety Yandao 8. The zygo1-3 mutant allele was used for data generation throughout our research. The pair1, Oscom1, Oszip4, and hei10 mutants used in this study were reported previously (Ji et al., 2012; Shen et al., 2012; Wang et al., 2012; Che et al., 2014). The japonica rice variety Yandao 8 was used for wild-type analysis. All plant materials were grown in paddy fields in Beijing or Hainan province of China.
Molecular Cloning
For map-based cloning of ZYGO1, heterozygous mutant plants of zygo1-1 were crossed with the indica variety Zhongxian 3037, and ∼240 sterile plants segregated from the F2 population were selected for analysis. Markers were developed based on sequence differences between the japonica variety Yandao 8 and the indica variety Zhongxian 3037. The primer sequences of these markers are listed in Supplemental Table 2. Total RNA was extracted from the root, internode, leaf, and young panicle (5–7 cm) of the wild type and reverse-transcribed into cDNA using the SuperScript III reverse transcriptase (Invitrogen; catalog no. 18080-044). The cDNA sequence of ZYGO1 was redefined using the 5′-Full RACE kit with TAP (TaKaRa; catalog no. 6107) and the 3′-Full RACE kit (TaKaRa; catalog no. 6106). The gene-specific primers for 5′ RACE were RACE1R and RACE2R and the gene-specific primers for 3′ RACE were RACE1F and RACE2F (Supplemental Table 2). The PCR products were sequenced and analyzed.
Complementation Test and CRISPR-Cas9 Targeting of ZYGO1
For the complementation test, an 8538-bp genomic sequence containing 3422 bp of promoter region and the entire ZYGO1 gene region was inserted into the binary vector pCAMBIA 1300. This construct was transformed into Agrobacterium tumefaciens strain EHA105, and then into zygo1-1+/− embryonic calli. For the CRISPR-Cas9 targeting of ZYGO1, the reported CRISPR-Cas9 binary vector pC1300-cas9 and the intermediate vector SK-gRNA were used in this study (Hu et al., 2017). The target sequences of ZYGO1 were “TGAGGAAGCGCCCGAGGCCG” and “GTACGAGGCGGACATGTGGA,” respectively. These two constructs were introduced into Agrobacterium strain EHA105 and then independently transformed into the japonica variety Yandao 8.
Neighbor-Joining Tree Construction and Multiple Alignments
Using MEGA5 software (http://www.megasoftware.net/), the neighbor-joining tree was constructed based on the full-length amino acid sequence of the related proteins. The pairwise deletion option and 1000 bootstrap replicates were applied. The protein domains were detected by SMART (http://smart.embl-heidelberg.de/) and drawn by IBS software (http://ibs.biocuckoo.org/). PSI-BLAST (NCBI) was used for homology searches. Multiple alignments were conducted using MAFFT (https://toolkit.tuebingen.mpg.de/mafft) and colored with ESPript (http://espript.ibcp.fr/ESPript/ESPript/). A text file of the alignment is provided as Supplemental File 1.
Library Screening and Y2H Assay
The library was constructed using the purified total RNA from the meiotic-stage wild-type anthers. The “make your own mate and plate library system” (Clontech; no. 630490) was used to generate the library. The full-length coding sequence of ZYGO1 was cloned into the pGBKT7 vector (Clontech) and transformed into yeast strain AH109. Transformed cells were further transformed with plasmids from the cDNA library. These transformants (2 × 106) were screened on SD –Trp –Leu –His –Ade medium. Surviving clones were further screened on SD –Trp –Leu –His –Ade medium with X-α-gal to examine the LacZ activity. Then, positive clones were sequenced and analyzed. For the confirmation assays, the full-length or truncated coding sequences of related genes were inserted into the vector pGADT7 or pGBKT7. The Y2H assays were conducted with a Matchmaker Gold Yeast Two-Hybrid system (Clontech; no. 630489) and the yeast strain Y2HGOLD was used in these confirmation assays. Detailed procedures from the Yeast Handbook (Clontech) were followed. All primers are listed in Supplemental Table 2. These assays were repeated at least three times.
BiFC Assay
To conduct BiFC assays, the full-length or truncated coding sequences of ZYGO1 and OSK1 were cloned into pUC-SPYNE and pUC-SPYCE, respectively. The vector pairs enable the expression of proteins of interest fused to the N-terminal 155 amino acids (nYFP) or to the C-terminal 86 amino acids (cYFP) of YFP (Walter et al., 2004). The proper plasmid pairs were cotransformed into rice stem protoplasts. After overnight incubation in the dark at 28°C, cells were collected for observation. Images were captured using a confocal laser scanning microscope (Leica TCS SP5). These assays were repeated three times.
