Plants have evolved to utilize the SCD complex, which is architecturally distinct from—but performs similar functions as—the structurally related exocytic yeast and mammalian Sec2/Rabin8 Rab GEFs.
Abstract
Although exocytosis is critical for the proper trafficking of materials to the plasma membrane, relatively little is known about the mechanistic details of post-Golgi trafficking in plants. Here, we demonstrate that the DENN (Differentially Expressed in Normal and Neoplastic cells) domain protein STOMATAL CYTOKINESIS DEFECTIVE1 (SCD1) and SCD2 form a previously unknown protein complex, the SCD complex, that functionally interacts with subunits of the exocyst complex and the RabE1 family of GTPases in Arabidopsis thaliana. Consistent with a role in post-Golgi trafficking, scd1 and scd2 mutants display defects in exocytosis and recycling of PIN2-GFP. Perturbation of exocytosis using the small molecule Endosidin2 results in growth inhibition and PIN2-GFP trafficking defects in scd1 and scd2 mutants. In addition to the exocyst, the SCD complex binds in a nucleotide state-specific manner with Sec4p/Rab8-related RabE1 GTPases and overexpression of wild-type RabE1 rescues scd1 temperature-sensitive mutants. Furthermore, SCD1 colocalizes with the exocyst subunit, SEC15B, and RabE1 at the cell plate and in distinct punctae at or near the plasma membrane. Our findings reveal a mechanism for plant exocytosis, through the identification and characterization of a protein interaction network that includes the SCD complex, RabE1, and the exocyst.
INTRODUCTION
Vesicular trafficking to and from the plasma membrane is paramount to plant growth and development, as it facilitates multiple important processes including cell wall biosynthesis, nutrient uptake, hormone signaling, and pathogen defense (Takano et al., 2005; Tanaka et al., 2006; Robatzek, 2007). Similarly, in dividing cells, exocytic and endocytic trafficking pathways are essential for the formation of the cytokinetic organelle known as the cell plate.
Proper regulation and function of biosynthetic secretory and endocytic membrane trafficking pathways depend on stage-specific Rab GTPases. In their GTP-bound form, Rabs recruit divergent effectors to coordinate the formation, transport, tethering, and fusion of transport vesicles and organelles. To function properly, Rabs must continually cycle between active and inactive forms through the exchange of GTP and GDP via interactions with Rab GEFs (guanine nucleotide exchange factors) and Rab GAPs (GTPase activating proteins) (Stenmark, 2009). Thus, the association of Rab GTPases with downstream effector proteins is inherently dependent upon their interactions with GEFs and GAPs. Surprisingly, of the 57 Arabidopsis thaliana Rabs, only a few of their GEFs/GAPs and downstream effector proteins are known (Preuss et al., 2006; Goh et al., 2007; Camacho et al., 2009; Thellmann et al., 2010; Qi and Zheng, 2011; Fukuda et al., 2013; Singh et al., 2014). Therefore, despite recent advances in our knowledge of Rab function in plants, a significant gap remains in our understanding of the molecular machinery involved in their regulation.
In yeast, communication between the GEF Sec2p and the Rab GTPases Ypt32p and Sec4p establishes a functional connection between cargo-containing Golgi-derived exocytic vesicles and the molecular machinery necessary for their targeting and fusion with the plasma membrane (Ortiz et al., 2002). Specifically, Ypt32p recruits Sec2p to the trans-Golgi network which in turn binds and activates the Sec4p, which facilitates interactions with the exocyst complex, an evolutionarily conserved eight-subunit complex consisting of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84, to promote docking with the plasma membrane prior to fusion (Hammer and Sellers, 2011; Wu and Guo, 2015; Vukašinovic and Žárský, 2016). A similar mechanism occurs in mammalian cells, in which a Rab cascade comprising Rab11, Rab8, and the Rab8 GEF Rabin8 promotes exocyst-dependent vesicle targeting to the plasma membrane (Knödler et al., 2010; Westlake et al., 2011; Mizuno-Yamasaki et al., 2012).
Similar to its function in yeast and mammalian cells (Heider and Munson, 2012; Wu and Guo, 2015), the exocyst plays an important role in the trafficking of materials to the plasma membrane and also functions in multiple steps during cytokinesis and cell plate formation in plants (McMichael and Bednarek, 2013; Rybak et al., 2014). Arabidopsis exocyst subunit mutants exhibit developmental phenotypes, including dwarfism, improper guard cell cytokinesis, and cell plate maturation defects (Fendrych et al., 2010; Drdová et al., 2013; Rybak et al., 2014; Wu and Guo, 2015). However, although homologs for Ypt32/Rab11, Sec4p/Rab8, and exocyst subunits are known to function in exocytosis and cytokinesis in plants, the molecular details of their interactions are not known.
Arabidopsis stomatal cytokinesis defective1 (scd1) and scd2 mutants exhibit strikingly similar phenotypes to exocyst mutants: Plants are dwarfed and have defects in cell division and expansion that result in guard cell cytokinesis and root hair morphogenesis defects (Falbel et al., 2003; Korasick et al., 2010; McMichael et al., 2013). In addition, SCD1 and SCD2 genetically interact and are associated with clathrin-coated vesicles (CCVs), suggesting a role for these proteins in membrane trafficking during cytokinesis and cell expansion including endocytosis (McMichael and Bednarek, 2013; McMichael et al., 2013). The SCD1 protein is defined by an N-terminal DENN (Differentially Expressed in Normal and Neoplastic cells) domain that in metazoans has been demonstrated to activate Rab GTPases (Marat and McPherson, 2010; Yoshimura et al., 2010; Marat et al., 2012); however, a connection between DENN domain proteins and Rab GTPases has not been established in plants.
Members of the plant RabE1 GTPase (RabE1a-e) family are most closely related to mammalian Rab8 and Saccharomyces cerevisiae Sec4p (Rutherford and Moore, 2002). Live-cell imaging has demonstrated that RabE1d and RabE1c localize to the Golgi stacks, plasma membrane, and the cell plate in dividing cells (Zheng et al., 2005; Chow et al., 2008; Speth et al., 2009). Moreover, functional studies have indicated that RabE1s act in post-Golgi trafficking to the plasma membrane and cell plate (Speth et al., 2009; Ahn et al., 2013). Similar to the scd1 and scd2 mutant phenotypes, silencing of RabE1 expression in Nicotiana benthamiana results in defective guard mother cell cytokinesis, and overexpression of dominant-negative mutant RabE1 in plants manifests in shoot and root growth defects (Speth et al., 2009; Ahn et al., 2013).
To date, only a few factors that function to regulate the exocyst complex have been identified in plants (Lavy et al., 2007; Hazak et al., 2010). Here, we show, through the use of proteomics, in vitro binding studies, cargo trafficking assays, and colocalization analysis, that SCD1 and SCD2 are subunits of a protein complex, which we refer to as the SCD complex, that functions together with RabE1s and the exocyst in a protein interaction network to mediate post-Golgi trafficking to the plasma membrane and cell plate.
RESULTS
SCD1 and SCD2 Are Subunits of an Oligomeric Protein Complex
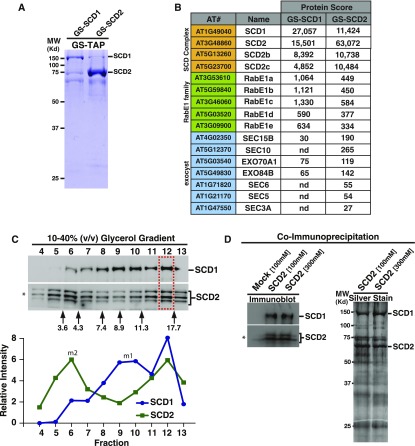
Previously we demonstrated that SCD1 and SCD2 function in membrane transport required for cytokinesis and cell expansion (McMichael et al., 2013). To further define the protein interaction network of SCD1 and SCD2, we generated Arabidopsis cell lines (PSB-d) that express N-terminal G-protein/Streptavidin binding peptide (GS)-tagged SCD1 (GS-SCD1), SCD2 (GS-SCD2), or GFP (GS-GFP) fusion proteins. GS-SCD1, GS-SCD2, and GS-GFP were purified from Arabidopsis cell extracts using tandem affinity purification (TAP) chromatography as described (Van Leene et al., 2011). TAP elutions were analyzed by SDS-PAGE (Figure 1A; Supplemental Figure 1) and liquid chromatography tandem mass spectrometry (LC/MS/MS) (Figure 1B; Supplemental Data Set 1). The major polypeptides detected in the TAP GS-SCD1 and GS-SCD2 elutions migrated in SDS-PAGE at the expected molecular mass of GS-SCD1 (∼140 kD) and GS-SCD2 (∼70 kD), respectively, with the G-protein removed during purification as described (Van Leene et al., 2011) (Figure 1A). In addition, polypeptides of the expected size for endogenous SCD1 (∼132 kD) and SCD2 (∼64 kD) were observed in the GS-SCD1 and GS-SCD2 TAP fractions, respectively. LC/MS/MS analysis confirmed that endogenous untagged SCD2 copurified with GS-SCD1 and vice versa (Figure 1B; Supplemental Data Set 1), indicating that SCD1 and SCD2 physically interact. The association of endogenous untagged SCD1 and SCD2 was further analyzed using independent methods. Immunoprecipitation of untransformed Arabidopsis cell lysate using anti-SCD2 antibodies demonstrated coimmunoprecipitation of SCD1 with SCD2 under standard (100 mM KCl) and higher salt (300 mM KCl) conditions, indicating that the association of SCD1 and SCD2 is salt stable (Figure 1D). Furthermore, SCD1 and SCD2 were found to cofractionate by velocity gradient sedimentation analysis, which separates proteins based on their native molecular mass and hydrodynamic properties (Harding, 1999). Specifically, Arabidopsis cell extracts were fractionated by glycerol gradient sedimentation and analyzed by immunoblotting with antibodies against both SCD1 and SCD2 (Figure 1C). Quantitative analysis of the immunoblots revealed that endogenous SCD1 and SCD2 fractionated in separate slower sedimenting peaks corresponding to their expected monomeric forms (molecular masses 132 and 64 kD, respectively), as well as in an overlapping ∼17S peak with an estimated molecular mass of ∼430 kD, which is larger than expected for a complex containing only single copies of SCD1 and SCD2, suggesting that SCD1 and SCD2 are subunits of a multimeric protein complex. In addition, LC/MS/MS analysis of proteins that copurified with GS-SCD1 and GS-SCD2 identified two SCD2-like polypeptides encoded by At5g23700 and At5g13260, which we have named SCD2c and SCD2b, respectively. SCD2c and SCD2b share 75% and 56% amino acid identity with SCD2, respectively (Figure 1B; Supplemental Data Set 1). Similar to SCD2, these two SCD2-like proteins are predicted to contain two centrally located coiled-coil domains as well as a ProDom-defined (Corpet et al., 1998), plant-specific domain of unknown function, PD147848, at their respective carboxyl termini. LC/MS/MS analysis of proteins that copurified with GS-GFP did not show similar enrichment profiles of interacting proteins (Supplemental Data Set 1 and Supplemental Table 1). A small amount of RabE1c copurified with GS-GFP, but its abundance in the GS-SCD1 and GS-SCD2 samples was 9- and 29-fold higher, respectively, based on exponentially modified protein abundance index as determined by Mascot (Supplemental Table 1).
