Using optimized protocols, nuclear photoreceptor genes were successfully modified or inactivated in the alga Chlamydomonas via directed gene targeting using zinc-finger nucleases or the nuclease Cas9.
Abstract
The fast-growing biflagellated single-celled chlorophyte Chlamydomonas reinhardtii is the most widely used alga in basic research. The physiological functions of the 18 sensory photoreceptors are of particular interest with respect to Chlamydomonas development and behavior. Despite the demonstration of gene editing in Chlamydomonas in 1995, the isolation of mutants lacking easily ascertained newly acquired phenotypes remains problematic due to low DNA recombination efficiency. We optimized gene-editing protocols for several Chlamydomonas strains (including wild-type CC-125) using zinc-finger nucleases (ZFNs), genetically encoded CRISPR/associated protein 9 (Cas9) from Staphylococcus aureus and Streptococcus pyogenes, and recombinant Cas9 and developed protocols for rapidly isolating nonselectable gene mutants. Using this technique, we disrupted the photoreceptor genes COP1/2, COP3 (encoding channelrhodopsin 1 [ChR1]), COP4 (encoding ChR2), COP5, PHOT, UVR8, VGCC, MAT3, and aCRY and created the chr1 chr2 and uvr8 phot double mutants. Characterization of the chr1, chr2, and mat3 mutants confirmed the value of photoreceptor mutants for physiological studies. Genes of interest were disrupted in 5 to 15% of preselected clones (∼1 out of 4000 initial cells). Using ZFNs, genes were edited in a reliable, predictable manner via homologous recombination, whereas Cas9 primarily caused gene disruption via the insertion of cotransformed DNA. These methods should be widely applicable to research involving green algae.
INTRODUCTION
Light provides both energy and information to guide or affect plant and algal growth, development, orientation, adaptation, and stress responses. Whereas energy conversion via photosynthesis is widely understood, our knowledge of the signaling processes initiated by light through sensory photoreceptors remains fragmentary, despite considerable research over the past three decades. The fast, synchronized growth of Chlamydomonas reinhardtii (hereafter Chlamydomonas), its ability to grow heterotrophically, and the extensive knowledge of its biochemistry and cellular and molecular biology that has accumulated over the past decades has rendered it an excellent organism to study complex networks of sensory photoreceptors. To date, 18 photoreceptor genes have been assigned in the Chlamydomonas genome (Figure 1). Some of these genes are more or less universal, such as UVR8, which encodes a homomultimeric UV-B photoreceptor that monomerizes upon UV-B reception to regulate gene expression (e.g., in UV-B acclimation and stress responses). However, the specific physiological functions are largely unknown for the light-sensitive cryptochrome proteins, of which Chlamydomonas contains four variants: aCRY, pCRY, CRY-DASH-1, and CRY-DASH-2 (Spexard et al., 2014; Beel et al., 2012). Nevertheless, there is evidence that aCRY modulates the expression of several genes in response to blue, orange, and red light. The unique red light response of aCRY has been linked to the neutral radical of the flavin chromophore acting as a sensor that absorbs light over almost the entire visible spectral range up to 680 nm (Spexard et al., 2014; Beel et al., 2012). Additionally, recent evidence points to a central role for pCRY in controlling the circadian clock and the algal life cycle (Müller et al., 2017) and to an involvement of CRY in the sexual life cycle (Zou et al., 2017).
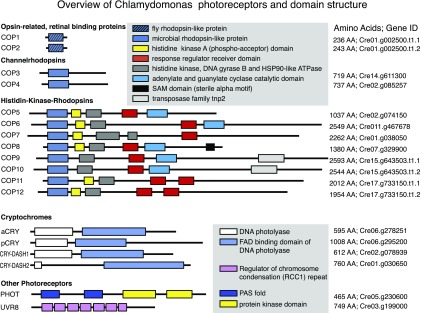
Figure 1.
Chlamydomonas Photoreceptors.
Overview of Chlamydomonas photoreceptors and domain structure. Gene IDs taken from Phytozome V12. AA, number of amino acids.
Although Chlamydomonas is widely used in basic and applied research and important discoveries including sensory photoreceptors have been made using this organism, functional studies involving Chlamydomonas have been limited by difficulties in modifying or inactivating genes by directed gene targeting. Homologous recombination (HR) between endogenous DNA and homologous DNA templates in Chlamydomonas is rare because double-strand breaks (DSBs)—if they occur—are in most cases repaired via error prone nonhomologous end joining (NHEJ) rather than high-fidelity homology-directed repair (HDR) (Zorin et al., 2005), thus impeding targeted gene modification. More recently, several attempts were made to improve the HDR frequency and suppress NHEJ in Chlamydomonas. A single-stranded DNA approach enabling marked suppression of random integration of template DNA allowed for the inactivation of the phototropin-encoding PHOT gene in the nonmotile Chlamydomonas strain CW15-302 (also named CC-4350) with an efficiency of ∼1% in coselected mutants (Zorin et al., 2009). The disruption of PHOT suppressed both eyespot size reduction and downregulation of channelrhodopsin at high light intensities (Trippens et al., 2012), as well as high-energy nonphotochemical quenching, which dissipates harmful excessive light energy in photosystem II as heat to prevent protein damage (Petroutsos et al., 2016).
Further advances in developing techniques for increasing the frequency of HDR in Chlamydomonas were realized by exploiting zinc-finger nuclease (ZFN) technology (Bibikova et al., 2003), which produces targeted DNA-DSBs suitable for the insertion of templates via HDR. COP3, which encodes channelrhodopsin 1 (ChR1), was inactivated by HDR using the ZFN approach (Sizova et al., 2013). ChR1 and ChR2 are the most well-characterized sensory photoreceptors in Chlamydomonas to date (Schneider et al., 2015; Deisseroth and Hegemann, 2017). The inactivation of COP3 and COP4 (which encodes ChR2) using antisense approaches revealed that both proteins are photoreceptors for phototactic and photophobic responses via previously described photocurrents (Harz et al., 1992; Holland et al., 1996; Braun and Hegemann, 1999; Sineshchekov et al., 2002; Govorunova et al., 2004; Berthold et al., 2008). Electrical studies in ChR1 and ChR2 expressing Xenopus laevis oocytes and HEK (human embryonic kidney) cells revealed that both proteins function as light-gated ion channels (Nagel et al., 2002, 2003). More detailed physiologic studies of ChR function and processing would require disruption of the genes encoding both ChR1 and ChR2 and replacement with genes encoding modified ChR variants. Moreover, neither of the above-described earlier approaches (single-stranded DNA or ZFN based) enabled a gene of interest (GOI) to be targeted in motile Chlamydomonas strains (Sizova et al. 2013).
Although the ZFN technology has proven to be useful in single knockout experiments and for promoting HDR, this technology was likely too limited to be used to investigate the four cryptochrome and eight enzyme rhodopsin photoreceptors (Figure 1) with strong sequence homology and (probably) functions. The latter family, named histidinkinase rhodopsins (HKR1–HKR8, encoded by the genes COP5–COP12) is characterized by the presence of microbial-type sensory rhodopsins connected C-terminally to a histidine kinase and a response regulator (Kateriya et al., 2004). HKR1, 2, and 4 to 6 contain a C-terminal guanylyl- or adenylyl-cyclase effector domain, suggesting that HKRs regulate cGMP and cAMP (Figure 1). Little is known about the HKRs aside from data regarding the photophysics and photochemistry of the bimodally switchable HKR1 photoreceptor (Luck et al., 2012; Penzkofer et al., 2014; Luck and Hegemann, 2017).
By far the most popular and widely used programmable nuclease system for gene modification in many organisms is clustered regularly interspaced short palindromic repeat (CRISPR)/associated protein 9 (Cas9) technology (Jinek et al., 2012, 2013; Mali et al., 2013; Cong et al., 2013). The CRISPR DNA-targeting system consists of three components: Cas9 nuclease protein; complementary base-pairing CRISPR RNA (crRNA), which includes the target sequence (protospacer); and transactivating CRISPR RNA (tracrRNA), which forms a secondary scaffold structure recognized by Cas9. By inserting a small linker sequence, the crRNA:tracrRNA duplex can be expressed as a single guide RNA (sgRNA) that binds to the Cas9 nuclease to form a Cas9/sgRNA ribonucleoprotein (RNP) complex. The only prerequisite for target DNA recognition by the Cas9 RNP is the protospacer adjacent motif (PAM), which varies among Cas9 variants and is located immediately downstream of the target sequence. In the first Cas9 discovered, SpCas9 (from Streptococcus pyogenes), the PAM motif is NGG, whereas in Cas9 from Staphylococcus aureus (SaCas9), the PAM motif is NNGRRT, where R indicates either adenine or guanine.
Three research groups have reported using the CRISPR/Cas9 system in Chlamydomonas (Jiang et al., 2013, 2014; Baek et al., 2016; Shin et al., 2016; Jiang and Weeks, 2017). Promising results were achieved when Cas9/sgRNA RNP complexes assembled in vitro were delivered into Chlamydomonas cells via electroporation (Baek et al., 2016; Shin et al., 2016). After targeting the photosynthesis-associated genes ZEP, CpFTSY, CpSRP43, and ChlM, some of the colonies (which were preselected with an antibiotic resistance marker) appeared pale due to reduced antenna size and/or chlorophyll synthesis (Kirst et al., 2012; Meinecke et al., 2010). The targeting efficiency (targeted colonies/selected colonies) obtained from visual clone identification in these studies was ∼1:600 (0.17%), 1:73 (1.4%) (Shin et al., 2016), and 0.56% (Baek et al., 2016). Earlier, another group established a plasmid-based CRISPR/Cas9 system in Chlamydomonas (Jiang et al., 2014). In their initial approach, the authors targeted the peptidyl-prolyl cis-trans isomerase gene FKB12, allowing for positive selection of cells with FKB12 gene knockouts on medium containing rapamycin. This resulted in the production of one mutant colony out of 16 transformations, or one colony with a modified target locus per 1.5 × 109 initial cells (Jiang et al., 2014). Later, using a hybrid Cas9/sgRNA expression construct, the efficiency was improved to yield 13 colonies out of four transformations (equivalent to ∼1 colony per 3 × 107 initial cells) (Jiang and Weeks, 2017). Additionally, positive selection of prototrophic strains after precise repair of the point mutation in the ARG gene in the auxotrophic mutant, arg7, was demonstrated using the hybrid Cas9/sgRNA gene coelectroporated with a short, 80-nucleotide, ssODN (oligodeoxynucleotide) with 5′ and 3′ phosphothioate (PTO)-protected ends. In this study, seven arginine prototroph clones were isolated from two transformation reactions (∼1 colony per 2.5 × 107 initial cells) (Jiang and Weeks, 2017). These results indicate that the CRISPR/Cas9 system is suitable for gene modification in Chlamydomonas. However, the efficiency of the reported methods is low, and the methods do not enable routine disruption of nonselectable or nonphenotypic genes.
Our goal in this study was to overcome the drawbacks associated with existing methods and to optimize gene targeting in Chlamydomonas by developing a technology that (1) allows for efficient gene disruption and ultimately precise and predictable gene editing via HR; (2) enables efficient isolation of non-phenotypic mutants; and (3) is applicable to numerous Chlamydomonas strains. Toward this end, we optimized ZFN technology to enable precise gene modification in Chlamydomonas. We then modified these protocols further for CRISPR/Cas9 to disrupt genes rapidly and in a cost-effective, routine manner.
RESULTS
A Selectable Marker Repair Assay to Evaluate Target-Specific Zinc-Finger Nucleases
To elucidate the roles of ChR1 and ChR2 in phototactic and photophobic responses in detail, single and double ChR1 and ChR2 knockouts are required. We previously reported the development and application of ChR1-specific ZFNs in the nonmotile Chlamydomonas strain CW15-302 (Sizova et al., 2013). In a continuation of this work, we used these nucleases in a motile Chlamydomonas strain to develop ChR2-specific ZFNs. We analyzed ChR2 for ZFN target sites using the ZiFiT (Zinc Finger Targeter) database (Sander et al., 2007). As predictions gave rise to only a poorly active ChR2-ZFN pair (hereafter designated ChR2-a-ZFNs), we ordered a set of ChR2-ZFNs from a commercial supplier (hereafter designated ChR2-b-ZFNs). To characterize both nuclease pairs, we used an optimized version (Figure 2A) of the previously reported mutated aphVIII (mut-aphVIII) repair gene targeting selection (GTS) system (Sizova et al., 2013) (explained in detail below) to qualify HDR of nuclear genes in Chlamydomonas.
