The CELLULOSE SYNTHASE INTERACTING1 protein directs secondary wall patterning during the early phases of xylem vessel development.
Abstract
The evolution of the plant vasculature was essential for the emergence of terrestrial life. Xylem vessels are solute-transporting elements in the vasculature that possess secondary wall thickenings deposited in intricate patterns. Evenly dispersed microtubule (MT) bands support the formation of these wall thickenings, but how the MTs direct cell wall synthesis during this process remains largely unknown. Cellulose is the major secondary wall constituent and is synthesized by plasma membrane-localized cellulose synthases (CesAs) whose catalytic activity propels them through the membrane. We show that the protein CELLULOSE SYNTHASE INTERACTING1 (CSI1)/POM2 is necessary to align the secondary wall CesAs and MTs during the initial phase of xylem vessel development in Arabidopsis thaliana and rice (Oryza sativa). Surprisingly, these MT-driven patterns successively become imprinted and sufficient to sustain the continued progression of wall thickening in the absence of MTs and CSI1/POM2 function. Hence, two complementary principles underpin wall patterning during xylem vessel development.
INTRODUCTION
The plant vasculature is one of the most important evolutionary innovations for terrestrial life, as it allowed plants to adapt and grow to significant stature (Myburg et al., 2013). The xylem tissue provides essential functions in the vasculature by distributing water throughout the plant and providing structural support to the plant body. The xylem cells are encased by thickened cell walls that reinforce them and therefore are essential for their function (Turner et al., 2007). The organization of the secondary cell walls differs between xylem vessel cell types and is typically described either as an annular/spiral pattern (called protoxylem) or a reticulate/pitted pattern (called metaxylem; Pesquet et al., 2011). Before these thickened secondary walls are assembled, the xylem cells, like all plant cells, are encased by a flexible but strong primary cell wall (Somerville et al., 2004). These walls largely comprise polysaccharides, of which cellulose, an unbranched, linear β-1,4-linked glucan, forms a significant constituent. Cellulose is synthesized at the plasma membrane by large cellulose synthase (CesA) complexes (CSCs; Schneider et al., 2016). The CSCs are composed of a heterotrimeric configuration of 18 to 24 CesAs where CesA1, CesA3, and CesA6-like (i.e., CesA2, 5, 6, and 9) CesAs produce primary wall cellulose in Arabidopsis thaliana, and CesA4, CesA7, and CesA8 comprise the CSCs necessary to make secondary wall cellulose (Persson et al., 2007; Desprez et al., 2007; Taylor et al., 2003; Atanassov et al., 2009).
The CSCs move along linear tracks at the plasma membrane (Paredez et al., 2006), likely due to the catalytic activity of the CSCs. Nascent cellulose microfibrils become entrapped in the cell wall and further synthesis therefore exerts a force on the CSCs that propels them forward through the plasma membrane. The movement of the CSCs is guided by cortical microtubules (MTs) during both primary and secondary wall cellulose synthesis (Paredez et al., 2006; Watanabe et al., 2015). The protein CELLULOSE SYNTHASE INTERACTING1 (CSI1), also called POM-POM2 (POM2), is necessary for the MT-based guidance of the primary wall CSCs, as lesions in the protein impaired coalignment between tracks of primary wall CSCs and cortical MTs (Bringmann et al., 2012; Li et al., 2012). However, reports on the function of CSI1/POM2 during secondary wall cellulose production differ. Gu and Somerville (2010) reported no defects on secondary walls nor decreased cellulose content in csi1/pom2 mutant Arabidopsis stems. By contrast, Derbyshire et al. (2015) showed that induction of tracheary elements in Arabidopsis cell cultures was impaired in cells with reduced CSI1/POM2 expression. The role of CSI1/POM2 in secondary wall cellulose production therefore remains unclear.
Secondary walls are typically produced around cells that are situated deep in tissues and that therefore are largely masked by other cells. This location makes it difficult to study secondary wall synthesis where it normally occurs. Instead, alternative systems have been developed for this purpose, including trans-differentiating cell cultures that can be induced by hormone cocktails (Kubo et al., 2005; Demura et al., 2002; Pesquet et al., 2010) and inducible transcription factor-based systems. The latter systems make use of the NAC-related transcription factors VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 that promote meta- and protoxylem-like cell wall structures, respectively (Kubo et al., 2005; Yamaguchi et al., 2010; Oda et al., 2010). By selectively controlling the activity of the VNDs with an inducible promoter system, it is possible to induce and explore secondary wall formation in cells that normally do not form these structures. VND7-inducible Arabidopsis seedlings have been used to evaluate the behavior of the secondary wall CSCs using a fluorescently tagged CesA7 (Watanabe et al., 2015) and to assess the coordination between transcripts and metabolites during this process (Li et al., 2016a).
Here, we investigated how protoxylem vessel wall patterns are controlled by analyzing the coordination of MTs and cell wall deposition in Arabidopsis and rice (Oryza sativa). We found that CSI1/POM2 orchestrates cell wall synthesis along MTs during the initial developmental phase of xylem vessel formation but that subsequent synthesis occurs via a CSI1/POM2 autonomous mechanism. Our results indicate that cell wall patterns are directed by two complementary principles during xylem vessel development.
RESULTS
CSI1/POM2 Influences Xylem Vessel Wall Patterning
To evaluate if defects in CSI1/POM2 function alter cell wall patterning during xylem vessel formation in Arabidopsis, we examined secondary wall formation in three different systems where the function of CSI1/POM2 was impaired. First, we confirmed that the downregulation of CSI1/POM2 caused aberrant secondary wall deposition in proto- and metaxylem trans-differentiating cell suspension cultures (Derbyshire et al., 2015; Supplemental Figures 1A and 1B). Using confocal microscopy, we quantified the occurrence of spiral, reticulate, and pitted secondary wall patterns and the percentage of calcofluor-stained irregular deposits in the secondary walls of nontransgenic and CSI1 downregulated cell lines (Supplemental Figures 1C and 1D). Although it was difficult to assess defects in cell wall patterning in these lines, downregulation of CSI1/POM2 caused a significant increase in irregular deposits along the secondary walls (Supplemental Figures 1B and 1D). This defect was irrespective of the patterning of the secondary walls (Supplemental Figure 1D).
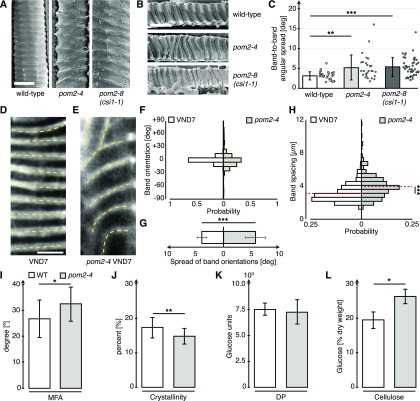
We next investigated if the xylem of mature stems of Arabidopsis plants showed structural defects when CSI1/POM2 function was impaired. We made longitudinal sections of the first internodes allowing structural characterization of intact and transected xylem vessels in the previously described csi1/pom2 mutants pom2-4 and csi1-1/pom2-8 (Bringmann et al., 2012) as well as wild-type plants (Figures 1A and 1B). We found that the secondary wall bands were significantly more disordered in the pom2-4 and csi1-1/pom2-8 mutants, as evident from measuring the spread in orientation angles of neighboring wall bands (Figure 1C).
Figure 1.
Defects in CSI1/POM2 Cause Aberrant Xylem Vessel Patterns.
(A) and (B) Scanning electron micrographs of longitudinal sections of mature wild-type stems. Exposed (A) and transected (B) xylem vessels of wild-type plants and pom2-4 and pom2-8 (csi1-1) mutants. Bar = 10 µm.
(C) Band-to-band orientations in pom2-4 (16 cells in 6 seedlings) and pom2-8 (44 cells in 6 seedlings) compared with wild-type xylem (27 cells in 6 seedlings) obtained from the images in (A) and (B).
(D) and (E) S4B staining of cellulose in VND7-induced hypocotyls. Dotted lines indicate highly ordered bands in the secondary walls of wild-type cells (D) and irregular bands in pom2-4 mutant cells (E). Bar = 5 μm.
(F) Distribution of the average band orientations (yellow lines in [A] and [B]).
(G) The spread of band orientations within individual cells of induced pom2-4 cells (602 bands in 115 cells in 5 seedlings) compared with wild-type cells (824 bands in 132 cells in 5 seedlings).
(H) Secondary wall band spacing in the pom2-4 mutant compared with wild-type cells.
(I) MFA (relative to growth axis) in the pom2-4 mutant compared with the wild type.
(J) Cell wall crystallinity in the pom2-4 mutant compared with the wild type.
(K) Degree of polymerization (DP) in the pom2-4 mutant compared with the wild type.
(L) Cellulose content (% fraction of dry weight) in the pom2-4 mutant compared with the wild type. All measurements ([I] to [L]) were done on ground stems of 10-week-old, fully senesced plants grown in 16-h-light/8-h-dark conditions. Data are means ± sd. Statistical significance was tested by Welch’s unpaired t test: *P < 0.05, **P < 0.005, and ***P < 0.0005.
We next used a VND7-inducible Arabidopsis line (Yamaguchi et al., 2010) to study protoxylem vessel secondary wall patterning. Here, we observed xylem-related wall synthesis as indicated by well-organized band patterns that were transversely and evenly distributed around induced hypocotyl cells (Figure 1D). We quantified the geometry of the bands and found that they were aligned tightly around an average angle of 0.6 ± 3.8° (mean ± sd, 132 cells from five seedlings) against the horizontal axis (Figures 1D and 1F).
To assess if the CSI1/POM2 function influenced the wall patterns, we introgressed the pom2-4 mutant into the VND7-inducible line. The xylem vessel wall patterns were less well aligned in the pom2-4 background (Figures 1E and 1F). Here, the bands displayed significantly wider and less uniform angles compared with control (−2.4 ± 22.7°; 136 cells from five seedlings; Figures 1E to 1G). In addition, the band spacing was substantially altered in the pom2-4 mutant compared with the control VND7-inducible line (Figure 1H). These results indicate that while CSI1/POM2 is not essential for the formation of secondary wall bands, confirming that xylem vessels are intact (Gu and Somerville, 2010), the protein influences the geometry and relative position of the deposition of the bands.
To investigate if the defects in secondary wall patterning were associated with changes in cell wall architecture and ultrastructure, we measured the microfibril angle (MFA), cell wall crystallinity, degree of cellulose polymerization, and cellulose content in pom2-4 mutant stems and compared the results with wild-type stems (Figures 1I to 1L). We found that the cellulose showed differences in both MFAs and crystallinity (Figures 1I and 1J), corroborating defects in cellulose synthesis. We also found a slight increase in glucose content, most likely due to an increase in amorphous cellulose due to the decreased levels of crystalline cellulose (Figure 1L). These data indicate that CSI1/POM2 influence the quality of secondary wall cellulose synthesis.