Figure 1.
Identification of the SCD Complex and Its Interactors.
(A) Coomassie-stained SDS-PAGE analysis of TCA-precipitated TAP-purified GS-SCD1 and GS-SCD2 fractions.
(B) Abridged list of proteins that copurified with GS-SCD1 and GS-SCD2 as identified by LC/MS/MS. Shown is the Mascot protein score.
(C) Cofractionation of SCD1 and SCD2. Glycerol gradient velocity sedimentation analysis of native SCD1 and SCD2 from Arabidopsis cell extracts. Glycerol gradient fractions were analyzed by quantitative immunoblotting using indicated antibodies. Arrows indicate the fractionation of molecular mass standards with their indicated Svedberg (S)-values. Graph depicts the intensity of the SCD1 and SCD2 immunoblot signal peaks. SCD1 and SCD2 fractionate in separate lower molecular weight peaks corresponding to their expected monomeric molecular masses 132 and 64 kD, respectively (labeled m1 and m2), as well as in an overlapping (∼430 kD) peak (fraction 12; boxed region).
(D) Co-IP of SCD2 and SCD1. Immunoblot and silver stain analysis of Arabidopsis cell extracts immunoprecipitated without (mock) or with anti-SCD2 antibodies in the presence of 100 or 300 mM KCl. Asterisk: affinity-purified anti-SCD2 antibodies detect multiple SCD2 bands as described (McMichael et al., 2013).
scd Mutants Display Post-Golgi Trafficking Defects
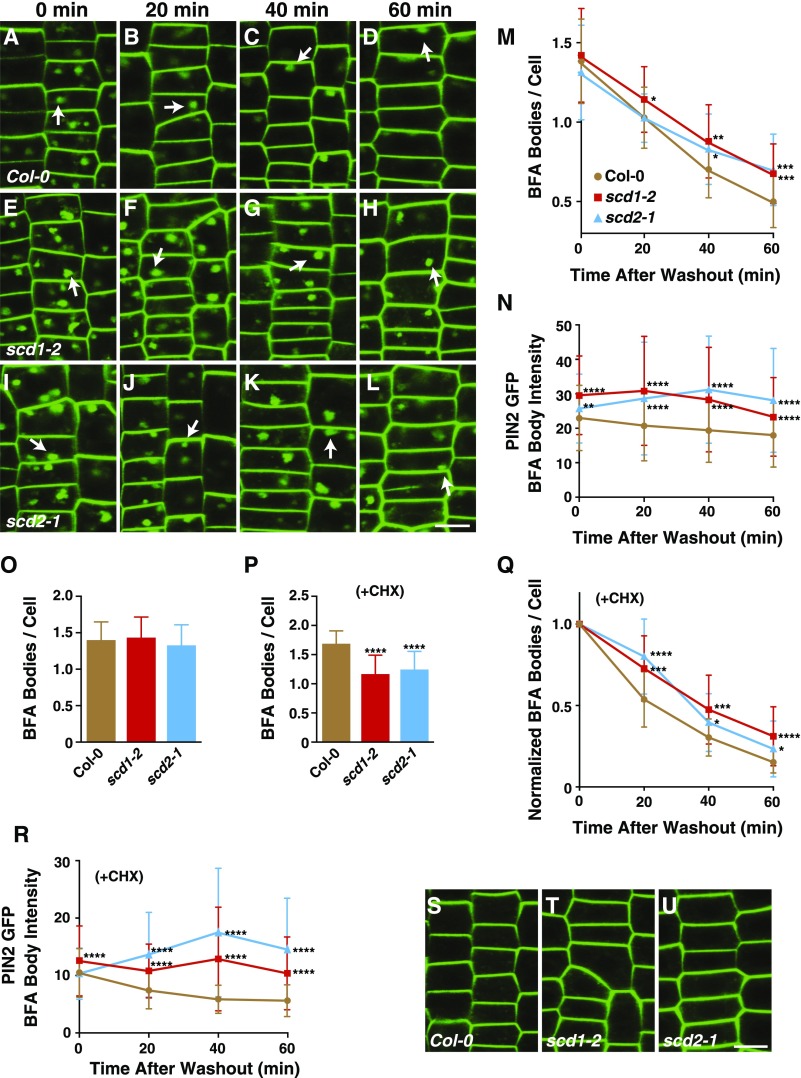
Arabidopsis scd1 and scd2 mutant plants exhibit cytokinesis and cell expansion defects, including impaired internalization of the endocytic tracer dye, FM4-64, indicative of potential roles in cell plate and plasma membrane trafficking (Falbel et al., 2003; McMichael et al., 2013). LC/MS/MS analysis of GS-TAP experiments have identified exocyst subunits (SEC3A, SEC5, SEC6, SEC10, SEC15B, EXO70A1, and EXO84B) as potential interactors with SCD1 and SCD2, suggesting that the SCD complex may function in exocytic vesicle trafficking to the cell plate and plasma membrane (Figure 1B; Supplemental Data Set 1). To test this, we assayed the trafficking of the plant plasma membrane protein PIN-FORMED2 fused to green fluorescent protein (PIN2-GFP) (Xu and Scheres, 2005) in scd1 and scd2 mutants. PIN2 is a member of the PIN family of transport proteins that functions to transport auxin, a crucial developmental hormone, across the plasma membrane (Krecek et al., 2009). PIN2-GFP is trafficked from the trans-Golgi network/early endosome to the plasma membrane where it can undergo rounds of constitutive endocytosis and recycling from endosomes to the plasma membrane or be transported to the vacuole for degradation (Löfke et al., 2013; Wang et al., 2013). To monitor PIN2-GFP trafficking, wild-type (Col-0) and loss-of-function scd1-2 and scd2-1 mutant plants expressing PIN2-GFP, under control of its native promoter (ProPIN2:PIN2-GFP), were treated with the reversible vesicle trafficking inhibitor Brefeldin A (BFA), which causes formation of intracellular endosomal and trans-Golgi compartment aggregates (BFA bodies) (Rosa et al., 1992; Ritzenthaler et al., 2002; Wang et al., 2013). Following 50 µM BFA treatment for 60 min, de novo-synthesized and internalized PIN2-GFP accumulated in one to two BFA bodies/root cell in scd1-2, scd2-1, and wild-type plants as monitored by confocal microscopy (Figure 2). Time-course assays demonstrated that the levels of intracellular PIN2-GFP decreased upon BFA removal in wild-type, scd1-2, and scd2-1 root cells, indicative of the restoration of protein trafficking to the plasma membrane (Dhonukshe et al., 2007). Relative to the wild type, however, scd1-2 and scd2-1 mutants displayed a significant delay in the rate of loss of PIN2-GFP-positive BFA bodies per cell and higher PIN2-GFP fluorescent signal intensity (Figures 2A to 2O).
Figure 2.
scd Mutants Display Post-Golgi Trafficking Defects.
Quantitative time-lapse analysis of the number and intensity of PIN2-GFP-labeled BFA bodies in wild-type, scd1, and scd2 seedling root cells pretreated with BFA ([A] to [O]) and BFA plus CHX ([P] to [R]) followed by removal of the inhibitors.
(A) to (L) Trafficking of intracellular PIN2-GFP following BFA washout in wild-type ([A] to [D]), scd1-2 ([E] to [H]), and scd2-1 ([I] to [L]) root cells. Seedlings were treated with 50 µM BFA for 60 min prior to washout with 0.5× MS and imaged at 0, 20, 40, and 60 min by CLSM. Arrowheads indicate PIN2-GFP-labeled BFA bodies. Bar = 10 µm.
(M) The number of PIN2-GFP-labeled BFA bodies per cell after BFA removal (minimum of 872 cells counted from a minimum of 27 roots each). Shown are means ± sd. **P < 0.01 and ***P < 0.001 (t test, compared with wild-type value).
(N) PIN2-GFP BFA body intensity/area. Total PIN2-GFP BFA body intensity was determined and divided by the BFA body area per cell (minimum of 163 cells from a minimum of 9 roots). Arbitrary intensity units. Shown are means ± sd. **P < 0.01 and ****P < 0.0001 (t test, compared with wild-type value). Seedlings were treated as described in (A) to (L).
(O) Number of BFA bodies prior to washout in Col-0 and scd mutants without CHX. Shown are means ± sd.
(P) Number of BFA bodies prior to washout in Col-0 and scd mutants in the presence of CHX. Shown are means ± sd. ****P < 0.0001 (t test, compared with wild-type value)
(Q) Relative number of PIN2-GFP-labeled BFA bodies per cell in the presence of CHX after BFA removal (minimum of 404 cells counted from a minimum of 13 roots each). Shown are means ± sd. *P < 0.05, ***P < 0.001, and ****P < 0.0001 (t test, compared with wild-type value). Seedlings were pretreated with 50 μM CHX for 30 min and then treated with 50 µM BFA and 50 μM CHX for 60 min prior to washout with 0.5× MS and imaged at 0, 20, 40, and 60 min using CLSM. Total number of BFA bodies per cell was normalized with the number of BFA bodies per cell at the start of BFA washout for each genotype.
(R) PIN2-GFP BFA body intensity/area in the presence of CHX. Total PIN2-GFP BFA body intensity was determined and divided by the total BFA body area per cell (minimum of 100 cells from a minimum of 9 roots each). Arbitrary intensity units. Shown are means ± sd. ****P < 0.0001 (t test, compared with wild-type value). Seedlings were treated as described in (Q).
(S) to (U) PIN2-GFP in Col-0, scd1-2, and scd2-1 in untreated root cells. Bar = 10 µm.
To distinguish between the trafficking of de novo-synthesized and internalized PIN2-GFP, BFA washout experiments were performed in the presence of cycloheximide (CHX), an inhibitor of eukaryotic translation (Schneider-Poetsch et al., 2010). In the presence of CHX and BFA, the number of PIN2-GFP-labeled BFA bodies/cell was found to be reduced in both scd mutants compared with the wild type (Figure 2P), indicative of an endocytosis defect and consistent with previous data showing that scd1 and scd2 mutants exhibit defects in the uptake of the endocytic tracer dye, FM4-64 (McMichael et al., 2013). In addition to a reduction in endocytosis, recycling of PIN2-GFP from BFA bodies was delayed in scd mutants. As shown in Figures 2P and 2Q, after normalization for the initial number of BFA bodies per root cell in the presence of CHX, we observed a decrease in the rate of PIN2-GFP-labeled BFA body disappearance in scd1-2 and scd2-1 mutants following removal of BFA. Similarly, scd mutants had increased PIN2-GFP intensity per BFA body area per cell after washout (Figure 2R). Taken together, our analysis of PIN2-GFP trafficking in the scd mutants indicates that the SCD complex functions in exocytosis and endosomal recycling of plasma membrane proteins.