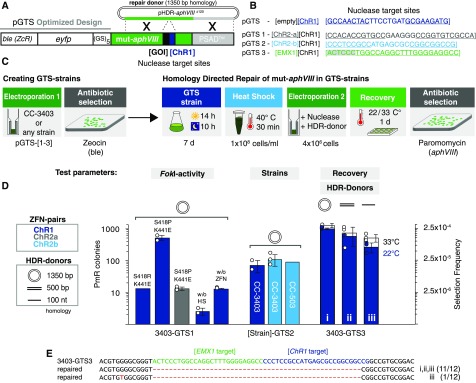
Figure 2.
Optimization of ZFNs and Transformation Parameters.
(A) Schematic drawing of a GTS construct. Transcription of the GTS cassette is controlled by the HSP70A/RBCS2 promoter and PSAD-terminator. ble (phleomycin resistance gene) was used as a selection marker to isolate the GTS strains; the nuclease target site for any GOI can be inserted into mut-aphVIII [GOI][ChR1]. ChR1-ZFNs serve as a control to test the HDR capabilities of GTS strains. eyfp, enhanced yellow fluorescent protein gene; GS5, glycine-serine linker; aphVIII, paromomycin resistance marker; pHDR-aphVIII∆120, HDR plasmid to restore aphVIII functionality.
(B) ZFN target site sequences inserted into mut-aphVIII (pGTS1-3) used to create the GTS-strains. The ChR1-ZFN target sequence served as a control in all pGTS plasmids. pGTS1 includes the ChR2-a-ZFN target site and pGTS2 includes the respective ChR2-b-ZFN target site. pGTS3 includes the target site from the human EMX1 homeobox protein gene and was created to test the HDR capability of Cas9. Binding sites of left and right ZF domains for ChR1, ChR2-a, and ChR2-b are underlined. Binding site of EMX1 gRNA is underlined. PAM is highlighted in gray.
(C) Workflow for the generation of a GTS strain followed by HDR experiments. Electroporation 1, pGTS-[1-3] is electroporated into a Chlamydomonas strain. Single colonies are isolated on Zeocin-containing agar plates. Next, isolated and synchronously cultured GTS strains are concentrated and heat shocked before Electroporation 2. Here, functional nucleases (plasmid-encoded or recombinant) may induce the formation of DNA-DSBs within their respective mut-aphVIII target site and trigger HDR with coapplied donor DNA, which results in selectable paromomycin resistant (PmR) cells. After electroporation and before paromomycin selection, the cells are allowed to recover for 24 h at 22°C or 33°C.
(D) GTS strain experiments. Bars indicate the number of PmR colonies per transformation reaction. Secondary y axis: selection frequencies (PmR colonies/4 × 106 electroporated cells). FokI-domain variants (mutations are indicated) were tested in their efficiency to introduce DNA-DSBs, thereby promoting HDR of mut-aphVIII in 3403-GTS1. Controls without heat shock before transformation (without HS) or without ZFN plasmids (without ZFN). Middle: Functionality of ChR2-ZFNs was tested in GTS-strain 3403-GTS2. The same results were obtained in the CC-503 background (503-GTS2). Right: ChR1-ZFNs (3403-GTS3) in combination with different HDR donors as indicated and recovery temperatures of 22°C and 33°C. Error bars indicate standard deviation and white circles reflect individual data points.
(E) Sequence alignment of amplified mut-aphVIII [EMX1][ChR1] (3403-GTS1) and repaired aphVIII loci from PmR colonies after HDR. Hyphens indicate deleted nucleotides. A point mutation found in one clone from (D) iii is indicated in red.
The GTS construct (ble:yfp:mut-aphVIII; Figure 2A; Supplemental Table 1) for creating the GTS test strains was improved by replacing rarely used (<15%) codons at the 5′ end of the ble (phleomycin [Zeocin]-resistance gene [ZcR]), substituting the previously used gfp gene (Fuhrmann et al., 1999) with the codon-optimized enhanced yfp gene (eyfp) and inserting a glycine-serine linker between eyfp and the mutated aphVIII selection marker. The linker was inserted to separate the ble and eyfp protein domains to potentially preserve their activities and stabilize the fusion protein by preventing premature protein degradation. The aphVIII (paromomycin resistance [PmR]) gene was mutated by inserting target sites for the previously functionally characterized ChR1-ZFNs (Sizova et al. 2013) and the newly designed but uncharacterized ChR2-a-ZFNs (pGTS1-mut-aphVIII [ChR2-a][ChR1]; Figure 2A). Cell wall-deficient, motile Chlamydomonas strain CC-3403 (RU-387 nit1 arg7 cw15 mt−) cells were transformed with pGTS1 via electroporation. Full-length integration of the GTS1 cassette was confirmed by PCR analysis of genomic DNA isolated from ZcR clones. The verified clonal cell line was designated the “GTS-strain.” After a second round of electroporation of the GTS strain with ZFN-encoding plasmids (Figure 2C), functional ZFNs recognized and cleaved their respective mut-aphVIII target sites. The resulting DNA-DSBs triggered the cellular DNA repair machinery to utilize a cosupplied repair-donor to restore aphVIII functionality (Figure 2C), thereby creating selectable PmR clones. The number of PmR clones served as an indicator of nuclease functionality. The GTS approach thus allows for testing and optimization of many parameters that could limit HDR-based modification of Chlamydomonas genes, such as nuclease activity, HDR donor design, and transformation conditions.
Highly Active FokI Enables HDR of Endogenous Genes in Strains CC-3403 and CC-503
The objective of our first experiments was to test the overall functionality of the GTS system in the CC-3403 background. Verified 3403-GTS1 (pGTS1 in CC-3403) cells were grown synchronously before electroporation of ChR1-ZFN plasmids and mut-aphVIII repair templates (pHDR-APHVIIIΔ120; Figures 2A and 2C). Employment of the heat shock protein (HSP) 70A promoter (triggered by a short heat shock of 30 min at 40°C before electroporation) was expected to allow for time-limited ZFN transcription and to minimize possible toxic effects resulting from long-term nuclease expression (Schroda et al., 2000; Sizova et al., 2013).
ZFNs are artificial restriction enzymes consisting of three to four zinc-finger DNA binding modules fused to the heterodimeric nuclease domain FokI (Miller et al., 2007). Using strain 3403-GTS1, we tested two different engineered FokI heterodimer variants (Guo et al., 2010; Sizova et al., 2013) in combination with ChR1-ZFNs. FokI variant S418R/K441E, originally optimized for CW15-302 cells (Sizova et al., 2013), generated only a limited number of PmR colonies after electroporation and selection, similar to the control. The more active FokI variant FokI-S418P/K441E (SP/KE) was more efficient with respect to mut-aphVIII repair, generating ∼500 colonies per electroporation (Figure 2D). For comparison, glass-bead transformation (Kindle, 1990) with ChR1-ZFNs (FokI-SP/KE) resulted in only 6 ± 2 PmR colonies. Thus, we used FokI-KE/SP and electroporation in subsequent ZFN experiments.
We then tested the two new pairs of ChR2-ZFNs. Using ChR2-a-ZFNs (with FokI-SP/KE) in strain 3403-GTS1, very few (n = 15) PmR colonies appeared. By contrast, ChR2-b-ZFNs were 50% more active than the ChR1-ZFNs in strain 3403-GTS2 with both ChR1 and ChR2-b target sites included (mut-aphVIII [ChR2-b][ChR1]) (Figure 2D). Similar repair efficiencies were observed for 503-GTS2 cells derived from strain CC-503 (Figure 2D). We concluded that the previously reported high level of transgene expression in CW15-302 is disadvantageous in combination with the use of highly active FokI-SP/KE ZFNs. The superior performance of the more active ZFN variants in CC-3403 and CC-503 cells compared with CW15-302 cells is thus most likely associated with lower levels of transgene expression (Sizova et al., 2013).
Short HDR Donors Enhance Mutant Screening Efficiency
To simplify donor generation and analysis of potential ChR1 and ChR2 (or any GOI) recombinants, we tested shorter templates in the mut-aphVIII repair assay. We compared circular plasmids (1350 bp), short double-stranded linear fragments with PTO-blocking groups (500 bp), and single-stranded oligodeoxynucleotides (ssODNs) with 5′- and 3′-PTO protections (100 nucleotides) as templates in the GTS system (Liang et al., 2017; Jiang et al., 2017). As expected, a shorter template homology region was correlated with fewer PmR colonies (Figure 2D, dark-blue bars; 994 ± 65, 570 ± 15, and 266 ± 93, respectively, for plasmids, short double-stranded linear fragments with 5′-PTO blocking groups, and ssODNs with 5′- and 3′-PTO protections). However, all tested small donors promoted mut-aphVIII HDR at reasonable frequencies. In our protocol, cells were allowed to recover for 24 h at room temperature (22°C) before plating. Increasing the recovery temperature from 22 to 33°C increased the number of colonies by 25 to 90%, depending on the template (Figure 2D, white bars; 1238 ± 190, 709 ± 427, and 508 ± 130). For each donor type, we sequenced four PmR clones. With the ssODN template, only one PmR clone contained an integrated silent, single point mutation, whereas the 11 other sequenced clones carried repaired aphVIII, as specified by the template (Figure 2E). With respect to the proportion of PmR colonies, we achieved editing efficiencies in the range of 2.5 × 10−5 to 2.5 × 10−4 (1 HDR event per 4000 cells).
ZFN-Mediated Disruption of Nonselectable Genes for ChR1 and ChR2
After these optimization experiments in GTS strains, we next targeted the endogenous ChR1 and ChR2 genes in strain CC-3403. Prior electrode voltage clamp studies revealed that ChR1 and ChR2 directly function as light-gated ion channels (Nagel et al., 2002, 2003). The light sensor and channel pore is confined to the seven-transmembrane helix fragment of the proteins, whereas the long C terminus is likely responsible for membrane targeting to the eyespot (Mittelmeier et al., 2011). First, we tested whether these proteins are the only photoreceptors mediating phototaxis by creating protein-coding disruptive HDR knockin templates for both genes (Figures 3A and 3B). As CC-3403 is an arginine auxotroph and therefore requires arginine supplementation in the medium, we used the argininosuccinate lyase gene ARG7 (pHR11) as a selection marker in cotransformation experiments with ZFNs and HDR donors. Correct donor integration introduced an additional 30 bp to the sequence, referred to as “FLAG” in the following experiments, that was used for unique ODN binding in our HDR-specific PCR screening strategy (Figure 3A, blue arrow). In an experiment targeting the ChR1 gene, eight out of 96 analyzed arginine prototroph clones exhibited donor integration (Figure 3C). In seven chr1 clones, the HDR donor had been correctly inserted (FLAG insert; Figures 3C and 3D, chr1 [1-7]). In one clone, the donor had been integrated via by 5′ HR of the cleavage site, but it also contained additional plasmid DNA fragments. For ChR2, we screened 96 clones each from three independent transformation experiments and found 22 “FLAG” insertions (7.6% on average; Figure 3C). Of the 16 sequenced chr2 clones, 15 contained a correctly inserted HDR donor (Figure 3D, chr2 [1-15]) and in only one clone, the template plus parts of plasmid DNA had been integrated by NHEJ at the 3′site (chr2 [16]). In controls without an HDR donor, we found two mutants out of 96 clones with small mutations at the target site (Figures 3C and 3D, chr2 [17+18]). To generate a strain where both ChRs are disrupted, we transformed strain 3403-chr1-1 with ChR2-ZFNs and ChR2-HDR donor and coselected them using an aphVIII marker gene. In six out of 96 analyzed clones, both ChR1 and ChR2 were disrupted (Figures 3C and 3D, chr1 chr2 [1-6]). Protein immunoblots using a ChR1-specific antiserum (Nagel et al., 2003) demonstrated the absence of ChR1 in the chr1-1 strains, and only very low amounts of ChR1:FLAG (ΔCT) of a reduced size were detected (Figure 3E). Unfortunately, our ChR2 antiserum cross-reacted with ChR1, and due to the similar molecular weights of both ChRs, ChR2 was only detectable by immunoblotting protein extracts from CC-3403-chr1 cells. In strain CC-3403-chr1-1, ChR2 comprised only a minor fraction (Figure 3F) and disappeared in the double knockout strain CC-3403-chr1-1/chr2-1 (Figure 3F). The single and double-knockout strains were motile and suitable for physiological studies.