CSI1/POM2 Mimics the Behavior of, and Can Interact with, the Secondary Wall CesA Proteins
To investigate how the CSI1/POM2 behaves during the transition from primary to secondary wall synthesis, we crossed plants expressing a functional, native promoter-driven triple (3x) YFP translational fusion with CSI1/POM2 (Worden et al., 2015) into the VND7-inducible Arabidopsis line. The 3xYFP-CSI1/POM2 can be seen as fluorescent foci that track together with the CSCs at the cell cortex along linear trajectories during primary wall synthesis (Worden et al., 2015). After induction of VND7, we observed a clear change in the cellular distribution of the 3xYFP-CSI1/POM2. Although the 3xYFP-CSI1/POM2 foci maintained linear movement, the pattern of movement changed following induction. The foci were initially evenly distributed across the plasma membrane; however, this pattern changed in favor of dense and regularly spaced banded patterns (Supplemental Movie 1).
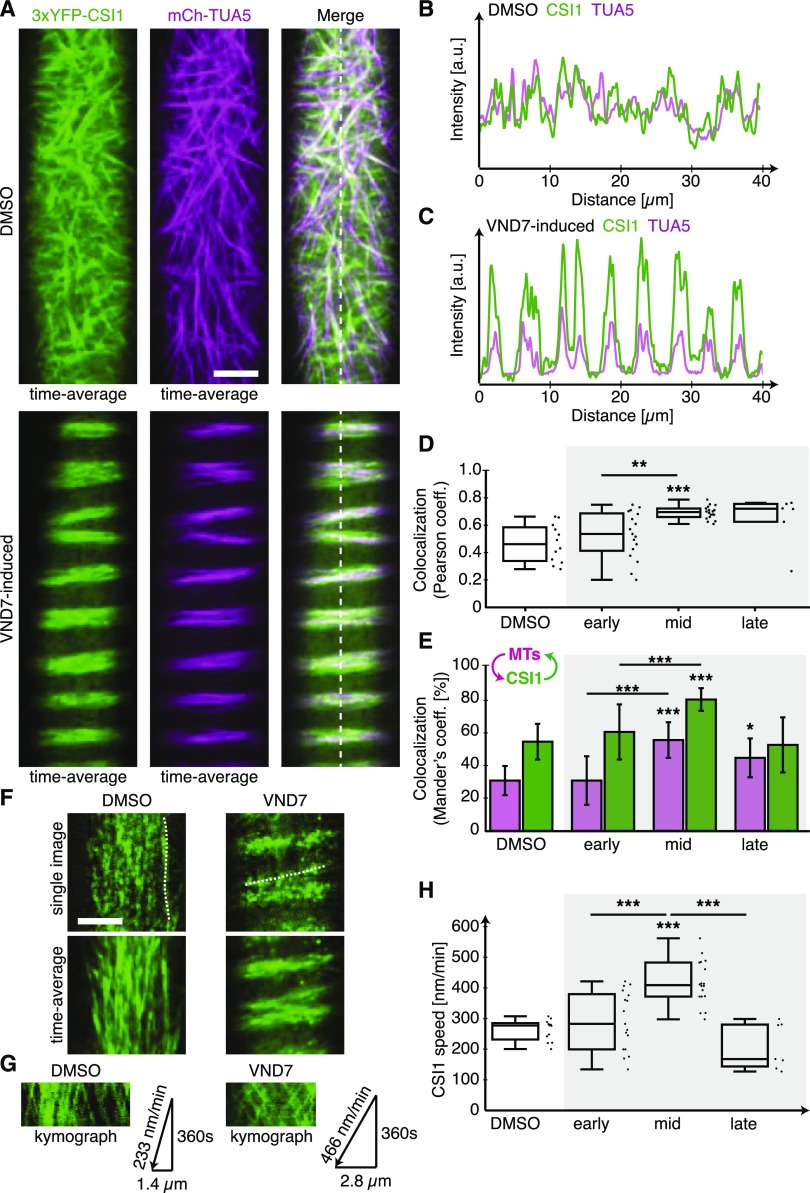
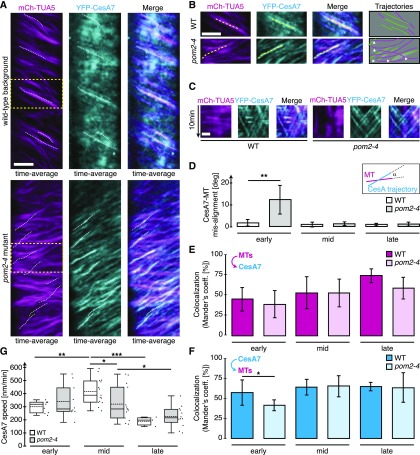
Cortical MTs change their distribution during the progression of xylem vessel production and form distinct banded or helical arrays (Watanabe et al., 2015), similar to what we observed for the 3xYFP-CSI1/POM2 foci. To see if the 3xYFP-CSI1/POM2 and MT re-distributions co-occurred during xylem vessel development, we crossed a mCherry-TUA5-expressing plant with the 3xYFP-CSI1/POM2 VND7-inducible plant and analyzed the progeny. We found that the changes in the 3xYFP-CSI1/POM2 patterns co-occurred with the rearrangement of the MT array during the transition from primary to secondary wall synthesis (Figures 2A to 2E), indicating that the CSI1/POM2 likely tracks with both primary and secondary wall CSCs.
Figure 2.
CSI1/POM2 Colocalizes with Cortical Microtubules and Behaves Similarly to Secondary Wall CesAs during Xylem Vessel Development.
(A) Time-averaged images of MTs and 3xYFP-CSI1 in DMSO-treated control cells (upper panel) and VND7-induced cells in mid stage (lower panel; see Supplemental Figure 3 for definition of stages). Bar = 5 µm.
(B) and (C) Fluorescence intensity plots of CSI1 (green) and MTs (lilac) in DMSO-treated (B) and VND7-induced (C) cells along the dashed lines in (A).
(D) and (E) Measurement of the Pearson’s (D) and Mander’s (E) correlation coefficient comparing CSI1 colocalization with MTs during secondary wall developmental program compared with DMSO-treated cells. The arrow scheme in (E) indicates intensity overlap of MTs with CSI1 (lilac) and CSI1 with MTs (green).
(F) Single and time-averaged images of 3xYFP-CSI1 in DMSO-treated (left) and VND7-induced (right) cells. Bar = 5 µm.
(G) Kymographs along the dotted lines in (F) comparing 3xYFP-CSI1 velocity in DMSO-treated and VND7-induced cells.
(H) 3xYFP-CSI1 migration speeds during secondary wall developmental compared with DMSO-treated cells. Statistical significance was tested by Welch’s unpaired t test relative to DMSO-treated controls: *P < 0.05, **P < 0.005, and ***P < 0.0005.
The primary wall CSCs typically track with a speed of ∼250 nm/min (Paredez et al., 2006), and CSI1/POM2 proteins track together with these CSCs (Bringmann et al., 2012). However, the secondary wall CSCs track significantly faster than the primary wall CSCs (Watanabe et al., 2015), and if the CSI1/POM2 proteins are associated with the secondary wall CSCs, one would anticipate an increase in speed of the CSI1/POM2s over time after VND7 induction. To test this, we measured the speed of the 3xYFP-CSI1/POM2 at early, mid, and late time points after VND7 induction. We selected these time points based on the MT reorganization status after induction (Supplemental Figure 2), and they roughly coincide with time points used by Watanabe et al. (2015). We found that the CSI1/POM2 proteins moved with a speed of ∼266 ± 35 nm/min (1015 foci in 12 cells from 3 seedlings) in DMSO-treated cells and at 293 ± 98 nm/min (905 foci in 17 cells in 15 seedlings) in cells in the early stages of the secondary wall synthesis, i.e., where MTs still exhibited a primary wall-like pattern (Figures 2F to 2H). However, during the middle stages of secondary wall synthesis, i.e., where MTs formed diffuse bands, we observed a significant increase in CSI1/POM2 speed (422 ± 72 nm/min; 1391 foci in 18 cells from 15 seedlings). Once secondary cell wall synthesis had progressed to late stages, we found that the speed of the CSI1/POM2 proteins declined (211 ± 75 nm/min; 234 foci in 7 cells from 15 seedlings), possibly related to the initiation of programmed cell death. Notably, the secondary wall CSCs underwent a very similar transition in speeds during early, mid, and late secondary wall stages (Watanabe et al., 2015). These data indicate that the CSI1/POM2 may track with the secondary wall CesA proteins, similar to what has been shown for the primary wall CesAs (Gu et al., 2010). To test whether CSI1/POM2 can interact with secondary wall CesAs, we performed bimolecular fluorescence complementation (BiFC) assays between the three secondary wall Arabidopsis CesAs and CSI1/POM2. We found that the proteins can interact when transiently expressed in tobacco (Nicotiana tabacum) epidermal leaf cells (Supplemental Figure 3). Hence, the CSI1/POM2 proteins behave similarly to the secondary wall CesAs and can interact with them.
The CSI1/POM2s Track with the Secondary Wall CesAs and Are Rapidly Recruited to Their Sites of Action
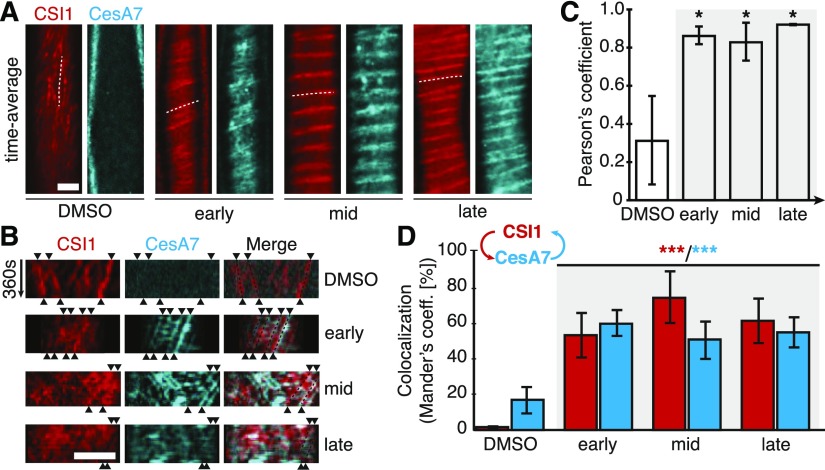
The CSI1/POM2 and secondary wall CesAs behave similarly and can interact, suggesting that the proteins also track together during xylem vessel development. To test this, we generated plants expressing a mCherry-tagged CSI1/POM2 fusion protein under control of the CSI1/POM2 promoter and introgressed these plants with VND7-inducible lines expressing the YFP-CesA7 construct. The two fluorescent proteins showed similar behavior and closely colocalized throughout the different stages of VND7 induction (Figures 3A to 3D). These observations are supported by close inspections of kymographs from movies of the fluorescent proteins, where the tracking of the proteins coincided (Figure 3B).