The SCD Complex Communicates with the Exocyst Complex
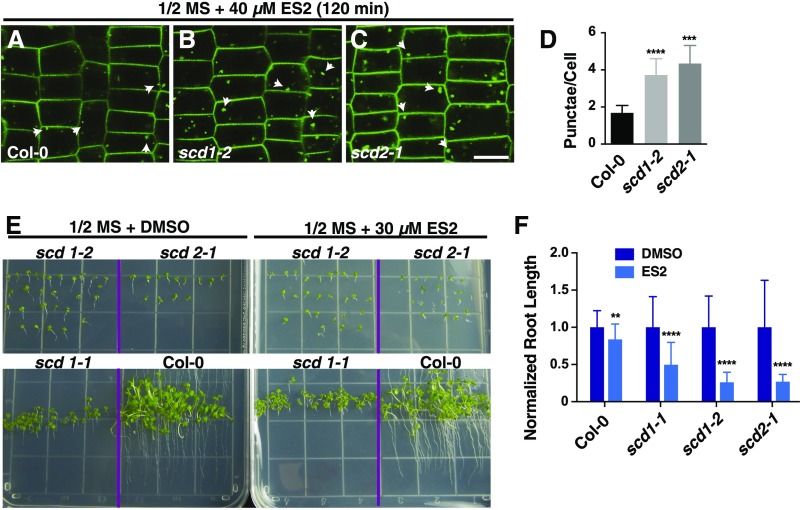
Given that the SCD complex functions in post-Golgi trafficking, and the identification of exocyst subunits as putative SCD complex interactors (Figure 1B; Supplemental Data Set 1), we further explored the relationship between the SCD complex and the exocyst. To determine if SCD complex-mediated post-Golgi trafficking to the plasma membrane is dependent on exocyst function, we performed pharmacological studies with the small molecule Endosidin2 (ES2), which selectively inhibits exocytosis by binding to exocyst subunit Exo70 (Zhang et al., 2016). As previously shown, ES2 treatment of wild-type seedlings expressing PIN2-GFP blocks exocytosis and endosomal recycling, resulting in the intracellular accumulation of PIN2-GFP in punctate structures (Figure 3A) that are distinct from BFA bodies (Zhang et al., 2016). Interestingly, ES2 was found to enhance the intracellular accumulation of PIN2-GFP in both scd1 and scd2 mutants compared with wild-type plants. Specifically, scd1-2 and scd2-1 mutant root cells treated with 40 µM ES2 showed 2.3-fold and 2.6-fold elevation, respectively, in the average number of PIN2-GFP-labeled punctae/cell (Figures 3A to 3D) relative to ES2-treated wild-type (Col-0) root cells. These intracellular PIN2-GFP punctae were not detectable in untreated Col-0 and scd mutants (Figures 2S to 2U). To confirm the presence of internalized PIN2-GFP in ES2 bodies, experiments were performed with CHX. Treatment with both ES2 and CHX together enhanced intracellular accumulation of PIN2-GFP, likely due to inhibition of recycling PIN2-GFP back to the plasma membrane by ES2 (Supplemental Figure 2).
Figure 3.
Inhibition of Exocyst Function Results in Growth Inhibition and PIN2-GFP Trafficking Defects in scd1 and scd2 Mutants.
(A) to (C) PIN2-GFP in wild-type (A), scd1-2 (B), and scd2-1 (C) roots from 5-d-old seedlings treated for 2 h in 0.5× MS plus 40 µM ES2. Arrowheads indicate PIN2-GFP-labeled intracellular accumulations. Bar = 10 µm.
(D) Quantitation of average PIN2-GFP-labeled ES2-induced punctae per cell. Shown are means ± sd. ***P < 0.001 and ****P < 0.0001 (t test, compared with wild-type value).
(E) Seedlings grown on agar plates with 0.5× MS or 0.5× MS plus 30 µM ES2.
(F) Quantitation of relative root length of wild-type and scd1-2 and scd2-1 mutants grown on 0.5× MS agar plates containing DMSO or 30 µM ES2. Shown are means ± sd. **P < 0.01 and ****P < 0.0001 (t test, compared with DMSO value).
Homozygous temperature-sensitive scd1-1 grown above the permissive temperature of 16°C and loss-of-function scd1-2 and scd2-1 plants are dwarfed and display numerous growth defects including stunted roots (Falbel et al., 2003; McMichael et al., 2013). To assess the effects of ES2 on root growth, wild-type (Col-0) and scd mutant seedlings were germinated and grown for 13 d in the presence of DMSO or 30 µM ES2 and root length was measured. Relative to DMSO-treated control seedlings, ES2-treated wild-type (Col-0) seedlings showed a ∼10% reduction in root length. By contrast, ES2 inhibited scd1-1, scd1-2, and scd2-1 seedling root growth by ∼50, ∼80, and ∼80%, respectively, relative to DMSO-treated scd mutant seedlings (Figures 3E and 3F). The observed enhancement of growth defects observed in ES2-treated scd1 and scd2 mutants provides further support for a functional connection between the SCD and exocyst protein complexes and suggests that the SCD complex functions in protein trafficking through downstream interactions with the exocyst.
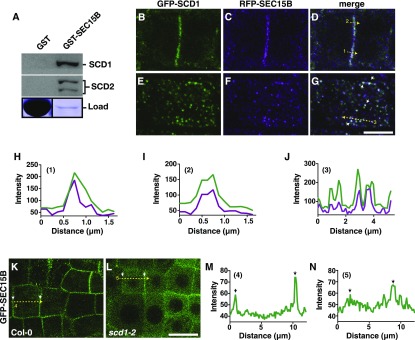
LC/MS/MS analysis of tandem affinity-purified GS-SCD2 from Arabidopsis cell extracts identified seven of the eight exocyst subunits, with the notable absence of Sec8 (Figure 1B; Supplemental Data Set 1). Interestingly, SEC15 and SEC10, which in yeast act as a linker between transport vesicles and the rest of the exocyst complex (Guo et al., 1999), were the most highly represented exocyst subunits that copurified with GS-SCD2 based on the number of spectral counts. To verify the physical connection between the SCD and exocyst complexes, we chose to further explore the interaction of the SCD complex with SEC15B using in vitro binding and colocalization studies. To determine if SCD1 and SCD2 interact with SEC15B, immobilized GST and GST-SEC15B were incubated with Arabidopsis cell extracts. Immunoblot analysis of proteins bound to GST and GST-SEC15B demonstrated that SCD1 and SCD2 interacted with GST-SEC15B and not with the GST negative control (Figure 4A).
Figure 4.
The SCD Complex Colocalizes in Vivo and Associates in Vitro with SEC15B.
(A) GST-SEC15B was incubated with Arabidopsis cell extract, and binding of candidate proteins was determined by immunoblot (top panel) analysis with indicated antibodies. Lower panel: Coomassie-stained SDS-PAGE of GST and GST-fusion protein load used in the binding experiment.
(B) to (D) CLSM images of a dividing root cell expressing GFP-SCD1 and RFP-SEC15B show colocalization at the cell plate (Costes P value, 1.00; Pearson’s R value, 0.65).
(E) to (G) GFP-SCD1 and RFP-SEC15B colocalize at distinct punctae in root cells. Arrowheads denote colocalization at distinct punctae (Costes P value, 1.00; Pearson’s R value, 0.61). Bar = 5 µm.
(H) and (I) Line scan measurements (1 and 2 respectively, yellow dotted arrows) of CLSM image (D) are plotted to show overlapping intensity profiles.
(J) Line scan measurement (yellow dotted arrow 3) of CLSM image in (G) plotted to show overlapping intensity profiles.
(K) and (L) CLSM images of GFP-SEC15B localization in wild-type (Col-0) and scd1-2 mutant plant roots. Bar = 10 µm. Arrowheads denote the plasma membrane.
(M) and (N) Line scan measurements (4 and 5 respectively, yellow dotted arrows) of CLSM images ([K] and [L]) are plotted to show intensity profiles. Arrowheads denote the plasma membrane.
Next, we utilized confocal laser scanning microscopy (CLSM) to assess whether GFP-SCD1 and red fluorescent protein (RFP)-SEC15B colocalize. As previously shown (Fendrych et al., 2010; Rybak et al., 2014), RFP-SEC15B predominantly localized to the cell plate and to punctate structures at or adjacent to the plasma membrane in dividing and nondividing cells, respectively (Figured 4C and 4F). Similarly, SCD1 was associated with punctae closely associated with the plasma membrane (McMichael et al., 2013) (Figure 4E) as well as the cell plate in dividing cells (Figure 4B), which had previously not been reported.
Fluorescence intensity line scans across the cell plate (Figure 4D) and punctate structures (Figure 4G) revealed overlapping intensity profiles of GFP-SCD1 and RFP-SEC15B consistent with colocalization (Figures 4H to 4J). Furthermore, colocalization was quantitatively verified for image pairs using the Costes randomization test (Costes et al., 2004). Images were analyzed with 100 Costes iterations returning a Costes P value of 1.00 for both punctae and cell plate localization with Pearson’s R values of 0.65 and 0.61, respectively.
Next, we utilized CLSM to assess whether SCD function is required for the localization of the exocyst. As shown previously (Fendrych et al., 2010), GFP-SEC15B was associated with the plasma membrane in nondividing cells. However, the subcellular distribution of GFP-SEC15B was altered in scd1-2 mutant root cells compared with the wild type (Figures 4K and 4L), with the most dramatic differences seen near the plasma membrane. Line scan intensity measurements also demonstrated an increase in the intracellular levels of GFP-SEC15B in scd1-2 mutants relative to the wild type (Figures 4M and 4N). These data, together with the ES2 inhibitor and in vitro binding studies, indicate that the SCD complex and exocyst function together to mediate post-Golgi vesicle trafficking to the plasma membrane.
The SCD Complex Selectively Interacts with RabE1 GTPases
Given that SCD1 contains a tripartite DENN domain that has been demonstrated in other systems to function as a GEF for Rab GTPases (Marat and McPherson, 2010; Yoshimura et al., 2010; Marat et al., 2012), the SCD complex may function through interactions with Rab GTPases to regulate exocytic vesicle trafficking. Consistent with this, LC/MS/MS analysis of proteins associated with SCD1 and SCD2 identified all members of the RabE1 family; specifically, RabE1a-e were identified by TAP and coimmunoprecipitation (co-IP) of GS-tagged and endogenous SCD proteins (Figure 1B; Supplemental Data Set 1). Previously published reports have implicated RabE1 GTPases in post-Golgi trafficking to the plasma membrane and cell plate (Zheng et al., 2005; Speth et al., 2009; Ahn et al., 2013), suggesting that the SCD complex’s role in membrane trafficking may be through its interaction with RabE1. We utilized colocalization analysis and in vitro binding studies to validate the interaction of the SCD complex with RabE1 GTPases.