Antibody Production
The primary antibodies against OsREC8, PAIR2, PAIR3, γ-H2AX, OsCOM1, OsDMC1, OsZIP4, and HEI10 were previously reported (Wang et al., 2011, 2012; Ji et al., 2012; Shen et al., 2012; Wang et al., 2016). The guinea pig antibody against ZEP1 was generated with the ZEP1 fusion protein (amino acids 426–612) based on the report of the mouse antibody against ZEP1 (Wang et al., 2010). To produce the rabbit polyclonal antibody against OsSAD1, the amino acid sequence “IRGESVLGKSKYPL” was selected for peptide synthesis. Then, the synthetic peptide was used for immunization and the resulting antiserum was affinity-purified against this peptide. Peptide synthesis, antibody production, and affinity purification were performed by GenScript.
Chromosome Preparation
The 1% acetocarmine solution used in these assays was prepared according to a research protocol (Cheng, 2013). In brief, 1 g carmine was added to 100 mL of 45% acetic acid in a boiling flask, which was thereafter boiled for 8 h on a hot plate with an attached reflux column. This solution was stored at room temperature until use. The FAA solution was prepared by adding 10 mL of formaldehyde (37–40%), 5 mL of glacial acetic acid, and 47.5 mL of ethyl alcohol to 37.5 mL of distilled water. To prepare chromosomes fixed in the ethanol-acetic acid (3:1) solution, meiotic stage anthers of the wild type or mutants were squashed in 1% acetocarmine solution and then washed with 45% acetic acid. Slides were frozen in liquid nitrogen for a few minutes, and then the cover slips were removed. After dehydration through an ethanol series (70, 90, and 100%), the slides were stained with DAPI in an antifade solution (Vector Laboratories). The anthers fixed in the FAA solution were also processed by following these procedures.
FISH Analysis
The FISH analysis was conducted based on a previous protocol (Yang et al., 2016). Two repetitive DNA elements were used as probes: the pAtT4 clone containing telomeric repeats (Richards and Ausubel, 1988) and the pTa794 clone containing 5S rRNA genes from wheat (Triticum aestivum; Cuadrado and Jouve, 1994). The bulked oligonucleotide probes (11S and 11L) were newly developed based on the procedures of a previous report (Han et al., 2015). First, thousands of independent oligonucleotides, specific to the short or long arm of chromosome 11 in rice, were selected by the bioinformatics pipeline. The selected oligonucleotides were then synthesized, amplified, and labeled with biotin or digoxigenin to make the probe for FISH assays. Biotin-labeled probes were detected by Alexa Fluor 488 streptavidin and digoxigenin-labeled probes were detected by rhodamine antidigoxigenin. We observed at least 50 meiocytes from three individual wild-type or mutant plants in each assay.
Immunofluorescence Assay
For immunofluorescence assays, the fresh panicles around meiotic stage were fixed in 4% (w/v) paraformaldehyde. The procedure was performed as previously described (Cheng, 2013). Fluorochrome-coupled secondary antibodies were used for fluorescence detection, including the fluorescein isothiocyanate-conjugated goat anti-mouse antibody (Southern Biotech), the rhodamine-conjugated goat anti-rabbit antibody (Southern Biotech), and the AMCA-conjugated goat anti-guinea pig antibody (Jackson Immunoresearch). We observed at least 50 meiocytes from three individual wild-type or mutant plants in each assay.
Immuno-FISH Analysis
The immuno-FISH analysis was performed as previously described (Han et al., 2007). First, immunofluorescence assays were conducted as described above. The rabbit antibody against PAIR2 and the guinea pig antibody against PAIR3 were used in this assay. Thereafter, slides were screened and the X-Y position of each collected image was recorded. Those slides with good spreads were washed with 1× PBS to remove cover slips and DAPI. Then, those slides were ready for FISH assays. The pAtT4 probe labeled with biotin was used to detect telomeres. After the FISH assay, those slides were screened again to collect further images. Finally, three-channel images were composed using Photoshop CS2 (Adobe).
Image Capture, Processing, and Statistical Analysis
The super-resolution images were captured using a DeltaVision microscope (GE Healthcare; OMX V4) and processed with SoftWoRx (Applied Precision) to generate projected images. Other images were captured under a Zeiss A2 fluorescence microscope with a micro CCD camera and viewed with ZEN software (Zeiss). To quantitate the length of SC, images were analyzed using IPLab 4.0 software (Scanalytics). Statistical significance was determined by unpaired two-tailed t test and graphs were drawn with GraphPad Prism 6 software (http://www.graphpad.com/).
