Silencing phosphoenolpyruvate carboxylase kinase in a CAM species more than halves dark period CO2 fixation and causes arrhythmia in some components of the central circadian clock.
Abstract
Phosphoenolpyruvate carboxylase (PPC; EC 4.1.1.31) catalyzes primary nocturnal CO2 fixation in Crassulacean acid metabolism (CAM) species. CAM PPC is regulated posttranslationally by a circadian clock-controlled protein kinase called phosphoenolpyruvate carboxylase kinase (PPCK). PPCK phosphorylates PPC during the dark period, reducing its sensitivity to feedback inhibition by malate and thus enhancing nocturnal CO2 fixation to stored malate. Here, we report the generation and characterization of transgenic RNAi lines of the obligate CAM species Kalanchoë fedtschenkoi with reduced levels of KfPPCK1 transcripts. Plants with reduced or no detectable dark phosphorylation of PPC displayed up to a 66% reduction in total dark period CO2 fixation. These perturbations paralleled reduced malate accumulation at dawn and decreased nocturnal starch turnover. Loss of oscillations in the transcript abundance of KfPPCK1 was accompanied by a loss of oscillations in the transcript abundance of many core circadian clock genes, suggesting that perturbing the only known link between CAM and the circadian clock feeds back to perturb the central circadian clock itself. This work shows that clock control of KfPPCK1 prolongs the activity of PPC throughout the dark period in K. fedtschenkoi, optimizing CAM-associated dark CO2 fixation, malate accumulation, CAM productivity, and core circadian clock robustness.
INTRODUCTION
Crassulacean acid metabolism (CAM) is one of several higher plant photosynthetic CO2-concentrating mechanisms. It is particularly noteworthy for its high water-use efficiency (WUE) relative to C3 and C4 photosynthesis (Borland et al., 2009; Cushman et al., 2015). The functional genomics and evolutionary biology of CAM species has recently received a dramatic upsurge in interest due to the potential of CAM crops, such as agaves and opuntias, as water-wise sources of biomass for biofuels, platform chemicals, and food (Cushman et al., 2015; Yang et al., 2015). In addition, efforts are underway to engineer CAM into C3 species using synthetic biology approaches (Borland et al., 2014, 2015). CAM into C3 aims to improve the WUE of key C3 crops, thereby adapting them to the increased frequency and intensity of droughts and extreme high temperatures predicted under future climate change scenarios. The further exploitation of CAM in these ways will require detailed characterization of CAM gene function, especially with respect to their daily regulation by the circadian clock (Yang et al., 2015; Hartwell et al., 2016).
CAM is a classic example of convergent evolution that is known from 35 families of plants spanning monocots, dicots, and ferns (Smith and Winter, 1996; Holtum et al., 2007; Tsen and Holtum, 2012; Christin et al., 2014; Silvera et al., 2014). Based on our currently incomplete understanding of the molecular-genetic blueprint for CAM (Hartwell et al., 2016), C3 species are believed to possess ancestral copies of all of the genes required for CAM (Cushman and Bohnert, 1999; Aubry et al., 2011; Ming et al., 2015). In principle, for CAM to evolve from the C3 ancestral state in a leaf succulent CAM species such as Kalanchoë fedtschenkoi, the corresponding metabolic enzymes, metabolite transporters, and their cognate regulatory proteins must increase in abundance in leaf mesophyll cells and come under tight temporal control such that certain metabolic steps are confined to the dark period, and others are confined to the light (Cushman and Bohnert, 1999; Borland et al., 2009; Abraham et al., 2016; Brilhaus et al., 2016; Hartwell et al., 2016). However, only limited work has been reported testing this principle using a genetic approach, which represents a powerful method for in planta functional testing of candidate CAM genes when combined with detailed characterization of CAM-associated phenotypes in the resulting transgenic lines (Hartwell et al., 2016; Dever et al., 2015).
During the daily CAM cycle, CO2 fixation occurs via two temporally separated steps. In the dark, atmospheric CO2 enters through open stomata and is converted to HCO3− either passively or via the reaction catalyzed by a β-carbonic anhydrase. Primary CO2 fixation is catalyzed by phosphoenolpyruvate carboxylase (PPC), which converts PEP and HCO3− to OAA and inorganic phosphate (Chollet et al., 1996). Malate dehydrogenase reduces OAA to malate, which is transported to and stored in the vacuole as malic acid (Borland et al., 2009). In the light, malate decarboxylation generates CO2 that is refixed by Rubisco in the Calvin cycle, referred to as secondary CO2 fixation. Both primary and secondary CO2 fixation occurs in each leaf mesophyll cell; therefore, strict temporal regulation is required to prevent futile cycling between carboxylation and decarboxylation (Hartwell, 2005, 2006).
In the model obligate CAM species K. fedtschenkoi (Hartwell et al., 2016), both PPC abundance and specific activity are stable over the light/dark cycle (Nimmo et al., 1984; Dever et al., 2015), but its allosteric properties are subject to tight temporal control (Nimmo et al., 1986, 1987). Several primary metabolites have been reported to influence the activity of PPC in vitro, including malate (allosteric inhibitor) and glucose 6-phosphate (allosteric activator) (Doncaster and Leegood, 1987; Vidal and Chollet, 1997). For a CAM leaf, as the dark period progresses, the vacuolar storage capacity for malic acid reaches its limit, and malate begins to accumulate in the cytosol where it feeds back to inhibit PPC activity. In K. fedtschenkoi, the Ki of PPC for malate was found to be increased by dark period phosphorylation of the enzyme on an N-terminal serine residue catalyzed by the activity of a specific, Ca2+-independent protein kinase, named PPC kinase (PPCK) (Carter et al., 1991; Hartwell et al., 1999; Nimmo, 2000, 2003). The circadian clock controls PPCK transcript levels and activity, such that they peak during the dark period (Hartwell et al., 1996, 1999; Taybi et al., 2000; Boxall et al., 2005; Theng et al., 2008). In the light, a protein phosphatase type 2A dephosphorylates PPC, leaving it up to 10 times more sensitive to malate inhibition (Carter et al., 1990). This phosphatase was found to be active throughout the light/dark cycle (Carter et al., 1991), supporting the proposal that the light/dark regulation of PPC is mainly a result of the circadian rhythm of PPCK levels (Hartwell et al., 1999), although this has yet to be demonstrated in planta.
PPCK remains the only well-defined, temporal/circadian control point for CAM (Hartwell, 2006, 2016). To further elucidate the role of PPC phosphorylation in the temporal control of CO2 fixation associated with CAM, and the extent to which this phospho-regulation step helps to prevent metabolic futile cycling during CAM, we generated stable transformants of K. fedtschenkoi expressing RNA interference (RNAi) constructs that targeted the degradation of endogenous KfPPCK1 transcripts. We recovered independent transgenic lines with reduced KfPPCK1 transcripts and reduced phosphorylation of PPC in the dark and found that phosphorylation was vital for optimizing and sustaining dark CO2 fixation associated with CAM. Furthermore, the loss of dark phosphorylation of PPC led to a loss of robust rhythmicity for certain components of the central molecular circadian clock and for the regulation of several distinct circadian clock outputs. Surprisingly, several key components of the central oscillator were induced in the KfPPCK1 RNAi lines and these genes maintained robust transcript oscillations under constant light and temperature conditions.
RESULTS
Initial Screening and Characterization of KfPPCK1 RNAi Lines of K. fedtschenkoi
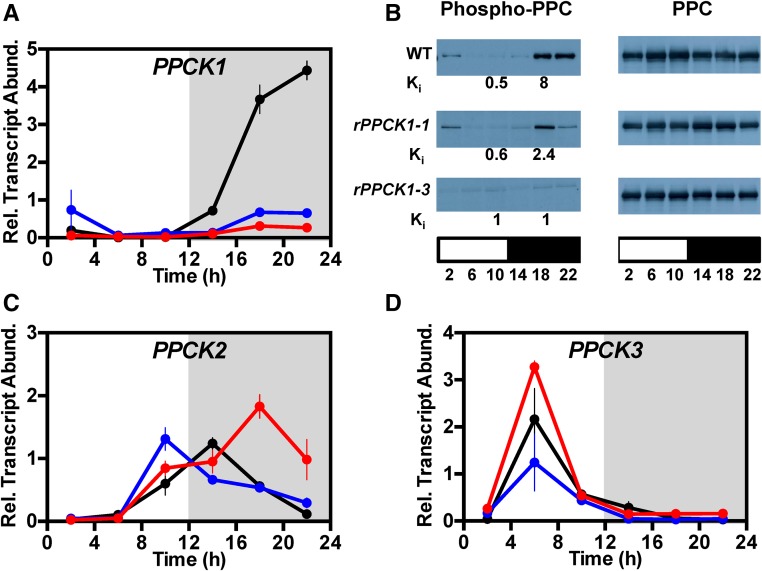
As K. fedtschenkoi does not produce viable seed (Garcês et al., 2007; Hartwell et al., 2016), data are presented for primary transformants that were propagated clonally via leaf plantlets and/or stem cuttings. Primary transformants were screened initially using high-throughput leaf disc tests for starch and acidity (Cushman et al., 2008; Dever et al., 2015). Leaf disc screening identified KfPPCK1 RNAi lines because they had higher leaf pH at dawn than the wild type due to reduced malic acid accumulation during the dark period. Transgenic lines that acidified less during the dark period were screened for the steady state abundance of KfPPCK1 transcript levels with RT-qPCR (Figure 1). Comparison of KfPPCK1 transcript levels over a 12-h-light/12-h-dark cycle revealed that, relative to the wild type, the two best RNAi lines (rPPCK1-1 and rPPCK1-3) had a large reduction in the transcript abundance of the target transcript (Figure 1A). While both lines displayed reduced levels of KfPPCK1 transcripts relative to the wild type, line rPPCK1-1 had a higher level of KfPPCK1 transcripts in the middle of the dark period than line rPPCK1-3, and neither line displayed a complete loss of KfPPCK1 transcript accumulation in the dark (Figure 1A). These results indicated that we had recovered independent lines with different degrees of silencing of the endogenous transcript level, with line rPPCK1-3 showing the greatest reduction (Figure 1A).
Figure 1.
Confirmation of Target Gene Silencing in Transgenic K. fedtschenkoi RNAi Lines rPPCK1-1 and rPPCK1-3.
Downregulation of the target endogenous KfPPCK1 gene was confirmed at the level of KfPPCK1 transcript abundance (A), and target protein PPC phosphorylation and the apparent Ki of PPC for l-malate in rapidly desalted leaf extracts, measured as l-malate (mM) required for a 50% reduction in extractable PPC activity (B). Gene transcript abundance was measured using RT-qPCR for the wild type, rPPCK1-1, and rPPCK1-3 for target genes: KfPPCK1 (A), KfPPCK2 (C), and KfPPCK3 (D). Mature leaves (leaf pair 6) were sampled from three individual, clonal, biological replicate plants collected every 4 h across the 12-h-light/12-h-dark cycle. A thioesterase/thiol ester dehydrase-isomerase superfamily gene (KfTEDI) was amplified from the same cDNAs as a reference gene. Gene transcript abundance data represent the mean of three technical replicates of each of three biological replicates and were normalized to reference gene KfTEDI; error bars represent the se of the mean calculated for each biological replicate. In all cases, plants were entrained under 12-h-light/12-h-dark cycles for 7 d prior to sampling. In (B), protein abundance (right panel) and the phosphorylation state of PPC (left panel) were determined by immunoblot analyses, and the apparent Ki of PPC for l-malate was measured using rapidly desalted leaf extracts (Ki values in mM below the corresponding left panel immunoblot time points). Total leaf protein (leaf pair 6) was isolated from leaves sampled every 4 h across the 12-h-light/12-h-dark cycle, separated using SDS-PAGE, and used for immunoblot analyses with antibodies raised to a phospho-PPC peptide (left panel). Sample loading was normalized according to total protein and confirmed using the immunoblot for total PPC protein, which is stable over the 12-h-light/12-h-dark cycle (right panel). The white bar below each panel represents the 12-h-light period, and the black bar below each panel represents the 12-h-dark period. For (A), (C), and (D), black data are for the wild type, blue for rPPCK1-1, and red for rPPCK1-3.