Figure 3.
CSI1/POM2 Comigrates with CesA7 and Recovers More Quickly Than CesA7 after Photobleaching during Xylem Vessel Formation.
(A) Average projections of mCherry-CSI1/POM2 and YFP-CesA7 in noninduced cells (DMSO) and early, mid, and late stages of secondary wall synthesis. Bar = 5 μm.
(B) Kymographs along the dotted lines in (A) showing the migration of CSI1/POM2 with YFP-CesA7 in early, mid, and late stages of secondary wall synthesis. Trajectories are indicated by arrowheads. Bar = 5 μm.
(C) and (D) CSI1/POM2 colocalization with CesA7 throughout secondary wall synthesis as shown by Pearson’s (C) and Mander’s coefficients (D). The arrow scheme in (D) indicates intensity overlap of CSI1 with CesA7 (red) and CesA7 with CSI1 (cyan). Statistical significance was tested by Welch’s unpaired t test: *P < 0.05, **P < 0.005, and ***P < 0.0005.
To assess the recruitment of CSI1/POM2 and CesA7 to their sites during secondary wall synthesis, we performed fluorescence recovery after photobleaching (FRAP) experiments. We first used the VND7-lines expressing mCherry-CSI1/POM2 and YFP-CesA7; however, the mCherry signal proved too weak to accurately assess fluorescence recovery. Instead, we used VND7-induced lines expressing either 3xYFP-CSI1/POM2 or YFP-CesA7 and counted the number of insertion events over time, which permitted measurement of the average insertion, or delivery, times (Supplemental Figures 4A to 4D). The 3xYFP-CSI1/POM2 signal rapidly repopulated the bleached area after FRAP (Supplemental Figures 4A and 4D; recovery time 32 ± 13 s, 46 bands in 7 cells in 3 seedlings), whereas the recovery of the YFP-CesA7 was significantly slower (106 ± 68 s, 32 bands in 6 cells in 4 seedlings). We calculated the ratio of the recovery of the two fluorescently labeled proteins to be 3.3 ± 2.5. As the secondary wall CSCs contain CesA4, CesA7, and CesA8, possibly in equal stoichiometry (Gonneau et al., 2014; Hill et al., 2014), it is likely that each CesA in the secondary wall CSC is associated with one CSI1/POM2 protein. However, it is important to note two things: First, the pom2-4 and irx3-4 mutations were not homozygous in the 3xYFP-CSI1/POM2 and the YFP-CesA7 lines, respectively. While one might assume that each CSC will contain both labeled and unlabeled CesA7, and thus that each CSC is tracked in our image analysis, it is possible that we underestimate the numbers of CSI1/POM2s associated with each CSC. Second, the analyses were done on seedlings with either the 3xYFP-CSI1/POM2 or YFP-CesA7, which alone may introduce experimental differences. While we therefore favor a ratio between the CSI1/POM2 and CesA as 1:1 at a given secondary wall CSC, further experiments are needed to firmly corroborate this hypothesis.
Optical Flow Analyses Support Global Bidirectionality, but Local Unidirectionality, of the CSI1/POM2
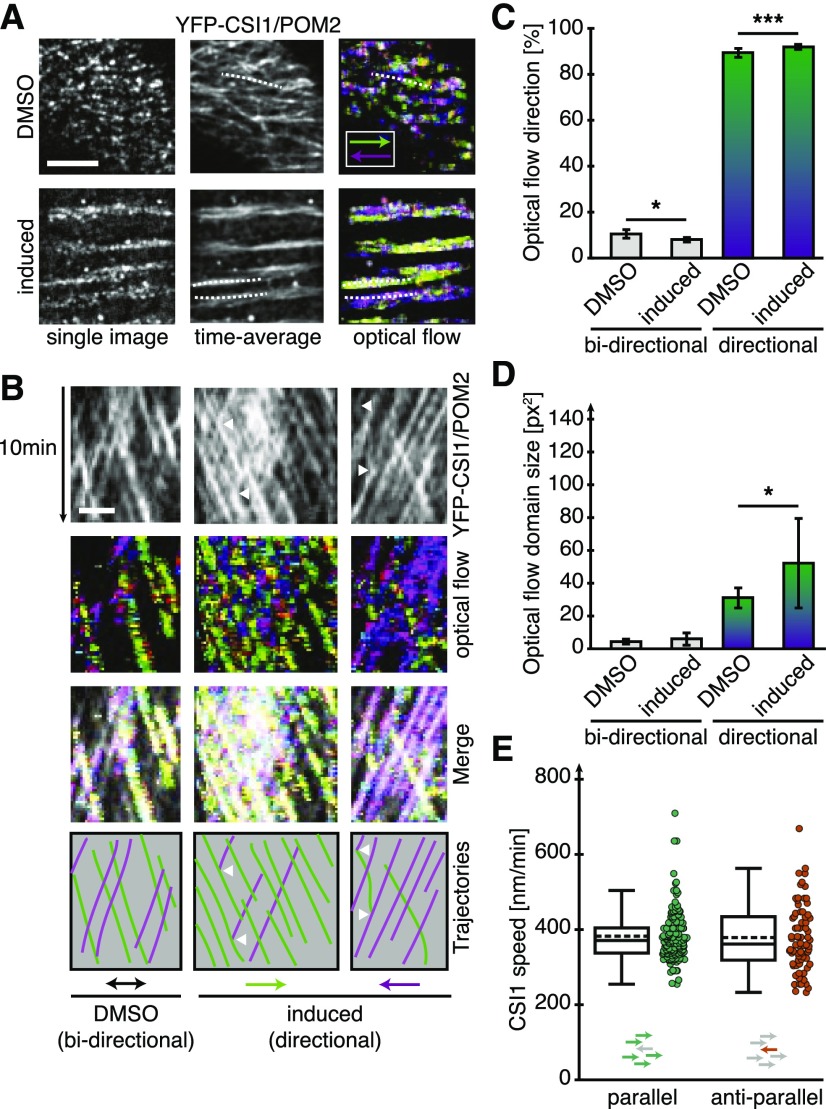
To assess, in more detail, the migratory patterns of CSI1/POM2 during xylem vessel development, we analyzed the behavior of the 3xYFP-CSI1/POM2 using optical flow analyses (Supplemental Figures 5A to 5C). This analysis can examine the patterns of apparent motion and size of fluorescent objects. We false-colored the motion of fluorescent objects based on direction, i.e., movement to the left or right were colored purple and green, respectively (Figure 4A). We detected clear bidirectional movement of the 3xYFP-CSI1/POM2 objects in both DMSO and VND7-induced cells, i.e., the YFP-CSI1/POM2 trajectories clearly overlapped along kymograph sections (Figure 4B), and the average optical flow images contained a significant number of white pixels (Figures 4B and 4C). However, domains of apparent unidirectional movement were significantly larger in the cells undergoing xylem differentiation (Figure 4D). These data indicate that the flow of the CSI1/POM2, and therefore most likely also the CSCs, is preferentially bidirectional on a cellular scale, but unidirectional on a local scale. Hence, in regions where one CSI1/POM2 migrated in a defined direction, the majority of the associated CSI1/POM2s were likely to follow the same direction (Figures 4B and 4C). To see if the movement of the 3xYFP-CSI1/POM2 foci depended on whether they were part of uni- or bidirectional domains, we measured the speeds of the foci from the different domains. We found that the CSI1/POM2 migrated with similar speeds independent of being part of a bi- or unidirectionally moving domain (Figure 4E). These data indicate that the secondary wall cellulose is preferentially produced in one direction at any given subregion of the cell wall bands.
Figure 4.
CSI1/POM2 Moves in Local Directional Patches along MT Bands during Xylem Vessel Development.
(A) Single, time-averaged, and time-averaged optical flow images of 3xYFP-CSI1/POM2 time-lapse series in DMSO-treated cells (upper panels) and in mid stages of secondary wall synthesis (lower panels). Inset: Color code for movements to the left and right. Overlapping bidirectional movements are displayed in white. Bar = 5 μm.
(B) Kymographs drawn along the dotted lines in (A) reveal predominantly bidirectional movement of individual CSI1/POM2s in DMSO-treated cells (left column) but local unidirectional movement of large patches of CSI1/POM2 toward the right (green, middle column) or the left (purple, right column) in induced cells. Individual CSI1/POM2 foci that moved against the dominant direction of the patch were able to change direction (arrowheads). Bar = 2 μm.
(C) Fraction of pixels in the time-averaged optical flow images that represent bidirectional (white pixels in [A]) and local unidirectional (green and purple pixels in [A]) movement for DMSO-treated and induced cells.
(D) Size of bi- and unidirectionally moving CSI1/POM2 patches in DMSO-treated and induced cells.
(E) Box plot of CSI1/POM2 speeds moving in groups with each other (parallel, left) or against the group (anti-parallel, right). Statistical significance was tested by Welch’s unpaired t test: *P < 0.05, **P < 0.005, and ***P < 0.0005.
We observed many 3xYFP-CSI1/POM2 foci that first moved in one direction but suddenly stopped and changed direction (white arrows in Figure 4B). Such events were detected only in xylem vessel-differentiating cells and not in the DMSO-treated cells. These observations suggest that CSI1/POM2 can be transferred between CSCs moving in opposite directions during secondary wall synthesis, perhaps to support tight associations between the secondary wall CSCs and underlying MTs.
Mutations in CSI1/POM2 Cause Misalignments of Secondary Wall CesAs and Microtubules
Lesions in CSI1/POM2 caused defects in the alignment of primary wall CesA trajectories and cortical MTs (Bringmann et al., 2012; Li et al., 2012). To investigate whether defects in CSI1/POM2 also affected the alignment of the secondary wall CesAs and the MTs, we generated YFP-CesA7 mCh-TUA5 dual-labeled VND7-inducible lines in wild-type or pom2-4 mutant backgrounds. The YFP-CesA7 trajectories coaligned with cortical MTs during all stages of xylem vessel development in the wild-type background (Supplemental Figure 6). However, we observed clear defects in the alignment of CesA7 trajectories and MTs in pom2-4 mutant cells (Figures 5A to 5F; Supplemental Movie 2). Notably, substantial misalignment was observed only during early stages of secondary wall synthesis (Figure 5D). These observations were corroborated by quantification of YFP-CesA7 and mCh-TUA5 colocalization, revealing a significant reduction in CesA7 overlap with MTs during the early developmental stage, but not during subsequent stages, in pom2-4 compared with the wild type (Figures 5E and 5F).
Figure 5.