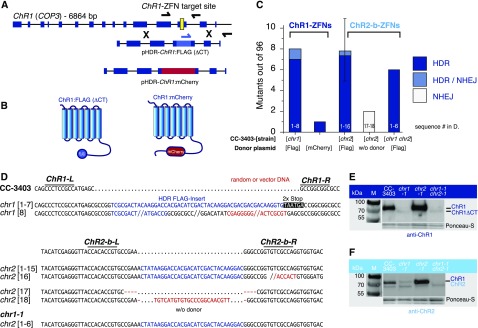
Figure 3.
Inactivation of Nonselectable Genes Using ZFNs.
(A) Schematic diagram of ChR1 (COP3) with exons shown as blue rectangles and the ZFN target site shown in yellow. HDR donors ChR1:FLAG and ChR1:mCherry are illustrated. Homology regions are identical for both constructs, and sequence insertions are centered on the ZFN cleavage site. For the FLAG insert, two stop codons were added in-frame at the 3′end, whereas mCherry was inserted in-frame without stop codons.
(B) 78 amino acids at the C terminus of ChR1:FLAG are missing due to the presence of stop codons after FLAG. The mCherry coding sequence is integrated in-frame.
(C) Generation of chr1 and chr2 mutants in strain CC-3403. Cells were transformed via electroporation of ChR1- or ChR2-ZFN plasmids and HDR donors as indicated. The ChR2-FLAG donor plasmid was constructed in a manner analogous to that of ChR1:FLAG, as shown in (A). The chr1 disruption strain was transformed again to obtain the chr1 chr2 double disruption strains. Bars reflect the number of HDR events found for 96 colonies analyzed. Conditions for ChR2-b-ZFNs were replicated. Error bar shows triplicates ± sd.
(D) Sequencing results of chr1 [1-8] and chr2 [1-18] target locus amplicons. chr1 [8] and chr2 [16] contain additional sequences from the HDR donor plasmid. The clones chr2 [17, 18] were generated without HDR donors, resulting in small InDels (NHEJ; +23, −8 bp) at the cleavage site.
(E) Protein immunoblotting of ChR1 mutants (chr1) using anti-ChR1 antiserum and secondary HRP-conjugated antibody for chemiluminescence detection. M, marker. Ponceau staining was used as a loading control.
(F) Protein immunoblotting of ChR2 mutants (chr2) using anti-ChR2 antiserum and secondary HRP-conjugated antibody for chemiluminescence detection. Total ChR2 abundance in CC-3403 is low (chr1 background). ChR2 depletion is visible in chr1 chr2 double knockouts. Ponceau staining was used as a loading control. The protein below the comigrating ChR1 and ChR2 proteins is labeled unspecifically and is not encoded by ChR1 or ChR2.
Photomovement Analysis of ChR1 and ChR2 Disruption Strains
We performed light scattering (LS) assays to quantify the phototactic sensitivity of the strains (Uhl and Hegemann, 1990). The LS signals from CC-3403 gamete cells were nearly identical to those of previously studied CW2 cells (Berthold et al., 2008). After lights-on, the LS signal linearly rose until it approached a stationary level (Figure 4A). Under low-intensity light (455 nm), the increase was graded with the light intensity over 1.5 log units. However, phototaxis was delayed at higher intensities due to a transient switch from forward to reverse swimming during the phobic response (Figure 4A) (Hegemann and Bruck, 1989). The behavior and light sensitivity in chr2 cells were almost indistinguishable from those of the parent cells at both 455- and 530-nm wavelengths (Figures 4A and 4B, light blue), confirming that ChR1, with an absorption maximum λmax = 495 nm at pH 6.5, is the dominant phototaxis photoreceptor in CC-3403 cells. In contrast, chr1 showed an almost 1000-fold lower sensitivity at 455 nm (Figures 4A and 4B, dark blue), where ChR1 and ChR2 absorb light equally well (Berthold et al., 2008). Photoorientation was almost undetectable in the chr1 chr2 double mutant (Figures 4A and 4B, light gray). These results confirm that while both ChRs are phototaxis photoreceptors, ChR2 has a low abundance and is only a minor contributor to the phototactic sensitivity and performance of CC-3404 cells.
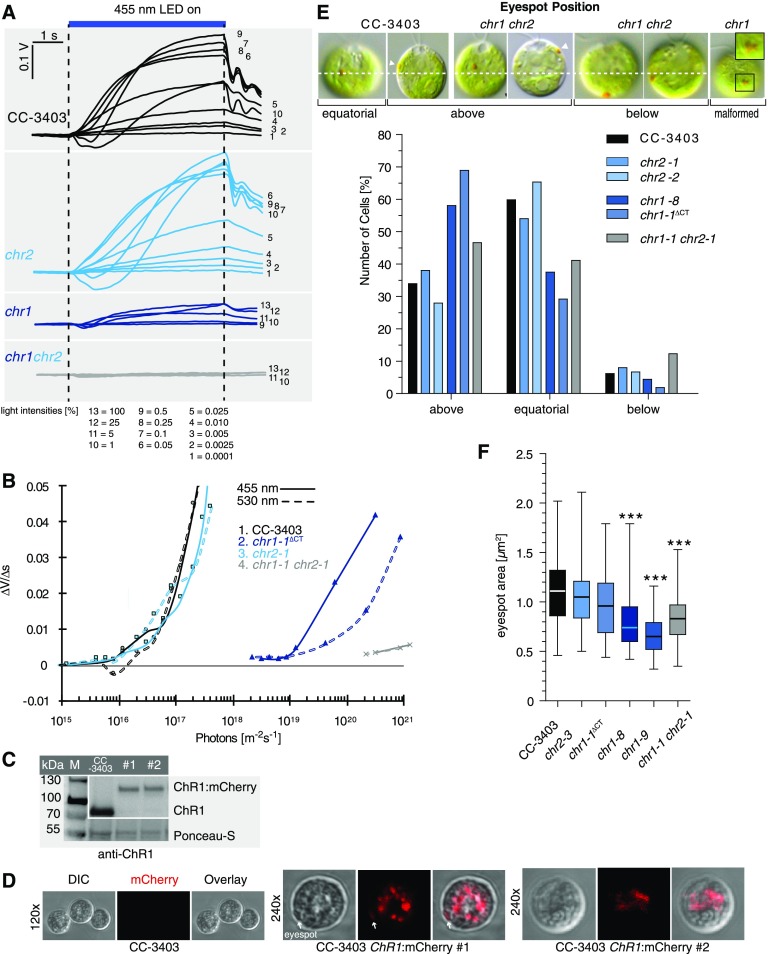
Figure 4.
Physiological Analysis of ChR1 and ChR2 Disruption Strains.
(A) Light-scattering assay of strains as indicated. Phototactic responses to different intensities of blue light (455 nm). The 100% light corresponds to 1.25 × 1021 photons m−2 s−1. Numbers below correspond to the various light intensities employed.
(B) Phototactic sensitivity of various Chlamydomonas lines. Initial linear slopes of the light-scattering responses at 455 nm (solid lines) and 530 nm (dashed lines) were calculated (∆V/∆s) for CC-3403 cells and cells containing chr1, chr2, or chr1 chr2 modifications and plotted against the normalized light intensity.
(C) Protein immunoblotting of two CC-3403 strains with mCherry inserted into ChR1 (#1; #2). Anti-ChR1 antibodies detected ChR1:mCherry (∼110 kD) fusion protein. M, marker; WT, CC-3403 crude extracts. Ponceau staining was used as a loading control.
(D) Confocal microscopy: Live-cell imaging of CC-3403 ChR1:mCherry strains. CC-3403 was used as a control. ChR1:mCherry is mainly located within the cell cytoplasm. Only minor fractions are found within the plasma membrane region of the eyespot (white arrow). The same settings and filters were used in all images. DIC, differential interference contrast.
(E) Eyespot position. DIC images of the indicated ChR1 and ChR2 disruption strains are shown at the top. Dashed line indicates equatorial position and arrowheads the eyespot. For statistical analysis, between 100 and 193 cells of each strain grown under identical conditions were analyzed.
(F) Box plot (whiskers min to max) of the eyespot area of the indicated strains. ANOVA analyses with Tukey's multiple comparison post-test revealed a significant difference (***P < 0.001; Supplemental Table 9) for the marked strains compared with other strains. n = 62 to 65 cells.
Analysis of Eyespots in ChR1 and ChR2 Disruption Strains
As a second physiological approach, we explored the C terminus of ChR1 and its role in the transport of ChR to the eyespot. All HDR templates designed to this point contained a short insert of only ∼30 bp. However, we were interested in labeling algal proteins with fluorescent tags. As proof of principle, we created a new donor that included the coding sequence for mCherry in-frame, interspacing the ChR1-ZFN target site located on the C terminus of the respective ChR1 protein (pHDR-ChR1:mCherry; Figures 3A and 3B). The mCherry sequence was correctly inserted and expressed in ∼1% of analyzed clones (3/288). Confocal microscopy confirmed ChR1 tagging (Figures 4C and 4D). However, only a minor fraction of the ChR1:mCherry protein was targeted to the eyespot (Berthold et al., 2008; Mittelmeier et al., 2011), whereas the main fraction was localized as patches within the cell (Figure 4D). Nevertheless, an eyespot was visible in the strains with an mCherry-tagged ChR1.
Next, we analyzed whether the different ChR-modified strains developed alterations in eyespot positioning and size, as measured by examining the carotenoid-rich eyespot globule layers visible via differential interference microscopy. All strains in nonsynchronous growing cultures had visible eyespots, as exemplified in the wild-type-like CC-3403, strain chr1 chr2, and chr1 in Figure 4E. However, the relative eyespot positions within the cell differed. In the majority of CC-3403 cells (∼60%), the eyespot localized to an equatorial position. Approximately 35% localized more anteriorly toward the flagella-bearing pole, while only ∼5% localized toward the posterior end. Both analyzed chr2 strains had an identical distribution. In the chr1 strains, however, a clear tendency toward a predominantly anterior position was evident (∼60%), and an equatorial position was seen in only ∼30% of the cells. Deletion of the last C-terminal 78 amino acids [Figure 3B, ChR1:FLAG (ΔCT)] of ChR1 (Figure 4E, chr1ΔCT) had an identical effect. In contrast, the chr1 chr2 population exhibited an almost equal distribution of the eyespots between these two positions and a slight increase in the posterior localization. Occasionally, two eyespots were visible in chr1 chr2 cells. Furthermore, malformed eyespots missing the typical roundish-ellipsoid form were sometimes evident in chr1 and chr1 chr2 cells (Figure 4E). In addition, the eyespot area was significantly smaller in the analyzed chr1 and chr1 chr2 strains compared with CC-3403 (Figure 4F). In the other strains, no significant difference from the wild type was evident. These data indicate that ChR1 has a larger impact on eyespot plate positioning and size/stability compared with ChR2. Furthermore, the observation that chr1 chr2 forms visible, albeit smaller, eyespots supports the idea that additional eyespot proteins (EYE2 and others) are required for the initial organization of the eyespot globule plate and that this process is ChR independent (Roberts et al., 2001; Boyd et al., 2011b; Mittelmeier et al., 2013).
Gene Disruption Using Plasmid-Encoded Cas9
Although our ZFN experiments enabled us to specifically modify target genes and respective proteins at acceptable frequencies, it is difficult to design new ZFN pairs, and the number of efficient target sites within a gene is limited. To develop a faster and more flexible gene-editing approach, we endeavored to use the CRISPR/Cas9 system for targeting nonselectable genes in Chlamydomonas. With functional Cas9 protein expression, only the guide RNA needs to be adapted for any new GOI. Low target site restrictions (PAM) and easy guide RNA design facilitate positional modifications.