PPC Phosphorylation and the Apparent Ki of PPC for Feedback Inhibition by l-Malate
In K. fedtschenkoi, PPC has been shown to be subject to a circadian rhythm of dark period phosphorylation/light period dephosphorylation as a result of rhythmic synthesis and degradation of PPCK (Hartwell et al., 1999). Immunoblotting using an anti-phospho-PPC antibody was used to investigate the phosphorylation state of PPC over a 12-h-light/12-h-dark cycle in CAM leaves of both the wild type and the rPPCK1 lines (Figure 1B, left panel). The abundance of PPC itself was measured on replicate immunoblots to check for even protein loading of the blots (Figure 1B, right panel). In the wild-type CAM leaves, PPC was phosphorylated to a high level both at 18:00 and 22:00, 6 and 10 h into the 12-h dark period (Figure 1B, left panel). Wild-type PPC phosphorylation declined rapidly in the light, with a low level of phospho-PPC detected at 2 h into the 12-h light period, but virtually no phosphorylation detected at both 6 and 10 h into the light period (Figure 1B, left panel). The phosphorylation of PPC in the dark was reduced in both of the rPPCK1 lines (Figure 1B, left panel). Relative to the wild type, line rPPCK1-1 had a reduced peak of phospho-PPC in the middle of the dark period (18:00), but there was a dramatic reduction relative to the wild type at the end of the dark period (22:00) (Figure 1B, left panel). Line rPPCK1-3 failed to achieve a detectable level of PPC phosphorylation above background at any of the sampled light/dark time points. Thus, rPPCK1-1 had intermediate levels of PPC phosphorylation in the dark, and rPPCK1-3 had the strongest phenotype, as it lacked detectable levels of dark period phosphorylation of PPC (Figure 1B, left panel). These perturbations of dark phosphorylation of PPC correlated well with the level of KfPPCK1 transcripts in the dark in the two lines, with rPPCK1-3 showing the greatest reduction in KfPPCK1 transcript abundance (Figure 1A), consistent with the loss of PPC phosphorylation at the same time points (Figure 1B, left panel). The relative transcript abundance of two other detectable KfPPCK genes, namely, KfPPCK2 and KfPPCK3, was higher in the rPPCK1-3 lines relative to the wild type (Figures 1C and 1D). Interestingly, the increased transcript levels for KfPPCK2 and KfPPCK3 in the rPPCK1-3 line (Figures 1C and 1D) did not rescue the dark period phosphorylation of PPC (Figure 1B, left panel). The fourth PPCK gene (KfPPCK4) in the K. fedtschenkoi genome could not be detected reliably using RT-qPCR, even after 40 cycles.
As the phosphorylation of PPC in the dark results in an increase in its apparent Ki for its feedback inhibitor l-malate (Nimmo et al., 1984, Carter et al., 1991), we also investigated the apparent Ki of PPC for l-malate using rapidly desalted extracts of LP6 sampled in the light at 10:00 (2 h before dusk) and in the dark at 18:00 (middle of the 12-h dark period) (Figure 1B, Ki values below corresponding time points on the left panel; Supplemental Figure 1). The wild type displayed the well-documented large increase in the apparent Ki of PPC for l-malate in the dark period, with the apparent Ki l-malate increasing from 0.5 mM for leaves sampled 2 h before the end of the light period up to 8 mM in the middle of the dark period, the latter coincident with high PPC phosphorylation detected at the same time point (Figure 1B, left panel; Supplemental Figure 1). Line rPPCK1-1 displayed a large reduction in the light to dark differential for the apparent Ki of PPC for l-malate, with the measured values being reduced to an average of 0.6 mM in the light samples, and 2.4 mM for the dark samples (Figure 1B; Supplemental Figure 1). This was consistent with the reduced level of PPC phosphorylation in rPPCK1-1 in the middle of the dark period (Figure 1B, left panel). In line rPPCK1-3, which displayed the largest reduction in PPC phosphorylation in the dark period, the apparent Ki of PPC for l-malate was 1 mM in both the light and the dark, confirming that the lack of measurable PPC phosphorylation in the dark period in line rPPCK1-3 resulted in a failure of the leaf mesophyll cells to convert the major, CAM-associated PPC isoform into the phosphorylated, malate-insensitive form that occurs in wild-type leaves in the dark period (Figure 1B, left panel; Supplemental Figure 1).
Gas Exchange Characteristics in Light/Dark Cycles
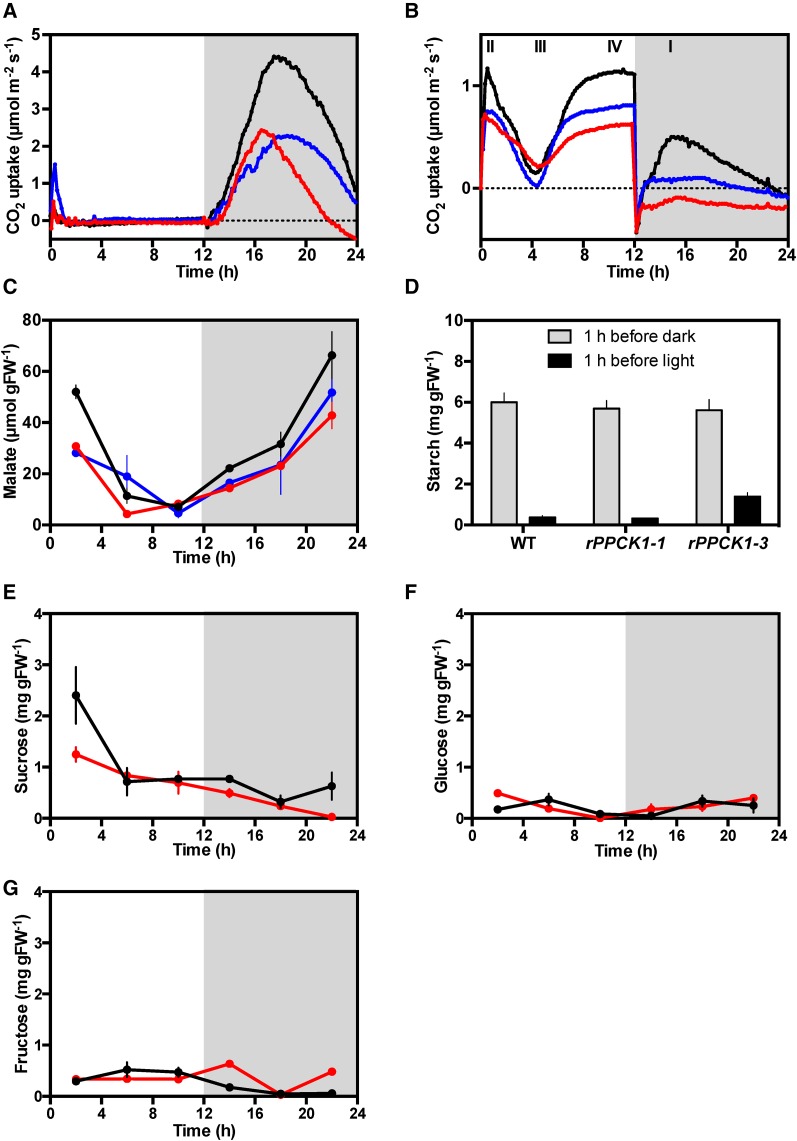
In K. fedtschenkoi, CAM is induced developmentally along the growing shoot, with the youngest leaves performing only daytime CO2 fixation via Rubisco and older leaves developing nocturnal CO2 fixation associated with increasing CAM (Jones, 1975; Borland et al., 2009; Dever et al., 2015; Hartwell et al., 2016). Using a multi-cuvette, gas switching, infrared gas analyzer system (described in detail in Dever et al., 2015), we investigated the 24 h pattern of CO2 exchange for detached, full-CAM leaves (leaf pair 6 [LP6]) from rPPCK1-1, rPPCK1-3, and the wild type that had been entrained in 12-h-light/12-h-dark cycles (Figure 2). Both the wild type and the transgenic lines with reduced KfPPCK1 fixed the majority of their CO2 at night (phase I of CAM: stomata open, primary atmospheric CO2 fixation via PPC; Figure 2A) (Osmond, 1978). Lines rPPCK1-1 and rPPCK1-3 fixed only 53% and 34%, respectively, of wild-type CO2 uptake in the dark (Figure 2A). rPPCK1-1 and rPPCK1-3 also fixed a small amount of CO2 at dawn (phase II; early hours of the light period, stomata remain open briefly, CO2 fixation via PPC and Rubisco), and this phase II peak of CO2 fixation was greater in rPPCK1-3 than in the wild type (Figure 2A). The wild type reached a maximum CO2 fixation rate in the dark period of 4.4 µmol m−2 s−1, whereas lines rPPCK1-1 and rPPCK1-3 reached a peak dark CO2 fixation rate of only 2.4 µmol m−2 s−1. It was noteworthy that the rPPCK1-3 line that had the greatest reduction in dark phosphorylation of PPC (Figure 1B) reached its peak dark CO2 fixation rate 1 h earlier in the dark period than either the wild type or rPPCK1-1 line (Figure 2A). Furthermore, the CO2 fixation of line rPPCK1-3 began to decline after only 4.5 h of the dark period, to such an extent that, from 2 h before dawn, this line was respiring an increasing amount of CO2 throughout the remainder of the dark period (Figure 2A). By contrast, the wild type and intermediate rPPCK1-1 line continued net atmospheric CO2 fixation for the whole of the dark period (Figure 2A).
Figure 2.
Impact of Silencing KfPPCK1 on 24-h-Light/Dark Gas Exchange Profiles, and Malate, Starch, and Soluble Sugar Levels for the Wild Type, rPPCK1-1, and rPPCK1-3 under Well-Watered Conditions.
(A) Gas exchange profile for CAM leaves (leaf pair 6) using plants pre-entrained for 7 d under 12-h-light/12-h-dark cycles.
(B) Gas exchange profile for well-watered whole young plants (8-leaf-pairs stage) using plants entrained for 7 d under 12-h-light/12-h-dark cycles.
(C) Malate content was determined from leaf pair 6 samples collected every 4 h using plants entrained under 12-h-light/12-h-dark cycles using methanol extracts of leaves from the wild type, rPPCK1-1, and rPPCK1-3.
(D) Starch content was determined from leaf pair 6 samples collected at 1 h before dawn and 1 h before dusk.
(E) to (G) Soluble sugars levels were determined separately for sucrose, glucose, and fructose using three biological replicates of leaf pair 6 sampled every 4 h using plants entrained under 12-h-light/12-h-dark cycles.
In (C) to (G), the error bars represent the se of the mean calculated for the three biological replicates measured at each time point. Black data traces represent the wild type, blue data rPPCK1-1, and red data rPPCK1-3.
Intact small plants (8-leaf-pair stage) with their root system in soil performed all four phases of CAM (labeled I–IV in Figure 2B). rPPCK1-1 and rPPCK1-3 fixed 72% and 62%, respectively, of wild-type light period CO2 fixation. In the dark, rPPCK1-1 fixed only 12%, and rPPCK1-3 respired −76%, of wild-type CO2 fixation (Figure 2B). Whole young plants of rPPCK1-3 respired CO2 throughout the dark period, but respiratory CO2 loss from the plants declined from an initial peak straight after the lights went off to a minimum 4 h into the dark period, before rising gently throughout the remainder of the dark (Figure 2B). However, whole plants of all three lines showed a clear phase III of CAM in the light, evidenced by the pronounced dip in CO2 fixation in the middle of the light period (Figure 2B).
Malate, Starch, and Soluble Sugar Levels
Malate and starch levels reciprocate over the light/dark cycle in starch-accumulating CAM species such as K. fedtschenkoi (Borland et al., 2016). Starch breakdown in the dark generates the substrate for glycolysis, which in turn supplies PEP as a substrate for PPC to use in dark CO2 fixation. As we had observed such large changes in the magnitude and duration of dark CO2 fixation in the rPPCK1 lines (Figures 2A and 2B), it was important to measure the levels and daily light/dark dynamics of the total leaf malate and starch pools.