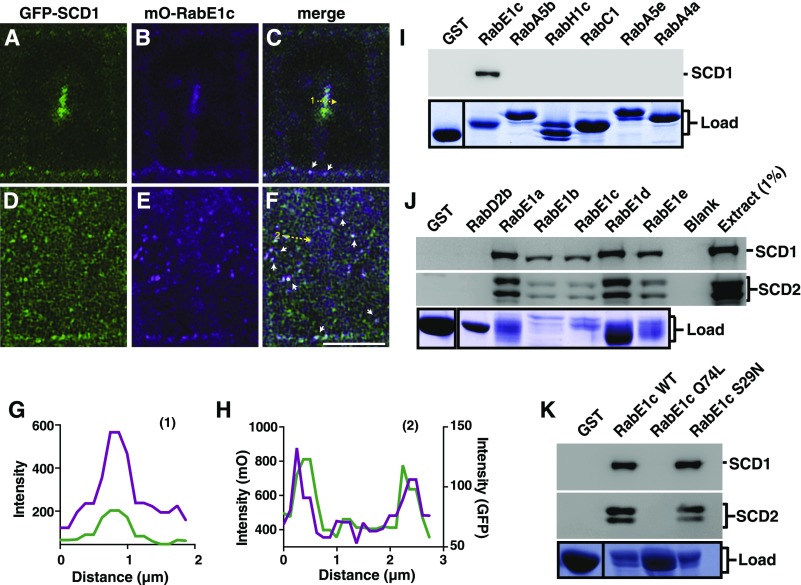
CLSM imaging of root cells from Arabidopsis lines that express GFP-SCD1 and mO-RabE1c showed colocalization between GFP-SCD1 and mO-RabE1c at distinct intracellular punctae, some of which appear at or near the plasma membrane, as well as at the cell plate (Figures 5A to 5H). Fluorescence intensity line scans across the cell plate (Figures 5C and 5G) and punctate structures (Figures 5F and 5H) revealed overlapping intensity profiles of GFP-SCD1 and mO-RabE1c consistent with colocalization. Additionally, colocalization was quantitatively verified for image pairs as described above, with 100 Costes iterations returning a Costes P value of 1.00 for both punctae and cell plate localization with Pearson’s R values of 0.50 and 0.61, respectively.
Figure 5.
RabE1 GTPases Colocalize in Vivo and Interact in Vitro with the SCD Complex.
(A) to (C) CLSM microscopy of a dividing root cell expressing GFP-SCD1 and mO-RabE1c show colocalization at the cell plate (Costes P value, 1.00; Pearson’s R value, 0.61).
(D) to (F) GFP-SCD1 and mO-RabE1c colocalize at distinct punctae in root cells (Costes P value, 1.00; Pearson’s R value, 0.50). Bar = 5 µm. Arrowheads denote colocalization at distinct subcellular punctae.
(G) Line scan measurement (yellow dotted arrow 1) of cell plate images is plotted to show overlapping intensity profiles. Bar = 5 µm.
(H) Line scan measurement (yellow dotted arrow 2) of punctae show overlapping intensity profiles.
(I) to (K) GST and GST-fusions of the indicated Rabs were incubated with Arabidopsis cell extract. Binding was determined by immunoblot analysis of elutions probed with indicated antibodies (top panel). Lower panel: Coomassie-stained SDS-PAGE of GST and GST-fusion protein loads used in the binding experiments.
To confirm the specificity of RabE1 GTPase interaction with the SCD complex, in vitro binding studies were performed with GST-tagged RabE1 and members of other Arabidopsis Rab GTPase families that function in various stages of the plant biosynthetic secretory and endocytic pathways. Following incubation of immobilized candidate GST-Rab fusion proteins with Arabidopsis cell extracts, bound proteins were eluted and analyzed by immunoblotting. As shown in Figure 5I, SCD1 selectively bound to RabE1c. Binding of SCD1 to GST alone, or to GST-tagged members of other Rab GTPases families including RabA5, A4, A2, C1, D2, G3, and H1, was not detected (Figures 5I and 5J). SCD complex association with any other Rab families was also not detected by LC/MS/MS (Supplemental Data Set 1). Furthermore, both SCD1 and SCD2 bound to all RabE1 family members in vitro, (Figure 5J) but not to the GST control.
To further define the interaction between RabE1 and the SCD complex, we tested the binding of SCD proteins from Arabidopsis cell extract to nucleotide state-specific RabE1c mutants in vitro. SCD1 and SCD2 bound to either wild-type RabE1c or to the S29N mutant RabE1c, RabE1cS29N, which is analogous to dominant inhibitory H-rasS17N mutant that displays reduced nucleotide affinity (Farnsworth and Feig, 1991; Nassar et al., 2010). By contrast, no binding was observed to the RabE1cQ74L mutant (Figure 5K), which is predicted to have a reduced intrinsic hydrolysis rate of GTP as demonstrated for other Rab proteins (Walworth et al., 1992). These data demonstrate that the SCD1 and SCD2 bind RabE1c in a nucleotide-dependent manner.
To test whether the SCD complex functions in the activation of RabE, we determined if overexpression of wild-type RabE1 would rescue the conditional growth and developmental defects of partial loss-of-function scd1-1 mutants. Precedence for this has been established as Rab Sec4p was shown to rescue the temperature-sensitive growth defects of mutant alleles of its GEF Sec2p, in S. cerevisiae (Nair et al., 1990; Walch-Solimena et al., 1997). To determine if RabE1 overexpression rescues the scd1 mutant, we generated multiple independent transgenic scd1-1 mutant lines that express wild-type N-terminal mOrange (mO)-tagged RabE1c (mO-RabE1c) under the control of the constitutive 35S CaMV promoter. Whereas partial loss-of-function scd1-1 mutant plants exhibit temperature-sensitive growth and stomatal cytokinesis defects at temperatures at or above 22°C (Falbel et al., 2003; McMichael et al., 2013), scd1-1 mO-RabE1c grown at 22°C were found to be phenotypically similar to wild-type (Col-0) plants at various stages of development including leaf expansion, inflorescence growth, and guard cell formation (Figures 6A to 6C). By contrast, overexpression of mO-RabD2b (scd1-1 mO-RabD2b), which is involved in ER to Golgi trafficking (Zheng et al., 2005), failed to suppress the growth defects of scd1-1 plants (Supplemental Figures 3A and 3B).
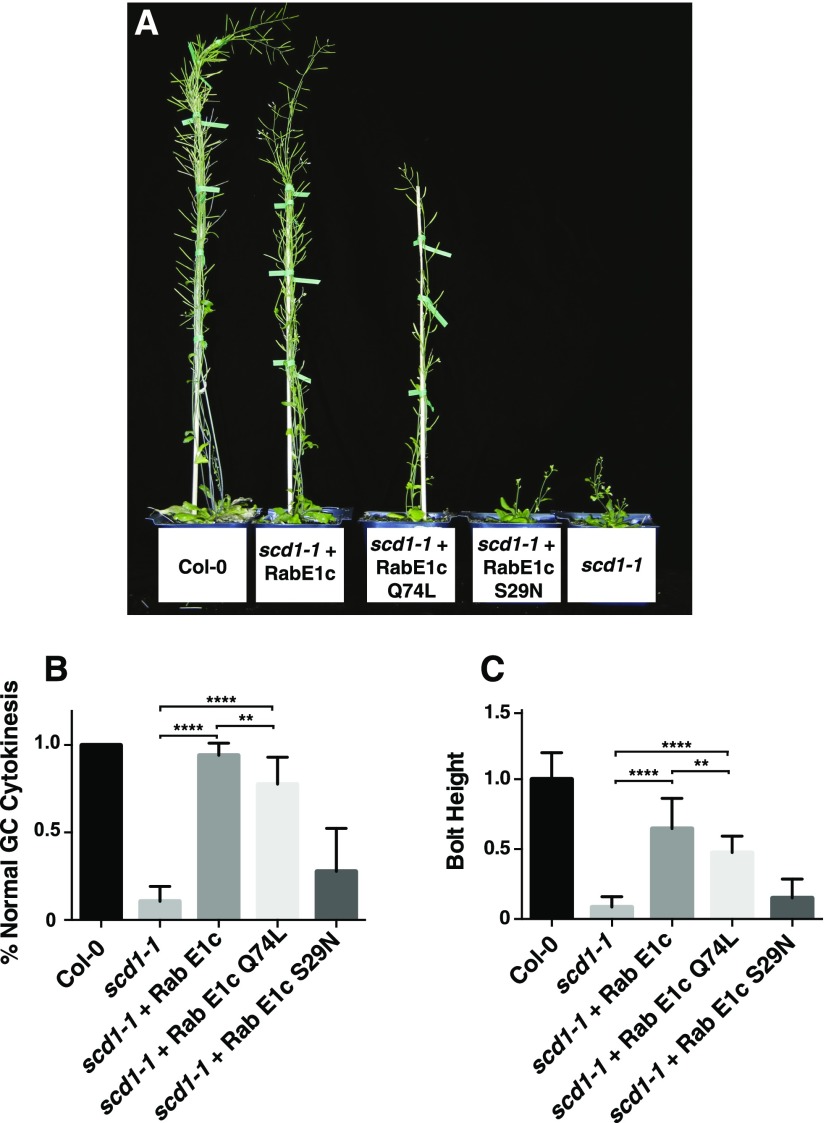
Figure 6.
Overexpression of RabE1c and RabE1c Q74L Rescues the Growth and Guard Cell Cytokinesis Defects of the Temperature-Sensitive Mutant scd1-1.
(A) Wild-type (Col-0), scd1-1, and scd1-1 plants expressing RaE1c, RabE1c Q74L, and RabE1c S29N grown at 22°C.
(B) Quantitation of stomatal cytokinesis defects seen in leaves of wild-type (Col-0), scd1-1, and scd1-1 plants expressing wild-type and mutant RabE1c. Shown are means ± sd. ****P < 0.0001 (t test).
(C) Inflorescence stem height of 60-d-old wild-type (Col-0), scd1-1, and scd1-1 plants expressing wild-type and mutant RabE1c. Shown are means ± sd. ****P < 0.0001 (t test).