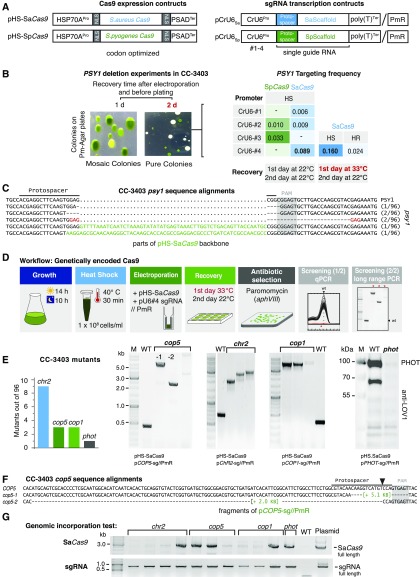
A first prerequisite for CRISPR/Cas9 experiments is sgRNA transcription under the control of an RNA polymerase III promoter (RNAPIII). In general, RNAPIII transcription factor binding sites are inherent to the transcribed sequences, making them unsuitable for sgRNA transcription. One of the few examples of sgRNAs with proximal RNAPIII promoter elements is U6snRNA. Therefore, we employed a previously characterized U6snRNA sequence from chromosome 8 (Jakab et al., 1997) and three other homologous CrU6snRNA sequences of Chlamydomonas. The respective amplified promoter regions (500 bp) were used to drive sgRNA transcription using the appropriate SaCas9 or SpCas9 scaffold sequences (pCrU6 #1-4//PmR; Figure 5A; Supplemental Table 2).
Figure 5.
Gene Disruption by Genetically Encoded Cas9.
(A) Schematic of DNA constructs. The promoter of heat shock protein HSP70A promotes the transcription of Chlamydomonas codon-adapted S. aureus or S. pyogenes Cas9. Guide RNA transcription under the control of different CrU6-promoters (#1-4). Guide sequences were inserted into vectors containing the appropriate SaCas9 or SpCas9 scaffold. The coding sequence of aphVIII (PmR) is also located on the pCrU6-plasmids to enable antibiotic selection using paromomycin. NLS, nuclear localization signal; T, poly-thymine terminator.
(B) The phytoene synthase gene, PSY1, was chosen as a target gene. sgRNA transcription driven by Chlamydomonas U6 promoters (CrU6 #1-4) was assayed. Photographs of a selective agar plates from PSY1 inactivation experiments. If the cells were allowed to recover for 1 d before antibiotic selection on paromomycin-containing agar plates, mixed colonies of wild-type and PSY1-inactivated mutants were obtained. If the cells were allowed to recover for 2 d, pure white colonies with inactivated PSY1 were found. Recovery at 33°C for the first 24 h increased targeting frequencies for HS by approximately one-third. Targeting frequencies (targeted colonies/selected colonies) are the mean of three independent experiments. HS, HSP70A promoter; HR, HSP70A/RBCS2 promoter.
(C) Sequence alignment of amplified psy1 loci from white colonies. Dots indicate spacers, hyphens indicate deleted nucleotides. “PAM” (gray) and “Protospacer” sequences are indicated. PSY1, wild-type gene sequence.
(D) Workflow diagram summarizing steps for generation of mutants using genetically encoded Cas9. Red letters highlight the key step enabling mutant generation and isolation. The use of this two-step screening strategy (qPCR + long-range PCR) enables the isolation of clones having long insertions within the target site.
(E) Genomic DNA of clones that failed in the initial qPCR locus amplification was column purified and subjected to long-range PCR (200-s elongation time). This step identified cop5, chr2, cop1, and phot mutants with target site insertions in the range of 2 to 5 kb. The absence of PHOT protein due to failed phot mutant target site amplification is shown by immunoblotting. Left: Graph summarizing the number of deletion mutants found per 96 analyzed clones. WT, target loci amplicons from CC-3403 genomic DNA.
(F) cop5 #1 and 2 amplicons from (E) were sequenced from both ends. In both cases, fragments of plasmid pCOP5-sg//PmR, used in the respective targeting experiment, had inserted into the target site by NHEJ. Hyphens indicate deleted nucleotides.
(G) Integration of the full-length SaCas9 coding sequence and sgRNA-DNA into the genomes of isolated photoreceptor mutants. WT, negative control; the pHS-SaCas9 and pCOP5-sg//PmR plasmid templates were used as positive controls for PCR.
Disruption of the phytoene synthase-1 gene (PSY1) produces white colonies that are easy to detect and count (Inwood et al., 2008; McCarthy et al., 2004). Therefore, we targeted the two Cas9 variants to PSY1 (PSY1-sg) to test promoter functionality. We chose a PSY1 target site for SaCas9 with a corresponding PAM (5′CGGAGT) that was also suitable for SpCas9 (5′CGG) recognition. The transcription of PSY1-sg from each of the four different CrU6 sgRNA promoters was tested in combination with either SaCas9 or SpCas9 expression (Figure 5A). CC-3403 cells were electroporated with two separate circular plasmids, one containing a codon-optimized Cas9 gene and a second plasmid containing a U6-driven sgRNA gene and a PmR gene to allow for selection of transformed cells. Cells were allowed to recover for 24 h at 22°C and selected on PmR. With transformation conditions optimized for ZFNs, no white colonies were observed. However, we detected a reasonable number of pale green colonies that were potentially derived from colonies mixed with wild-type and psy1 cells; such colonies were absent in the controls (Figure 5B). Furthermore, extending the recovery time after electroporation from 24 to 48 h allowed us to select white colonies. Among the CrU6 promoters examined, most white psy1 colonies were derived from the SaCas9/CrU6#4 electroporation (5 out of 56 PmR colonies; 9%). Increasing the recovery temperature from 22°C to 33°C for 24 h, followed by 24 h at 22°C, further improved the targeting efficiency (n = 3; 16% ± 4%; Figure 5B). Exchanging the heat-shock-inducible HSP70A promoter with the constitutively active HSP70A/RBCS2 promoter (HR) led to the production of fewer psy1 clones (Figure 5B). SpCas9 created white colonies with reduced targeting frequencies (2 out of 61 PmR colonies; 3.3%) compared with SaCas9. To characterize the modifications, the PSY1-sg target site was PCR amplified and the product sequences were compared with wild-type PSY1. In contrast to ZFN modifications, most Cas9-modified psy1 target loci were impossible to amplify, possibly due to the large insertion size of plasmid DNA, as found by Shin et al. (2016). Of the white clones for which we obtained PCR products (7/96), three had 5- to 7-bp deletions within the cleavage site, two potential siblings contained identical 25-bp deletions of PSY1 resulting from microhomology-mediated end joining, and two contained inserted short fragments of pHS_SaCas9 (Figure 5C). Based on these results, we determined that translated Cas9 was active and that sufficient sgRNA was transcribed in CC-3403 cells to obtain clones with GOI inactivation. The final transformation protocol for gene targeting using genetically encoded Cas9 and optimized conditions is illustrated in Figure 5D.
Using the optimized transformation conditions, we next disrupted photoreceptor genes that were not expected to produce an immediate phenotype. For the four targeted photoreceptor genes tested, ChR2 (ChR2-sg), chlamyopsin-1/2 (COP1/2-sg), chlamyopsin-5 (COP5-sg), and phototropin (PHOT-sg), we found target site insertions in the range of 2 to 10 kb (Figure 5E). The insertion for the only phototropin disruption clone could not be amplified, but protein immunoblotting confirmed the absence of PHOT protein in this mutant (Figure 5E). The two cop5 mutants (cop5-1 and cop5-2) contained large but amplifiable fragments of pCOP5-sg (Figure 5F). For SpCas9, the prolonged expression of the genome-integrated Cas9 coding sequence was hypothesized to cause cytotoxic effects (Jiang et al., 2014). We successfully amplified full-length SaCas9 DNA in 5 out of the 11 SaCas9 modified strains tested (Figure 5G). For two of the SaCas9 amplicons from cop5 clones, correct integration of the full-length SaCas9 was confirmed by sequencing. sgRNA sequences, including the CrU6#4 promoter, were supplied on the same plasmid as the aphVIII selection marker and had integrated in all clones tested.
In summary, we disrupted four different photoreceptor genes in strain CC-3403 cells using plasmid-based, genetically encoded SaCas9, and disruption efficiencies reached levels up to 9% in preselected colonies.
Gene Disruption Using Preassembled Cas9/gRNA RNP Complexes
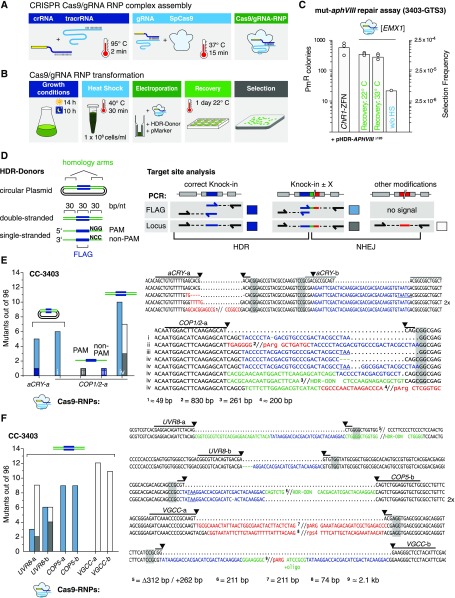
An alternative way to ensure the presence of functional CRISPR/Cas9 in cells is through direct delivery of am RNP complex with the Cas9 protein and synthetic CRISPR RNA (Kim et al., 2014). In Chlamydomonas, this delivery method has previously been used to obtain mutants for phenotype screening (Baek et al., 2016; Shin et al., 2016). To enable easier disruption of Chlamydomonas genes whose knockout would not create selectable or easily scorable phenotypes, we modified the Cas9gRNA RNP technology to allow use of our mut-aphVIII gene repair assay. CC-3403-GTS3 (Figure 2B) contains target sites for the well-characterized human EMX1 homeobox protein gene (Ran et al., 2015) and for ChR1-ZFNs. We transformed the mut-aphVIII strain with EMX1 SpCas/gRNA RNPs and HDR-repair donor (pHDR-APHVIII∆120) (Figures 6A and 6B). On average, we obtained ∼300 PmR colonies with EMX1 SpCas9/gRNA RNPs per 4 × 106 electroporated cells compared with ∼500 PmR colonies from the ChR1-ZFN controls (Figure 6C). Omitting the pretransformation heat shock step reduced the number of PmR colonies 15-fold (21 PmR colonies). Increasing the temperature from 22°C to 33°C for recovery after transformation did not alter the efficiency, unlike in the plasmid-based system (Figure 6C).
Figure 6.
Gene Targeting Using Exogenously Supplied Recombinant Cas9/gRNA RNP Complexes.
(A) Schematic illustrating Cas9/gRNA RNP assembly detailed in Methods.
(B) Schematic workflow of the Cas9/gRNA RNP transformation procedure.
(C) Number of PmR colonies obtained with the GTS mut-aphVIII repair assay from three independent transformations. Strain 3403-GTS3, with mut-aphVIII [EMX1] and [ChR1] target sites, was transformed with HDR donor plasmids (pHDR-APHVIII∆120) and either ChR1-ZFN plasmids or Cas9/gRNA[EMX1] RNP. Selection frequency is calculated as PmR colonies/electroporated cells. The experiments were performed as depicted in (A) and (B) except where indicated. Controls without heat shock before transformation (without HS).
(D) Left: HDR donors contain a 30-bp target site insertion sequence (blue box, “FLAG”) surrounded by two target gene homology arms (equal length, square brackets). Right: Possible mutations and underlying repair mechanisms are drawn schematically. Arrows indicate ODN bindings sites used during PCR analysis. Colored boxes are mutation variants and refer to color-coded sequences in (E) to (G), and squares indicate the type of modification: dark blue, HDR on both sides; light blue, single side HDR detected by flag-PCR; dark gray, single side HDR detected by locus PCR; open square, dual side NHEJ detected by locus PCR.
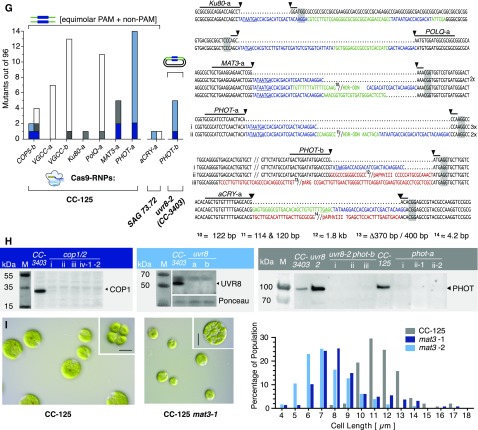
(E) to (G) Left: number of identified mutants (out of 96 tested). The target site, HDR donor, and strain are indicated for each experiment. Cells were cotransformed with a selection marker (ARG7 for CC-3403, aphVII for CC-125, and aphVIII for 3403-uvr8). Sequence alignment of target site amplicons from mutant strains with the corresponding wild-type sequence. Lines indicate 20-bp gRNA binding sites. Predicted cutting sites are shown (black arrowheads), and NGG PAM sequences are highlighted by gray boxes. Gaps are marked with dots, deletions with minus signs. FLAG sequences are shown in blue, HDR donor sequences inserted in a non-HDR manner are shown in green, and other inserting DNA e.g., genomic DNA fragments or DNA from marker plasmids, are shown in red. Premature STOP codons are underlined.