The wild type, rPPCK1-1, and rPPCK1-3 all accumulated malate in the dark (Figure 2C). Wild-type plants accumulated the most malate by dawn (75.76 µmol gFW−1 se 4.65). Line rPPCK1-1 accumulated 78% and rPPCK1-3 accumulated 64%, of the wild-type level of malate by dawn (Figure 2C). At the end of the light period, ∼90% of the dark accumulated malate had been decarboxylated in the wild-type and rPPCK1 lines. The wild type had a Δ malate of 59.19 ± 8.82 µmol gFW−1, rPPCK1-1 had a Δ malate of 47.16 ± 5.23 µmol gFW−1, and PPCK1-3 had a Δ malate of 38.54 ± 4.89 µmol gFW−1.
At the end of the light period, wild-type and rPPCK1 CAM leaves (LP6) had synthesized similar amounts of starch (Figure 2D). Wild-type and rPPCK1-1 leaves broke down 94% of the accumulated starch during the dark period, whereas rPPCK1-3 only broke down 82% of its starch, consistent with the reduced CO2 fixation and malate accumulation achieved by this line (Figures 2A and 2C). The wild type had a Δ starch of 5.63 ± 0.47 mg gFW−1, PPCK1-1 had a Δ starch of 5.37 ± 0.38 mg gFW−1, and PPCK1-3 had a Δ starch of 4.23 ± 0.67 mg gFW−1.
CAM leaves of the closely related obligate species Kalanchoë daigremontiana have been reported previously to display a characteristic postdawn peak of sucrose peaking at the end of phase II of CAM (Wild et al., 2010). At the beginning of the light period, wild-type K. fedtschenkoi had a peak of sucrose phased to 2 h after dawn that was reduced to around half the wild-type level in line rPPCK1-3 (Figure 2E). Fructose accumulation peaked slightly later than the wild type in rPPCK1-3 at 2 h into the dark period (Figure 2G), whereas the temporal pattern of glucose accumulation was similar between rPPCK1-3 and the wild type (Figure 2F).
Gas Exchange Characteristics under Constant Light and Temperature Free-Running Conditions
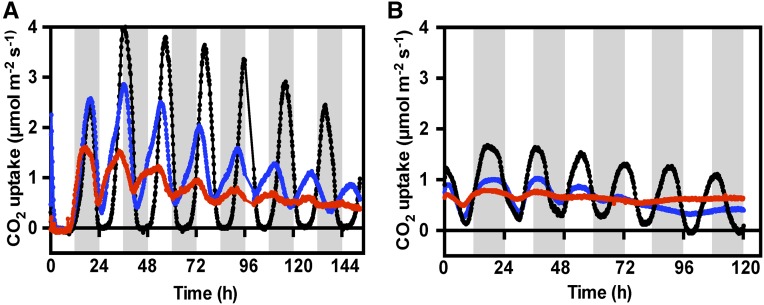
In constant light, free-running circadian conditions (LL; constant light 100 µmol m−2 s−1, and temperature, 15°C), detached LP6 from wild-type plants displayed the characteristic, robust, and persistent CAM circadian rhythm, with oscillating peaks and troughs of CO2 fixation (Figure 3) (Wilkins, 1992; Hartwell, 2006). rPPCK1-1 and rPPCK1-3 showed a dampening of the CO2 fixation rhythm in constant conditions, with a dramatic reduction in rhythm amplitude (Figure 3A). The rPPCK1-1 CO2 fixation rhythm maintained clear oscillations throughout the 6.5-d experiment, but the period of the rhythm was shorter than the wild type such that, after 6.5 d, the wild-type CO2 fixation trough or minimum coincided with the rPPCK1-1 peak of CO2 fixation (Figure 3A). The rPPCK1-3 CO2 fixation rhythm dampened more rapidly than rPPCK1-1, so much so that it was virtually arrhythmic after 6.5 d. The weak LL rhythm in line rPPCK1-3 also had an even shorter period than rPPCK1-1 (Figure 3A).
Figure 3.
Effects of Silencing KfPPCK1 on CAM CO2 Exchange Rhythms Measured under LL Conditions.
(A) Gas exchange profile for CAM leaves (leaf pair 6) was measured using leaves entrained under a 12-h-light/12-h-dark cycle followed by release into constant LL conditions (100 µmol m−2 s−1 light at 15°C).
(B) Gas exchange profile for well-watered whole young plants (8-leaf-pair stage) using plants entrained under 12-h-light/12-h-dark cycles followed by release into constant LL conditions (100 µmol m−2 s−1 light at 15°C). Black data traces represent the wild type, blue data rPPCK1-1, and red data rPPCK1-3.
When whole young plants at the 8-leaf-pair stage were measured in the gas exchange system under LL free-running conditions, the CO2 exchange rhythms in the rPPCK1-1 and rPPCK1-3 lines both collapsed to arrhythmia after 2 to 3 d (Figure 3B). The amplitude of the rPPCK1-1 and rPPCK1-3 CO2 fixation rhythms was even lower for whole plants than for detached LP6 (compare Figure 3B with 3A). In particular, comparing the first peak and trough of its CO2 fixation rhythm, rPPCK1-3 achieved a maximum amplitude of ∼0.25 µmol m−2 s−1. This contrasted strongly with the amplitude of the wild-type rhythm for whole plants during the first peak and trough when the amplitude reached over 1.5 µmol m−2 s−1 (Figure 3B).
Phenotypic Characterization under Well-Watered and Drought-Stressed Conditions
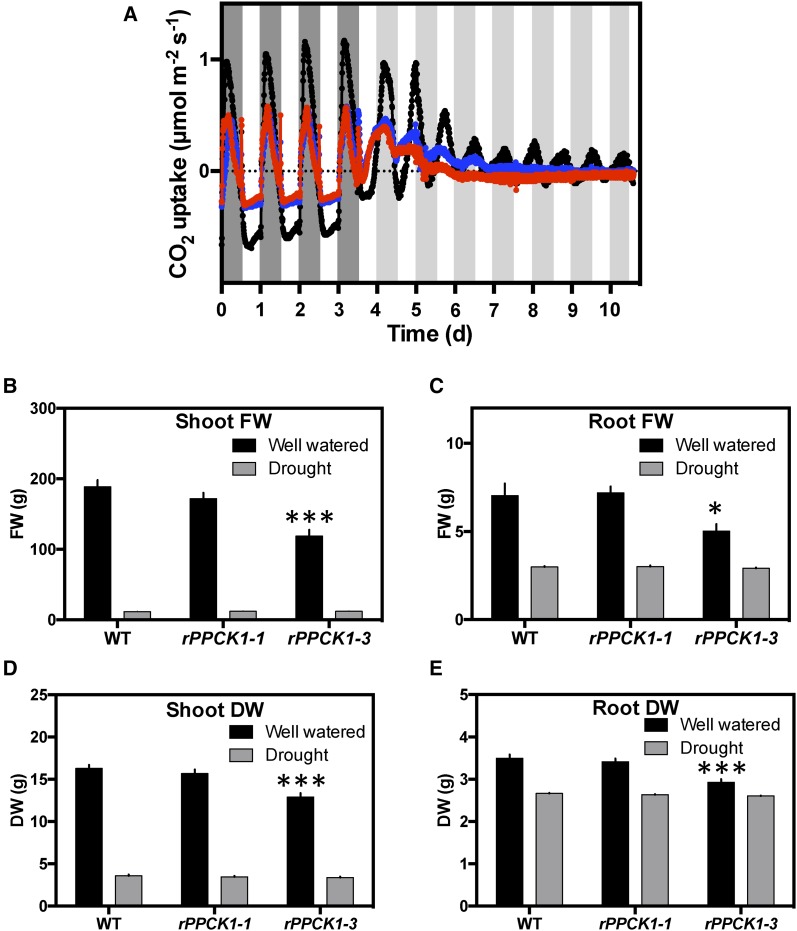
CAM is widely recognized and characterized as a water-conserving adaptation to seasonally dry environments. It was therefore important to investigate the physiological responses and performance of the rPPCK1 lines under drought-stress conditions. CO2 exchange was measured for drought-stressed whole plants. Wild-type and rPPCK1 plants were grown from plantlets under identical conditions to the LP6 stage. Water was then withheld for 25 d. Wild-type drought-stressed plants displayed strong CAM in light/dark cycles, with a large nocturnal phase I period of dark CO2 fixation and a net loss of CO2 in the light period (Figure 4). Phase II CO2 uptake at dawn was present in both wild-type and RNAi lines, but phase IV CO2 fixation at the end of the light period, which is characteristically observed in well-watered wild-type plants (Figure 2B), was absent in both the RNAi lines and the wild type.
Figure 4.
Impact of Loss of KfPPCK1 Activity on Gas Exchange Profiles and Vegetative Yield for Plants Subjected to Drought Stress.
(A) Drought-stressed whole plants (8-leaf-pair stage) were entrained under 12-h-light/12-h-dark cycles followed by release into constant LL conditions (100 µmol m−2 s−1 light at 15°C). The black data trace represents the wild type, blue rPPCK1-1, and red rPPCK1-3.
(B) to (E) Fresh ([B] and [C]) and dry ([D] and [E]) weight of aboveground biomass (shoot; [B] and [D]) and belowground tissues (roots; [C] and [E]) at maturity (138 d under greenhouse conditions) for the wild type, rPPCK1-1, and rPPCK1-3 under well-watered and drought-stressed conditions. n = 7 plants; error bars represent se of the mean. Asterisks indicate significant difference from the wild type based on Student’s t test: *P < 0.05 and ***P < 0.001.
As observed in well-watered conditions (Figure 3B), the LL circadian rhythm of CO2 exchange persisted robustly in the wild-type drought-stressed plants, but collapsed rapidly toward arrhythmia in both rPPCK1 lines (Figure 4A). In particular, after 2.5 d of LL, drought-stressed rPPCK1-3 moved into net respiratory loss of CO2, and this continued throughout the remaining 4.5 d of the LL experimental run (Figure 4A). This contrasted strongly with the LL gas exchange for line rPPCK1-3 under well-watered conditions, where it maintained positive net CO2 fixation throughout the LL experiment, both for detached LP6 and young whole plants (Figures 3A and 3B).
Growth Performance under Well-Watered and Drought-Stressed Conditions
The high WUE of CAM plants allows them to grow productively in arid and semi-arid environments (Borland et al., 2009; Davis et al., 2017). Multiple clones of all lines were grown from plantlets under identical conditions to the LP6 stage. Half of the plants were maintained well-watered as controls, and half were subjected to drought stress for 138 d. Both sets of plants were maintained in the same growth cabinet such that the only environmental variable was water supply.
In well-watered conditions, all plants grew well. However, weighing aboveground shoots and belowground roots, both as fresh weight and dry weight, revealed that the average fresh and dry weight of the shoots and roots was significantly less than the wild type for line rPPCK1-3 under well-watered conditions (Figures 4B to 4E). The intermediate line rPPCK1-1 also had a lower average fresh and dry weight for its shoots and roots under well-watered conditions, although the reductions were not significantly different from the wild type based on Student’s t tests (Figures 4B and 4D). The wet weight of the shoots of rPPCK1-1 weighed 91% of the wild type, whereas the shoots of rPPCK1-3 weighed 55% of the wild type (n = 7). Differences in shoot dry weight were smaller, with rPPCK1-1 shoots weighing 96% of the wild type, and rPPCK1-3 weighing 76% of the wild type shoot dry weight (n = 7). Belowground, root growth differences were less pronounced, although line rPPCK1-3 achieved an average root wet weight that was only 71% of the wild type (Figure 4C).