To determine if RabE1 suppression of the scd1-1 phenotype was dependent on nucleotide state, we examined the effect of overexpression of GTPase deficient “constitutively active” RabE1cQ74L and “inactive” RabE1cS29N mutants. Previous studies have reported that expression of GFP-RabE1dQ74L in wild-type plants did not significantly affect plant growth and development (Speth et al., 2009; Ahn et al., 2013). Similar to scd1-1 plants that expressed wild-type mO-RabE1c, the scd1-1 mO-RabE1cQ74L lines showed significant rescue of the scd1-1 mutant growth and development defects (Figures 6A to 6C). However, whereas bolt height and stomatal development were restored in the scd1 mO-RabE1cQ74L lines, the level of suppression of the scd1-1 phenotype was significantly reduced relative to scd1-1 plants expressing wild-type mO-RabE1c (Figures 6A to 6C). In contrast to scd1-1 mO-RabE1c and scd1-1 mO-RabE1cQ74L lines, scd1-1 mO-RabE1cS29N plants were phenotypically indistinguishable from the parental scd1-1 lines (Figures 6A to 6C). Overexpression of mO-RabE1cS29N did not suppress or enhance growth and guard cell cytokinesis defects associated with the scd1-1 mutation, providing further evidence that the scd1-1 phenotype is likely due to defects in its nucleotide state-specific interaction with RabE1. Expression of mO-RabD2b, mO-RabE1c, mO-RabE1cQ74L, and mO-RabE1cS29N was confirmed by RT-PCR and immunoblot analysis (Supplemental Figures 3C and 3D). Taken together, the biochemical interaction and genetic suppression studies suggest that the SCD complex is required in some manner for RabE1 activation.
DISCUSSION
Here, we demonstrate that the SCD1 and SCD2 proteins are members of the SCD complex that functions in concert with the exocyst and RabE1s in post-Golgi trafficking to the plasma membrane and cell plate (Figure 7). This conclusion is supported by the results presented in this work as well as the previous phenotypic characterization of scd, rabE1, and exocyst mutants (Speth et al., 2009; Fendrych et al., 2010; Ahn et al., 2013; Drdová et al., 2013; Rybak et al., 2014; Wu and Guo, 2015). Exocyst mutants, including exo84b, sec6, and scd mutants, show similar developmental phenotypes, including dwarfism, reduced leaf pavement cell expansion, and aberrant guard mother cell cytokinesis (Falbel et al., 2003; Fendrych et al., 2010; McMichael et al., 2013; Wu et al., 2013). At the cellular level, scd1 and scd2 share similar defects as exocyst subunit loss-of-function mutants and ES2-treated cells (Zhang et al., 2016) in exocytosis and endosomal recycling of the plasma membrane protein, PIN2-GFP, in the presence of BFA (Drdová et al., 2013) (Figure 2) and show an accumulation of secretory vesicles, indicating that the SCD and exocyst complexes function in post-Golgi vesicle targeting and fusion (Falbel et al., 2003; Fendrych et al., 2010). Importantly, the SCD and exocyst complexes were found to colocalize at the cell plate and in punctae adjacent to the plasma membrane (Figures 4B to 4J) and to biochemically interact by proteomic analysis of the SCD complex (Figure 1; Supplemental Data Set 1) and by in vitro binding studies (Figure 4A). The interaction of the SCD and exocyst complexes is further supported by the finding that ES2 inhibition of exocyst function enhances growth and trafficking defects of scd1 and scd2 mutants (Figure 3). Given the large number of EXO70 subunits, 23 in the Arabidopsis genome (Cvrčková et al., 2012), ES2 experiments cannot distinguish whether the SCD complex functions in concert solely with EXO70A1 and/or other EXO70-containing exocyst complexes in the trafficking of proteins to the plasma membrane.
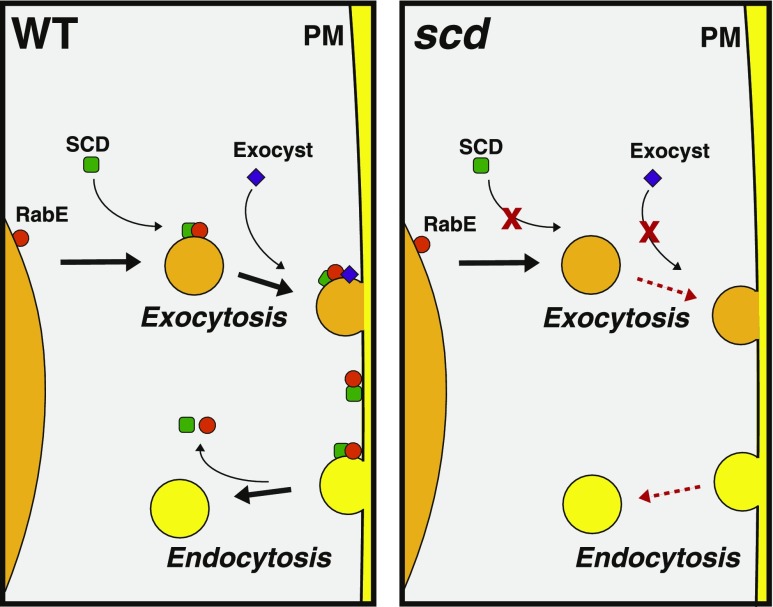
Figure 7.
SCD Function in Post-Golgi Trafficking.
In wild-type cells (left panel), the SCD complex (green square) associates with RabE1 (orange circle) and the exocyst complex (purple diamond) on exocytic post-Golgi vesicles. Following vesicle fusion, the SCD complex and RabE1 are deposited at the plasma membrane. In addition to its role in exocytosis, the SCD complex may function directly or indirectly in endocytosis as scd mutants exhibit exocytic as well as endocytic trafficking defects (Figure 2; McMichael et al., 2013). Our working model is that SCD-dependent exocytosis may deliver factors to the plasma membrane required for endocytosis and/or that following delivery to the cell surface, the SCD complex and activated RabE1 serve a role in the initiation of endocytosis. In scd mutants (right panel), activation and/or recruitment of RabE1 and the exocyst to post-Golgi secretory vesicles is impaired, leading to defects in exocytosis and endocytosis. Black arrows denote the direction of normal membrane trafficking. Red dashed arrows represent inhibition in membrane trafficking. PM, plasma membrane.
In addition to its association with the exocyst, the SCD complex was found to interact with RabE1 GTPases, which have previously been shown to localize to the Golgi, plasma membrane, and the cell plate (Zheng et al., 2005; Chow et al., 2008; Speth et al., 2009). Similar to scd1 and scd2 mutants, silencing of NbRabE1 expression or expression of dominant-negative NbRabE1S29N results in defects in protein trafficking from the Golgi to the plasma membrane and inhibition of N. benthamiana plant growth accompanied by defects in guard cell cytokinesis, polarized root hair expansion, and pathogen defense responses (Falbel et al., 2003; Zheng et al., 2005; Speth et al., 2009; Korasick et al., 2010; Ahn et al., 2013; McMichael et al., 2013). In this study, we show that RabE1s bind to the SCD complex in a nucleotide-state specific manner and colocalize at the cell plate and in punctae at or near the plasma membrane (Figures 1 and 5; Supplemental Data Set 1).
The SCD Complex Biochemically and Genetically Interacts with RabE1
The SCD1 subunit of the SCD complex contains an N-terminal tripartite DENN domain, which in other systems has been demonstrated to possess GEF activity for specific Rab GTPases (Allaire et al., 2010; Yoshimura et al., 2010), defining the DENN domain as a protein module characteristic of vesicle-trafficking regulators. In particular, DENN-domain-containing proteins connecdenn 1 to 3 are CCV-associated GEFs for Rab35, which functions in CCV trafficking, endosomal recycling, actin regulation, and cytokinesis in animals (Allaire et al., 2006, 2010; Kouranti et al., 2006; Patino-Lopez et al., 2008; Marat et al., 2012). Our data showing that SCD complex binds to “inactive” RabE1cS29N and not the “constitutively active” RabE1cQ74L (Figure 5K) is intriguing and suggests that the SCD complex functions to activate members of the RabE1 family. This hypothesis is further supported by the genetic evidence that overexpression of wild-type RabE1c and constitutively active RabE1cQ74L, but not the inactive RabE1cS29N mutant protein, suppressed the growth and developmental defects in scd1-1 mutants (Figure 6). In a similar manner, overexpression of Sec4p in S. cerevisiae rescues the temperature-sensitive growth defects of mutant alleles of its GEF, Sec2p (Nair et al., 1990; Walch-Solimena et al., 1997), and expression of “constitutively active” mutant RAB-A1cQ72L partially rescues the growth and development of attrs130 mutant plants (Qi et al., 2011), which have defects in the plant homolog of the yeast and mammalian Ypt32/Rab11 TRAPPII complex. In all these cases, it is likely that when overexpressed the levels of GTP-bound or constitutive-active Rab GTPase are sufficient to overcome the need for GEF-catalyzed activation to support vesicle trafficking.
In S. cerevisiae and mammalian cells, Sec4p and Rab8 are activated through the structurally related GEFs Sec2p and Rabin8, respectively (Guo et al., 2013). Vesicle-associated GTP-bound Sec4p/Rab8 GTPases and their cognate GEFs interact with the exocyst Sec15 subunit to promote docking and fusion of exocytic vesicles at the plasma membrane (Huber et al., 1993; Guo et al., 1999; Feng et al., 2012). In contrast to Sec4p- and Rab8-dependent trafficking, our understanding of the effectors involved in RabE1 activation and post-Golgi vesicle docking/fusion is limited, and unlike the Sec4p/Rab8 GEFs, Sec2p and Rabin 8, and other DENN GEFs (Dong et al., 2007; Allaire et al., 2010; Yoshimura et al., 2010; Marat et al., 2012; Vetter et al., 2015), the architecture of the multisubunit SCD complex, with the exception of the SCD1 DENN domain, is fundamentally distinct. Of interest are future experiments to determine if the SCD complex functions as a GEF for RabE1. A complication in testing its GEF activity is the multisubunit nature of the SCD complex as we are only able to purify relatively low quantities of the complex from tissue-cultured cells, and individual subunits and domains including the SCD1 DENN domain are not stable when individually purified, thereby necessitating reconstitution of the intact complex. In addition, it will be important to determine the molecular function(s) of the plant-specific protein SCD2. Furthermore, the presence of two SCD2-like proteins (AT5G13260 and AT5G23700) in GS-SCD1, GS-SCD2, and anti-SCD2 co-IPs provides interesting insight for other subunits in the SCD complex. While more work is required to fully delineate the subunit identity and stoichiometry of the complex, it is tempting to speculate, given the cofractionation of these two proteins with SCD1 and SCD2, that they may be additional subunits of the SCD complex.
Although we favor a model in which the SCD complex functions in the activation of RabE1, we cannot rule out potential alternative roles for the SCD complex in RabE1-dependent membrane trafficking. In addition to GEFs and GAPs, Rab GTPases are regulated by other factors including guanine nucleotide release factors, GDP dissociation inhibitors (GDIs), and GDI displacement factors (GDF), which serve to deliver Rabs to membranes (Stenmark, 2009). However, the selective interaction of SCD complex with bacterially expressed (i.e., nonprenylated) RabE1 (Figure 5) appears to be distinct from that of GDIs, which bind with high affinity exclusively to the prenylated GDP form of Rabs (Araki et al., 1990; Pylypenko et al., 2006) and display limited specificity in Rab binding (Grosshans et al., 2006). Similarly, the GDF, Yip3 (Sivars et al., 2003), and the mammalian guanine-nucleotide release factor MSS4, which has been proposed to function as a chaperone for nucleotide-free Rabs (Itzen et al., 2006), bind to a number of distinct Rabs, unlike the SCD complex, which binds selectively to RabE1s but not to other early or late secretory pathway Rabs (Figure 5I; Supplemental Data Set 1).