(H) Protein immunoblotting using protein-specific antisera and anti-rabbit HRP-conjugated secondary antibodies for chemiluminescent visualization. Left: Immunoblotting of cop1 mutants obtained in (E), i to iv, anti-VOP rabbit antiserum, 1:2000; middle: uvr8 mutants described in (F), anti-UVR8 rabbit antiserum, 1:2000; right: phot mutants described in (G), i to iii, anti-LOV1 rabbit antiserum, 1:2000. M, marker.
(I) Left: DIC images of unfixed CC-125 and CC-125 mat3-1 cells and division clusters (upper right corner). Right: cell size distribution of CC-125 (n = 167 cells), CC-125 mat3-1 (n = 170 cells), and CC-125 mat3-2 (n = 136 cells). Bars = 10 µm.
Next, we targeted photoreceptor genes for which disruption strains were not yet available. Based on our ZFN experiments, we created HDR donors with 30-bp unique sequence information (FLAG) flanked by arms with homology to the target sequence, either as double-stranded fragments (500–750 bp) cloned into plasmids or as ssODNs of 90 nucleotides total length. The use of short donors simplifies target site analysis because they allow for the detection of FLAG-PCR-based integration or changes in target locus size by Locus PCR (Figure 6D). For our first candidate gene, aCRY, we used a single donor spanning two adjacent target sites (Figure 6E). The aCRY-a target site is located 24 bp upstream of the FLAG integration site and is not destroyed by correct FLAG integration, whereas aCRY-b is disrupted by the FLAG insertion. After transformation of CC-3403 cells (which contain the arg7 gene mutation) with Cas9/gRNA RNPs, plasmid donors, and a wild-type ARG7 marker gene, we analyzed 96 prototrophic clones for aCRY-a and aCRY-b target site modifications. The aCRY-b gRNA apparently had little or no activity because no arginine prototrophic colonies containing aCRY gene modifications were obtained. However, we successfully derived five mutants using a aCRY-a gRNA (Figure 6E). All five clones contained a FLAG sequence integrated into the genome, but only one correct HDR mutant was found. The four other clones had additional deletions within the remaining aCRY-a target site, possibly due to multiple nuclease digestions (Figure 6E).
Next, we compared different donor species for the same target site within the COP1/2 gene, including plasmids, 90-nucleotide ssODNs in PAM (NGG) or non-PAM (NCC) orientation, and 90-bp linear double-stranded templates (dsODNs) of complementary PAM and non-PAM ssODNs. After transformation, 96 clones for each cosupplied HDR donor species were first screened by FLAG-PCR. We found six HDR/NHEJ hybrid mutations with incorrect donor integration from plasmid transformations (Figure 6E, i) and 10 hybrid mutations for 90-bp dsODNs (Figure 6E, iv) with FLAG inserts plus other mutations or insertions (e.g., duplications of the homology arms or marker plasmid DNA fragments). Using Locus PCR, which is especially suited for detecting insertions or deletions by NHEJ, we reanalyzed the same 96 clones and identified seven mutant clones. Three of the HDR/NHEJ hybrids were also found by FLAG-PCR, but Locus PCR identified four additional NHEJ mutants created by insertions of donor DNA or selection marker plasmid fragments (Figure 6E, iv) (pHR11-Arg7). For PAM and non-PAM ssODN donors, we found only a single mutant in each case, and only one of them contained a flawless FLAG integration (Figure 6E, COP1/2 ii and iii).
Our data demonstrate that plasmids and ssODN HDR substrates create low amounts of errorless knockin mutants in conjunction with Cas9/gRNA RNPs, whereas short double-stranded donors create higher numbers of unpredictable but valuable gene disruptions/modifications. Since HDR integration of dsODNs could depend on the sequence context, we designed two gRNAs each for UVR8, COP5 (Histidine-kinase rhodopsin-1), and VGCC (voltage-gated calcium channel) and used 90-bp dsODN templates. We analyzed 566 arginine prototroph colonies and identified between 3 and 12 disruptions per target and 67 mutants in total (Figure 6F). However, high gene disruption rates for dsODNs were only produced with HDR/NHEJ hybrids or NHEJ mutations, disqualifying dsODNs for use in HDR in Chlamydomonas.
For further in-depth characterization of the disruption mutants, we intend to perform physiological assays and several imaging techniques, including super-resolution microscopy and cryo-electron tomography (Engel et al. 2015), all techniques for which cell wall-containing strains are preferable. We repeated targeting of COP5 and VGCC using the wild-type strain 137c (CC-125) and aCRY using the wild-type strain SAG73.72. Additionally, we targeted PHOT and three nonphotoreceptor genes (MAT3, KU80, and POLQ) involved in cell cycle regulation and DNA repair (Figure 6G). DSBs can be repaired by HR, by error-prone canonical NHEJ (supported by KU80), or by alternative end joining mediated by the polymerase Theta POLQ (altEJ). Our data demonstrated that the precise repair of Cas9-induced DSBs is a rare event and that mutated clones that had undergone HR on one side of the break carried NHEJ/AltEJ-induced inserts of donor DNA or random fragments of Chlamydomonas chromosome at the other side of the break. To better understand the contribution of the NHEJ/AltEJ routes, especially for Cas9-induced gene targeting, suppression of random insertions, and stimulation of HR, we generated KU80 and POLQ-deficient mutants for future analysis.
For an HDR template, we mixed two complementary PAM and non-PAM ssODNs without annealing. We transformed CC-125 and SAG73.72 cells using adjusted electroporation conditions and used the aphVII marker to preselect transformed cells for hygromycin resistance (HygR). Locus and/or FLAG-PCR revealed disruptions in all seven genes with frequencies ranging from 1 (for KU80-a) to 14 (PHOT-a) out of 96 analyzed HygR colonies (Figure 6G). Notably, for three target sites (COP5-b, MAT3-a, and PHOT-a), we found one to two “clean” FLAG insertions (Figure 6G, dark-blue bars). Finally, we created a double knockout by transforming an uvr8 disruption strain with a Cas9/gRNA RNP and a plasmid bearing the PHOT template sequence using aphVIII as a coselection marker (Figure 6G). We confirmed the absence of UVR8, COP1/2, and PHOT proteins from the respective deletion strains via protein immunoblotting using available antibodies for these proteins (Figure 6H). Strain phot-1 was motile and tested for mating competence. phot-1 cells mated reliably with CC-124 [137c(-)] cells (Jun Minagawa, personal communication), challenging the previously drawn conclusion that PHOT is needed for gamete formation (Huang and Beck, 2003). The mat3 mutant is defective in a gene with strong homology to an animal retinoblastoma cancer gene involved in cell cycle control. Our mutant showed the same small-size phenotype as related mutants originally created by Gillham and colleagues via insertional mutagenesis (Gillham et al., 1987; Umen and Goodenough, 2001) (Figure 6I).
In conclusion, we optimized a method for direct transformation of preassembled Cas9/gRNA RNP complexes into Chlamydomonas and disrupted eight different genes (COP1/2, COP5, aCRY, PHOT, UVR8, VGCC, MAT3, KU80, and POLQ) using four different strains (CC-3403, CC-125, SAG73.72, and 3403-uvr8-2) with various HDR donors (plasmids, ssODNS, and dsODNs) and three marker genes (ARG7, aphVII, and aphVIII). Figure 7 summarizes the workflows for all three described nuclease systems with references to detailed descriptions in Method.
Figure 7.
Protocols for Gene Editing in Chlamydomonas.
The stepwise diagram serves as a guide for the application of ZFNs or CRISPR-Cas9 followed by mutant screening procedures. Details for every section can be found in Methods.
DISCUSSION
In this study, we optimized several experimental parameters for more efficient application of ZFN and Cas9-based gene-editing technology in motile Chlamydomonas cells, including the wild-type strain CC-125 (Figure 7). The GTS system that we used for optimization has advantages over most other monitoring assays, such as targeting selectable genes or phenotypic markers, because it creates antibiotic-resistant clones after aphVIII repair by HDR. This allowed us to test and quantify the activity of new nucleases and to compare different HDR donor designs and transformation conditions.
Essentials for ZFN- and Cas9-Mediated Gene Targeting
One of the most effective improvements to the ZFN approach is the change from glass bead transformation to electroporation. Although the overall DNA concentration for each plasmid used in transformation remained constant (∼30 ng/μL cell suspension), electroporation generated ∼50-fold more PmR colonies in the GTS system. Considering that 25 times fewer cells (4 × 106 cells) were employed for electroporation compared with glass beads (1 × 108 cells), the overall efficiency with respect to cell number increased by 1250-fold, with a value of up to 1 colony per 4000 transformed cells, which is a factor of ∼100-fold above the previously reported efficiency (Jiang and Weeks 2017).
In both plasmid-encoded nuclease systems (ZFNs and Cas9), increasing the recovery temperature to 33°C was beneficial for targeting events, but it did not affect the efficiency of recombinant Cas9/gRNA RNPs. We used the HSP70A promoter, which was previously characterized using a limited heat shock period of 30 min at 40°C, yielding high levels of protein expression after 60 min (Schroda et al., 2000). Our studies demonstrated that increasing the recovery temperature to 33°C for 24 h posttransformation allowed for continued transcription from this promoter and was a good compromise between high protein expression and survival rate. Most critically, we expect that the 30 min heat shock treatment applied pretransformation is one (if not the only) essential parameter for the superior efficiency compared with previous reports (Shin et al., 2016; Jiang and Weeks, 2017). Heat shock treatment prior to transformation induces unknown physiological changes that are favorable to Cas9/gRNA activity and/or to processes involved in DNA repair (Figure 6C), DNA integration, and/or homologous recombination that merit future investigation.
Activity of Genetically Encoded Cas9
We demonstrated that Cas9 can be expressed in Chlamydomonas in sufficient amounts to generate mutants with or without integration of the Cas9 coding DNA into the genome (Figure 5G). There are two main factors that may have prevented earlier establishment of this plasmid-based CRISPR-Cas9 system. First, the use of standard transformation conditions, including a recovery time of 24 h at 22°C before plating, results in the production of mixed nonhomogenous colonies due to delayed Cas9 expression (Kouranova et al., 2016), as we demonstrated in the PSY1 disruption experiments. Second, Chlamydomonas exhibits atypical gene modification preferences. Although small insertions/deletions are predominant in other systems (Zheng et al., 2016; Zhu et al., 2017), most Chlamydomonas mutants have acquired large insertions that are difficult to analyze and might have remained undetected or disregarded during analysis (Shin et al., 2016). Additional experiments are needed to determine if any HDR donors (e.g., PTO-protected HDR substrates) can remain undegraded long enough for Cas9 translation to peak and further increase the number of mutants obtained. Irrespective of the gene-editing method used, the adaptation of this technology to Chlamydomonas can serve as a basis for improving other Cas9 applications, such as gene activation/repression (Kao and Ng, 2017).
ZFN and Cas9 Favor Different Repair Mechanisms
HDR is approximately 10 times less likely after DNA cleavage with Cas9 compared with ZFN. Moreover, the ratio of HDR to NHEJ/alternative end joining (altEJ; Ceccaldi et al., 2016) differed significantly, even though ZFNs and recombinant Cas9 created nearly equal numbers of PmR colonies. ZFNs created DSBs that were repaired via HDR in most isolated clones with all donor types we tested. In contrast, in the Cas9 experiments, we found that short double-stranded HDR donors tended to create multiple insertions. This observation is in accordance with the creation of blunt cuts, a longer dwell time on DNA, and a delayed release of one DNA strand after cleavage by Cas9 (Sternberg et al., 2014). These differences might prevent perfect integration of symmetric HDR donors and promote nonhomologous integration of double-stranded templates. Nevertheless, HDR via Cas9 is possible in Chlamydomonas. Suppression of NHEJ was achieved using single-stranded DNA templates, but in this case, the number of clones with HDR-based GOI modification was again small. Alternatively, the disruption of KU70/KU80 or POLQ or the inhibition of Ligase IV by SCR7 could be used to suppress NHEJ/altEJ of double-stranded donors (Chu et al., 2015). The ku80 and polq strains are currently available for such experiments (Figure 6G).