The replicate plants subjected to 138 d of drought stress had fresh weights for both shoots and roots that were much lower than the fresh weights for the well-watered plants. For example, the average fresh weight of wild-type shoots from well-watered plants was 188 g, whereas the average fresh weight for the drought-stressed wild type shoots was only 11 g. This demonstrated that the 138-d drought-stress treatment was a severe drought-stress regime, as might be experienced by K. fedtschenkoi during the dry season in its native Madagascan environment. After drought stress, the fresh and dry weights of both the shoots and roots of the rPPCK1 lines did not differ significantly from the wild-type values. The very low dry weights of the drought-stressed shoots of all lines relative to the dry weights achieved by all of the well-watered plants indicated that all lines performed very little shoot growth under such a long-term drought-stress regime (Figure 4D). However, the reduction in the dry weight of drought-stressed roots relative to the well-watered values was less dramatic (Figure 4E). Drought-stressed root dry weights were between 76 and 89% of the corresponding well-watered values, whereas drought-stressed shoots weighed less than 22 to 26% of the well-watered shoots for all lines. It is clear that all plants put more of their available fixed carbon into belowground root growth during the prolonged drought-stress treatment used in this experiment. Furthermore, the root fresh weights were almost the same as the root dry weights for the drought-stressed plants, suggesting that, by the end of the 138-d drought treatment, the roots of all lines were almost completely dry, even before being placed in a drying oven.
Diurnal Regulation of the Transcript Abundance of Circadian Clock Genes
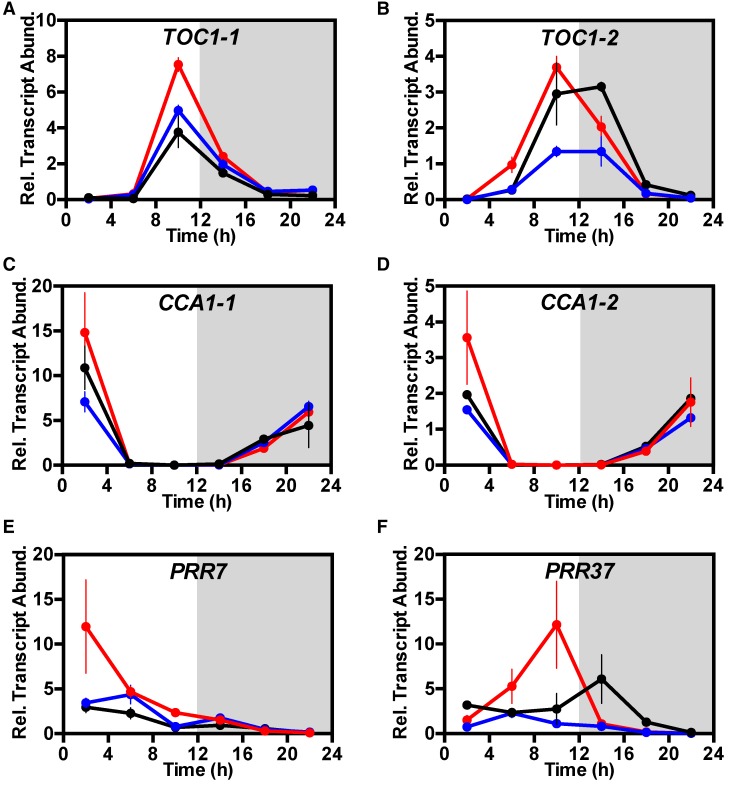
Having established that plants lacking KfPPCK1 had lost the rhythm of CO2 uptake in free-running conditions (Figures 3A and 3B), it was also important to investigate whether the regulation of genes in the core molecular circadian clock was affected under both light/dark cycles (Figure 5) and free-running LL conditions (Figure 6). K. fedtschenkoi has two copies of the circadian clock gene TIMING OF CHLOROPHYLL A/B BINDING PROTEIN1 (KfTOC1-1 and KfTOC1-2) and two CIRCADIAN CLOCK-ASSOCIATED1/LATE ELONGATED HYPOCOTYL-related genes (KfCCA1-1 and KfCCA1-2). Both KfTOC1 genes were upregulated under light/dark conditions in the rPPCK1-3 line, with the phasing of the KfTOC1-2 peak occurring 4 h earlier in rPPCK1-3 (Figures 5A and 5B). In rPPCK1-3, both KfCCA1-1 and KfCCA1-2 were slightly upregulated at the time of their daily peak 2 h after dawn, whereas transcript levels of these genes in line rPPCK1-1 were similar to or slightly lower than those in the wild type (Figures 5C and 5D). A K. fedtschenkoi PSEUDO RESPONSE REGULATOR7 ortholog (KfPRR7) was markedly upregulated at 2 h after dawn in the rPPCK1-3 line (Figure 5E), and a KfPRR37 gene was also upregulated and peaked 4 h earlier than the wild type at 10:00 (2 h before dusk) in line rPPCK1-3 (Figure 5F).
Figure 5.
Impact of the Loss of KfPPCK1 Activity on the Light/Dark Regulation of the Transcript Abundance of Central Circadian Clock Genes KfTOC1-1, KfTOC1-2, KfCCA1-1, KfCCA1-2, KfPRR7, and KfPRR37.
Mature leaves (leaf pair 6) were sampled from three biological replicates every 4 h across the 12-h-light/12-h-dark cycle, RNA isolated, and used for real-time RT-qPCR. TOC1-1 (A), TOC1-2 (B), CCA1-1 (C), CCA1-2 (D), PRR7 (E), and PRR37 (F). A thioesterase/thiol ester dehydrase-isomerase superfamily (KfTEDI) gene was amplified from the same cDNAs as a reference gene. Gene transcript abundance data represents the mean of three technical replicates for each of three biological replicates and was normalized to the reference gene (KfTEDI); error bars represent the se of the mean calculated for the three biological replicates. In all cases, plants were entrained under 12-h-light/12-h-dark cycles for 7 d prior to sampling. Black data are for the wild type, blue data rPPCK1-1, and red data rPPCK1-3.
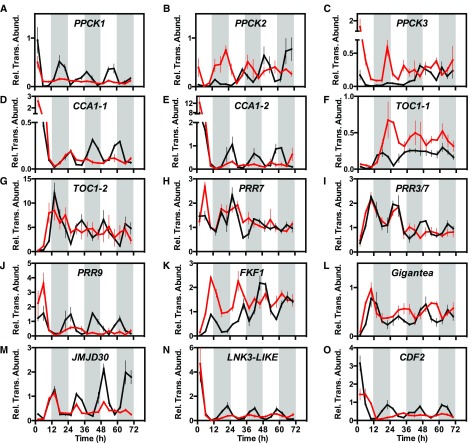
Figure 6.
Impact of the Loss of KfPPCK1 Activity on Circadian Clock-Controlled Gene Transcript Abundance during Constant Light and Temperature Free-Running Conditions.
(A) Circadian rhythm of KfPPCK1 transcript abundance under constant LL conditions (100 µmol m−2 s−1 light at 15°C) for the wild type (black line) and rPPCK1-3 (red line).
(B) to (O) KfPPCK2 (B), KfPPCK3 (C), KfCCA1-1 (D), KfCCA1-2 (E), KfTOC1-1 (F), KfTOC1-2 (G), KfPRR7 (H), KfPRR37 (I), KfPRR9 (J), KfFKF1 (K), KfGIGANTEA (L), KfJMJD30 (M), KfLNK3-like (N), and KfCDF2 (O). Mature leaves (leaf pair 6) were sampled from three biological replicates every 4 h under constant conditions (100 µmol m−2 s−1 light at 15°C) for the wild type and rPPCK1-3. RNA was isolated and used for real-time RT-qPCR. A thioesterase/thiol ester dehydrase-isomerase superfamily gene (KfTEDI) was amplified as a reference gene from the same cDNAs. Gene transcript abundance data represent the mean of three technical replicates for each of three biological replicates and were normalized to the reference gene (KfTEDI); error bars represent the se of the mean calculated for the three biological replicates. In all cases, plants were entrained under 12-h-light/12-h-dark cycles prior to release into LL free-running conditions. Black data are for the wild type and red data rPPCK1-3.
Regulation of Circadian Clock Genes under Free-Running Conditions
The robust circadian rhythm of CAM CO2 exchange in LL conditions for the wild type (Figures 3A and 3B) has been shown to correlate tightly with circadian clock control of PPCK and the phosphorylation state of PPC and its Ki for malate (Hartwell, 2005, 2006; Dever et al., 2015). Since CO2 exchange rhythms in LL conditions for rPPCK1-1 and rPPCK1-3 showed, at best, low-amplitude rhythmicity in well-watered plants (Figures 3A and 3B), it was important to also investigate the circadian regulation of the transcript abundance of KfPPCK1 under LL conditions. In wild-type plants, KfPPCK1 transcript levels oscillated with a robust circadian rhythm throughout 3 d under LL (Figure 6A). The period of the rhythmic peaks and troughs matched the well-documented, temperature-dependent, short-period rhythm of gas exchange observed in K. fedtschenkoi (Anderson and Wilkins, 1989; Dever et al., 2015). Consistent with the RNAi-mediated silencing of the target gene, KfPPCK1 transcript abundance was very low in rPPCK1-3 in free-running conditions and did not oscillate (Figure 6A). The upregulation of KfPPCK2 and KfPPCK3 in plants lacking KfPPCK1 under LD conditions (Figures 1C and 1D) led us to investigate whether these genes remained rhythmic in constant conditions in line rPPCK1-3. Both KfPPCK2 and KfPPCK3 transcript oscillations remained rhythmic and were dramatically upregulated at the start of the LL time course in rPPCK1-3 (Figures 6B and 6C).
As these results revealed that certain circadian clock-controlled output genes were subject to robust oscillations under LL free-running conditions, it was also important to investigate the rhythmicity of core circadian oscillator genes. The rhythmicity of both KfCCA1-1 and KfCCA1-2 was dampened in rPPCK1-3 (Figures 6D and 6E). Interestingly, the two KfTOC1 genes were differentially regulated in rPPCK1-3 under constant conditions with KfTOC1-1 being upregulated and rhythmic, whereas KfTOC1-2 was downregulated and its rhythm collapsed toward arrythmia (Figures 6F and 6G). This was surprising as it indicated that the two KfTOC1 genes may be involved in distinct circadian clocks having potentially divergent roles and responding to different entrainment signals, possibly in different tissues/cell types of the CAM leaf.
KfPRR7, KfPRR37, and KfPRR9 all displayed dampening of their transcript rhythms toward arrhythmia in rPPCK1-3 under constant conditions (Figures 6H to 6J). K. fedtschenkoi FLAVIN BINDING KELCH-REPEAT F-BOX PROTEIN (KfFKF1) and Kf GIGANTEA (KfGI) were both upregulated and rhythmic in LL (Figures 6K and 6L), whereas Kf JUMONJI C-DOMAIN CONTAINING PROTEIN30 (KfJMJ30), Kf NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED3 (KfLNK3-like), and Kf CYCLING DOF-TRANSCRIPTION FACTOR2 (KfCDF2) were all downregulated but rhythmic in rPPCK1-3 (Figures 6M to 6O). KfGI was particularly noteworthy for the fact that the phasing of its transcript peaks and troughs shifted late relative to the wild type in line rPPCK1-3 on the second and third 24-h period of the LL experimental run (Figure 6L), which contrasted strikingly with the earlier phase/ shorter period of the LL CO2 fixation rhythm for rPPCK1-3 (Figure 3A).