Defects in CSI1/POM2 Cause Misalignment of YFP-CesA7 Trajectories and Cortical Microtubules during Early Stages of Xylem Vessel Development.
(A) Time-averaged images of YFP-CesA7 mCh-TUA5 in VND7-induced wild-type (upper panel) and pom2-4 mutant (lower panel) seedlings during early stages of secondary wall synthesis. The YFP-CesA7 trajectories (dashed lines) fail to align with the MTs in the pom2-4 mutant (lower panel). Bar = 5 µm.
(B) Enlarged regions of yellow, dashed rectangles in (A) for wild type (upper panel) and the pom2-4 mutant (lower panel). Prominent YFP-CesA7 trajectories (dashed lines) are highlighted in the MT and YFP-CesA7 images. Schematic illustrations of MTs (lilac lines) and CesA7 trajectories (green lines) in the wild type and pom2-4 reveal clear misalignment as indicated by the white arrow heads (right column). Bar = 5 µm.
(C) Kymographs along the yellow, dashed lines in (B) confirming linear movement of YFP-CesA7 along the selected lines. Bar = 2 µm.
(D) Misalignment between YFP-CesA7 trajectories and MTs during early, mid, and late stages of secondary wall synthesis. Note: Pronounced misalignment is only present during early stages of secondary wall synthesis.
(E) and (F) Quantification of colocalization of YFP-CesA7 and MTs in VND7-inducible wild-type and the pom2-4 mutant background using Mander’s coefficient describing the intensity overlap of MTs with YFP-CesA7 (upper panel) and YFP-CesA7 with MTs (lower panel).
(G) YFP-CesA7 speeds during early, mid, and late stages of secondary wall synthesis in wild-type and pom2-4 mutant background, respectively. Statistical significance was tested by Welch’s unpaired t test: *P < 0.05, **P < 0.005, and ***P < 0.0005.
To further assess whether defects in CSI1/POM2 influenced the behavior of the secondary wall CSC, we measured insertion rates and speeds of YFP-CesA7 in either VND7-induced wild-type or pom2-4 mutant backgrounds. We found that the insertion times of YFP-CesA7 in pom2-4 were not significantly different from the wild type (Supplemental Figures 4C and 4D; 84 ± 27 s, 39 bands in 6 cells in 5 seedlings). By contrast, we observed changes in the distribution of YFP-CesA7 speeds during the progression of secondary cell wall synthesis (Figure 5G). In the pom2-4 mutant background, YFP-CesA7 moved with higher speeds during early stages of secondary wall synthesis, but showed significantly slower speeds than the wild type during mid stages. In addition, during late secondary wall synthesis stages, the YFP-CesA7 speeds appeared less tightly controlled in the pom2-4 mutant compared with the wild type. These findings indicate that CSI1/POM2 is involved in regulating the speed of secondary wall CesAs, possibly by maintaining the CesAs in close vicinity of the MTs.
Xylem Vessel Cell Wall Patterns Can Be Maintained in the Absence of Microtubules
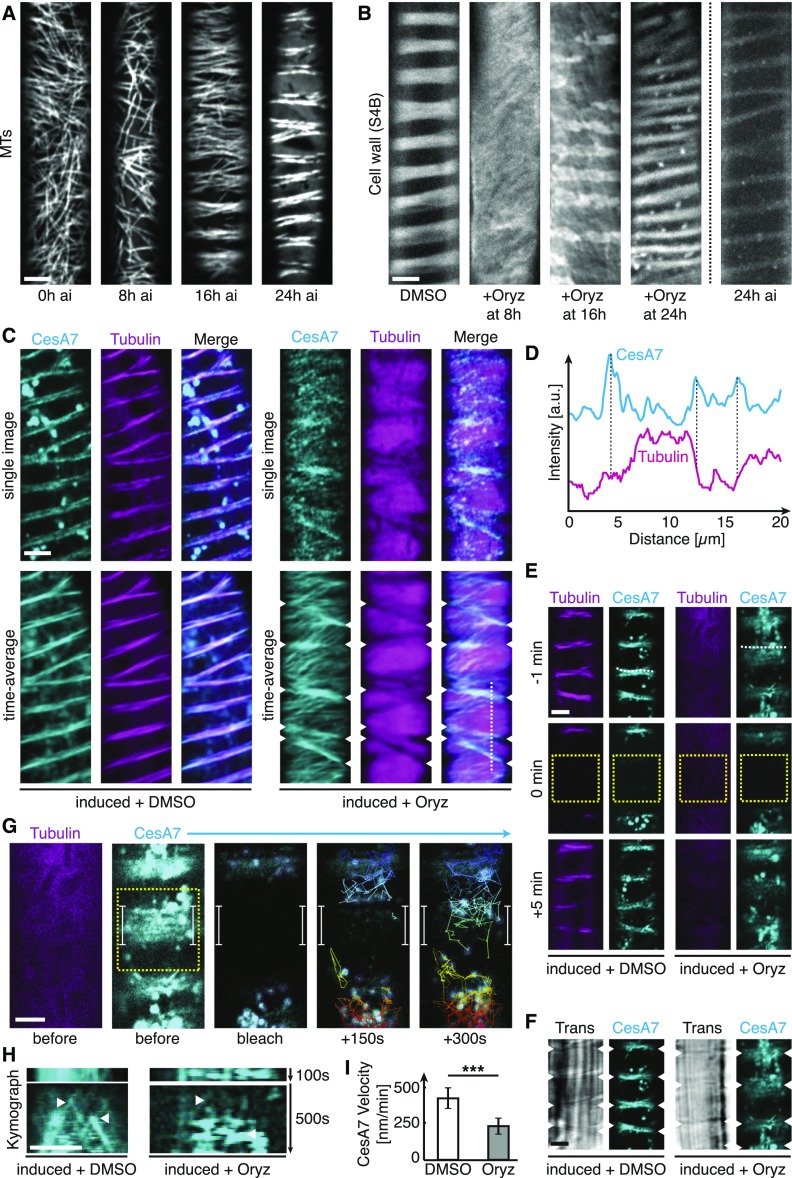
CSI1/POM2 is regarded as the component that guides CSCs along cortical MTs during primary wall synthesis (Bringmann et al., 2012; Li et al., 2012). The observation that the CSI1/POM2 is not essential for alignment of the CSCs and MTs during the mid and late stages of xylem vessel development indicated that these stages do not depend on MT-based guidance to maintain cell wall patterning. To test this conclusion, we first established time points when the MT array was reorganized during VND7-induced xylem vessel formation. In our hands, diffuse bands of MTs were not established until around 16 h after the VND7 induction (Figure 6A), and the bands became progressively more condensed during the subsequent 8 h. To assess the influence of MTs on cell wall pattern maintenance, we treated VND7-induced seedlings with the MT-depolymerizing drug oryzalin (Morejohn et al., 1987; 20 µM) at different time points after induction, and then investigated the ensuing wall patterns 48 h after VND7 induction. Seedlings treated with oryzalin 8 h after induction lacked cell wall bands entirely. By contrast, cell wall bands were evident in seedlings treated with oryzalin 16 and 24 h after VND7 induction (Figure 6B, third and fourth image from left). These wall bands were not as well defined and evenly spaced as the control seedlings (DMSO-treated; Figure 6B, left image). However, when comparing the wall patterns with the typical MT array organization after 16 and 24 h VND7 induction (Figure 6A, third and fourth images), the 16 and 24 h wall bands showed very similar distributions (Figures 6A and 6B). In addition, the wall bands in the oryzalin-treated seedlings (treated 16 and 24 h, and imaged at 48 h, after VND7 induction) were substantially more pronounced compared with the wall patterns in seedlings at 24 h after VND7 induction (Figure 6B). These data indicate that the xylem vessel wall patterns become reinforced despite removal of the MT array.
Figure 6.
Secondary Wall Patterning Can Be Maintained also in the Absence of MTs.
(A) Representative images of mCherry-TUA5-labeled MT arrays at 0, 8, 16, and 24 h after VND7 induction. Bar = 5 μm.
(B) Representative images of cellulose-stained secondary walls of VND7-induced seedlings 48 h after induction. We applied DMSO or 20 μM oryzalin 8, 16, and 24 h after induction until the final imaging at 48 h after induction to the seedlings. As comparison, a representative image of the secondary wall 24 h after induction is shown on the right. Bar = 5 μm.
(C) Single and time-averaged images of dual-labeled YFP-CesA7 (cyan) and mCherry-TUA5 (magenta) after being treated with DMSO (left panel) or 20 μM oryzalin (right panel) for 4 h. Arrowheads indicate YFP-CesA7 trajectories still localized to distinct bands despite complete depolymerization of the cortical MT array. Bar = 5 μm.
(D) Intensity cross section along dotted line in (C). YFP-CesA7 fluorescence accumulates at distinct bands (indicated by dashed lines) where mCherry-TUA5 fluorescence is absent.
(E) Single images of YFP-CesA7 and mCherry-Tubulin before, during, and after photobleaching a rectangular region (dotted box) in absence (left) and presence (right) of oryzalin (40 µM for 2 h). Bar = 5 µm.
(F) Trans-illumination image in absence (left) and presence (right) of oryzalin showing overlap of secondary wall thickenings and YFP-CesA7 bands (arrowheads).
(G) Fluorescence recovery of secondary wall bands after photobleaching of YFP-CesA7. Bright objects (Golgi, purple circles) were tracked and found accumulating at and moving quickly between the previously bleached bands (see colored tracks coding for the vertical position of the Golgi). Bar = 5 µm.
(H) Kymographs along the dotted lines in (E) show the insertion of individual CesAs to the secondary cell wall band (left) also in absence of MTs (right). Bar = 5 µm.
(I) Velocity of CesA7 inserted into bands in the presence or absence of MTs. Statistical significance was tested by Welch’s unpaired t test: *P < 0.05, **P < 0.005, and ***P < 0.0005.
Secondary Wall CesAs Remain Preferentially Delivered to Sites of Secondary Cell Wall Bands in Absence of Microtubules
To determine the behavior of the CSCs and the CSI1/POM2 in the absence of MTs during the VND7 induction, we used the dual-labeled YFP-CesA7 mCherry-TUA5 and 3xYFP-CSI1/POM2 mCherry-TUA5 lines. We treated the seedlings with oryzalin for 4 h after 24-h VND7 induction, confirming effective MT depolymerization, and assessed the behavior of the YFP-tagged proteins. While some YFP-CesA7 puncta clearly were not associated with distinct bands, many were, despite complete depolymerization of MTs (Figure 6C). These observations were confirmed with fluorescence intensity values along transects from time average images (Figures 6C and 6D). Similar observations were made using the 3xYFP-CSI1/POM2 (Supplemental Figure 7), indicating that the protoxylem vessels need MTs to establish the wall patterns but that the patterns can be maintained in the absence of MTs.