Accession Numbers
Sequence data used in this article can be found in the NCBI databases under the following accession numbers: ZYGO1, NP_001042418; OSK1, XP_015617535; and OsSAD1, NP_001055057. Accession numbers of the homologs of ZYGO1 used in the neighbor-joining tree construction are as follows: Brachypodium distachyon, XP_003565503.1; Sorghum bicolor, XP_002457119.1; Arabidopsis thaliana, NP_001319817.1, NP_198449.1, and NP_191809.1; Solanum lycopersicum, XP_01032704.1; Physcomitrella patens, XP_001784016.1; Zea mays, NP_001168584.1 and XP_008673124.1; Vitis vinifera, XP_003633984.1 and XP_002281745.1; and FBL12, NP_038939.2.
Supplemental Data
Supplemental Figure 1. ZYGO1 Is Required for Rice Fertility.
Supplemental Figure 2. Linkage and Physical Map of zygo1-1.
Supplemental Figure 3. Meiotic Chromosome Behaviors of the Complemented Lines of zygo1-1 and the Three Other zygo1 Alleles.
Supplemental Figure 4. The cDNA Sequence of ZYGO1.
Supplemental Figure 5. Alignment of the Full-Length Amino Acid Sequences of ZYGO1 and the Mouse F-Box Protein FBL12.
Supplemental Figure 6. Meiocytes Fixed with Formalin-Acetic Acid-Alcohol Solution in the Wild Type and zygo1.
Supplemental Figure 7. Quantification of the Number of pAtT4 foci in zygo1.
Supplemental Figure 8. Full-length Synapsis Is Severely Affected in zygo1.
Supplemental Figure 9. Chromosome Behaviors at Metaphase I in hei10, Oszip4, hei10 zygo1, and Oszip4 zygo1.
Supplemental Table 1. Positive Clones from the Yeast Two-Hybrid Screening.
Supplemental Table 2. List of Primers Used in This Study.
Supplemental File 1. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 1.
AUTHOR CONTRIBUTIONS
Z.C. and F.Z. conceived the research plans. Y.S. and Y.L. supervised the experiments. F.Z., D.T., Z.X., W.S., L.R., and G.D. performed most of the experiments. F.Z. and D.T. analyzed the data. F.Z. wrote the article. Z.C. supervised and completed the writing.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100901) and the National Natural Science Foundation of China (31230038 and 31360260).
Footnotes
Articles can be viewed without a subscription.
References
- Ahuja J.S., Sandhu R., Mainpal R., Lawson C., Henley H., Hunt P.A., Yanowitz J.L., Börner G.V. (2017). Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 355: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che L., Wang K., Tang D., Liu Q., Chen X., Li Y., Hu Q., Shen Y., Yu H., Gu M., Cheng Z. (2014). OsHUS1 facilitates accurate meiotic recombination in rice. PLoS Genet. 10: e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Vezon D., Chambon A., Gendrot G., Pereira L., Lemhemdi A., Vrielynck N., Le Guin S., Novatchkova M., Grelon M. (2012). The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8: e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. (2013). Analyzing meiotic chromosomes in rice. Methods Mol. Biol. 990: 125–134. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. (2006). Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Yamane M., Okamasa K., Tsutsumi C., Kojidani T., Sato M., Haraguchi T., Hiraoka Y. (2009). Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 187: 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.N., Dominguez A.M., Dresser M.E. (1997). Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276: 1252–1255. [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Jouve N. (1994). Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6x-triticale. Chromosome Res. 2: 331–338. [DOI] [PubMed] [Google Scholar]
- Dawe R.K., Sedat J.W., Agard D.A., Cande W.Z. (1994). Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76: 901–912. [DOI] [PubMed] [Google Scholar]
- De Muyt A., Vezon D., Gendrot G., Gallois J.L., Stevens R., Grelon M. (2007). AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 26: 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Pereira L., Vezon D., Chelysheva L., Gendrot G., Chambon A., Lainé-Choinard S., Pelletier G., Mercier R., Nogué F., Grelon M. (2009). A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. (2007). SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12: 863–872. [DOI] [PubMed] [Google Scholar]
- Golubovskaya I.N., Harper L.C., Pawlowski W.P., Schichnes D., Cande W.Z. (2002). The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162: 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., del Pozo J.C., Walker L., Hobbie L., Risseeuw E., Banks T., Crosby W.L., Yang M., Ma H., Estelle M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13: 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Gao Z., Yu W., Birchler J.A. (2007). Minichromosome analysis of chromosome pairing, disjunction, and sister chromatid cohesion in maize. Plant Cell 19: 3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang T., Thammapichai P., Weng Y., Jiang J. (2015). Chromosome-specific painting in cucumis species using bulked oligonucleotides. Genetics 200: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L., Golubovskaya I., Cande W.Z. (2004). A bouquet of chromosomes. J. Cell Sci. 117: 4025–4032. [DOI] [PubMed] [Google Scholar]
- He Y., Wang C., Higgins J.D., Yu J., Zong J., Lu P., Zhang D., Liang W. (2016). MEIOTIC F-BOX is essential for male meiotic DNA double-strand break repair in rice. Plant Cell 28: 1879–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Wang C., Liu Q., Fu Y., Wang K. (2017). Targeted mutagenesis in rice using CRISPR-Cpf1 system. J. Genet. Genomics 44: 71–73. [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334. [DOI] [PubMed] [Google Scholar]