Delayed Fluorescence Rhythms Revealed That a Further, Distinct Circadian Clock Output inside the Chloroplast Was Perturbed in the Absence of PPCK Activity
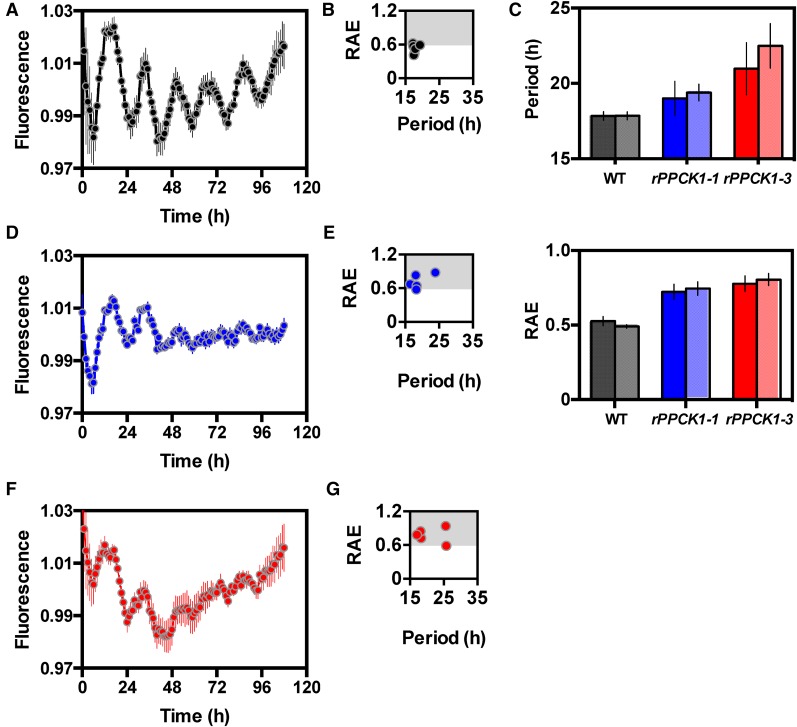
The level of delayed fluorescence (DF) has previously been reported to be under robust circadian control in K. fedtschenkoi and provides a simple high-throughput assay for measuring circadian period, robustness, and accuracy (Gould et al., 2009). DF was measured in the wild type, rPPCK1-1, and rPPCK1-3 under LL free-running conditions (Figure 7), and the data were analyzed to calculate key circadian rhythm statistics using Biodare (Moore et al., 2014; Zielinski et al., 2014). The wild type had a robust DF oscillation (Figure 7A). The wild-type relative amplitude error (RAE) plot (Figure 7B), and summary bar chart for the computed period length for the wild type (Figure 7C, top panel), demonstrated a short period (mean 17.8 h) from both fast Fourier transform (nonlinear least squares method) and spectral resampling analysis. The mean RAE plot revealed that the values were below the 0.6 cutoff (Figure 7C, bottom panel), supporting statistically the visibly robust and high-amplitude wild-type short period DF rhythm (Figure 7A). Biodare analysis demonstrated that both rPPCK1-1 (Figures 7D and 7E) and rPPCK1-3 (Figures 7F and 7G) lost rhythm robustness through time, particularly in the later stages of the LL free-running conditions. Line rPPCK1-1 had an intermediate effect with dampening of the rhythm occurring over several cycles (Figure 7D). The period length plot for rPPCK1-1 suggested that its average free-running period was slightly longer than that of the wild type, but the larger error suggested this period estimate was not reliable (Figure 7C, top panel). The corresponding RAE values showed a much higher average for rPPCK1-1 than the wild type due to its dampening, reflected in the wide spread of period lengths from short to long (Figure 7E). The analysis outputs for the DF results from rPPCK1-3 revealed a slightly more severe rhythm phenotype than rPPCK1-1 (Figures 7F and 7G). Although the Biodare graph summarizing period length for DF rhythms in rPPCK1-1 and rPPCK1-3 indicated longer DF period length for the transgenic lines relative to the wild type (Figure 7C, top panel), which was at odds with the shorter period for the gas exchange LL rhythms (Figures 3A and 3B), visual inspection of the DF rhythms revealed that there may have been a shorter period length for the very low amplitude DF rhythms in rPPCK1-1 and rPPCK1-3 that were observed throughout the latter stages of the LL time course (Figures 7D and 7F). The RAE values above the 0.6 cutoff for rPPCK1-1 and rPPCK1-3 also indicated that the rhythms were not robust and thus that the period estimates in Figure 7C should not be overinterpreted due to their high associated RAE values.
Figure 7.
DF Rhythms Collapsed toward Arrythmia in Lines rPPCK1-1 and rPPCK1-3.
Plants were entrained under 12-h-light/12-h-dark cycles before being transferred to constant red/ blue light under the CCD imaging camera (35 µmol m−2 s−1). DF was assayed with a 1-h time resolution for 108 h. The plots represent normalized averages for DF measured for six leaf discs sampled from three biological replicates of leaf pair 6 for each line. Error bars indicate se of the mean calculated from three biological replicates. Black data are for the wild type, blue data rPPCK1-1, and red data rPPCK1-3.
(A) Wild-type DF rhythm under LL.
(B) RAE plot for wild-type DF rhythm.
(C) Mean period length (upper graph) and RAE plots (lower graph) for the wild type (black/gray), rPPCK1-1 (blue/pale blue), and rPPCK1-3 (red/pale red); the plotted values were calculated using the Biodare package for circadian rhythm analysis. In both graphs, the dark shade of the color represents data from fast Fourier transform-nonlinear least squares analysis and the paler shade represents data from spectral resampling analysis.
(D) rPPCK1-1 DF rhythm.
(E) RAE plot for rPPCK1-1.
(F) rPPCK1-3 DF rhythm.
(G) RAE plot for rPPCK1-3.
DISCUSSION
Physiological Consequences of Loss of Clock-Controlled PPC Phosphorylation in an Obligate CAM Plant
Previous work had shown that the PPCK gene responsible for light period phosphorylation and in vivo activation of photosynthetic PPC in the C4 species Flaveria bidentis was not essential for high C4 photosynthetic rates in this species (Furumoto et al., 2007). This work on a C4 species surprised the wider community because decades of prior work had argued that PPC phosphorylation by PPCK was vital for alleviating malate/aspartate inhibition of PPC in planta and thus for optimizing photosynthetic CO2 fixation in C4 and CAM species (Vidal and Chollet, 1997; Nimmo, 2003). These findings have become all the more surprising in the light of recent work on the evolution of the PPCK gene family in C3 and C4 species of Flaveria (Aldous et al., 2014). A specific, light-induced, C4-recruited PPCKA gene was found in each C4 species of Flaveria and was found to have evolved rapidly since its recruitment to a C4-specific function. All Flaveria species also possessed a PPCKB gene that displayed peak transcript abundance in the dark period and that was downregulated in the C4 species relative to the C3 species. Aldous et al. (2014) suggested that the C4-recruited ppcA and PPCKA formed a coevolving substrate-kinase pair, and yet the findings of Furumoto et al. (2007) argued that PPCKA in F. bidentis was dispensable for optimal CO2 fixation via the C4 pathway. Why would there be strong evolutionary selection acting on the C4 ppcA/PPCKA substrate-kinase pair if the kinase and its phosphorylation of PPC were dispensable for optimal CO2 fixation via the C4 pathway? It was thus vital to investigate the physiological role, if any, of PPCK and PPC phosphorylation in relation to dark period CO2 fixation through to malate in a CAM plant.
Our findings revealed that, in the CAM species K. fedtschenkoi, phosphorylation of PPC during the dark period by the circadian clock-controlled KfPPCK1 both maximized and sustained primary atmospheric CO2 fixation via PPC. Overall, our data showed that dark phosphorylation of PPC during CAM in K. fedtschenkoi is not a prerequisite for dark CO2 fixation, as 34% of dark CO2 fixation and 64% of nocturnal malate accumulation happened even in the absence of detectable levels of dark PPC phosphorylation in rPPCK1-3 (Figure 2). However, KfPPCK1 does play a critical role in fine-tuning the amount and duration of dark CO2 fixation, leading to greater malate accumulation and growth in the wild type relative to rPPCK1-3.
The high WUE of CAM plants is regarded as one of their key adaptive strengths and the main trait of interest for future exploitation in CAM crops and CAM biodesign for climate-resilient agriculture (Borland et al., 2009, 2014). It was thus important to compare the performance of the rPPCK1 lines between well-watered and drought-stressed conditions. Gas exchange analysis of the rPPCK1 lines subjected to drought stress demonstrated that both lines showed similar reductions in dark CO2 fixation relative to the wild type in comparison to the differences observed under well-watered conditions (Figure 4A). However, the rPPCK1 lines did continue to fix atmospheric CO2 in the dark after drought stress, which contrasted strikingly with our previous findings for transgenic K. fedtschenkoi with reduced CAM and dark CO2 fixation due to reduced activity of NAD-ME or PPDK, both of which failed to fix CO2 for the majority of the light/dark cycle following drought stress (Dever et al., 2015).
Drought stress had a dramatic impact on both the wet and dry weight of plants for both the wild-type and the rPPCK1 lines, but there was no significant difference between the fresh or dry weight of both the shoots and roots of wild-type and rPPCK1 lines following drought stress (Figure 4). Thus, under our growth conditions, the increased dark CO2 fixation during drought stress achieved by the wild type relative to the rPPCK1 lines (Figure 4A) did not lead to a measurable increase in yield for the wild type. Instead, all lines achieved little if any net CO2 fixation after prolonged drought stress, as their respiratory loss of CO2 in the light period largely balanced their fixation of CO2 in the dark period. This was reflected in the very low dry weight yields, which were >3-fold lower for drought-stressed shoots relative to the well-watered shoots (Figure 4D). It would be interesting to monitor the gas exchange of the wild-type and rPPCK1 lines continuously throughout the drought-stress treatment, so that their gas exchange during the transition from fully hydrated to severely drought stressed could be investigated in detail. This may reveal whether or not the wild type is able to maintain net daily fixation of CO2 for longer into the drought-stress period. However, if this does occur, it was clearly not sufficient to generate a measurable difference in growth performance after the 138 d of drought treatment (Figure 4). It may be that all lines were locked down into a CAM-idling survival mode after 138 d of drought stress and that their wet and dry weights were indistinguishable because they had, by that time, respired away the small but significant differences in their shoot and root dry biomass that were evident under well-watered conditions.
Molecular and Biochemical Phenotypes of rPPCK1 Lines
Both rPPCK1 lines possessed lower KfPPCK1 transcript levels than the wild type throughout the dark period (Figure 1A). rPPCK1-1 and rPPCK1-3 had similar KfPPCK1 transcript levels at both the beginning and end of the light period, but possessed a large difference in transcript abundance in the middle of the dark period, when line rPPCK1-1 achieved a significantly higher transcript level than line rPPCK1-3 (18:00, 6 h into the 12-h dark period; Figure 1A). This was consistent with the higher PPC phosphorylation and apparent Ki of PPC for l-malate detected in line rPPCK1-1 relative to line rPPCK1-3, specifically in the middle of the dark period at 18:00 (Figure 1B), and the greater dark period CO2 fixation, malate accumulation, and starch turnover achieved by this line (Figures 2A to 2D). Overall, our biochemical measurements revealed a dosage-dependent impact of the level of KfPPCK1 transcripts each night on the biochemical correlates of CAM.
Impact of Silencing KfPPCK1 on Circadian Rhythms of CAM CO2 Fixation and the Operation of the Central Circadian Oscillator
Since the first discovery of PPCK activity and its circadian control in 1991 (Carter et al., 1991), through the subsequent cloning and characterization of the gene encoding this remarkable protein kinase in 1999 (Hartwell et al., 1999), there has been a long-held assumption that this circadian clock-controlled kinase is crucial for the temporal coordination and optimization of nocturnal atmospheric CO2 fixation in CAM species, and the associated persistent circadian rhythm of CO2 exchange observed under constant conditions in CAM species such as K. fedtschenkoi and K. daigremontiana (Wilkins, 1992; Nimmo, 2003; Wyka et al., 2004; Hartwell, 2006). However, it has not been possible previously to perturb genetically the level of PPCK activity and dark period PPC phosphorylation in a CAM species; thus, it has not been possible to test the validity of this long held assumption about the pivotal role of PPCK in the circadian control of CAM.
By silencing the CAM-associated KfPPCK1 gene with an RNAi construct, we have been able to test in planta the importance of KfPPCK1 for the circadian rhythm of CO2 exchange. The loss of dark period PPC phosphorylation in line rPPCK1-3 led to a rapid decline in the circadian rhythm of CO2 exchange when leaves or whole plants pre-entrained under 12-h-light/12-h-dark cycles were released into LL free-running conditions (Figures 3A, 3B and 4A). After 25 d of drought stress, the weak CO2 exchange rhythms displayed by the well-watered rPPCK1 lines (Figure 3B) collapsed to arrhythmia more rapidly (Figure 4A). In particular, line rPPCK1-3 became arrhythmic and continued a steady rate of respiratory loss of CO2 for the majority of the LL time course (Figure 4A), whereas the drought-stressed wild type continued with a persistent circadian rhythm of CO2 fixation throughout the LL experiment (Figure 4A). Furthermore, silencing KfPPCK1 also led to DF rhythms collapsing toward arrhythmia, revealing that a distinct circadian clock output that originates inside the chloroplast was also perturbed in the rPPCK1 lines (Figure 7). These results revealed that wild-type levels of circadian clock controlled PPCK are essential for the amplitude, robustness, and persistence of the CO2 fixation rhythm associated with CAM and the DF rhythms in the chloroplast in K. fedtschenkoi.