In future experiments in Chlamydomonas and related green algae, our results suggest that Cas9 DNA or protein should be used in combination with small PTO-protected double-stranded HDR donors for rapid, efficient gene disruption. However, for clean and predictable gene modification, ZFNs may be preferable for use in combination with larger plasmid donors (≥750 bp).
Advantage of Direct Gene Modification in Chlamydomonas
For the generation of gene modifications, direct gene targeting is an efficient alternative to screening of insertion libraries, a current widely used technology in cases where deletion mutants are required or requested (Galván et al., 2007; Li et al., 2016; Cheng et al., 2017). Insertion mutagenesis has the advantage that thousands of mutants are generated simultaneously. However, marker DNA is inserted randomly and most frequently into introns; genes are not necessarily inactivated, and insertions within the GOI are still laborious to detect in most cases. By contrast, gene deletion using Cas9 is fast, occurs specifically at the site of interest, and is cost effective. A handful of mutants can be retrieved from a single 96-well plate, which supports the interpretation of subsequent physiological experiments. This indicates that mutants can be produced on demand, preventing the need for expensive library maintenance. The technology can be scaled up or multiplexed, as shown for mammalian cell lines, where >18,000 genes were inactivated in parallel using >64,000 gRNAs in a single experiment (Shalem et al., 2014). Finally, direct gene targeting by ZFN or Cas9 separates the selection marker from the targeting site, enabling outcrossing of the marker and sequential inactivation of many genes applicable to any strain of choice.
Channelrhodopsin Physiology
By disrupting ChR1 and ChR2 in CC-3403, we demonstrated that precise modification of nonselectable genes in wild-type-like, motile Chlamydomonas cells is possible. We performed careful physiological characterization of single and double knockout lines of ChR1 and ChR2 and confirmed that ChR1 and ChR2 are the sole photoreceptors for phototaxis in Chlamydomonas, as previously determined using antisense experiments (Sineshchekov et al., 2002; Berthold et al., 2008). The availability of clean disruption mutants allowed us to quantify the relative contribution of ChR1 and ChR2, which is ∼100:1 in CC-3404 gametes. The mislocalization of ChR1 after tagging with mCherry (Figure 4D) suggests that an undisturbed C terminus is necessary for its targeting and/or transport to the eyespot along the four-membered microtubule rootlet (D4) (Mittelmeier et al., 2011; Awasthi et al., 2016). Using the ChR1 and ChR2 mutants, we determined that the ChR patch and eyespot globule layers can assemble independently. In addition, assembly of a functional eyespot is dependent on the chloroplast envelope protein EYE2, which nucleates the formation of the carotenoid-rich eyespot globule layers (Boyd et al., 2011a, 2011b; Mittelmeier et al., 2013). Our data demonstrate that the EYE2 patch and visible eyespots form independently of the ChRs. However, the reduction in eyespot size and the observed malformed eyespots in chr1 and chr1 chr2, but not in chr2 or chr1Δct, suggest that ChR1 may stabilize the eyespot globule plate. Furthermore, low levels of ChRs or ChR1 without a full C terminus were not sufficient for the retention of a normal dominating equatorial eyespot position, supporting the hypothesis of Mittelmeier et al. (2013) that eyespots will be more anterior in the absence of ChRs compared with wild-type cells. The ChR mutants generated here lay the foundation for further characterization of phototaxis and behavioral physiology. Moreover, our optimized methods can be used to generate additional ChR mutants with altered absorption, kinetics, ion selectivity, and adaptation to variable light conditions (Schneider et al., 2015). These ChR mutants will be of great value to the Chlamydomonas field for understanding ChR transport, as well as eyespot assembly, positioning, and size/stability control.
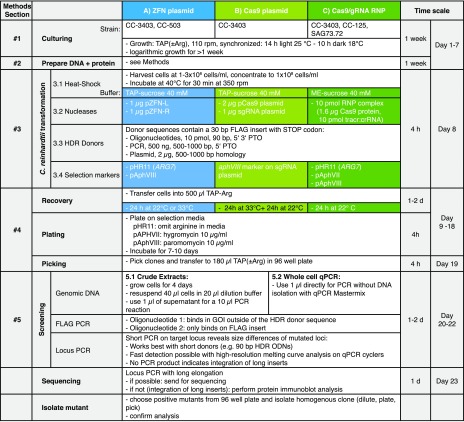
METHODS
The sections below are arranged in accordance with the workflow scheme in Figure 7. Methods used for preliminary experiments of specific analytic studies not necessarily recommended for users are described at the end of this section.
1. Strain and Culture Conditions
Motile Chlamydomonas reinhardtii strains CC-3403 (RU-387 nit1 arg7 cw15 mt−), CC-503 (cw92 mt+), and CC-125 (137c nit1 nit2 agg1+ mt+) were obtained from the Chlamydomonas Resource Center (http://www.chlamycollection.org), and SAG73.72 (mt+) was obtained from Maria Mittag (Friedrich Schiller University, Jena). Cells were grown in standard Tris-acetate-phosphate (TAP) medium (Gorman and Levine, 1965), optionally supplemented with 100 µg/mL l-arginine (TAP-Arg) under continuous cool fluorescent white light of 40 to 60 µE m−2 s−1 at 110 rpm at 22°C or alternatively for synchronized cultures in cycles of 14 h at 25°C under light and 10 h at 18°C in darkness.
2. DNA Preparation and Cas9 Protein Purification
Circular plasmid DNA used for Chlamydomonas transformation was isolated from XL-1 blue Escherichia coli cells and column purified according to the manufacturer’s instructions (Machery-Nagel NucleoSpin Plasmid EasyPure).
Streptococcus pyogenes Cas9 protein was expressed and purified as described (Gagnon et al., 2014). Briefly, the Cas9 expression plasmid pET-28b-Cas9-His (Addgene plasmid 47327) was transformed into E. coli strain Rosetta2(DE3). The Cas9-expressing clone was grown in 500 mL LB medium with 100 mg/mL ampicillin at 37°C for 2 h before induction with 1 mM IPTG. Expression was performed overnight at 25°C. E. coli cells were resuspended in lysis buffer (20 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole, DNaseI, and 0.1 mM PMSF) and lysed using an EmulsiFlex-B15 homogenizer (Avestin). The lysate was purified by immobilized affinity chromatography (5 mL nickel column FF-Crude, Desalt 16-60; GE Healthcare). Protein concentration was determined by A260/A280 absorption, diluted to 10 µM (∼3.1 µg/µL) in 1× Buffer O (BO5; Thermo Fisher Scientific), and filter sterilized, and aliquots were shock frozen in liquid nitrogen and stored at −80°C.
3. Transformation of Chlamydomonas Cells
3.1. Cell Growth, Heat Shock, and Transformation
Cells were grown under a synchronized light (14 h, 25°C)/dark (10 h, 18°C) cycle for at least 10 d and kept in exponential growth phase by diluting 1:50 every 3 to 4 d with fresh TAP(-Arg) medium. However, strictly speaking, whether or not synchronization of the algal culture has an effect on gene targeting efficiency has not been determined for the final recipe, and transformation of nonsynchronized cells might work equally well.
Cells at a density of 1 to 3 × 106 cells/mL were harvested by centrifugation at 2000g for 10 min at room temperature (RT) and resuspended in TAP medium supplemented with 40 mM sucrose (TAP-Suc) for ZFN and Cas9 plasmid-based experiments. For Cas9 protein experiments, cells were resuspended in MAX Efficiency transformation medium (A24229; Thermo Fisher Scientific) supplemented with 40 mM sucrose (ME-Suc) to a density of 108 cells/mL. Prior to transformation, concentrated cells were heat shocked at 40°C for 30 min in a thermomixer (Eppendorf) operated at 350 rpm. Procedures were timed in such a way that the cells were actually electroporated with nucleases (see below) −2 h to +2 h after entering the dark phase. Transformation was performed by electroporation using a NEPA21 electroporator (Nepa Gene Co.) according to the manufacturer’s instructions and Yamano et al. (2013). The impedance of a 40-μL cell suspension (4 × 106 cells) should be 400 to 550 ohms; if not, the volume was adjusted by withdrawing or adding 5 μL cell suspension. CC-3403 and CC-503 cells were electroporated using two 8-ms/200-V poring pulses at 50-ms intervals and a decay rate of 40%, followed by five 50-ms/20-V polarity-exchanged transfer pulses at 50-ms intervals and a decay rate of 40%. CC-125 and SAG73.72 cells were electroporated using one 8-ms/300-V poring pulse followed by five 50-ms/20-V polarity-exchanged transfer pulses at 50-ms intervals and a decay rate of 40%.
3.2. Nucleases
ZFNs (Figure 7A). For targeting the ChR1 gene, ZFNs were used as described previously (Sizova et al., 2013). To target ChR2, designed ChR2-a-ZFNs were designed using the ZiFit database. Sigma-Aldrich (CompoZr) designed and functionally evaluated the ChR2-b-ZFNs. An N-terminal Simian Virus 40 nuclear localization signal (MAPKKKRKVGIHG) was added and the ZF binding domains were fused to the FokI nuclease domain. ZFN expression was controlled by the HSP70A promoter (Schroda et al., 2000) and transcription termination regulated by the 3′ region of RBCS2. Sequences were codon-optimized for expression in Chlamydomonas and synthesized by Genescript. Plasmid maps are provided in Supplemental Table 1. One microgram of each ZFN plasmid (left and right, pZFN-L and pZFN-R), 0.3 µg of marker plasmid pHR11(ARG7), and 2 µg of pHDR donor plasmids (all circular) were used per transformation.
Plasmid-Encoded Cas9 (Figure 7B). Staphylococcus aureus Cas9 (SaCas9) and S. pyogenes Cas9 (SpCas9) coding sequences were codon optimized to the average Chlamydomonas codon bias and ordered from a commercial supplier (GenScript). Sequences 3′ of the HSP70A or HSP70A/RBCS2 promoter were cloned (Schroda et al., 2000). Fully annotated plasmid maps are provided in Supplemental Table 2, including nuclear localization signals. Chlamydomonas U6 promoters for in vivo sgRNA transcription were identified by BLAST searching the published U6 small nuclear RNA (snRNA) sequence (Jakab et al. 1997) against the Chlamydomonas genome (Phytozome v11). Four promoters were chosen, and synthetic double-stranded DNA fragments (Integrated DNA Technologies; gblocks) were ordered for the U6 promoter sequences, followed by two Esp3I restrictions sites and the corresponding SaCas9 or SpCas9 sgRNA scaffold. The Esp3I restriction sites were inversely positioned following the protocol of Ran et al. (2013) to create a specific 4-bp overhang after cleavage to allow for insertion of the protospacer sequence as annealed oligos in a cut-ligation reaction (for details, see Supplemental Tables 3 and 4). The immediate 4-bp sequence upstream of the transcriptional start site was changed to ACTT in all U6 constructs to simplify the cloning procedures. For the electroporation transformation reactions, 2 µg of nonlinearized Cas9 plasmid and 1 µg of sgRNA coding plasmid were used. For experiments involving the use of a donor DNA, 1 µg of plasmid was added. All plasmids and corresponding maps used for the Cas9 experiments are provided in Supplemental Table 2.
Recombinant Cas9 RNP (Figure 7C). Recombinant S. pyogenes Cas9 protein was complexed with guide RNA (gRNA), forming an RNP. gRNAs were ordered as two RNAs, the scaffold RNA (tracrRNA) with constant sequence, and the target sequence (crRNA), which differed for each target site, according to the guidelines of a commercial supplier (Alt-R CRISPR-Cas9 system; Integrated DNA Technologies). All RNAs and target (protospacer) sequences are shown in Supplemental Table 5. Equimolar amounts of tracrRNA and crRNA (Figure 6A) were annealed in DUPLEX buffer (100 mM potassium acetate and 30 mM HEPES, pH 7.5; Integrated DNA Technologies) to a final concentration of 10 µM by heating to 95°C for 2 min, followed by cooling at a rate of 0.1°C/min. Purified 10 µM Cas9 protein was mixed with equimolar amounts of annealed gRNA in 1× Buffer O (BO5; Thermo Fisher Scientific) to a final concentration of 3 µM each and incubated for 15 min at 37°C. Assembled Cas9/gRNA RNPs were kept on ice until the next experiment or stored at 4°C for up to 1 week. Cells were mixed with 2 to 5 μL of 3 µM Cas9 RNP, 10 to 20 pmol of ss- or ds-HDR ODNs, or 2 µg of pHDR donor plasmid, and 0.3 µg selection marker plasmid pHR11(ARG7), pAphVII, or pAphVIII. The resistance of transformation mixtures in electroporation cuvettes was carefully monitored and maintained at 0.3 to 0.7 kOhm by adding or withdrawing 5 μL to the cell-RNP-DNA mixture.