To investigate the dynamic behavior of the secondary wall CSCs in more detail, we first looked at the behavior of YFP-CesA7-containing Golgi bodies. We observed that the Golgi moved erratically at the cell cortex and that they preferentially associated with regions that coincided with MT bands (Supplemental Movie 3). We next investigated the behavior of the Golgi in cells where MTs had been depolymerized by oryzalin treatment. Golgi followed very similar patterns of movement, i.e., they preferentially populated regions where MT bands had been before the oryzalin treatment (Figures 6E to 6G; Supplemental Movie 4). To confirm these observations, we analyzed the number of Golgi localized to wall bands versus gaps using TrackMate. We found that 85% ± 16% (mean ± sd, n = 7 bands in 3 cells) of the Golgi were localized beneath wall bands, with the rest localized to gaps, in the presence of MTs. After oryzalin treatment 75% ± 18% of the Golgi were localized beneath wall bands. This difference was not significant (P = 0.39, Welch’s unpaired t test), indicating that Golgi movement is independent of MT bands. Golgi positions typically correspond to sites of delivery of CesAs (Crowell et al., 2009; Sampathkumar et al., 2013). To see if CesAs were inserted to the plasma membrane mainly above areas with Golgi movement, we applied FRAP and assessed how the YFP-CesA7 signal repopulated the bleached area. Indeed, the CesAs were preferentially inserted at regions where the bulk of Golgi was evident and, thus, in proximity of the wall bands both in presence and absence of MTs (Figures 6E to 6G). To investigate if the delivered CesAs moved in any direction after delivery, or if they followed the tracks of previous CesAs, we analyzed the CesA behavior at the plasma membrane. While we found that the YFP-CesA7 foci moved slower in the absence of MTs compared with cells with MTs (Figures 6H and 6I), the majority of CesAs moved parallel to the cell wall bands. In all, 88% ± 4% and 93% ± 8% of the newly inserted CesAs moved parallel to cell wall bands before and after oryzalin treatment, respectively (means ± sd, 101 and 35 CesAs in 3 cells; Figure 6F). Given the Golgi behavior, our data indicate that the cellular regions where cell wall bands are made are different in their molecular composition compared with interband regions.
Defects in CSI1/POM2 Affect Patterning of Cell Walls in Rice Xylem Vessels
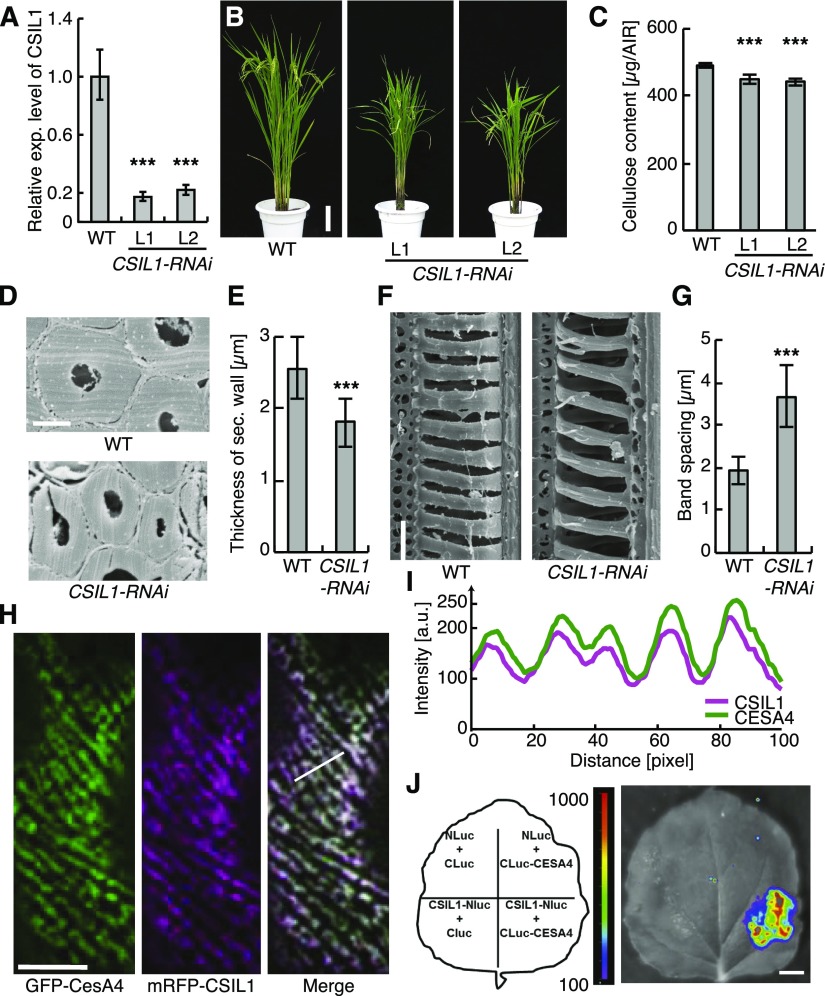
The importance of CSI1/POM2 in cellulose synthesis has been supported by data from only Arabidopsis. To see if the protein is also important for secondary wall synthesis in other plant species, we investigated the role of CSI1/POM2 in xylem vessel wall formation in rice. CSI1/POM2 was in part discovered based on coexpression of the corresponding gene with the primary wall CesA genes in Arabidopsis (Gu et al., 2010). We therefore explored what rice CSI1/POM2 homolog displayed the closest coexpression with the rice primary and secondary wall CesAs using FamNet (Ruprecht et al., 2016). We found that the most likely candidate for this function was Os06g11990, which we referred to as CSI-like 1 (CSIL1; Supplemental Figure 8A). These data were corroborated by phylogenetic analyses, which revealed that the rice CSIL1 was closely related to the Arabidopsis CSI1/POM2, and through expression analyses that showed ubiquitous expression of the gene and very low expression of the other CSIL genes (Supplemental Figures 8B to 8D and Supplemental Data Set 1). To assess whether this protein affects rice growth, we generated RNAi-mediated suppression constructs to downregulate CSIL1. Several independent homozygous T3 progeny of the transformants had substantially decreased CSI1L transcript abundance, as estimated by quantitative RT-PCR (Figure 7A), and showed stunted growth with reduced cellulose content (Figures 7B and 7C). Notably, when estimating the secondary cell wall thickness, we found that the CSIL1 RNAi plants had considerably thinner walls compared with control plants (Figures 7D and 7E). While these effects were more pronounced than what we observed in Arabidopsis, they clearly support a function of CSIL1 in secondary wall synthesis in rice. In addition, when we assessed the xylem vessel wall patterns, we found that the spacing between the bands was significantly changed (Figures 7F and 7G). These changes were in close agreement with the phenotypes we observed in the Arabidopsis pom2-4 and pom2-8 mutants (Figure 1).
Figure 7.
Rice CSIL1 Can Interact with Secondary Wall CESA4 and Affects Secondary Wall Cellulose Synthesis.
(A) RT-qPCR analysis of CSIL1 expression in rice wild-type and CSIL1 RNAi plants. The relative expression level was estimated by normalizing the expression of CSIL1 with that of TP1 (control). Mean ± sd (n = 3).
(B) Four-month-old wild-type and two lines of CSIL1 RNAi transgenic plants. Bar = 12 cm.
(C) Quantification of the cellulose content in the internodes of wild-type and RNAi plants. Mean ± sd (n = 4; biological replicates by using alcohol-insoluble residues from eight plants).
(D) Scanning electron micrographs of the sclerenchyma cells in cross sections of matured wild-type and CSIL1 RNAi internodes. Bar = 4 μm.
(E) Quantification of the thickness of sclerenchyma cell walls shown in (D). Mean ± sd (n = 40 cells from five plants).
(F) Scanning electron micrographs of tracheary elements in wild-type and the CSIL1 RNAi internodes. Bar = 10 μm.
(G) Quantification of the band space between wall spirals shown in (F). Mean ± sd (n = 218 bands from five plants).
(H) GFP-CESA4 and mRFP-CSIL1 are colocalized at foci at the plasma membrane of N. benthamiana epidermal cells. Bar = 10 μm.
(I) Intensity plot of GFP-CESA4 and mRFP-CSIL1 from transects in ([H], right panel).
(J) Split-luciferase complementation assay showing interactions between CESA4 and CSIL1. Bar = 1 cm. Statistical significance was tested by Welch’s unpaired t test: *P < 0.05, **P < 0.005, and ***P < 0.0005.
To assess if the CSIL1 can also interact with the rice secondary wall CesAs, we performed split-luciferase assays of the rice secondary wall CesAs, i.e., CesA4, CesA7, and CesA9, and the CSIL1. All the rice secondary wall CesAs could interact with the CSIL1 (Figure 7J; Supplemental Figure 8E), corroborating a function of CSIL1 in rice secondary wall cellulose production. In addition, transient coinfiltration of mRFP-CSIL1 and GFP-CesA4 into Nicotiana benthamiana leaves revealed tight colocalization of the proteins at plasma membrane focal planes (Figures 7H and 7I). The patterns of colocalization were reminiscent of cortical MTs, supporting a related function of the rice CSIL1 and the Arabidopsis CSI1/POM2. Hence, we conclude that CSI1/POM2 also contributes to xylem vessel patterns in rice.
DISCUSSION
Cell wall patterning has been attributed to MT-based guidance of CSCs (Oda and Fukuda, 2013; Schneider et al., 2016). While the guiding principles have been largely resolved for primary cell wall cellulose synthesis, the corresponding mechanisms for secondary wall deposition have remained ill defined. We show that secondary wall patterning depends on MT-based cell wall deposition. However, once the wall patterns are established, they can also be sustained in the absence of MTs, as hypothesized in Zinnia elegans cell suspensions (Roberts et al., 2004). The reorganization of the MT array therefore represents a critical initial establishment phase for the xylem vessel bands to form, whereas the patterns can be maintained in the absence of MTs during the subsequent phases.