In light of the weakened rhythmicity of CO2 exchange in the rPPCK1 lines, it was important to investigate the robustness of transcript abundance oscillations for components of the central molecular oscillator that underpin plant circadian rhythms (Figures 5 and 6). Wild-type K. fedtschenkoi displayed robust oscillations in the transcript abundance of KfCCA1-1, KfCCA1-2, KfTOC1-1, and KfTOC1-2, but rhythms in the transcript abundance of KfCCA1-1, KfCCA1-2, and KfTOC1-2 collapsed rapidly toward arrhythmia under LL in the rPPCK1-3 line (Figures 6D, 6E, and 6G). This revealed that multiple components of the core molecular oscillator failed to maintain wild-type levels of robust rhythmicity in the absence of KfPPCK1. In contrast, KfTOC1-1, KfPRR7, KfFKF1, and KfGI were induced and displayed a rhythm with a greater amplitude than the wild type under LL in line rPPCK1-3 (Figures 6F, 6H, 6K, and 6L), revealing that certain subcomponents of the core molecular circadian oscillator continued to function with a robust circadian rhythm in the absence of KfPPCK1 activity. These are paradigm-changing findings because KfPPCK1 has previously been regarded as a circadian clock-controlled output pathway component that is vital for coupling the central clock to CAM. However, our current findings reveal that some components of the central clock itself stop working correctly in the absence of KfPPCK1. This suggests either that KfPPCK1 is itself part of the central oscillator in K. fedtschenkoi, although this seems highly unlikely considering the known function of PPCK1 in PPC phosphorylation, or, more likely, that the failure to phosphorylate PPC in the dark leads to metabolic changes that influence the operation of the core oscillator, perhaps via a mechanism not dissimilar to that proposed for sugars acting as signals from photosynthesis that can entrain the central clock in Arabidopsis (Haydon et al., 2013).
It will be important to investigate the wider metabolic and/or gene expression changes associated with the loss of PPCK and PPC phosphorylation in these lines. Such investigations should provide valuable insights into the types of signals that could link CAM-associated metabolites to the central clock during the daily operation of the CAM pathway. It is particularly noteworthy in this respect that we also saw a loss of central clock gene TOC1 transcript oscillations in K. fedtschenkoi transgenic RNAi lines with reduced activity of NAD-ME (Dever et al., 2015). Those lines turned over only a small proportion of their malate each day and had a much greater reduction in starch accumulation and turnover than the rPPCK1 lines reported here. This suggests that the metabolic and/or gene regulatory changes that led to dampening of oscillations for a subset of central clock genes in these two distinct sets of K. fedtschenkoi transgenic lines may be more complex than a direct impact of malate or carbohydrate levels on clock operation. However, when we examined sucrose, fructose, and glucose levels over the 12-h-light/12-h-dark cycle in the rPPCK1 lines reported here, we discovered that line rPPCK1-3 displayed an almost 50% reduction in the 2 h after dawn-phased peak of sucrose detected in the wild type (Figure 2E).
Taking into consideration the fact that sucrose has been demonstrated as a central clock-entraining metabolite in the C3 species Arabidopsis (Haydon et al., 2013), our findings suggest that sucrose is a strong candidate for a metabolite linking the perturbations in CAM and core clock rhythms reported here for the rPPCK1 lines (Figure 2E). It is particularly noteworthy in this context that KfPRR7 and KfPRR37 transcript levels were increased in rPPCK1-3 at 2 and 10 h after dawn, respectively, relative to the wild type (Figures 5E and 5F). For KfPRR7, this change in transcript level was coincident with the decrease in the post-dawn sucrose peak (Figure 2E). Haydon et al. (2013) proposed that PRR7 could be a key clock gene functioning in the entrainment of the central oscillator by sucrose signals. However, it is noteworthy that our KfPRR7 ortholog displayed a transcript peak phased to 2 h after dawn, coincident with the daily sucrose peak, whereas the Arabidopsis PRR7 transcript level peaked at 8 h into the 12-h light period and was also strongly correlated with the daily sucrose peak, which occurs in the latter part of the light period in Arabidopsis (Haydon et al., 2013). Transcript levels of our KfPRR37 transcript did peak at an equivalent time (10:00, 2 h prior to dusk) to PRR7 in Arabidopsis and reached its daily peak in rPPCK1-3 some 4 h earlier than the wild type, which peaked at 14:00, 2 h into the dark (Figure 5F). These striking similarities in the timing and strong correlation of peak sucrose and peak PRR7 transcript levels in both K. fedtschenkoi and Arabidopsis provide support for the proposal that sucrose is a key clock entraining signal in K. fedtschenkoi and that the decrease in the 2 h post-dawn sucrose peak in rPPCK1-3 (Figure 2E) feeds back to perturb core clock gene regulation (Figure 6), potentially via KfPRR7 and KfPRR37 (Figures 5E and 5F).
Our data support the proposal that, in the CAM leaf, the circadian clock is set by both light and sucrose. Sucrose has been demonstrated to be a product of CAM-derived starch degradation during the hours at the end of the phase I dark period, although sucrose was also shown to accumulate further during phase II after dawn as a direct result of Rubisco-mediated fixation of atmospheric CO2, as is classically observed in the late afternoon in C3 plants such as Arabidopsis (Wild et al., 2010; Haydon et al., 2013). Both of the KfCCA1 genes and one of the two KfTOC1 genes were downregulated and displayed dampening rhythmicity in the rPPCK1-3 plants under LL conditions (Figure 6), and this may be due to the rPPCK1-3 plants being in a state of metabolic starvation after dawn. In C3 leaves of Arabidopsis during starvation, PRR7 represses CCA1 and derepresses the TOC1/GI loop (Dalchau et al., 2011; Haydon et al., 2013, 2015). The rPPCK1-3 transgenic lines of K. fedtschenkoi displayed derepression and rhythmicity for KfPRR7, KfTOC1-1, KfGI, and KfFKF1, supporting the proposal that the reduction in the post-dawn sucrose peak and associated increase in KfPRR7 and KfPRR37 may lead to the derepression of a TOC1-1/ GI loop in K. fedtschenkoi. However, in K. fedtschenkoi, while KfTOC1-1 was derepressed, which is similar to the TOC1 regulation reported in Arabidopsis, KfTOC1-2 was repressed, suggesting that in CAM leaves of Kalanchoë there are distinct suboscillators of the central circadian clock involving KfTOC1-1 and KfTOC1-2. We cannot at this stage rule out the possibility that these striking differences in the regulation of core circadian clock genes in line rPPCK1-3 were a result of differential regulation of the two TOC1 genes occurring in different cell types of the leaf.
We have previously demonstrated the utility of K. fedtschenkoi as a model system for elucidating the in planta functioning of candidate core metabolic enzymes associated with CAM (Dever et al., 2015; Hartwell et al., 2016). Our findings here provide in planta confirmation of the importance of the regulatory protein kinase PPCK and its circadian rhythm for optimal and maximal dark CO2 fixation. Plants lacking KfPPCK1 also failed to maintain robust rhythmicity of the transcript oscillations of several core clock components, including both isogenes of KfCCA1 and one isogene of KfTOC1. A key criterion for a gene to be classified as a core clock component is that misregulation of the gene should cause arrhythmia within the oscillator itself. Our data fulfill this criterion to some extent for KfPPCK1; thus, our data could be interpreted as evidence that KfPPCK1 operates as a core component of the central circadian clock in K. fedtschenkoi. However, we do not favor that explanation. Instead, our data for daily sucrose oscillations and KfPRR7, KfPRR37, KfTOC1-1, and KfGI transcript regulation support the proposal that the pathway of metabolic regulation suggested to integrate photosynthetic sugar metabolism with the clock in Arabidopsis plays a role in the partial loss of rhythmicity observed in this K. fedtschenkoi rPPCK1-3 line (Haydon et al., 2013). Alternatively, malate changes in the rPPCK1-3 line may influence the clock, as malate has been demonstrated to function to influence gene regulation (Finkemeier et al., 2013).
This work extends the categories of CAM-associated genes for which valuable functional insights have been gained using transgenic K. fedtschenkoi, moving beyond core CAM enzymes to include the circadian clock-controlled posttranslational regulatory step catalyzed by PPCK1. Based on these findings, we predict that transgenic approaches applied in K. fedtschenkoi will also prove useful for demonstrating the functional role of other proposed CAM regulators, such as PPDK regulatory protein (Dever et al., 2015), or pyruvate dehydrogenase kinase (Thelen et al., 2000), or as yet unknown transcription factors and other regulatory proteins that likely function in the circadian clock output pathway that couples the core molecular oscillator to the regulation of CAM enzymes via proteins such as PPCK (Hartwell, 2006; Borland et al., 2009; Hartwell et al., 2016).
Relevance of These Findings to Efforts to Engineer CAM into C3 Species
Our findings are vitally important to current and ongoing efforts to engineer the CAM pathway into C3 crops using plant synthetic biology approaches because they demonstrate that expressing high levels of PPC alone in leaf mesophyll cells will not be sufficient to achieve optimized operation of CAM dark CO2 fixation. Expressing elevated levels of PPC may yield plants that can fix some atmospheric CO2 in the dark and accumulate a certain level of malate by dawn, which would be a significant advance in and of itself. However, our data for the KfPPCK1 RNAi lines demonstrate that in order for an engineered CAM system to work with the full efficiency demonstrated by existing CAM species, C3 crops will need to be engineered to express both high levels of PPC in leaf mesophyll cells and a circadian clock-controlled PPCK that peaks in the dark period and achieves a 10-fold increase in the Ki of the PPC for malate.
We propose that the PPCK and PPC used for this forward engineering work should ideally be a pre-evolved substrate-kinase pair, as has been suggested to have coevolved in the C4 Flaveria system (Aldous et al., 2014). For example, KfPPC1 (GenBank KM078709) and KfPPCK1 (GenBank AF162662) from K. fedtschenkoi would be an ideal substrate-kinase pair for the forward engineering of CAM into C3 species, although care would need to be taken with promoter choice for the transgenes in order to ensure that the KfPPC1 gene was expressed to very high levels in the leaf mesophyll cells, and KfPPCK1 was expressed under circadian clock control to peak in the middle of the dark in leaf mesophyll cells.
METHODS
Plant Materials
Unless otherwise stated, Kalanchoë fedtschenkoi Hamet et Perrier plants were propagated clonally from stem cuttings or leaf margin adventitious plantlets using the same clonal stock originally obtained from the Royal Botanic Gardens, Kew, by Malcolm Wilkins (Wilkins, 1959). Plants were grown and entrained to light/dark cycles as described by Dever et al. (2015).
Time-Course Experiments
For light/dark time-course experiments, opposite pairs of LP6 (where leaf pair 1/LP1 are the youngest leaf pair flanking the shoot apical meristem and leaf pairs are counted down the stem) were collected every 4 h over a 12-h-light (450 µmoles m−2 s−1, 25°C, 60% humidity)/12-h-dark (15°C, 70% humidity) cycle, starting 2 h (02:00) after the lights came on at 00:00 h using plants raised and entrained as described by Dever et al. (2015). For constant light, constant temperature, constant humidity (LL) free-running circadian time-course experiments, plants were entrained in 12-h-light/12-h-dark conditions for 7 d and switched to LL after a dark period as described by Dever et al. (2015). For LL, the constant conditions were as follows: light 100 µmol m−2 s−1, temperature 15°C, and humidity 70%. LP6 were sampled every 4 h from three individual (clonal) plants, starting at 02:00 (2 h after lights on). All sampling involved immediate freezing of leaves in liquid nitrogen. Samples were stored at −80°C until use.