The SCD Complex Functions in Exocytosis, Recycling, and Endocytosis
The highly dynamic process of cell plate formation involves vesicle-mediated delivery and retrieval of membrane from the division plane. Likewise, in nondividing cells, the coordination of exocytosis and endocytosis is critical for polarized cell expansion and the polarized distribution of plasma membrane proteins. Interestingly, scd1 and scd2 mutants display defects not only in exocytosis but also in the uptake of the endocytic tracer dye FM4-64 (McMichael et al., 2013) and PIN2-GFP endocytosis (Figures 2P and 2R). While the SCD complex interaction network is composed of proteins involved in exocytosis, scd mutants exhibit defects in both endocytosis and exocytosis (Figure 7). Whether the SCD proteins function directly or indirectly in endocytosis remains to be determined. Consistent with the latter, defects in retrograde trafficking have been demonstrated to affect anterograde trafficking; for example, in human cells, defects in GARP and VAMP4-dependent endosome to trans-Golgi network trafficking result in an inhibition of recycling of factors necessary for the trafficking of proteins to the plasma membrane (Hirata et al., 2015). By contrast, however, treatment of plant cells with the small molecule ES16, which inhibits RabA-dependent exocytosis, does not affect endocytosis (Li et al., 2017). Nevertheless, it cannot be excluded that the SCD/RabE1 and ES16-sensitive/RabA trafficking pathways are distinct and that the SCD-dependent exocytic pathway is responsible for transporting factors required for endocytosis and/or that the SCD complex functions directly in endocytosis (Figure 7). Indeed, in S. cerevisiae, Sec4p and Sec2p have also been shown to function in exocytosis and endocytosis (Riezman, 1985; Johansen et al., 2016) and work in diverse systems, including yeast, metazoans, and Trypanosoma brucei has demonstrated a role for the exocyst in endocytosis (Riezman, 1985; Sommer et al., 2005; Jose et al., 2015; Boehm et al., 2017).
Although scd1 and scd2 mutants exhibit strong growth and developmental defects, PIN2-GFP exocytosis and/or endosomal recycling were found to be only partially impaired in mutant root cells (Figure 2). This may reflect the existence of RabE1-independent post-Golgi trafficking pathways including RabA-dependent pathways that could compensate for the loss, in scd mutants, of RabE1-dependent trafficking to the cell plate and/or plasma membrane. An alternative, but not mutually exclusive, possibility is that the severe developmental defects observed in scd mutants could be due to the lowered efficiency, as opposed to total inhibition, of the trafficking of critical cell-type-specific factors necessary at distinct times in plant development.
Our work demonstrates that SCD1 and SCD2 form a complex that functions in the post-Golgi trafficking to the plasma membrane and cell plate, a process that is mediated through interactions with RabE1 and the exocyst complex.
METHODS
Plant Materials
Columbia-0 lines used in this study were obtained from ABRC. scd1-1, scd1-2, and scd2-1 lines have been described previously (Falbel et al., 2003; McMichael et al., 2013). PIN2-GFP lines are described by Xu and Scheres (2005).
Transgenic Plants/scd1-1 Mutant Rescue
Transgenic plants created in this study were generated using the floral dip method, using Agrobacterium tumefaciens strain EHA105 (Clough and Bent, 1998). Transgenic scd1-1 mutant plants were transformed with the vector 35Spro:mO-RabE1c (or 35Spro:mO-RabD2b) (pSITE-II-5c; Martin et al., 2009) and the indicated point mutant constructs (Q74L or S29N). Following selection with kanamycin on 0.5× MS +1% agar (w/v), T3 plants were transferred to soil and grown at 22°C under 16-h light (T12 fluorescent bulb ∼160 µmol m−2 s−1) and 8-h dark cycle and phenotypically analyzed for defects in bolt height 60 d after germination. For the guard cell cytokinesis screen, stomata from seedling leaves were analyzed in 14- to 20-d-old plants grown under continuous light (T8 fluorescent bulb ∼120 µmol m−2 s−1 grown on 0.5× MS + 0.6% agar [w/v]). Measurements included at least three independent transformants for each overexpression line. Overexpression of mO-RabE1c and mutants was confirmed by immunoblot analysis using anti-RabE1 antibodies (Speth et al., 2009), with anti-CDC48 served as a loading control (Rancour et al., 2002).
For GFP-SCD1 colocalization with RabE1c or SEC15B, scd1-1 ProSCD1:GFP-SCD1 (McMichael et al., 2013) lines were transformed as described above with either 35Spro:mO-RabE1c or UBQ10pro:RFP-SEC15B. Initial transformants were selected on selective media (kanamycin or Basta on 0.5× MS +0.6% agar) and propagated on soil. Imaging of seedling roots was performed as described below.
Cloning/RT PCR
All PCR-generated cloning fragments used in this study were produced using Phusion DNA polymerase (Thermo Fisher). All oligonucleotide primers were synthesized by Integrated DNA Technologies. Detailed cloning information and primer sequences can be found in Supplemental Tables 2 and 3. All clones were sequence verified by Sanger Sequencing (UW-Madison Biotech Center).
Expression of mOrange-tagged Rabs in scd1-1 lines was verified by RT-PCR with primers JM15 and JM16 specific for the mOrange tag, and isolation of mRNA was verified in each sample using primers against UBQ10 (JM17 and JM18). Briefly, RNA from seedlings was isolated using the RNeasy plant mini kit (Qiagen). Following isolation, 1 μg of RNA was used in a reverse transcriptase reaction with M-MuLV reverse transcriptase (New England Biolabs) according to the manufacturer’s instructions. Following RT-PCR, first-strand cDNA was diluted 25-fold in a PCR reaction using primers JM15 and JM16 (mOrange) or JM17 and JM18 (UBQ10).
GST Binding Experiments
GST-fusion proteins were purified using standard techniques. Briefly, frozen Escherichia coli pellets (containing IPTG induced overnight expression of GST-fusion proteins) were thawed in GST buffer (PBS, pH 7.4, 500 mM NaCl, 0.1% Tween 20, and 1 mM BME) in the presence of 1 mM lysozyme plus PMSF (1 mM) and Benzamadine (1 mM) and then subjected to sonication for 2 min of total on time (20 s on, 40 s off). Pellets were clarified by centrifugation at 23,700g before incubation with glutathione resin for 1 h at 4°C and batch washed three times with GST buffer. For GST pull-down experiments, immobilized GST fusion proteins (50 μL) were incubated with 1.5 mL of T87 tissue culture extract (3-d-old T87 cells were lysed in lysis buffer [50 mM HEPES, pH 7.6, 100 mM KCl, 0.5 mM EDTA, 0.5 mM MgCl, 0.05% Nonidet P-40, and 10% glycerol + protease and/or phosphatase inhibitors] as a 50:50 slurry of pelleted cells to lysis buffer with depressurized nitrogen [1500 p.s.i. for 20 min on ice in a cell disruption vessel; Parr Instruments] and rotated for 15 min at 4°C after addition of 0.5% Triton X-100 followed by clarification with ultracentrifugation at 134,878g for 20 min in a TLA100.3 rotor] for 1 h at 4°C then washed with lysis buffer (three times) before elution with 10 mM glutathione in elution buffer (50 mM Tris, pH 8.1, and 150 mM NaCl). Eluants were analyzed via SDS-PAGE and immunoblotting as described in figure legends. Affinity-purified antibodies against SCD1 and SCD2 were generated as described (Falbel et al., 2003; McMichael et al., 2013).
Co-IP/TAP Purification
Co-IPs were performed from cell extract prepared as described above (for each panel, the same extract is used for each condition). For co-IP, anti-SCD2 antibodies were covalently linked to Protein A beads (Bio-Rad) using dimethylpimelimidate (Sigma-Aldrich). To covalently link beads, Protein A beads were washed in PBS with 0.1% Tween 20 (PBS-t) three times. Beads were then resuspended in PBS-t plus 55 μg of SCD2 antibody (McMichael et al., 2013) and incubated for 1 h at room temperature. Following incubation, beads were washed three times with PBS-t followed by three washes with 0.2 M sodium borate, pH 9.0. Dimethylpimelimidate (22 mM) was added and the beads were incubated for 30 min at room temperature. Beads were then washed three times with 0.2 M ethanolamine, pH 8.5, and 200 mM NaCl. For immunoprecipitation, antibody-linked beads (50 μL) were incubated with ∼1.5 mL of plant cell extract (prepared as described above) and washed with lysis buffer (described above) three times, and bound proteins were eluted with 100 mM glycine and trichloroacetic acid (TCA) precipitated. TCA pellets were analyzed using mass spectroscopy (described below) or immunoblot techniques.
GS-TAP fusions to SCD1 and SCD2 (or GS-TAP GFP) were generated using gateway cloning techniques into pKNGSTAP (Van Leene et al., 2011). Transgenic PSBd cell lines were generated, and TAP of GS-SCD1 and GS-SCD2 was performed as described (Van Leene et al., 2011), with some modifications (see below). Briefly, 6-d-old transgenic PSBd cell lines expressing GS-SCD1, GS-SCD2, or GS-GFP were lysed using depressurized nitrogen in lysis buffer (as described in GST binding experiment above). Following lysis, extracts were incubated with 0.5% Triton X-100 and clarified (as described in GST binding experiment above). After clarification, extracts were incubated with human IgG Sepharose 6 (GE Healthcare) for 1 h at 4°C. Following incubation, beads were batch washed with wash buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 0.1% [v/v] Nonidet P-40, and 5% [v/v] ethylene glycol) three times, and incubated with ∼200 units of TEV protease for 60 min at 16°C. The supernatant was collected and incubated with preequilibrated Streptavidin Sepharose Resin (GE Healthcare) for 1 h at 4°C. Following incubation, beads were batch washed with wash buffer three times and eluted with desthiobiotin in wash buffer. Eluants were TCA precipitated and analyzed by immunoblot or mass spectroscopy analysis.
Mass Spectroscopy
Enzymatic “in Liquid” Digestion
Purified protein samples were TCA/acetone precipitated (10% TCA and 28% acetone final) and then were pellets resolubilized and denatured in 7.5 μL of 8 M urea/50 mM NH4HCO3 (pH 8.5)/1 mM Tris-HCl for 5 min. Samples were subsequently diluted to 30 μL for a reduction step with 1.25 μL of 25 mM DTT, 2.5 μL methanol, and 18.75 μL 25 mM NH4HCO3 (pH 8.5). Samples were incubated at 50°C for 15 min and cooled on ice to room temperature and then 1.5 μL of 55 mM IAA was added for alkylation. Samples were incubated in darkness at room temperature for 15 min. Reactions were quenched by adding 4 μL of 25 mM DTT. Subsequently, 2 μL of trypsin/LysC solution (100 ng/μL trypsin/LysC Mix from Promega in 25 mM NH4HCO3) and 12.5 μL of 25 mM NH4HCO3 (pH 8.5) were added to generate a 50 μL final volume. Digestion was conducted for 2 h at 42°C and then an additional 1 μL of trypsin/LysC solution was added. Digestion proceeded overnight at 37°C. Reactions were terminated by acidification with 2.5% trifluoroacetic acid added to a 0.3% final concentration.