Several MT-associated proteins have been implicated in the MT reorganization during xylem vessel development, including MAP65-1, AIR9, MAP70-1, and MAP70-5 (Pesquet et al., 2010; Derbyshire et al., 2015). These proteins contribute to the MT-bundling/stabilization and are important to achieve the MT array rearrangements during secondary wall synthesis. In addition, several small GTPases, MICROTUBULE DEPLETION DOMAIN1, a KINESIN 13A, and a recently described member of the IQD family (IQD13) are involved in depleting MTs from the areas between thickenings (Oda and Fukuda, 2012; Sugiyama et al., 2017). Nevertheless, the mechanism for how the MTs guide cellulose synthesis during this important developmental process has remained elusive. Our results indicate that CSI1/POM2 is necessary for MT-based CSC guidance during the initial phase of xylem vessel development but that it is not needed during the subsequent stages. Time-course experiments using oryzalin corroborate that cell wall patterns, and the tracking of CSCs along defined bands, can be maintained also in the absence of MTs. These data indicate that other mechanisms, perhaps cell wall-mediated CSC guidance, may play significant roles during these stages. It is plausible that xylans and/or other cell wall constituents that are deposited along the MT bands may serve this function in the absence of CSI1/POM2. Computational modeling and NMR experiments suggest a tight interplay between xylans and cellulose microfibrils (Busse-Wicher et al., 2014; Simmons et al., 2016). Such interactions could influence the direction of the CSCs and therefore cause them to successively align along the secondary wall bands. Apart from potential cell wall polymers directing cellulose synthesis, the observation that Golgi movement and CesA delivery are different at regions that underlie cell wall bands indicates that other cellular features may also influence the cell wall band progression. We speculate that the membrane environment is different in these regions compared with membrane regions that lie between the bands and that these differences influence the movement of the Golgi bodies and, thus, the delivery of CesAs. Another possibility is that the movement of the Golgi is restricted due to physical constraints. The secondary wall bands lead to deformation of the plasma membrane, i.e., the membrane is slightly indented below the bands. These deformations could make it difficult for Golgi to pass through these regions and perhaps could trap Golgi beneath the bands. While there is much left to explore about this process, we find it unlikely that the maintenance of the band patterning is solely due to the cell wall polymers directing cellulose synthesis.
The primary wall CSCs typically track with a uniform speed and bidirectionality along cortical MTs (Paredez et al., 2006). The speed of the primary wall CSCs is reliant on CSI1/POM2 function, as CSC speeds were significantly reduced in csi1/pom2 mutants (Gu et al., 2010). During the secondary wall synthesis, we also see clear alterations in CesA7 speeds in pom2-4; however, these changes appear to be largely due to a wider spread of speeds rather than a uniform reduction and are tied to particular developmental stages of xylem vessel development. The increased variance in the CSC speeds was primarily observed during the mid stages of secondary wall synthesis (Figure 5G). While we observed major misalignment between the MTs and CesA7 trajectories only during the early stages, it is possible that the lack of direct engagement of the CSCs with the MTs causes difficulties in maintaining the speeds. It is worth highlighting that although we observed clear secondary wall bands in the csi1/pom2 lines, these bands were less well ordered compared with the wild type. We speculate that CSI1/POM2 proteins provide a feedback function to the formation of MT bands and that this may be compromised in the csi1/pom2 lines, which in turn may affect the final cell wall band patterns. This is in line with observations during primary wall synthesis where MT organization is perturbed when CSI1/POM2 is mutated (Bringmann et al., 2012; Landrein et al., 2013). If the CSCs are indeed also guided by cell wall components and membrane environment during this development stage, as discussed above, it is plausible that such guidance is not optimal and that it can manifest in changes in the quality and quantity of cellulose that is produced. These data are in agreement with our recorded changes in the MFA, cellulose microfibril crystallinity, and amorphous cellulose in the pom2-4 mutant.
The CSI1/POM2 speeds changed during the different stages of xylem vessel development. For example, the speeds significantly increased during the mid stages of transition (Figure 2H). These data are very similar to what has independently been reported for the secondary wall CesA7 (Watanabe et al., 2015), but contrast those of Li et al. (2016b). Assuming that the speed of the CSCs represents catalytic activity, these findings support a scenario in which an increase in speed of tracking and CesA abundance leads to a major boost in cellulose synthesis, which is compatible with the rapid development and subsequent death of the xylem vessels. Li et al. (2016b) also studied secondary wall CesA behavior in the VND7-inducible system and concluded that the CSCs moved unidirectionally as “swarms” (referred to as “directionally coherent movement”; Li et al., 2016b) during xylem vessel development. Our data support this report, but it is important to note that the unidirectional movement apparent in CSI1/POM2 was observed both during primary wall synthesis (DMSO treatment; Figure 4) and xylem vessel development and appears to depend on local versus global cell wall synthesis.
In summary, CSI1/POM2 directs xylem vessel patterning by coordinating the secondary wall CSCs and MTs during the transition from primary to secondary wall synthesis. However, the banding patterns can largely be maintained in absence of CSI1/POM2 and MTs during later stages of development. We therefore conclude that the wall patterning during proto-xylem development is initiated and more importantly sustained by two complementary mechanisms.
METHODS
More detailed descriptions of some procedures are provided in Supplemental Methods.
Plant Material
We used the previously described Arabidopsis thaliana lines pom2-4 and pom2-8/csi1-1 (SALK_136239; Bringmann et al., 2012). To generate multiple marker lines in the VND7 background, we crossed seeds of pom2-4 and native promoter-driven triple (3x)YFP-CSI1/POM2 (Worden et al., 2015) into the VND7-inducible Arabidopsis line proCaMV35s::VND7::VP16::GR (Yamaguchi et al., 2010). The F3 progeny was used for analysis. The pom2-4 mutant was used as the main allele, as it produces seeds more readily than some of the more severe csi1/pom2 lines. Note that the pom2-4 was generated from a T-DNA population between Nossen and Columbia. We have backcrossed the pom2-4 extensively to Col-0 and we used segregating progeny from crosses of pom2-4 and different markers to assure the best possible genetic homogeneity between samples. To generate dual-labeled plants for MTs, we crossed 3xYFP-CSI1/POM2 in the VND7 background into mCherry-TUA5 (Gutierrez et al., 2009) and used the F2 progeny of that cross. To visualize secondary wall CesAs, we crossed YFP-CesA7 in the irx3-4 background (Watanabe et al., 2015) into the wild-type and pom2-4-mutated mCherry-TUA5-VND7 background. We used the F2 generation for experiments. We confirmed the homozygosity of the pom2-4 mutation by growing seedlings on solid (0.8% agar) half-strength Murashige and Skoog (MS) medium (pH 5.7) supplemented with 5% sucrose allowing the identification of the obvious stunted root phenotype and confirmed via PCR (Supplemental Table 1). We further checked for the presence of YFP and mCherry markers using a fluorescence stereomicroscope prior to further treatment.
For generation of CSIL1-RNAi plants, the targeted fragments were amplified from the cDNA of rice (Oryza sativa) CSIL1 and inserted into PKANNIBAL vectors (see below). The construct was transfected into Agrobacterium tumefaciens EHA105 and introduced into the wild-type variety Nipponbare.
Plant Growth Conditions and Treatments
Arabidopsis plants were germinated and grown essentially as described by Liu et al. (2016). More specifically, seeds of the VND7-inducible Arabidopsis lines described above were surface-sterilized by washing for 10 min in 1.25% sodium-hypochlorite solution supplemented with 0.05% Tween 20. Sterilized seeds were washed excessively with sterile water. Seeds were plated on solid (0.8% agar) half-strength MS medium (pH 5.7) supplemented with 1% sucrose for normal growth and 5% sucrose for pom2-4 genotyping purposes, respectively. Plates were stratified for at least 2 d in a dark cold room (4°C). Germination was triggered by exposing plates for 8 h to light (100 µE m−2 s−1). Subsequently, the plates were wrapped with aluminum foil and placed vertically in a growth room air-conditioned to 60% relative humidity and 21°C temperatures. Seedlings used for spinning disc confocal microscopy were grown for 3 d in the dark, transferred to 24-well plates containing DMSO (control), or 10 to 100 µM dexamethasone (induction) to induce VND7. Subsequently, plates were wrapped with aluminum foil and placed on a slowly rotating orbital shaker in the growth room.
The plants used for determination of cell wall alterations were sterilized for 3 min in 70% ethanol followed by 10 min in 10% bleach, then rinsed six times in sterile distilled water, plated on solid (0.8% agar) half-strength MS plates, and stratified for 2 d in a dark cold room (4°C) before being incubated at 21°C at a 16/8-h light/dark cycle. Ten-day-old seedlings were transferred to soil and grown under the same conditions for ∼9 weeks through maturity to full senescence.
Arabidopsis cell suspension cultures were generated and tracheary element formation induced, as previously described (Pesquet et al., 2010).
Rice plants, including the wild-type plants and CSIL1-RNAi plants, were grown in experimental fields at the Institute of Genetics and Developmental Biology in Beijing and in Linshui, Hainan province during the natural growing seasons.
Nicotiana benthamiana plants were grown in soil in a glasshouse with continuous cool white fluorescent lights (100 µE m−2 s−1) and natural daylight at 20 to 26°C, as previously described (Lampugnani et al., 2016).
Live-Cell Imaging
Imaging was done essentially as described by Liu et al. (2016). Seedlings were observed under the microscope between 10 and 30 h after VND7 induction. Induced 3xYFP-CSI1/POM2 seedlings were imaged using the CSU-X1 spinning disk head (Yokogawa) mounted to an inverted Nikon Ti-E microscope equipped with a 100× oil-immersion objective (Plan Apo TIRF, NA 1.45). Fluorescence detection was achieved using an Evolve EM-CCD camera (Photometrics Technology). Induced YFP-CesA7 seedlings were imaged using the CSU-W1 spinning disk head (Yokogawa) mounted to an inverted Nikon Ti-E microscope equipped with a 100× oil-immersion objective (Apo TIRF, NA 1.49). Fluorescence detection was achieved using a deep-cooled iXon Ultra 888 EM-CCD (Andor Technology). Both setups were controlled via PC using MetaMorph (Molecular Devices). Photobleaching was achieved using either the iLas laser illumination system (Roper Scientific) or the Andor FRAPPA scanning instrument.
Seedlings were mounted on 1.5 grade glass cover slips and covered by 1-mm-thick agarose pads made from water supplemented with 1% agarose. We imaged 3xYFP-CSI1/POM2, mCherry-TUA5, and YFP-CesA7 using time-lapse recordings with typical exposure times between 200 and 400 ms, time intervals of 10 s, and total durations between 5 and 10 min. Fluorescence recovery was recorded in intervals of 2 to 5 s for 3xYFP-CSI1/POM2 and 10 s for YFP-CesA7.
For analysis of colocalization of rice CesA4-CSIL1, Agrobacterium EHA105 harboring GFP-CesA4 and mRFP-CSIL1 were coinjected into the lower epidermis of 4-week-old N. benthamiana leaves. After cultivation for two more days, the leaves were observed with oil immersed objective on the spinning-disc confocal microscope (Perkin-Elmer UltarVIEW VoX). To obtain the GFP and mRFP fluorescence images, the 488- and 561-nm lines of laser were used for excitation, and emission was detected at 500 to 540 nm and 600 to 640 nm, separately.