Generation of Transgenic K. fedtschenkoi Lines
An intron containing hairpin RNAi construct was designed to target the silencing of the CAM-induced and clock-controlled KfPPCK1 gene (GenBank accession AF162661) (Hartwell et al., 1999). A 359-bp fragment was amplified from CAM leaf cDNA using high fidelity PCR with KOD Hot Start DNA Polymerase (Merck) and the primers KfPPCK1 RNAi F 5′-CACCGCGCAGGACATATTTGAGG-3′ and KfPPCK1 RNAi R 5′-GGCCGAATTCCCTCACTCTGCTTC-3′. The amplified fragment spanned the 3′ end of the coding sequence and extended into the 3′ untranslated region to ensure specificity of the silencing to a single member of the K. fedtschenkoi PPCK gene family. Alignment of the 359-bp region with the homologous region from the three other PPCK genes in the K. fedtschenkoi genome demonstrated that none of the other PPCK genes shared any 21 nucleotide stretches that were an exact match for KfPPCK1. PCR products were cloned into the pENTR/D Gateway-compatible entry vector via directional TOPO cloning (Life Technologies) and recombined into the intron containing hairpin RNAi binary vector pK7GWIWG2(II) (Karimi et al., 2002) using LR Clonase II enzyme mix (Life Technologies). Constructs were confirmed by DNA sequencing and introduced into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method (Höfgen and Willmitzer, 1988).
Agrobacterium-mediated stable transformation of K. fedtschenkoi was achieved using the tissue culture based method described by Dever et al. (2015).
High-Throughput Leaf Acidity and Starch Content Screens
Leaf acidity (as a proxy for leaf malate content) and leaf starch content were screened with leaf disc stains using chlorophenol red and iodine solution, respectively, at both dawn and dusk, as described by Cushman et al. (2008). For each transgenic line, leaf discs were sampled in triplicate at 1 h before dawn and 1 h before dusk and stained in a 96-well plate format.
Net CO2 Exchange
Gas exchange measurements were performed using a six-channel, custom-built infrared gas analyzer system (PP Systems), which allowed the individual environmental control (CO2/H2O) and measurement of rates of CO2 uptake for each of six gas exchange cuvettes with measurements collected every 10 min (described in full in Dever et al., 2015). All experiments were repeated at least three times using three separate individual young plants (eight leaf pairs), or detached LP6 from three separate clonal plants, and representative gas exchange traces are shown. Wild-type and rPPCK1 lines were compared in neighboring gas exchange cuvettes during each run, such that the data are directly comparable between each line. For example, many runs included two cuvettes containing the wild type, two cuvettes containing rPPCK1-1, and two with rPPCK1-3.
Drought-Stress Experiments
For the drought-treatment gas exchange experiments, clonal leaf margin plantlets from wild-type and the rPPCK1 lines were grown to the 8-leaf-pair stage using 150 mL of our standard compost/perlite mix (Dever et al., 2015) and maintained well-watered throughout their development. Water was withheld for 25 d under our standard 12:12 LD growth conditions according to Dever et al. (2015).
Leaf Malate, Starch, and Sucrose Content
Leaf pair 6 (LP6; full CAM in the wild type) from mature plants was sampled into liquid nitrogen at the indicated times and stored at −80°C until use. The frozen leaf samples were prepared and assayed for malate and starch as described by Dever et al. (2015) using the standard enzyme-linked spectrophotometric methods for assaying malate (Möllering, 1985) and starch (Smith and Zeeman, 2006).
The concentrations of sucrose, d-fructose, and d-glucose were determined using enzyme-linked spectrophotometric assays according to the manufacturer’s instructions (K-SUFRG sucrose/d-fructose/d-glucose assay kit; Megazyme International). This involved the stepwise enzymatic conversion of each sugar to glucose-6-phosphate (G6P) and subsequent quantification by measuring NADPH production at 340 nm by following oxidation of the G6P in the presence of NADP+ and G6P-dehydrogenase.
Total RNA Isolation and RT-qPCR
Total RNA was isolated from 100 mg of frozen, ground leaf tissue using the Qiagen RNeasy kit following the manufacturer’s protocol with the addition of 13.5 μL 50 mg mL−1 PEG 20,000 to the 450 μL RLC buffer used for each extraction. cDNA was synthesized from the total RNA using the Qiagen Quantitect RT kit according to the manufacturer’s instructions (Qiagen). The resulting cDNA was diluted 1:4 with molecular biology grade water prior to use in RT-qPCR. Transcript levels were determined using three technical replicates and three biological replicates with SensiFAST SYBR No Rox kit (Bioline) in an Agilent MX3005P qPCR system cycler using the following program: 95°C for 2 min, 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 10 s, and primers designed with Primer3. Primer specificity was checked using BLAST searches, and primer efficiency was checked using a melting curve profile standard curve. In every PCR plate an interplate calibrator (made from a pool of RNAs assembled from K. fedtschenkoi wild-type leaf pair 6 samples taken every 4 h over a light/dark cycle) was run in triplicate in order to correct any interplate variation. In addition, nontemplate controls were included in every plate to confirm the absence of contamination. Each biological replicate was assayed in triplicate, and the relative abundance of each gene was determined using Agilent MxPRO QPCR software using the manufacturer’s instructions (Agilent Technologies). The results were normalized to a K. fedtschenkoi ortholog of a Thioesterase/thiol ester dehydrase-isomerase (TEDI) superfamily protein (Phytozome Kaladp0068s0118.1; Arabidopsis ortholog AT2G30720.1). Primers for RT-qPCR analyses are listed in Supplemental Table 1.
Immunoblotting
Total protein extracts of K. fedtschenkoi leaves were prepared according to Dever et al. (2015). One-dimensional SDS-PAGE and immunoblotting of leaf proteins was performed following standard methods. Blots were developed using the ECL system (GE Healthcare). Immunoblot analysis was performed using antisera to PPC (raised against PPC from K. fedtschenkoi, kindly supplied by H.G. Nimmo, University of Glasgow) (Nimmo et al., 1986) and the phosphorylated form of PPC (raised against a phospho-PPC peptide from barley [Hordeum vulgare] and kindly supplied by Cristina Echevarría, Universidad de Sevilla, Spain) (González et al., 2002; Feria et al., 2008).
Determination of the Apparent Ki of PPC for l-Malate in Rapidly Desalted Leaf Extracts
Rapidly desalted leaf extracts were prepared as described by Carter et al. (1991) using LP6 sampled from three biological replicates of the wild type, rPPCK1-1, and rPPCK1-3 at 10:00 (2 h before the end of the 12-h light period) and 18:00 (middle of the 12-h dark period). The apparent Ki of PPC activity for feedback inhibition by l-malate was determined for each rapidly desalted extract according to the method described by Nimmo et al. (1984), except that the range of l-malate concentrations added to the different PPC assays was 0, 0.2, 0.5, 0.7, 1.0, 2.0, 3.0, 5.0, 8.0, and 10.0 mM. The concentration of malate needed to achieve 50% inhibition of the initial PPC activity (apparent Ki) was determined using the plots shown in Supplemental Figure 1.
Dry Weight Growth Measurements
Mature plants were grown from developmentally synchronized clonal leaf plantlets in greenhouse conditions for 138 d, harvested as separated aboveground and belowground tissues, and dried in an oven at 60°C until they reached a constant weight. Weights were measured to two decimal places in grams using a fine balance. Seven individual clonal plants were grown for each line (Dever et al., 2015).
DF Measurements
The imaging system for DF was identical to the luciferase and delayed fluorescence imaging system described previously (Gould et al., 2009). DF was quantified using Imaris (Bitplane) to measure mean intensity for specific regions within an image. Background intensities were calculated for each image and subtracted, to give a final DF measurement (Gould et al., 2009).
DF Rhythm Analysis
K. fedtschenkoi plants were grown in greenhouse conditions for 4 months and then entrained in 12-h light/25°C: 12-h dark/15°C cycles in a Snijders Microclima MC-1000 (Snijders Scientific) growth cabinet for 7 d as described previously (Dever et al., 2015). At dawn on the 8th day, 1.5-cm leaf discs were punched from each of leaf pair 6 for three biological replicates (i.e., two leaf discs from each biological replicate, totaling six leaf discs per line) and placed with a few drops of distilled water in one of 25 2 × 2-cm wells in a 10 × 10-cm square Petri dish. The drops of distilled water were sufficient to keep the leaf discs hydrated but were not enough for the discs to float and thus move, which would make the DF image analysis extremely challenging. The Petri dish was placed in the imaging system at 22°C in constant red-blue light (LL). DF images were collected every hour for 108 h as described previously (Gould et al., 2009). The DF images were processed as described by Gould et al. (2009). The luminescence was normalized by subtracting the Y value of the best straight line from the raw Y value. Biodare was used to carry out fast Fourier transform (nonlinear least-square) analysis and spectral resampling on each DF time-course series using the time window from 24 to 108 h in order to generate period estimates and calculate the associated RAE (Moore et al., 2014; Zielinski et al., 2014). The first 24 h of the data were excluded as this represents a bedding-in period for the leaves when transition effects, resulting from the leaves moving from light/dark cycles to constant light and temperature (LL), may be influencing the data.
Accession Numbers
Sequence data associated with this article have been deposited in the GenBank/EMBL data libraries under the following accession numbers: KfPPCK1, AF162662; KfTOC1-1, KM078716; KfCCA1-1, KM078717; KfCCA1-2, KM078718; KfPPCK2, KM078720; KfGI, KM078724; and KfTOC1-2, KM078726. Other accession numbers and gene IDs are presented in Supplemental Table 1.
Supplemental Data
Supplemental Figure 1. Impact of silencing KfPPCK1 on the apparent Ki of PPC for l-malate in rapidly desalted leaf extracts.
Supplemental Table 1. Primers used for reverse transcription-quantitative PCR.
Acknowledgments
We thank Hugh Nimmo (University of Glasgow, UK) and Cristina Echevarria (Universidad de Sevilla, Spain) for providing the antibodies used in this study. This work was supported in part by the Biotechnology and Biological Sciences Research Council, UK (BBSRC grant no. BB/F009313/1 awarded to J.H.) and in part by the U.S. Department of Energy (DOE) Office of Science, Genomic Science Program under Award Number DE-SC0008834. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the DOE.