NanoLC-MS/MS
Digests were cleaned up using OMIX C18 SPE cartridges (Agilent) per the manufacturer’s protocol and eluted in 20 μL of 60/40/0.1% acetonitrile /water/trifluoroacetic acid, dried to completion in the SpeedVac, and finally reconstituted in 20 μL of 0.1% formic acid. Peptides were analyzed by nanoLC-MS/MS using the Agilent 1100 nanoflow system connected to a new generation hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap Elite; Thermo Fisher Scientific) equipped with an EASY-Spray electrospray source. Chromatography of peptides prior to mass spectral analysis was accomplished using capillary emitter column (PepMap C18, 3 µM, 100 Å, 150 × 0.075 mm; Thermo Fisher Scientific) onto which 3 μL of extracted peptides was automatically loaded. The NanoHPLC system delivered solvents A, 0.1% (v/v) formic acid, and B, 99.9% (v/v) acetonitrile, 0.1% (v/v) formic acid, at 0.50 µL/min to load the peptides (over a 30-min period) and 0.2 µL/min to elute peptides directly into the nano-electrospray with gradual gradient from 3% (v/v) B to 30% (v/v) B over 77 min and concluded with 5 min fast gradient from 30% (v/v) B to 50% (v/v) B, at which time a 5-min flash-out from 50 to 95% (v/v) B took place. As peptides eluted from the HPLC-column/electrospray source survey, MS scans were acquired in the Orbitrap with a resolution of 120,000 followed by MS2 fragmentation of the 20 most intense peptides detected in the MS1 scan from 300 to 2000 m/z; redundancy was limited by dynamic exclusion.
Data Analysis
Raw MS/MS data were converted to mgf file format using MSConvert (ProteoWizard: Open Source Software for Rapid Proteomics Tools Development) for downstream analysis. Resulting mgf files were used to search against Arabidopsis thaliana TAIR10 amino acid sequence database with a decoy reverse entries and a list of common contaminants (70,857 total entries) using in-house Mascot search engine 2.2.07 (Matrix Science) with variable methionine oxidation with asparagine and glutamine deamidation. Peptide mass tolerance was set at 15 ppm and fragment mass at 0.6 D. Protein annotations, significance of identification, and spectral-based quantification were done with help of Scaffold software (version 4.3.2; Proteome Software). Peptide identifications were accepted if they exceeded specific database search engine thresholds. Mascot identifications required that at least ion scores must be greater than both the associated identity scores and 20. Protein identifications were accepted if they contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Glycerol Gradient Velocity Sedimentation
T87 tissue culture extract generated (as described in GST pull-down above) and then separated on a 10 to 40% (v/v) glycerol gradient by centrifugation at 35,000 rpm for 16 h at 4°C in an SW 50.1 rotor. Following centrifugation, fractions were collected and analyzed by immunoblot analysis following by SDS-PAGE as described in figure legends. Densitometry was measured using Photoshop, and graphs were generated in GraphPad Prism. For protein standards, the following proteins were used: ovalbumin (Sigma-Aldrich), BSA (Sigma-Aldrich), yeast alcohol dehydrogenase (Sigma-Aldrich), sweet potato β-amylase (Sigma-Aldrich), catalase (Sigma-Aldrich), apoferritin (MP Biomedicals), and thyroglobulin (Sigma-Aldrich). Following centrifugation, SDS-PAGE gels were Coomassie stained and quantified as described above and used to generate standard curves from which experimental S-values were determined to make native molecular weight determinations.
Microscopy
All colocalization experiments, ES2-treated PIN2-GFP experiments, and GFP-SEC15B experiments were conducted on a confocal laser scanning microscope (Nikon A1R-Si+). For colocalization studies, roots of 5-d-old seedlings were grown on 0.5× MS plus 1% agar (continuous light T8 fluorescent bulb ∼120 µmol m−2 s−1; w/v) and imaged. For ES2 treatments, 5-d-old seedlings grown on 0.5× MS 1% agar (w/v) plates were placed in 0.5× MS plus 40 µM ES2 for 2 h at room temperature, and roots were imaged by CLSM. A single focal plane was used to determine the number of punctae per cell. For CHX plus ES2 experiments, seedlings were grown as described above and left untreated or pretreated with 50 µM CHX for 30 min before additional treatment with 50 µM CHX with and without 40 µM ES2 for 120 min. To verify our colocalization analysis, images were processed using ImageJ Coloc 2 plug-in. Briefly, and background subtraction from candidate images was performed using rolling ball subtraction with a 200-pixel ball size. ROIs were selected and run through Coloc 2 plug-in with 100 Costes randomizations using a PSF of 3. All graphs were generated in GraphPad Prism. All line scans were performed using NIS-Elements software with a 2-pixel width, and intensity over distance of the line scan was plotted using GraphPad Prism.
For PIN2-GFP plus BFA trafficking experiments, seedlings were pretreated with BFA for 60 min in 0.5× MS liquid media and followed by washout with 0.5× MS liquid medium for different lengths of time (0, 20, 40, and 60 min). For PIN2-GFP BFA+CHX trafficking experiments, seedlings were pretreated with CHX for 30 min in 0.5× MS liquid media, followed by washout with CHX and BFA for 60 min in 0.5× MS liquid media, and/or finally by washout with 0.5× MS liquid medium for different lengths of time (0, 20, 40, and 60 min). Following treatment, seedlings were imaged by CLSM (Leica TCS SP5 AOBS).
Plant Growth with ES2
Wild-type (Col-0), scd1-1, scd1-2, and scd 2-1 mutant plants were grown vertically on 0.5× MS 1% (w/v) agar with either DMSO or 30 µM ES2, under constant light (T8 fluorescent bulb ∼120 µmol m−2 s−1) at 22°C. Seedling images were taken using a digital camera (Nikon CoolPix S8100) and the root length of 13-d-old seedlings was measured using available tools in Photoshop. Graphs were generated using GraphPad Prism.
Quantification and Statistical Analysis
All statistically significance calculations were performed using the Student’s t test using GraphPad Prism software and are shown in Supplemental Data Set 2. For PIN2-GFP trafficking in the presence of BFA and/or CHX, sample sizes and P values are indicated in the figure legends. For ES2-treated seedling growth experiments, P values are indicated in the figure legend, and sample sizes for DMSO treated are as follows: Col-0 (26 roots), scd1-1 (26 roots), scd1-2 (28 roots), and scd2-1 (10 roots). Sample sizes for ES2 treated are as follows: Col-0 (26 roots), scd1-1 (34 roots), scd1-2 (26 roots), and scd2-1 (17 roots). For PIN2-GFP ES2 treatment experiments, the P values are indicated in the figure legends, and sample sizes are Col-0 (8 roots, 367 cells), scd1-2 (5 roots, 210 cells), and scd2-1 (4 roots, 221 cells). Significance calculations of colocalization experiments are described in the “Microscopy” section.
Accession Numbers
The accession numbers for DNA sequences used in this study are as follows: SCD1, AT1G4940; SCD2, AT3G48860; RabE1a, AT3G53610; RabE1b, AT5G59840; RabE1c, AT3G46060; RabE1d, AT5G03520; RabE1e, AT3G09900; RabA5b, AT3G07410; RabH1c, AT4G98890; RabC1, AT1G43890; RabA5e, AT1G05810; RabA4a, AT5G65270; RabD2b, AT5G47200; and SEC15B, AT4G02350.
Supplemental Data
Supplemental Figure 1. Tandem Affinity Purified GS-GFP.
Supplemental Figure 2. PIN2-GFP Trafficking in the Presence of CHX and ES2.
Supplemental Figure 3. RabD2b Overexpression Does Not Rescue scd1-1.
Supplemental Table 1. Mascot Protein Scores and Exponentially Modified Protein Abundance Index for GS-TAP Purifications.
Supplemental Table 2. Constructs Used in This Study
Supplemental Table 3. Primers Used in This Study
Supplemental Data Set 1. Detailed LC/MS/MS Data Sheet of Proteins Identified with GS-TAP and Co-IP Experiments Performed in This Study.
Supplemental Data Set 2. t Test Tables
AUTHOR CONTRIBUTIONS
J.R.M., J.J.C., J.P., and S.Y.B. designed the research. J.R.M., T.H., C.W., J.J.C., and Y.T. performed experiments. J.R.M., T.H., C.W., Y.T., J.P., and S.Y.B. analyzed data. J.R.M. and S.Y.B. wrote the article.
Acknowledgments
We thank members of the Bednarek lab and Richard Amasino for helpful discussions and critically reading the manuscript. We thank Elle Kielar Grevstad (UW-Madison Biochemistry) for assistance with confocal microscopy and data analysis. We thank Grzegorz Sabat for performing mass spectroscopy (UW-Madison Biotechnology Center) and for help with data analysis. We thank Glenn Hicks for providing ES2 and Christopher Grefen for pUBQN:XFP-Dest vectors. We also thank Sheng Yang He for the kind gift of RabE1 antibody. J.M. is supported by a fellowship from the National Institutes of Health (F32 GM112446-02). This work was supported by funding to S.Y.B. from the UW-Madison College of Agriculture and Life Sciences and the National Science Foundation (Awards 1121998 and 1614915), and to J.P. from the National Natural Science Foundation of China (31670283, 31370313, and 91317304).