Scanning Electron Microscopy
For xylem defect analysis, the first internodes of 8-week-old wild-type, pom2-4, and pom2-8 (csi1-1) mutant plants were cut into longitudinal sections and immediately fixed in 2.5% glutaraldehyde in PBS buffer for 30 min. Sections were washed three times in PBS and subsequently three times in water. Dehydration was achieved by washing the sections for minimum 1 h each in an ethanol series from 10% to 100% in 10% steps. After several washes with 100% ethanol, critical point drying was performed and the dried samples were gold-coated. Examination of the samples was performed using an XL30 field-emission scanning electron microscope from Phillips.
The second internodes of mature wild-type and CSIL1-RNAi plants were fixed in 4% paraformaldehyde (Sigma-Aldrich). To view the wall thickness of sclerenchyma cells, the internodes were transversely cut to expose the epidermal sclerenchyma cells. To observe the secondary pattern of vessel cells, the internodes were longitudinally cut under the stereoscope. After critical-point drying, the samples were sprayed with gold particles and observed with a scanning electron microscope (S-3000N; Hitachi).
Cell Wall Staining
To label the secondary walls in VND7-induced wild-type and pom2-4 mutants, DirectRed23 (Anderson et al., 2010; Sigma-Aldrich) was added to a final concentration of 0.06% to six-well plates containing 3-d-old seedlings 24 h after induction. The samples were washed with ultrapure water to reduce the amount of unbound dye. Subsequently, samples were observed under the spinning disk microscope by recording z-stacks using a 561-nm laser and 610/40-nm emission filters.
Image Analyses
The velocity of CSI1 foci was measured using the open-source software FIESTA (Ruhnow et al., 2011). Briefly, the velocity of moving foci is determined by measuring their slope in kymograph projections. We measured 1015 trajectories for noninduced cells and 905, 1391, and 367 trajectories for induced cells in early, mid, and late stages of the secondary wall program, respectively. Colocalization of CSI1 with MTs and CesA7 was measured using the JaCoP plug-in of Fiji. To increase the reliability of the colocalization measurements, we used a dual approach of measuring Pearson’s and Mander’s coefficients. Furthermore, we used Costes randomization to validate the significance of the determined Pearson’s coefficients. Costes-randomized image series always had a Pearson’s coefficient of at least a factor of 50 lower than the original image series.
To quantify the insertion rate of 3xYFP-CSI1 and YFP-CesA7 after photobleaching, we used the ThunderSTORM plug-in of Fiji to detect the appearance of foci in the bleached areas. We analyzed the recovery in areas slightly smaller than the bleached area to avoid migrating complexes in the plasma membrane to be included in the recovery signal. We plotted the number of detected foci over time and analyzed the recovery using a monoexponential growth model (reaction-limited case).
The misalignment between CesA trajectories and MTs was measured in dual-color average projections of the time series using Fiji. We measured the angle of short stretches of clearly visible CesA7 trajectories and compared them to the angle of the underlying MTs. For each cell, at least 10 trajectory-MT pairs were measured.
Orientation and spacing of secondary wall bands in VND7-induced seedlings were measured using Fiji. Z-stacks were smoothed and average-projected using inbuilt Fiji plug-ins. Subsequently, individual cells were cropped and aligned with the growth axis of the seedling. A total of 136 and 132 VND7-induced cells were captured for wild-type and the pom2-4 mutant, respectively. We quantified the average orientation of secondary wall bands and the variability of band orientations within each cell, termed dispersion, using the Fiji plug-in “Directionality” with default settings. Band spacing was analyzed using a custom-made Matlab (Mathworks) program. Briefly, the program displayed the intensity profile along the long axis of the cell and a graphical user interface subsequently allowed for the determination of band positions in a point-and-click manner.
Optical Flow Analysis
The optical flow was analyzed using the Fiji plug-in PIV analyzer using 4-by-4 pixel averaging, interpolation, and a mask of 0.1. The image series were preprocessed using subtract background (50 pixel sliding paraboloid) and four-frame walking averaging. The resulting optical flow image series was average projected to obtain images displaying the mean optical flow of intensity. The direction of the optical flow was determined using Fiji by decomposing the mean optical flow images into hue (H), saturation (S), and brightness (B) with the following thresholds: for movement to the right (H between 34 and 94, S between 50 and 255, and B between 1 and 255), for movement to the left (H between 161 and 221, S between 50 and 255, and B between 1 and 255), for movement into both directions (H between 0 and 255, S between 0 and 50, and B between 1 and 255).
Biochemical Analyses
The MFA of at least 18 Arabidopsis stems from VND7 and the pom2-4 mutant in VND7 was measured using an x-ray diffraction technique (Ukrainetz et al., 2008). The bottom 3 cm of mature, senesced plant stems was used for analysis. The 002 diffraction spectra of each stem were screened for T-value distribution and symmetry on a Bruker D8 discover x-ray diffraction unit equipped with an area array detector (GADDS). Wide-angle diffraction was used in the transmission mode, and measurements were made with CuKα1 radiation (λ = 1.54 Å). The x-ray source was fit with a 0.5-mm collimator and a GADDS detector collected the scattered photons. The x-ray source and the detector were both set at a theta angle of 0°. The diffraction data were integrated using GADDS software and further analyzed to estimate MFA values.
Cell wall crystallinity was determined on the same stems used for measuring MFA, using the same x-ray unit and parameters as the MFA measurements, except the source theta was set at 17°. The diffraction data were integrated using GADDS software and the output data further analyzed using a crystallinity calculation program based on the Vonk method (Vonk, 1973).
Cellulose Content
After x-ray data collection was complete, the same stems were then pooled by genotype and ground on a Thomas Wiley Mini Mill to pass through a #60 mesh (250 µm). The powdered sample was then dried for 24 h at 50°C and 15 mg of tissue was weighed into each preweighed tube. At least three technical replicates were done on each pooled genotype. First, the alcohol insoluble residue (AIR) was prepared as described by Pattathil et al. (2012). The AIR was then subjected to a series of extractions in a procedure modified from the AIR fractionation method also described by Pattathil et al. (2012). The modifications involved completing the chlorite extraction first as well as the removal of both the 1 M and the postchlorite 4 M potassium hydroxide extractions. The resulting cellulose residue was then predried in a vacuum centrifuge and finished in a 50°C oven for 48 h before the final weights were measured.
Degree of Polymerization
The resulting cellulose was dissolved at 5 mg/mL in 9% lithium chloride (LiCl)/N,N-dimethylacetamide (DMAc) through a four-step solvent exchange of nanopure water, anhydrous ethanol, DMAc, and 9% LiCl/DMAc. Once dissolved, the samples were diluted to 0.5 mg/mL cellulose in 0.9% LiCl/DMAc and each was separated on an Agilent 1100 SEC system containing Waters Styragel HR4 and HR6 columns coupled to a Wyatt Dawn Heleos II light scattering detector. The average molecular weight was calculated from the output using Wyatt’s Astra 6 software before converting to degree of polymerization.
Expression and Phylogenetic Analyses
Rice CSI1 homologous genes were identified based on the annotation of the rice genome database (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/). The phylogenetic tree of CSI1 and its like proteins in rice and Arabidopsis was generated using maximum likelihood with the MEGA5 software with 1000 bootstrap replications (Supplemental Data Set 1; Tamura et al., 2011). The spatio-temporal expression profiles of rice CSIL1 were from the expression data in RiceXPro database (http://ricexpro.dna.affrc.go.jp/).
To examine the expression of CSIL1 in the wild-type and transgenic plants, total RNA was extracted from young internodes using the Plant RNA Purification Reagent (Invitrogen), and cDNA was synthesized from RNA using the Reverse Transcription system kit (Takara). The expression level of CSIL1 was examined by qPCR with a CFX96 real-time system (Bio-Rad) using rice HNR as internal control. The primers for the RNAi construct and qPCR analyses are listed in Supplemental Table 1.
Constructs
The CSIL1-RNAi construct was generated by amplifying CSIL1 from a rice cDNA library, and the cDNA was inserted into a PKANNIBAL vector (Wesley et al., 2001) using BamHI and XbaI. The construct was transformed into the wild-type variety Nipponbare ecotype using Agrobacterium. The expression level of CSIL1 was quantified by qPCR with a CFX96 real-time system (Bio-Rad). Coding sequences of Arabidopsis EH1, CSI1/POM2, CesA4, CesA7, and CesA8 were amplified from cDNA and cloned into pAMON and pSUR using the Gibson assembly method to generate N-terminal fusions to VN155 (I152L) and VC155, respectively (Lee et al., 2014), for BiFC analyses (see below). All primers are listed in Supplemental Table 1.
Transgenic cell suspensions were produced by coculture with Agrobacterium transformed with 35S:CSI1-RNAi construct (Derbyshire et al., 2015) as previously described by Pesquet et al. (2010). Expression levels of CSI1 were measured using RT-qPCR on five independent biological repeats (primer sequences and method described in Derbyshire et al., 2015) and expressed as percentage of CSI1-to-UBIQUITIN gene ratio. Downregulated lines showed a residual CSI1 expression of 57% ± 17% (mean ± sd) compared with wild-type cells (100% ± 13%, P < 0.005, Welch’s unpaired t test). The expression of the different CSIs and primary and secondary CesAs during the TE differentiation time course have been checked using macroarray data (GEO GSE73146; Derbyshire et al., 2015).
Interaction Analyses
Coding sequences of EH1, CSI1/POM2, CesA4, CesA7, and CesA8 were amplified from cDNA using the primers defined in Supplemental Table 1 and cloned into linearized BiFC vectors pURIL [GENE-V(I152L)N], pDOX (GENE-VC), pAMON [V(I152L)N-GENE], or pSUR (VC-GENE) using the Gibson assembly method as previously described (Lampugnani et al., 2016). The pURIL and pDOX were linearized using KpnI and SfoI, while pAMON and pSUR were linearized using BamHI and SfoI. EH1 was cloned into pURIL and pDOX, while CSI1/POM2, CesA4, CesA7, and CesA8 were cloned into pAMON and pSUR to generate C-terminal and N-terminal fusions respectively. Constructs were introduced into Agrobacterium and combinations of BiFC constructs, together with Agrobacterium strains carrying 35S::CFP-N7 (Kaplan-Levy et al., 2014) and P19 , were introduced into N. benthamiana leaves by infiltration following the procedure described by Zhang et al. (2016). Leaves were examined for fluorescence 3 d postinfiltration on an inverted Nikon Ti-E microscope equipped with a CSU-W1 spinning disk head (Yokogawa). Detection occurred using a 100× oil-immersion objective (Apo TIRF, NA 1.49) and an iXon Ultra 888 EM-CCD (Andor Technology). All BiFC combinations were imaged under the same conditions. Specifically, a 445-nm laser line was used to excite CFP, while a 515-nm laser line was used to excite YFP. Emissions were detected with 470/40 and 535/30 band-pass filters. Z-stacks of images were collected using exposure times of 100 ms.