AUTHOR CONTRIBUTIONS
J.H., S.F.B., L.V.D., and P.D.G. designed the research. S.F.B. performed all experiments except the stable transformations, immunoblots, starch determinations, growth yield drought experiment, and delayed fluorescence measurements. L.V.D. performed the immunoblots, starch determinations, and the growth yield drought experiment. J.K. generated the binary constructs and carried out the stable transformation, regeneration, and initial screening of the K. fedtschenkoi rPPCK1 transgenic lines, and also collaborated with L.V.D. on the growth yield drought experiment. P.D.G. carried out the delayed fluorescence experiments in collaboration with S.F.B. S.F.B., L.V.D., P.D.G., and J.H. analyzed the data. S.F.B. and J.H. wrote the manuscript.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Abraham P.E., et al. (2016). Transcript, protein and metabolite temporal dynamics in the CAM plant Agave Nat. Plants 2: 1–10. [DOI] [PubMed] [Google Scholar]
- Aldous S.H., Weise S.E., Sharkey T.D., Waldera-Lupa D.M., Stühler K., Mallmann J., Groth G., Gowik U., Westhoff P., Arsova B. (2014). Evolution of the phosphoenolpyruvate carboxylase protein kinase family in C3 and C4 Flaveria spp. Plant Physiol. 165: 1076–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.M., Wilkins M.B. (1989). Period and phase control by temperature in the circadian rhythm of carbon dioxide fixation in illuminated leaves of Bryophyllum fedtschenkoi. Planta 177: 456–469. [DOI] [PubMed] [Google Scholar]
- Aubry S., Brown N.J., Hibberd J.M. (2011). The role of proteins in C(3) plants prior to their recruitment into the C(4) pathway. J. Exp. Bot. 62: 3049–3059. [DOI] [PubMed] [Google Scholar]
- Borland A.M., Griffiths H., Hartwell J., Smith J.A.C. (2009). Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J. Exp. Bot. 60: 2879–2896. [DOI] [PubMed] [Google Scholar]
- Borland A.M., Guo H.B., Yang X., Cushman J.C. (2016). Orchestration of carbohydrate processing for crassulacean acid metabolism. Curr. Opin. Plant Biol. 31: 118–124. [DOI] [PubMed] [Google Scholar]
- Borland A.M., Wullschleger S.D., Weston D.J., Hartwell J., Tuskan G.A., Yang X., Cushman J.C. (2015). Climate-resilient agroforestry: physiological responses to climate change and engineering of crassulacean acid metabolism (CAM) as a mitigation strategy. Plant Cell Environ. 38: 1833–1849. [DOI] [PubMed] [Google Scholar]
- Borland A.M., Hartwell J., Weston D.J., Schlauch K.A., Tschaplinski T.J., Tuskan G.A., Yang X., Cushman J.C. (2014). Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci. 19: 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxall S.F., Foster J.M., Bohnert H.J., Cushman J.C., Nimmo H.G., Hartwell J. (2005). Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol. 137: 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhaus D., Bräutigam A., Mettler-Altmann T., Winter K., Weber A.P.M. (2016). Reversible burst of transcriptional changes during induction of Crassulacean acid metabolism in Talinum triangulare. Plant Physiol. 170: 102–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P.J., Nimmo H.G., Fewson C.A., Wilkins M.B. (1990). Bryophyllum fedtschenkoi protein phosphatase type-2A can dephosphorylate phosphoenolpyruvate carboxylase. FEBS Lett. 263: 233–236. [Google Scholar]
- Carter P.J., Nimmo H.G., Fewson C.A., Wilkins M.B. (1991). Circadian rhythms in the activity of a plant protein kinase. EMBO J. 10: 2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R., Vidal J., O’Leary M.H. (1996). Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 273–298. [DOI] [PubMed] [Google Scholar]
- Christin P.A., Arakaki M., Osborne C.P., Bräutigam A., Sage R.F., Hibberd J.M., Kelly S., Covshoff S., Wong G.K.-S., Hancock L., Edwards E.J. (2014). Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. J. Exp. Bot. 65: 3609–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman J.C., Bohnert H.J. (1999). Crassulacean acid metabolism: Molecular genetics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 305–332. [DOI] [PubMed] [Google Scholar]
- Cushman J.C., Davis S.C., Yang X., Borland A.M. (2015). Development and use of bioenergy feedstocks for semi-arid and arid lands. J. Exp. Bot. 66: 4177–4193. [DOI] [PubMed] [Google Scholar]
- Cushman J.C., Agarie S., Albion R.L., Elliot S.M., Taybi T., Borland A.M. (2008). Isolation and characterization of mutants of common ice plant deficient in crassulacean acid metabolism. Plant Physiol. 147: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N., Baek S.J., Briggs H.M., Robertson F.C., Dodd A.N., Gardner M.J., Stancombe M.A., Haydon M.J., Stan G.B., Gonçalves J.M., Webb A.A.R. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 108: 5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.C., Kuzmick E.R., Niechayev N., Hunsaker D.J. (2017). Productivity and water use efficiency of Agave americana in the first field trial as bioenergy feedstock on arid lands. Glob. Change Biol. Bioenergy 9: 314–325. [Google Scholar]
- Dever L.V., Boxall S.F., Kneřová J., Hartwell J. (2015). Transgenic perturbation of the decarboxylation phase of Crassulacean acid metabolism alters physiology and metabolism but has only a small effect on growth. Plant Physiol. 167: 44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncaster H.D., Leegood R.C. (1987). Regulation of phosphoenolpyruvate carboxylase activity in maize leaves. Plant Physiol. 84: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feria A.B., Alvarez R., Cochereau L., Vidal J., García-Mauriño S., Echevarría C. (2008). Regulation of phosphoenolpyruvate carboxylase phosphorylation by metabolites and abscisic acid during the development and germination of barley seeds. Plant Physiol. 148: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I., König A.C., Heard W., Nunes-Nesi A., Pham P.A., Leister D., Fernie A.R., Sweetlove L.J. (2013). Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in Arabidopsis leaves. Plant Physiol. 162: 239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto T., Izui K., Quinn V., Furbank R.T., von Caemmerer S. (2007). Phosphorylation of phosphoenolpyruvate carboxylase is not essential for high photosynthetic rates in the C4 species Flaveria bidentis. Plant Physiol. 144: 1936–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcês H.M., Champagne C.E., Townsley B.T., Park S., Malhó R., Pedroso M.C., Harada J.J., Sinha N.R. (2007). Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc. Natl. Acad. Sci. USA 104: 15578–15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M.-C., Echevarria C., Vidal J., Cejudo F.J. (2002). Isolation and characterisation of a wheat phosphoenolpyruvate carboxylase gene. Modelling of the encoded protein. Plant Sci. 162: 233–238. [Google Scholar]
- Gould P.D., Diaz P., Hogben C., Kusakina J., Salem R., Hartwell J., Hall A. (2009). Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 58: 893–901. [DOI] [PubMed] [Google Scholar]
- Hartwell J. (2005). The co-ordination of central plant metabolism by the circadian clock. Biochem. Soc. Trans. 33: 945–948. [DOI] [PubMed] [Google Scholar]
- Hartwell J. (2006). The circadian clock in CAM plants. In Endogenous Plant Rhythms, Hall A.J.W., McWatters H.G., eds (Oxford, UK: Blackwell Publishing; ), pp. 211–236. [Google Scholar]
- Hartwell J., Dever L.V., Boxall S.F. (2016). Emerging model systems for functional genomics analysis of Crassulacean acid metabolism. Curr. Opin. Plant Biol. 31: 100–108. [DOI] [PubMed] [Google Scholar]
- Hartwell J., Smith L.H., Wilkins M.B., Jenkins G.I., Nimmo H.G. (1996). Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J. 10: 1071–1078. [Google Scholar]
- Hartwell J., Gill A., Nimmo G.A., Wilkins M.B., Jenkins G.I., Nimmo H.G. (1999). Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J. 20: 333–342. [DOI] [PubMed] [Google Scholar]
- Haydon M.J., Roman A., Arshad W. (2015). Nutrient homeostasis within the plant circadian network. Front. Plant Sci. 6: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon M.J., Mielczarek O., Robertson F.C., Hubbard K.E., Webb A.A.R. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R., Willmitzer L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtum J.A.M., Winter K., Weeks M.A., Sexton T.R. (2007). Crassulacean acid metabolism in the ZZ plant, Zamioculcas zamiifolia (Araceae). Am. J. Bot. 94: 1670–1676. [DOI] [PubMed] [Google Scholar]
- Jones M.B. (1975). The effect of leaf age on leaf resistance and CO2 exchange of the CAM plant Bryophyllum fedtschenkoi. Planta 123: 91–96. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Ming R., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47: 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllering H. (1985). L-malate: Determination with malate dehydrogenase and aspartate aminotransferase. In Methods in Enzymatic Analysis, Bermeyer H.U., Bermeyer J., Grasstl M., eds (Weinheim, Germany: Verlag Chemie; ), pp. 39–47. [Google Scholar]
- Moore A., Zielinski T., Millar A.J. (2014). Online period estimation and determination of rhythmicity in circadian data, using the BioDare data infrastructure. In Plant Circadian Networks: Methods and Protocols, Methods in Molecular Biology, Vol. 1158, D. Staiger, ed (New York: Springer Science and Business Media), pp. 13–44. [DOI] [PubMed] [Google Scholar]
- Nimmo G.A., Nimmo H.G., Fewson C.A., Wilkins M.B. (1984). Diurnal changes in the properties of phosphoenolpyruvate carboxylase in Bryophyllum leaves - a possible covalent modification. FEBS Lett. 178: 199–203. [Google Scholar]
- Nimmo G.A., Wilkins M.B., Fewson C.A., Nimmo H.G. (1987). Persistent circadian rhythms in the phosphorylation state of phosphoenolpyruvate carboxylase from Bryophyllum fedtschenkoi leaves and in its sensitivity to inhibition by malate. Planta 170: 408–415. [DOI] [PubMed] [Google Scholar]
- Nimmo G.A., Nimmo H.G., Hamilton I.D., Fewson C.A., Wilkins M.B. (1986). Purification of the phosphorylated night form and dephosphorylated day form of phosphoenolpyruvate carboxylase from Bryophyllum fedtschenkoi. Biochem. J. 239: 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo H.G. (2000). The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci. 5: 75–80. [DOI] [PubMed] [Google Scholar]
- Nimmo H.G. (2003). How to tell the time: the regulation of phosphoenolpyruvate carboxylase in Crassulacean acid metabolism (CAM) plants. Biochem. Soc. Trans. 31: 728–730. [DOI] [PubMed] [Google Scholar]
- Osmond C.B. (1978). Crassulacean acid metabolism - Curiosity in context. Annu. Rev. Plant Physiol. Plant Mol. Biol. 29: 379–414. [Google Scholar]
- Silvera K., Winter K., Rodriguez B.L., Albion R.L., Cushman J.C. (2014). Multiple isoforms of phosphoenolpyruvate carboxylase in the Orchidaceae (subtribe Oncidiinae): implications for the evolution of crassulacean acid metabolism. J. Exp. Bot. 65: 3623–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Zeeman S.C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1: 1342–1345. [DOI] [PubMed] [Google Scholar]
- Smith J.A.C., Winter K. (1996). Taxonomic distribution of crassulacean acid metabolism. In Crassulacean acid metabolism - biochemistry, ecophysiology and evolution, Winter K., Smith J.A.C., eds (Berlin, Heidelberg: Springer Verlag; ), pp. 427–436. [Google Scholar]
- Taybi T., Patil S., Chollet R., Cushman J.C. (2000). A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol. 123: 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen J.J., Miernyk J.A., Randall D.D. (2000). Pyruvate dehydrogenase kinase from Arabidopsis thaliana: a protein histidine kinase that phosphorylates serine residues. Biochem. J. 349: 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theng V., Agarie S., Nose A. (2008). Regulatory phosphorylation of phosphoenolpyruvate carboxylase in the leaves of Kalanchoe pinnata, K. daigremontiana and Ananas comosus. Biol. Plant. 52: 281–290. [Google Scholar]
- Tsen E.W.J., Holtum J.A.M. (2012). Crassulacean acid metabolism (CAM) in an epiphytic ant-plant, Myrmecodia beccarii Hook.f. (Rubiaceae). Photosynth. Res. 113: 311–320. [DOI] [PubMed] [Google Scholar]
- Vidal J., Chollet R. (1997). Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci. 2: 230–237. [Google Scholar]
- Wild B., Wanek W., Postl W., Richter A. (2010). Contribution of carbon fixed by Rubisco and PEPC to phloem export in the Crassulacean acid metabolism plant Kalanchoe daigremontiana. J. Exp. Bot. 61: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins M.B. (1959). An endogenous rhythm in the rate of carbon dioxide output of Bryophyllum. 1. Some preliminary experiments. J. Exp. Bot. 10: 377–390. [Google Scholar]
- Wilkins M.B. (1992). Circadian rhythms: Their origin and control. New Phytol. 121: 347–375. [DOI] [PubMed] [Google Scholar]
- Wyka T.P., Bohn A., Duarte H.M., Kaiser F., Lüttge U.E. (2004). Perturbations of malate accumulation and the endogenous rhythms of gas exchange in the Crassulacean acid metabolism plant Kalanchoë daigremontiana: testing the tonoplast-as-oscillator model. Planta 219: 705–713. [DOI] [PubMed] [Google Scholar]
- Yang X., et al. (2015). A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 207: 491–504. [DOI] [PubMed] [Google Scholar]
- Zielinski T., Moore A.M., Troup E., Halliday K.J., Millar A.J. (2014). Strengths and limitations of period estimation methods for circadian data. PLoS One 9: e96462. [DOI] [PMC free article] [PubMed] [Google Scholar]