References
- Ahn C.S., Han J.A., Pai H.S. (2013). Characterization of in vivo functions of Nicotiana benthamiana RabE1. Planta 237: 161–172. [DOI] [PubMed] [Google Scholar]
- Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., Ritter B. (2010). The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37: 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaire P.D., Ritter B., Thomas S., Burman J.L., Denisov A.Y., Legendre-Guillemin V., Harper S.Q., Davidson B.L., Gehring K., McPherson P.S. (2006). Connecdenn, a novel DENN domain-containing protein of neuronal clathrin-coated vesicles functioning in synaptic vesicle endocytosis. J. Neurosci. 26: 13202–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S., Kikuchi A., Hata Y., Isomura M., Takai Y. (1990). Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J. Biol. Chem. 265: 13007–13015. [PubMed] [Google Scholar]
- Boehm C.M., Obado S., Gadelha C., Kaupisch A., Manna P.T., Gould G.W., Munson M., Chait B.T., Rout M.P., Field M.C. (2017). The trypanosome exocyst: A conserved structure revealing a new role in endocytosis. PLoS Pathog. 13: e1006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L., Smertenko A.P., Pérez-Gómez J., Hussey P.J., Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J. Cell Sci. 122: 4383–4392. [DOI] [PubMed] [Google Scholar]
- Chow C.M., Neto H., Foucart C., Moore I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corpet F., Gouzy J., Kahn D. (1998). The ProDom database of protein domain families. Nucleic Acids Res. 26: 323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. (2004). Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86: 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrčková F., Grunt M., Bezvoda R., Hála M., Kulich I., Rawat A., Zárský V. (2012). Evolution of the land plant exocyst complexes. Front. Plant Sci. 3: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Dong G., Medkova M., Novick P., Reinisch K.M. (2007). A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol. Cell 25: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drdová E.J., Synek L., Pečenková T., Hála M., Kulich I., Fowler J.E., Murphy A.S., Zárský V. (2013). The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 73: 709–719. [DOI] [PubMed] [Google Scholar]
- Falbel T.G., Koch L.M., Nadeau J.A., Sack F.D., Bednarek S.Y. (2003). SCD1 is required for cytokinesis and polarized cell expansion in Arabidopsis thaliana [corrected]. Development 130: 4011–4024. [DOI] [PubMed] [Google Scholar]
- Farnsworth C.L., Feig L.A. (1991). Dominant inhibitory mutations in the Mg(2+)-binding site of RasH prevent its activation by GTP. Mol. Cell. Biol. 11: 4822–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M., Synek L., Pecenková T., Toupalová H., Cole R., Drdová E., Nebesárová J., Sedinová M., Hála M., Fowler J.E., Zársky V. (2010). The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22: 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Knödler A., Ren J., Zhang J., Zhang X., Hong Y., Huang S., Peränen J., Guo W. (2012). A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J. Biol. Chem. 287: 15602–15609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., et al. (2013). A guanine nucleotide exchange factor for Rab5 proteins is essential for intracellular transport of the proglutelin from the Golgi apparatus to the protein storage vacuole in rice endosperm. Plant Physiol. 162: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T., Uchida W., Arakawa S., Ito E., Dainobu T., Ebine K., Takeuchi M., Sato K., Ueda T., Nakano A. (2007). VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans B.L., Ortiz D., Novick P. (2006). Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103: 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., Novick P. (1999). The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 18: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Hou X., Goody R.S., Itzen A. (2013). Intermediates in the guanine nucleotide exchange reaction of Rab8 protein catalyzed by guanine nucleotide exchange factors Rabin8 and GRAB. J. Biol. Chem. 288: 32466–32474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J.A. III, Sellers J.R. (2011). Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13: 13–26. [DOI] [PubMed] [Google Scholar]
- Harding S. (1999). Protein hydrodynamics. In Protein: A Comprehensive Treatise, Vol. 2, Allen G., ed (JAI Press), pp. 271–305. [Google Scholar]
- Hazak O., Bloch D., Poraty L., Sternberg H., Zhang J., Friml J., Yalovsky S. (2010). A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol. 8: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider M.R., Munson M. (2012). Exorcising the exocyst complex. Traffic 13: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T., Fujita M., Nakamura S., Gotoh K., Motooka D., Murakami Y., Maeda Y., Kinoshita T. (2015). Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol. Biol. Cell 26: 3071–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber L.A., Pimplikar S., Parton R.G., Virta H., Zerial M., Simons K. (1993). Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J. Cell Biol. 123: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A., Pylypenko O., Goody R.S., Alexandrov K., Rak A. (2006). Nucleotide exchange via local protein unfolding--structure of Rab8 in complex with MSS4. EMBO J. 25: 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J., Alfaro G., Beh C.T. (2016). Polarized exocytosis induces compensatory endocytosis by Sec4p-regulated cortical actin polymerization. PLoS Biol. 14: e1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose M., Tollis S., Nair D., Mitteau R., Velours C., Massoni-Laporte A., Royou A., Sibarita J.B., McCusker D. (2015). A quantitative imaging-based screen reveals the exocyst as a network hub connecting endocytosis and exocytosis. Mol. Biol. Cell 26: 2519–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA 107: 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D.A., McMichael C., Walker K.A., Anderson J.C., Bednarek S.Y., Heese A. (2010). Novel functions of Stomatal Cytokinesis-Defective 1 (SCD1) in innate immune responses against bacteria. J. Biol. Chem. 285: 23342–23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I., Sachse M., Arouche N., Goud B., Echard A. (2006). Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16: 1719–1725. [DOI] [PubMed] [Google Scholar]
- Krecek P., Skupa P., Libus J., Naramoto S., Tejos R., Friml J., Zazímalová E. (2009). The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Bloch D., Hazak O., Gutman I., Poraty L., Sorek N., Sternberg H., Yalovsky S. (2007). A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17: 947–952. [DOI] [PubMed] [Google Scholar]
- Li R., Rodriguez-Furlan C., Wang J., van de Ven W., Gao T., Raikhel N.V., Hicks G.R. (2017). Different endomembrane trafficking pathways establish apical and basal polarities. Plant Cell 29: 90–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C., Luschnig C., Kleine-Vehn J. (2013). Posttranslational modification and trafficking of PIN auxin efflux carriers. Mech. Dev. 130: 82–94. [DOI] [PubMed] [Google Scholar]
- Marat A.L., Ioannou M.S., McPherson P.S. (2012). Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell 23: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., McPherson P.S. (2010). The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J. Biol. Chem. 285: 10627–10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- McMichael C.M., Bednarek S.Y. (2013). Cytoskeletal and membrane dynamics during higher plant cytokinesis. New Phytol. 197: 1039–1057. [DOI] [PubMed] [Google Scholar]
- McMichael C.M., Reynolds G.D., Koch L.M., Wang C., Jiang N., Nadeau J., Sack F.D., Gelderman M.B., Pan J., Bednarek S.Y. (2013). Mediation of clathrin-dependent trafficking during cytokinesis and cell expansion by Arabidopsis stomatal cytokinesis defective proteins. Plant Cell 25: 3910–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E., Rivera-Molina F., Novick P. (2012). GTPase networks in membrane traffic. Annu. Rev. Biochem. 81: 637–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J., Müller H., Peterson M., Novick P. (1990). Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol. 110: 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N., Singh K., Garcia-Diaz M. (2010). Structure of the dominant negative S17N mutant of Ras. Biochemistry 49: 1970–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75: 4646–4658. [DOI] [PubMed] [Google Scholar]
- Ortiz D., Medkova M., Walch-Solimena C., Novick P. (2002). Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 157: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino-Lopez G., Dong X., Ben-Aissa K., Bernot K.M., Itoh T., Fukuda M., Kruhlak M.J., Samelson L.E., Shaw S. (2008). Rab35 and its GAP EPI64C in T cells regulate receptor recycling and immunological synapse formation. J. Biol. Chem. 283: 18323–18330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M.L., Schmitz A.J., Thole J.M., Bonner H.K., Otegui M.S., Nielsen E. (2006). A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J. Cell Biol. 172: 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O., Rak A., Durek T., Kushnir S., Dursina B.E., Thomae N.H., Constantinescu A.T., Brunsveld L., Watzke A., Waldmann H., Goody R.S., Alexandrov K. (2006). Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J. 25: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Kaneda M., Chen J., Geitmann A., Zheng H. (2011). A specific role for Arabidopsis TRAPPII in post-Golgi trafficking that is crucial for cytokinesis and cell polarity. Plant J. 68: 234–248. [DOI] [PubMed] [Google Scholar]
- Qi X., Zheng H. (2011). Arabidopsis TRAPPII is functionally linked to Rab-A, but not Rab-D in polar protein trafficking in trans-Golgi network. Plant Signal. Behav. 6: 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour D.M., Dickey C.E., Park S., Bednarek S.Y. (2002). Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 130: 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. (1985). Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell 40: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C., Nebenführ A., Movafeghi A., Stussi-Garaud C., Behnia L., Pimpl P., Staehelin L.A., Robinson D.G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S. (2007). Vesicle trafficking in plant immune responses. Cell. Microbiol. 9: 1–8. [DOI] [PubMed] [Google Scholar]
- Rosa P., Barr F.A., Stinchcombe J.C., Binacchi C., Huttner W.B. (1992). Brefeldin A inhibits the formation of constitutive secretory vesicles and immature secretory granules from the trans-Golgi network. Eur. J. Cell Biol. 59: 265–274. [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5: 518–528. [DOI] [PubMed] [Google Scholar]
- Rybak K., Steiner A., Synek L., Klaeger S., Kulich I., Facher E., Wanner G., Kuster B., Zarsky V., Persson S., Assaad F.F. (2014). Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Dev. Cell 29: 607–620. [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. (2010). Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.K., Krüger F., Beckmann H., Brumm S., Vermeer J.E.M., Munnik T., Mayer U., Stierhof Y.D., Grefen C., Schumacher K., Jürgens G. (2014). Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr. Biol. 24: 1383–1389. [DOI] [PubMed] [Google Scholar]
- Sivars U., Aivazian D., Pfeffer S.R. (2003). Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425: 856–859. [DOI] [PubMed] [Google Scholar]
- Sommer B., Oprins A., Rabouille C., Munro S. (2005). The exocyst component Sec5 is present on endocytic vesicles in the oocyte of Drosophila melanogaster. J. Cell Biol. 169: 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth E.B., Imboden L., Hauck P., He S.Y. (2009). Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol. 149: 1824–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10: 513–525. [DOI] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Dhonukshe P., Brewer P.B., Friml J. (2006). Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell. Mol. Life Sci. 63: 2738–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellmann M., Rybak K., Thiele K., Wanner G., Assaad F.F. (2010). Tethering factors required for cytokinesis in Arabidopsis. Plant Physiol. 154: 720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Eeckhout D., Persiau G., Van De Slijke E., Geerinck J., Van Isterdael G., Witters E., De Jaeger G. (2011). Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol. Biol. 754: 195–218. [DOI] [PubMed] [Google Scholar]
- Vetter M., Stehle R., Basquin C., Lorentzen E. (2015). Structure of Rab11-FIP3-Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11. Nat. Struct. Mol. Biol. 22: 695–702. [DOI] [PubMed] [Google Scholar]
- Vukašinović N., Žárský V. (2016). Tethering complexes in the Arabidopsis endomembrane system. Front. Cell Dev. Biol. 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C., Collins R.N., Novick P.J. (1997). Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137: 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N.C., Brennwald P., Kabcenell A.K., Garrett M., Novick P. (1992). Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 2017–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yan X., Chen Q., Jiang N., Fu W., Ma B., Liu J., Li C., Bednarek S.Y., Pan J. (2013). Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25: 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C.J., et al. (2011). Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA 108: 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Guo W. (2015). The exocyst at a glance. J. Cell Sci. 128: 2957–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Tan X., Wu C., Cao K., Li Y., Bao Y. (2013). Regulation of cytokinesis by exocyst subunit SEC6 and KEULE in Arabidopsis thaliana. Mol. Plant 6: 1863–1876. [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., Barr F.A. (2010). Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191: 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., et al. (2016). Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. USA 113: E41–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Camacho L., Wee E., Batoko H., Legen J., Leaver C.J., Malhó R., Hussey P.J., Moore I. (2005). A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17: 2020–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]