The split-luciferase complementation assay was performed as described (Chen et al., 2008). In brief, the cDNA of CSIL1, CESA4, CESA7, and CESA9 were amplified (Supplemental Table 1) and inserted into the binary vectors for expression fused with N- or C-terminal luciferase. The resulting constructs were transfected into Agrobacterium strain C58 and infiltrated with the leaves of 4-week-old N. benthamiana. Interaction was determined based on the fluorescent signal intensity harvested by IndiGO software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CSI1/POM2, At2g22125; CesA4, At5g44030; CesA6, At5g64740; CesA7, At5g17420; CesA8, At4g18780; VND7, At1g71930; TUA5, At5g19780; EH1, At1g20760; OsCSIL1, Os06g11990; OsCesA4, Os01g54620; OsCesA7, Os10g32980; OsCesA9, Os09g25490; and GEO GSE73146.
Supplemental Data
Supplemental Figure 1. Defects in CSI1/POM2 Cause Aberrant Secondary Wall Patterns.
Supplemental Figure 2. Representative Images of Primary Wall Synthesis and Different Stages (Early, Mid, and Late) of Xylem Vessel Development.
Supplemental Figure 3. BiFC Assay Demonstrating Interactions between CSI1/POM2 and Secondary Wall CesA4, CesA7, and CesA8 Transiently Expressed in Epidermal Cells of N. benthamiana Leaves.
Supplemental Figure 4. CSI1/POM2 Recovers More Quickly after Photobleaching Than CesA7 during Xylem Vessel Formation.
Supplemental Figure 5. Schematic Workflow of Optical Flow Analyses.
Supplemental Figure 6. Secondary Wall CesA7 Tracks along Microtubules throughout All Stages of Xylem Vessel Development in Wild-Type Background, but Not in the pom2-4 Mutant.
Supplemental Figure 7. YFP-CSI1/POM2 Can Maintain Tracks along Bands in the Absence of MTs.
Supplemental Figure 8. Rice CSIL1 Is a Homolog of CSI1/POM2 and Can Interact with Secondary Wall Rice CesAs.
Supplemental Table 1. Primers used for BiFC constructs.
Supplemental Movie 1. Cellular Distribution of 3xYFP-CSI1/POM2 in Noninduced Cells and Early, Mid, and Late Stages of Secondary Wall Formation.
Supplemental Movie 2. YFP-CesA7 Trajectories Are Not Aligned with Cortical MTs in the pom2-4 Mutant during Early Stages of Secondary Wall Formation.
Supplemental Movie 3. YFP-CesA7 Quickly Recycles at MT Bands after Fluorescence Photobleaching.
Supplemental Movie 4. YFP-CesA7 Quickly Recycles to Sites of Secondary Wall Formation also in the Absence of MTs.
Supplemental Data Set 1. Multiple Protein Sequence Alignment of CSI Proteins in Arabidopsis and Rice.
Supplemental Data Set 2. ANOVA Tables.
Acknowledgments
S.P. was funded by a R@MAP Professorship at University of Melbourne. This work was in part supported by an ARC Discovery grant (DP150103495), a Future Fellowship grant (FT160100218), and the National Natural Science Foundation of China (Grant 31530051). S.D.M. acknowledges funding from the NSERC Discovery program. We thank Taku Demura for sharing the VND7-line. The P19 plasmid was a kind gift from David Baulcombe.
Footnotes
Articles can be viewed without a subscription.
References
- Anderson C.T., Carroll A., Akhmetova L., Somerville C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov I.I., Pittman J.K., Turner S.R. (2009). Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J. Biol. Chem. 284: 3833–3841. [DOI] [PubMed] [Google Scholar]
- Bringmann M., Li E., Sampathkumar A., Kocabek T., Hauser M.T., Persson S. (2012). POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse-Wicher M., Gomes T.C., Tryfona T., Nikolovski N., Stott K., Grantham N.J., Bolam D.N., Skaf M.S., Dupree P. (2014). The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 79: 492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T., et al. (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. USA 99: 15794–15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P., Ménard D., Green P., Saalbach G., Buschmann H., Lloyd C.W., Pesquet E. (2015). Proteomic analysis of microtubule interacting proteins over the course of xylem tracheary element formation in Arabidopsis. Plant Cell 27: 2709–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M., Desprez T., Guillot A., Vernhettes S., Höfte H. (2014). Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol. 166: 1709–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Kaplinsky N., Bringmann M., Cobb A., Carroll A., Sampathkumar A., Baskin T.I., Persson S., Somerville C.R. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 107: 12866–12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Somerville C. (2010). Cellulose synthase interacting protein: a new factor in cellulose synthesis. Plant Signal. Behav. 5: 1571–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Hill J.L. Jr., Hammudi M.B., Tien M. (2014). The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan-Levy R.N., Quon T., O’Brien M., Sappl P.G., Smyth D.R. (2014). Functional domains of the PETAL LOSS protein, a trihelix transcription factor that represses regional growth in Arabidopsis thaliana. Plant J. 79: 477–491. [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani E.R., Ho Y.Y., Moller I.E., Koh P.L., Golz J.F., Bacic A., Newbigin E. (2016). A glycosyltransferase from Nicotiana alata pollen mediates synthesis of a linear (1, 5)-α-L-arabinan when expressed in Arabidopsis. Plant Physiol. 170: 1962–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B., Lathe R., Bringmann M., Vouillot C., Ivakov A., Boudaoud A., Persson S., Hamant O. (2013). Impaired cellulose synthase guidance leads to stem torsion and twists phyllotactic patterns in Arabidopsis. Curr. Biol. 23: 895–900. [DOI] [PubMed] [Google Scholar]
- Lee J.E., Lampugnani E.R., Bacic A., Golz J.F. (2014). SEUSS and SEUSS-LIKE 2 coordinate auxin distribution and KNOXI activity during embryogenesis. Plant J. 80: 122–135. [DOI] [PubMed] [Google Scholar]
- Li S., Lei L., Somerville C.R., Gu Y. (2012). Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc. Natl. Acad. Sci. USA 109: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Bashline L., Zheng Y., Xin X., Huang S., Kong Z., Kim S.H., Cosgrove D.J., Gu Y. (2016b). Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc. Natl. Acad. Sci. USA 113: 11348–11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Omranian N., Neumetzler L., Wang T., Herter T., Usadel B., Demura T., Giavalisco P., Nikoloski Z., Persson S. (2016a). A transcriptional and metabolic framework for secondary wall formation in Arabidopsis. Plant Physiol. 172: 1334–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Schneider R., Kesten C., Zhang Y., Somssich M., Zhang Y., Fernie A.R., Persson S. (2016). Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev. Cell 38: 305–315. [DOI] [PubMed] [Google Scholar]
- Morejohn L.C., Bureau T.E., Molè-Bajer J., Bajer A.S., Fosket D.E. (1987). Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172: 252–264. [DOI] [PubMed] [Google Scholar]
- Myburg A.A., Lev‐Yadun S., Sederoff R.R. (2013). Xylem structure and function. In eLS, 10.1002/9780470015902.a0001302.pub2.
- Oda Y., Iida Y., Kondo Y., Fukuda H. (2010). Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr. Biol. 20: 1197–1202. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2012). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2013). Spatial organization of xylem cell walls by ROP GTPases and microtubule-associated proteins. Curr. Opin. Plant Biol. 16: 743–748. [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Pattathil S., Avci U., Miller J.S., Hahn M.G. (2012). Immunological approaches to plant cell wall and biomass characterization: Glycome profiling. Methods Mol. Biol. 908: 61–72. [DOI] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E., Korolev A.V., Calder G., Lloyd C.W. (2010). The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr. Biol. 20: 744–749. [DOI] [PubMed] [Google Scholar]
- Pesquet E., Korolev A.V., Calder G., Lloyd C.W. (2011). Mechanisms for shaping, orienting, positioning and patterning plant secondary cell walls. Plant Signal. Behav. 6: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.W., Frost A.O., Roberts E.M., Haigler C.H. (2004). Roles of microtubules and cellulose microfibril assembly in the localization of secondary-cell-wall deposition in developing tracheary elements. Protoplasma 224: 217–229. [DOI] [PubMed] [Google Scholar]
- Ruhnow F., Zwicker D., Diez S. (2011). Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 100: 2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C., Mendrinna A., Tohge T., Sampathkumar A., Klie S., Fernie A.R., Nikoloski Z., Persson S., Mutwil M. (2016). FamNet: A framework to identify multiplied modules driving pathway expansion in plants. Plant Physiol. 170: 1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A., Gutierrez R., McFarlane H.E., Bringmann M., Lindeboom J., Emons A.M., Samuels L., Ketelaar T., Ehrhardt D.W., Persson S. (2013). Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 162: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Hanak T., Persson S., Voigt C.A. (2016). Cellulose and callose synthesis and organization in focus, what’s new? Curr. Opin. Plant Biol. 34: 9–16. [DOI] [PubMed] [Google Scholar]
- Simmons T.J., Mortimer J.C., Bernardinelli O.D., Pöppler A.C., Brown S.P., deAzevedo E.R., Dupree R., Dupree P. (2016). Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat. Commun. 7: 13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Wakazaki M, Toyooka K, Fukuda H, Oda Y. (2017). A novel plasma membrane-anchored protein regulates xylem cell-wall deposition through microtubule-dependent lateral inhibition of Rho GTPase domains. Curr Biol. 27: 2522–2528. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.G., Howells R.M., Huttly A.K., Vickers K., Turner S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S., Gallois P., Brown D. (2007). Tracheary element differentiation. Annu. Rev. Plant Biol. 58: 407–433. [DOI] [PubMed] [Google Scholar]
- Ukrainetz N.K., Ritland K., Mansfield S.D. (2008). Identification of quantitative trait loci for wood quality and growth across eight full-sib coastal douglas-fir families. Tree Genet. Genomes 4: 159. [Google Scholar]
- Vonk C. (1973). Computerization of Ruland’s X-ray method for determination of the crystallinity in polymers. J. Appl. Crystallogr. 6: 148–152. [Google Scholar]
- Watanabe Y., Meents M.J., McDonnell L.M., Barkwill S., Sampathkumar A., Cartwright H.N., Demura T., Ehrhardt D.W., Samuels A.L., Mansfield S.D. (2015). Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203. [DOI] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590. [DOI] [PubMed] [Google Scholar]
- Worden N., et al. (2015). CESA TRAFFICKING INHIBITOR inhibits cellulose deposition and interferes with the trafficking of cellulose synthase complexes and their associated proteins KORRIGAN1 and POM2/CELLULOSE SYNTHASE INTERACTIVE PROTEIN1. Plant Physiol. 167: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Ohtani M., Nakano Y., Mortimer J.C., Nishikubo N., Kubo M., Katayama Y., Kakegawa K., Dupree P., Demura T. (2010). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153: 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., et al. (2016). Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 7: 11656. [DOI] [PMC free article] [PubMed] [Google Scholar]