3.3. HDR Donors
Three classes of HDR donors were used: linear single-stranded oligonucleotides, linear double-stranded DNA, and plasmids. These classes all had the integration of a 28- to 32-bp FLAG insert interspacing the cutting site in common. This “FLAG” sequence contains an in-frame stop codon and alters the reading frame. The “FLAG” sequences used in the study are listed in Supplemental Table 6. Oligonucleotides: single-stranded (ss) 90-bp oligos were ordered with two to three PTO bonds at the 3′ and/or 5′ end bases. In the case of short double-stranded (ds) HDR donors, 100 μL 20 μM equimolar sense and antisense oligonucleotide were annealed in 1× duplex buffer (100 mM potassium acetate and 30 mM HEPES, pH 7.5; Integrated DNA Technologies) by incubating at 95°C for 2 min, followed by cooling at a rate of 0.1°C/min. A total of 10 pmol of single-stranded or annealed double-stranded oligos were used per transformation. PCR products are as follows (Figure 2D): Donor sequences of 500 bp were amplified with 5′PTO-protected oligonucleotides. A total of 500 to 1000 ng of purified PCR product was used per transformation. Plasmids are as follows: Donor sequences of 500 to 3000 bp with a FLAG insert were ordered as gBlocks or generated via overlapping PCR and cloned blunt-ended into pBluescript KS(−) vectors. A total of 1 to 2 µg circular donor plasmid was used per transformation. All donor sequences are listed in Supplemental Tables 7 and 8.
3.4. Selection Markers
The Streptomyces aminoglycoside-5′-phosphotranspherase aphVIII selection marker (Sizova et al., 2001) was used in all experiments involving plasmid-encoded Cas9 and in double knockout experiments. The marker was either cloned into the sgRNA plasmid or cotransformed as a separate plasmid along with other plasmids. pHR11(ARG7), a gift kindly provided by Hussam Hassan Nour-Eldin (Chlamydomonas Resource Center, pHR11), was used to reconstitute arginine auxotrophy in ZFN and Cas9 RNP experiments with strain CC-3403. pAphVII (Berthold et al., 2002) gives resistance to hygromycin B and was used to transform CC-125 cells. If GTS strains were complemented with donor DNA, no additional marker was used. Plasmids and corresponding maps are listed in Supplemental Table 1.
4. Recovery, Plating, and Picking
After electroporation, the cells were diluted in 500 μL of TAP(-Arg) and incubated at 22°C or 33°C for 24 h for ZFNs, 33°C for 24 h plus 22°C for another 24 h for plasmid Cas9, or 22°C for 24 h prior to plating for Cas9/gRNA RNP. After incubation, the cell suspensions were transferred to agar plates. In the case of paramomycine selection via aphVIII, plates contained 10 µg/mL of paromomycin; for hygromycin selection via aphVII, plates contained 10 µg/mL of hygromycin B, and for ARG7 selection, arginine-free agar plates were used. Colonies appeared after 7 to 10 d and were picked with sterile toothpicks and transferred to 96-well plates containing 180 μL of TAP-Arg.
5. Screening Procedures
5.1. Crude Cell Extracts
PCR amplification of genomic Chlamydomonas DNA was performed in a 96-well format using Phire Plant Direct PCR Master Mix (Thermo Fisher Scientific; dilution buffer protocol). Cells were grown in 180 μL of TAP-Arg medium in 96-well plates until all wells turned uniformly green. A 40-μL aliquot of each cell culture was then transferred to a 96-well V-bottom culture plate and centrifuged at 2000g for 10 min at RT (22°C). The supernatant was removed and the pellet thoroughly resuspended in 20 μL of dilution buffer, incubated for 5 min at RT (22°C), and centrifuged again at 4000g for 10 min at RT; 80 μL of double-distilled water was added carefully to the supernatant, avoiding cell pellet resuspension. This step is optional, but we found higher dilution of genomic DNA from dilution buffer extractions work more reliably possibly due to lower amounts of inhibitory compounds from plant material. PCR was performed according to the manufacturer’s instructions. A total of 0.5 to 2 μL of DNA extract solution was used for a 10-μL PCR reaction, containing 10 pmol oligonucleotides, 1 M betaine, and 1× Phire Plant Master Mix. Typically, an initial denaturation (5 min, 98°C) was followed by 30 to 35 cycles of denaturation (10 s, 98°C), annealing (10 s, 60–72°C), and elongation (20–200 s, 72°C) with a final elongation (2 min, 72°C). Oligonucleotides with 25 to 30 bp were designed with a melting temperature of 60 to 65°C and tested before screenings. A PerfectBlue Maxi ExW electrophoresis system (Peqlab; VWR) was used to analyze the amplicons from the 96-well PCR plates with 1 to 3% Tris-borate-EDTA agarose gels. For PCR products of 300 to 2000 bp, crude cell extracts were used instead of whole cells. All oligonucleotide sequences used for screening are listed in Supplemental Table 6.
5.2. Whole-Cell qPCR
Due to the high failure rate in psy1 locus amplification, we reasoned that most Cas9 target sites contain large insertions from electroporated plasmid DNA. Therefore, our screening strategy was changed from a single-stage conventional PCR to a double-stage qPCR/long range PCR protocol. First, qPCR was applied to genomic DNA from selected clones using short elongation times of 20 s. All corresponding clones that failed in qPCR target locus amplification were subjected to a conventional long-range PCR elongation time of 300 s. Isolated colonies (1–2 mm diameter) were picked and transferred into 180 μL of TAP medium in a 96-well plate. The plates were immediately processed for qPCR, or the cells were allowed to grow overnight. However, further growth should be avoided, as most qPCR mixes are sensitive to inhibitors from plant materials. In preliminary tests, we found SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) or SsoFast Evagreen Supermixes (Bio-Rad) were most reliable for amplification of genomic Chlamydomonas template DNA; 1.5 μL of the 180 μL cell suspension was used for a 10-μL qPCR reaction mixture including 10 pmol of each oligonucleotide, 1 M betaine (final concentration), and 1× Mastermix. Cycling protocol is as follows: initial denaturing step of 5 min at 98°C; 40 amplification cycles of 98°C for 10 s, 60 to 65°C for 20 s. Oligonucleotides for locus PCR were chosen to create a product in the range of 150 to 300 bp. COP1 forward and reverse oligonucleotides (COP1-F and COP1-R; Supplemental Table 4) were used as a control in different GOI targeting experiments because they facilitate reliable amplification over the range of 60 to 72°C for annealing/elongation. PCR was performed using a CFX-Connect cycler (Bio-Rad). Melting curves were recorded using 0.2 to 0.5°C steps, depending on the expected product size differences. For FLAG PCRs and Locus PCR, the respective oligonucleotide pairs must be used, as explained in Figure 7.
Creating GTS Strains
A 40- to 45-μL volume of concentrated cell culture (1*108 cell/mL; CC-3403) was mixed with 500 ng of linearized pGTS plasmid DNA. After transformation, the cells were allowed to recover overnight in 0.5 mL of TAP medium under continuous light at RT (GRO LUX WIDE, 280 μE m−2 s−1, 22°C). The next day, cells were plated on TAP agar plates containing 10 µg/mL of Zeocin (Invitrogen). After 10 to 14 d, colonies were picked and analyzed under a microscope for signs of motility. Complete integration of the GTS cassette was tested in motile cells with oligonucleotide Ble-Starfw and 3′Psa-467rev. For further experiments, two to eight potential GT strains were tested using the aphVIII repair assay with ChR1-ZFNs, which are known to be functional.
mut-aphVIII Repair Assay Transformations
One microgram of each ZFN plasmid (left and right) or 6 pmol EMX1 Cas9/gRNA RNP and 2 µg of pHDR-APHVIII∆120 (all circular) were used per transformation. Unless specified differently, a heat shock (40°C, 30 min) was applied before transformation and cells were recovered at RT (22°C) for 24 h. Cells were plated and selected on paromomycin (10 µg/mL). Colonies appeared after 8 to 10 d and were counted from photographs of plates using OpenCFU software (Geissmann, 2013).
PSY1, COP1, VGCC, COP5, and PHOT Deletion Experiments
The sequence of each gene was examined for suitable Cas9 target sites using Benchling software, in which the Chlamydomonas genome is integrated to evaluate off-targeting. In the case of SaCas9 experiments, the NNGRRT PAM was always chosen, except for phototropin, which had an NNGRR PAM (Ran et al., 2015). All protospacers had on-target scores of ≥60 (Doench et al., 2014). Detailed protospacer sequences, plasmid maps, and gene IDs are provided in Supplemental Table 3. Because PSY1 deletion mutants are light sensitive, these cells were always kept in the dark. The preplating incubation temperature was either 33 or 22°C for 24 h, followed by 24 h at 22°C. After incubation, the cells were plated on TAP agar plates containing 10 µg/mL of paromomycin. In the case of PSY1, 0.3% (w/v) yeast extract and 0.2% (w/v) tryptone was added to the agar plates to enhance growth in darkness. Single colonies were obtained after ∼2 weeks. PHOT-deletion mutants had to be selected under dim light conditions of 20 µE m−2 s−1. After colony picking, the clones only grew under a light/dark regime.
Protein Analysis
Proteins were separated by SDS-PAGE using 4 to 15% Mini Protean TGX precast protein gels (Bio-Rad) and transferred onto low-fluorescence polyvinylidene difluoride membranes using a Trans-Blot Turbo transfer system (Bio-Rad). Blots were incubated overnight with anti-LOV1 (1:2000), anti-VOP (1:2000), or anti-UVR8 (1:2000), or affinity-purified anti-ChR1 (1:500) or anti-ChR2 (1:500) rabbit polyclonal antibodies. Secondary antibodies were allowed to bind for 2 h (horseradish peroxidase-conjugated ECL anti-rabbit IgG [donkey]; GE Healthcare; NA934V). Clarity ECL Western substrate-induced luminescence was detected using the ChemieDoc MP system (Bio-Rad).
Phototaxis Assay
Phototactic orientation was measured in a custom-made light-scattering apparatus (Berthold et al., 2008; Uhl and Hegemann, 1990). In brief, cells were transferred to NMM medium (80 µM MgSO4, 100 µM CaCl2, 3.1 mM K2HPO4, and 3.4 mM KH2PO4, pH 6.8) 2 d before experiments to induce gametogenesis. Infrared measuring light was scattered by a Chlamydomonas suspension (1 × 106/mL) onto infrared sensitive photodiodes. The current produced by the IR diode is proportional to the intensity of the scattered light and gives a measure of its orientation toward a perpendicular monochromatic light. For excitation, LEDs of 455 nm (10 µE m−2 s−1) and 530 nm (10 µE m−2 s−1) were used and light intensities were adjusted using neutral-density filters.
Microscopy
Color photographs of unfixed cells were taken with an Axioimager M2 (Zeiss; 63× PlanApoChromat 1.4-numerical aperture oil immersion objective; differential interference contrast [DIC]) equipped with an AxioCam ERc5S camera (Zeiss). Cells for eyespot area measurements, position and cell length determinations were analyzed as described in detail by Trippens et al. (2012) using an Eclipse 800 microscope (Nikon; Plan Apo 100×, 1.4-numerical aperture oil immersion objective) and DIC microscopy. NIS-Elements software (Nikon) was used for measurements, and statistical analyses were done using GraphPad Prism 5 software. Parameters and results of ANOVA and multiple comparison test are listed in Supplemental Table 9. Live-cell imaging of mCherry-expressing strains was performed using an Olympus FV1000 confocal microscope equipped with an Upslapo 60× W (numerical aperture: 1.20) objective and appropriate filter sets.
Accession Numbers
The accession numbers for all Chlamydomonas genes are provided in Figure 1 and Supplemental Tables 3 and 5.
Supplemental Data
Supplemental Table 1. Plasmids used for zinc-finger nuclease experiments.
Supplemental Table 2. Plasmids used for the CRISPR-Cas9 experiments.
Supplemental Table 3. Protospacer sequences and target gene information.
Supplemental Table 4. Oligos used for protospacer insertion.