- Jantsch V., Tang L., Pasierbek P., Penkner A., Nayak S., Baudrimont A., Schedl T., Gartner A., Loidl J. (2007). Caenorhabditis elegans prom-1 is required for meiotic prophase progression and homologous chromosome pairing. Mol. Biol. Cell 18: 4911–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45: 123–132. [DOI] [PubMed] [Google Scholar]
- Ji J., Tang D., Wang K., Wang M., Che L., Li M., Cheng Z. (2012). The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 72: 18–30. [DOI] [PubMed] [Google Scholar]
- Kahloul S., HajSalah El Beji I., Boulaflous A., Ferchichi A., Kong H., Mouzeyar S., Bouzidi M.F. (2013). Structural, expression and interaction analysis of rice SKP1-like genes. DNA Res. 20: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Nakamoto T., Nishimori S., Tanaka K., Chiba T. (2008). A new ubiquitin ligase involved in p57KIP2 proteolysis regulates osteoblast cell differentiation. EMBO Rep. 9: 878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C., Osman K., Nuntasoontorn K., West A., Higgins J.D., Copenhaver G.P., Yang J., Armstrong S.J., Mechtler K., Roitinger E., Franklin F.C. (2015). Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11: e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Villeneuve A.M. (2001). Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.P., Bass H.W. (2012). The maize (Zea mays) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation, linking nuclear morphology, telomere distribution and synapsis. J. Cell Sci. 125: 3681–3690. [DOI] [PubMed] [Google Scholar]
- Murphy S.P., Simmons C.R., Bass H.W. (2010). Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biol. 10: 269–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.P., Gumber H.K., Mao Y., Bass H.W. (2014). A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.). Front. Plant Sci. 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., Jantsch V. (2009). Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139: 920–933. [DOI] [PubMed] [Google Scholar]
- Peoples T.L., Dean E., Gonzalez O., Lambourne L., Burgess S.M. (2002). Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 16: 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L., Funabiki H. (2013). An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle 12: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H.B., Qiao H., Bhatt S.K., Bailey L.R., Tran H.D., Bourne S.L., Qiu W., Deshpande A., Sharma A.N., Beebout C.J., Pezza R.J., Hunter N. (2017). A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J., Ausubel F.M. (1988). Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136. [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz P., Jones G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4: 507–516. [DOI] [PubMed] [Google Scholar]
- Scherthan H. (2001). A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Shen Y., Tang D., Wang K., Wang M., Huang J., Luo W., Luo Q., Hong L., Li M., Cheng Z. (2012). ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 125: 2581–2591. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Ishiguro K., Watanabe Y. (2014). The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 16: 145–156. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Hernández-Hernández A., Morimoto A., Negishi L., Höög C., Watanabe Y. (2015). MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163: 1252–1266. [DOI] [PubMed] [Google Scholar]
- Starr D.A., Fridolfsson H.N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26: 421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E., Dresser M.E., Scherthan H. (2000). Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas J., Graumann K., Osman K., Pradillo M., Evans D.E., Santos J.L., Armstrong S.J. (2015). Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 81: 329–346. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang H., Hu Q., Tang D., Liu X., Du G., Shen Y., Li Y., Cheng Z. (2016). OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol. 171: 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Qin B., Li M., Cheng Z. (2011). PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Miao C., Hu Q., Lu T., Cheng Z. (2012). The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang K., Tang D., Wei C., Li M., Shen Y., Chi Z., Gu M., Cheng Z. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Liu P., Inuzuka H., Wei W. (2014). Roles of F-box proteins in cancer. Nat. Rev. Cancer 14: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Hu Y., Lodhi M., McCombie W.R., Ma H. (1999). The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96: 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Li Y., Su Y., Shen Y., Tang D., Luo Q., Cheng Z. (2016). A functional centromere lacking CentO sequences in a newly formed ring chromosome in rice. J. Genet. Genomics 43: 694–701. [DOI] [PubMed] [Google Scholar]
- Yao R., et al. (2016). DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473. [DOI] [PubMed] [Google Scholar]
- Yoshida M., et al. (2013). Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J. Cell Biol. 200: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Yu Q., Chen M., Ma H. (2001). The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128: 2735–2746. [DOI] [PubMed] [Google Scholar]
- Zhao D., Yang X., Quan L., Timofejeva L., Rigel N.W., Ma H., Makaroff C.A. (2006). ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol. Biol. 62: 99–110. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1998). The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (2015). Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]