Supplemental Table 5. crRNAs used for recombinant Cas9 transformations.
Supplemental Table 6. Oligonucleotides used for cloning, screening, and HDR donor amplification.
Supplemental Table 7. Oligonucleotides used as HDR donors.
Supplemental Table 8. gBlocks ordered as HDR templates.
Supplemental Table 9. Statistical analysis for Figure 4F.
Acknowledgments
We thank Margret Michalsky, Thi Bich Thao Nguyen, Mirjam Langenegger, and Francisca Boehning for excellent technical assistance; Wenshuang Li and Patrick Segelitz for isolating the aCry disruption mutants; Suneel Kateriya and Lina Sciesielski for fruitful discussions; Roman Ulm for supplying the UVR8 antibody; Richard Chappuis for immunoblot analysis of our UVR8 knockouts; and Jun Minagawa for the phot-1 mating assay. We also thank the Benchling team for integrating the Chlamydomonas genome into the CRISPR/Cas9 guide RNA design tool upon request. This work was supported by the Deutsche Forschungsgemeinschaft (FOR1261 and Leibniz grant) and by the Russian Foundation for Basic Research (Grant 17-54-45088 to I.S.). P.H. is a Hertie Senior Professor for Neuroscience supported by the Hertie Foundation.
AUTHOR CONTRIBUTIONS
P.H., I.S., and A.G. designed the experiments. I.S. and A.G. developed the GTS system. I.S. designed the ZFNs. S.K. established the ZFN protocols. A.G. established the plasmid-based Cas9 and screening protocols. S.K. established the recombinant Cas9 protocols. H.E. made the DNA constructs and selected and characterized the clones. G.K. analyzed cell shape and eyespot positioning by microscopy. A.G. and P.H. wrote the manuscript, with input from I.S., S.K., and G.K.
References
- Awasthi M., Ranjan P., Sharma K., Veetil S. K., Kateriya S. (2016). The trafficking of bacterial type rhodopsins into the Chlamydomonas eyespot and flagella is IFT mediated. Scientific Reports 6: 34646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K., Kim D.H., Jeong J., Sim S.J., Melis A., Kim J.S., Jin E., Bae S. (2016). DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 6: 30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B., Prager K., Spexard M., Sasso S., Weiss D., Müller N., Heinnickel M., Dewez D., Ikoma D., Grossman A.R., Kottke T., Mittag M. (2012). A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24: 2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W. (2002). An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412. [DOI] [PubMed] [Google Scholar]
- Berthold P., Tsunoda S.P., Ernst O.P., Mages W., Gradmann D., Hegemann P. (2008). Channelrhodopsin-1 initiates phototaxis and photophobic responses in chlamydomonas by immediate light-induced depolarization. Plant Cell 20: 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Beumer K., Trautman J.K., Carroll D. (2003). Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Boyd J.S., Mittelmeier T.M., Dieckmann C.L. (2011a). New insights into eyespot placement and assembly in Chlamydomonas. Bioarchitecture 1: 196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.S., Mittelmeier T.M., Lamb M.R., Dieckmann C.L. (2011b). Thioredoxin-family protein EYE2 and Ser/Thr kinase EYE3 play interdependent roles in eyespot assembly. Mol. Biol. Cell 22: 1421–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F.J., Hegemann P. (1999). Two light-activated conductances in the eye of the green alga Volvox carteri. Biophys. J. 76: 1668–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Rondinelli B., D’Andrea A.D. (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Liu G., Ke W., Zhao L., Lv B., Ma X., Xu N., Xia X., Deng X., Zheng C., Huang K. (2017). Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33: 543–548. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K., Hegemann P. (2017). The form and function of channelrhodopsin. Science 357: eaan5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J.M., Baumeister W. (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M., Oertel W., Hegemann P. (1999). A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19: 353–361. [DOI] [PubMed] [Google Scholar]
- Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9: e98186 Erratum. PLoS One 9: e106396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván A., González-Ballester D., Fernández E. (2007). Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv. Exp. Med. Biol. 616: 77–89. [DOI] [PubMed] [Google Scholar]
- Geissmann Q. (2013). OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS One 8: e54072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham N.W., Boynton J.E., Johnson A.M., Burkhart B.D. (1987). Mating type linked mutations which disrupt the uniparental transmission of chloroplast genes in chlamydomonas. Genetics 115: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman D.S., Levine R.P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54: 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E.G., Jung K.H., Sineshchekov O.A., Spudich J.L. (2004). Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys. J. 86: 2342–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Gaj T., Barbas C.F. III (2010). Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 400: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harz H., Nonnengasser C., Hegemann P. (1992). The photoreceptor current of the green alga Chlamydomonas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 338: 39–52. [Google Scholar]
- Hegemann P., Bruck B. (1989). Light-induced stop response in Chlamydomonas reinhardtii - Occurrence and adaptation phenomena. Cell Motil. Cytoskeleton 14: 501–515. [Google Scholar]
- Holland E.M., Braun F.J., Nonnengässer C., Harz H., Hegemann P. (1996). The nature of rhodopsin-triggered photocurrents in Chlamydomonas. I. Kinetics and influence of divalent ions. Biophys. J. 70: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Beck C.F. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 100: 6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inwood W., Yoshihara C., Zalpuri R., Kim K.S., Kustu S. (2008). The ultrastructure of a Chlamydomonas reinhardtii mutant strain lacking phytoene synthase resembles that of a colorless alga. Mol. Plant 1: 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G., Mougin A., Kis M., Pollák T., Antal M., Branlant C., Solymosy F. (1997). Chlamydomonas U2, U4 and U6 snRNAs. An evolutionary conserved putative third interaction between U4 and U6 snRNAs which has a counterpart in the U4atac-U6atac snRNA duplex. Biochimie 79: 387–395. [DOI] [PubMed] [Google Scholar]
- Jiang W., Brueggeman A.J., Horken K.M., Plucinak T.M., Weeks D.P. (2014). Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 13: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.Z., Dumm S., Knuth M.E., Sanders S.L., Weeks D.P. (2017). Precise oligonucleotide-directed mutagenesis of the Chlamydomonas reinhardtii genome. Plant Cell Rep. 36: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Jiang WZ, Weeks DP (2017). A gene-within-a-gene Cas9/sgRNA hybrid construct enables gene editing and gene replacement strategies in Chlamydomonas reinhardtii Algal Res. 10.1016/j.algal.2017.04.001. [DOI]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. (2013). RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao P.H., Ng I.S. (2017). CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour. Technol. 245: 1527–1537.. [DOI] [PubMed] [Google Scholar]
- Kateriya S., Nagel G., Bamberg E., Hegemann P. (2004). “Vision” in single-celled algae. News Physiol. Sci. 19: 133–137. [DOI] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S.W., Kim J., Kim J.S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H., Garcia-Cerdan J.G., Zurbriggen A., Ruehle T., Melis A. (2012). Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol. 160: 2251–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranova E., Forbes K., Zhao G., Warren J., Bartels A., Wu Y., Cui X. (2016). CRISPRs for optimal targeting: Delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Hum. Gene Ther. 27: 464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang R., Patena W., Gang S.S., Blum S.R., Ivanova N., Yue R., Robertson J.M., Lefebvre P.A., Fitz-Gibbon S.T., Grossman A.R., Jonikas M.C. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Potter J., Kumar S., Ravinder N., Chesnut J.D. (2017). Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J. Biotechnol. 241: 136–146. [DOI] [PubMed] [Google Scholar]
- Luck M., Hegemann P. (2017). The two parallel photocycles of the Chlamydomonas sensory photoreceptor histidine kinase rhodopsin 1. J. Plant Physiol. 217: 77–84. [DOI] [PubMed] [Google Scholar]
- Luck M., Mathes T., Bruun S., Fudim R., Hagedorn R., Tran Nguyen T.M., Kateriya S., Kennis J.T., Hildebrandt P., Hegemann P. (2012). A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J. Biol. Chem. 287: 40083–40090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S.S., Kobayashi M.C., Niyogi K.K. (2004). White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics 168: 1249–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke L., Alawady A., Schroda M., Willows R., Kobayashi M.C., Niyogi K.K., Grimm B., Beck C.F. (2010). Chlorophyll-deficient mutants of Chlamydomonas reinhardtii that accumulate magnesium protoporphyrin IX. Plant Mol. Biol. 72: 643–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Mittelmeier T.M., Boyd J.S., Lamb M.R., Dieckmann C.L. (2011). Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J. Cell Biol. 193: 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelmeier T.M., Thompson M.D., Öztürk E., Dieckmann C.L. (2013). Independent localization of plasma membrane and chloroplast components during eyespot assembly. Eukaryot. Cell 12: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N., Wenzel S., Zou Y., Künzel S., Sasso S., Weiß D., Prager K., Grossman A., Kottke T., Mittag M. (2017). A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol. 174: 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G., Ollig D., Fuhrmann M., Kateriya S., Musti A.M., Bamberg E., Hegemann P. (2002). Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296: 2395–2398. [DOI] [PubMed] [Google Scholar]
- Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., Bamberg E. (2003). Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 100: 13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzkofer A., Luck M., Mathes T., Hegemann P. (2014). Bistable retinal schiff base photodynamics of histidine kinase rhodopsin HKR1 from Chlamydomonas reinhardtii. Photochem. Photobiol. 90: 773–785. [DOI] [PubMed] [Google Scholar]
- Petroutsos D., Tokutsu R., Maruyama S., Flori S., Greiner A., Magneschi L., Cusant L., Kottke T., Mittag M., Hegemann P., Finazzi G., Minagawa J. (2016). A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537: 563–566. [DOI] [PubMed] [Google Scholar]
- Ran F.A., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.G.W., Lamb M.R., Dieckmann C.L. (2001). Characterization of the EYE2 gene required for eyespot assembly in Chlamydomonas reinhardtii. Genetics 158: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D (2007). Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 35: W599–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F., Grimm C., Hegemann P. (2015). Biophysics of Channelrhodopsin. Annu. Rev. Biophys. 44: 167–186. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck CF (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21: 121–131. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.E., et al. (2016). CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci. Rep. 6: 27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov O.A., Jung K.H., Spudich J.L. (2002). Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 99: 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229. [DOI] [PubMed] [Google Scholar]
- Sizova I, Greiner A, Awasthi M, Kateriya S, Hegemann P (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 73: 873–882. [DOI] [PubMed] [Google Scholar]
- Spexard M., Thöing C., Beel B., Mittag M., Kottke T. (2014). Response of the Sensory animal-like cryptochrome aCRY to blue and red light as revealed by infrared difference spectroscopy. Biochemistry 53: 1041–1050. [DOI] [PubMed] [Google Scholar]
- Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippens J., Greiner A., Schellwat J., Neukam M., Rottmann T., Lu Y., Kateriya S., Hegemann P., Kreimer G. (2012). Phototropin influence on eyespot development and regulation of phototactic behavior in Chlamydomonas reinhardtii. Plant Cell 24: 4687–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl R., Hegemann P. (1990). Probing visual transduction in a plant cell: Optical recording of rhodopsin-induced structural changes from Chlamydomonas reinhardtii. Biophys. J. 58: 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Iguchi H., Fukuzawa H. (2013). Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 115: 691–694. [DOI] [PubMed] [Google Scholar]
- Zheng X., Yang S., Zhang D., Zhong Z., Tang X., Deng K., Zhou J., Qi Y., Zhang Y. (2016). Effective screen of CRISPR/Cas9-induced mutants in rice by single-strand conformation polymorphism. Plant Cell Rep. 35: 1545–1554. [DOI] [PubMed] [Google Scholar]
- Zhu C., Bortesi L., Baysal C., Twyman R.M., Fischer R., Capell T., Schillberg S., Christou P. (2017). Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. 22: 38–52. [DOI] [PubMed] [Google Scholar]
- Zorin B., Hegemann P., Sizova I. (2005). Nuclear-gene targeting by using single-stranded DNA avoids illegitimate DNA integration in Chlamydomonas reinhardtii. Eukaryot. Cell 4: 1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorin B., Lu Y., Sizova I., Hegemann P. (2009). Nuclear gene targeting in Chlamydomonas as exemplified by disruption of the PHOT gene. Gene 432: 91–96. [DOI] [PubMed] [Google Scholar]
- Zou Y., Wenzel S., Müller N., Prager K., Jung E.M., Kothe E., Kottke T., Mittag M. (2017). An animal-like cryptochrome controls the Chlamydomonas sexual cycle. Plant Physiol. 174: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]