Abstract
High throughput genomic and molecular profiling of tumors is emerging as an important clinical approach. Molecular profiling is increasingly being utilized to guide cancer patient care, especially in advanced and incurable cancers. However, navigating the scientific literature to make evidenced-based clinical decisions based on molecular profiling results is overwhelming for many oncology clinicians and researchers. The Personalized Cancer Therapy website (www.personalizedcancertherapy.org) was created to provide an online resource for clinicians and researchers to facilitate navigation of available data. Specifically, this resource can be used to help identify potential therapy options for patients harboring oncogenic genomic alterations. Herein, we describe how content on www.personalizedcancertherapy.org is generated and maintained. We end with case scenarios to illustrate the clinical utility of the website. The goal of this publically-available resource is to provide easily-accessible information to a broad oncology audience, as this may help ease the information retrieval burden facing participants in the precision oncology field.
Keywords: Precision Oncology, Targeted Therapy, Bioinformatics
Introduction
With the increasing number of potential therapeutic targets and novel therapeutic agents (1), clinicians and researchers may be overwhelmed by the amount of information available in public repositories and in the published literature. At The University of Texas MD Anderson Cancer Center, we have established a team of scientists, clinicians, and bioinformatics experts whose mission is to “provide the scientific support and rationale for all cancer patients and physicians to use as a resource in guiding potential personalized cancer therapy options” (2–4). Our Precision Oncology Decision Support (PODS) team has developed the Personalized Cancer Therapy website (www.personalizedcancertherapy.org) to help clinicians and researchers navigate the growing amount of available information. We developed the website with the intention of being gene-centric and therefore “patient tumor type-agnostic,” and our goal was to facilitate clinical use of this complex information. Specifically, we envision that patient care team members would use this resource during patient encounters, particularly to identify available genotype-matched therapies.
Methodology
Gene Information
Individual gene sheets form the basis for the layout of the website. To be fully annotated and published on www.personalizedcancertherapy.org, a gene must fulfill an initial set of actionability criteria: 1) The gene must be a cancer-associated gene (e.g., associated with cancer development or progression, or with cancer cell proliferation or survival); 2) There must be evidence that targeting the gene or its pathway therapeutically may result in tumor suppression; and 3) Drugs targeting the gene product or its downstream consequence (e.g., downstream signaling) are either FDA-approved or are being clinically investigated. Members of the PODS team select genes for inclusion on the website based upon the strength of available scientific data, with input from MD Anderson clinicians and researchers. Once we select a gene for annotation, a systematic scientific literature review is performed to generate content for the sections of the gene page on the website (Figure 1). The majority of this content is generated using manual literature reviews of PubMed or internet search engines, but we are also developing and using semi-automated approaches (5).
Figure 1. PODS Database.
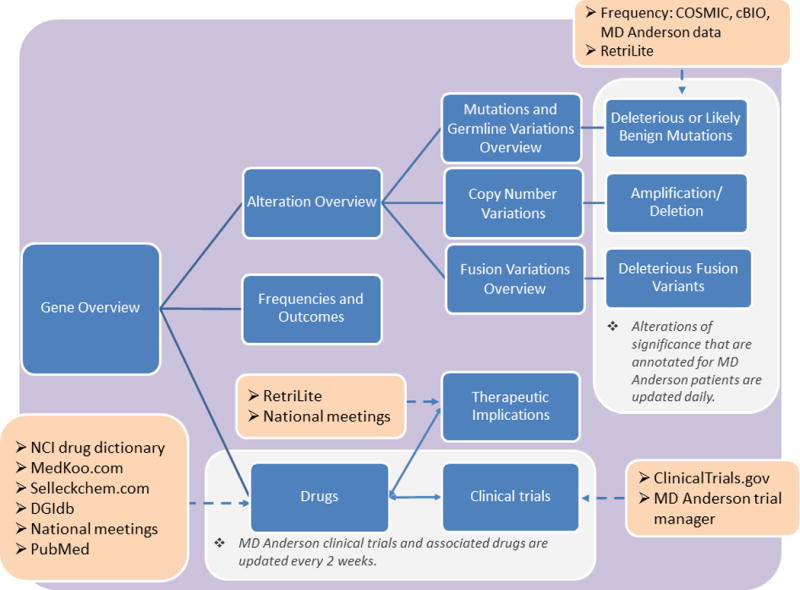
Workflow overview for the development of gene pages. Blue boxes indicate sections included on the website for each gene. Orange boxes indicate resources outside of the Precision Oncology Decision Support (PODS) database that are used to help generate gene page content. Solid lines indicate workflow progression; bidirectional arrows indicate processes that flow in both directions; dashed arrows indicate resources and databases used during that step.
Pathway and Gene Image
The MD Anderson medical graphics department assists with the generation of gene and pathway images. For gene images, we retrieve chromosomal location, transcript information from NM_transcript, and protein domain information from Uniprot (6). Information is then aggregated automatically from the Entrez database (7). For creation of condensed signaling pathway images, we use manual review of the scientific literature.
Functional Annotation of Single Nucleotide or Gene Fusion Variants
For functional annotation of variants, we compile unique alterations within the PODS database. This includes somatic or germline single-nucleotide alterations, and small insertions or deletions. Resources used for functional annotation include cBIO (8), COSMIC (9), alterations included on institutional next-generation sequencing platforms, alterations encountered during annotation of individual patient reports, and published findings. We use manual review of the scientific literature for annotation of copy number variations (i.e., amplifications and deletions) and gene fusion variants. Relevant literature is identified and retrieved using a specially designed information retrieval tool called RetriLite (10), which we developed to streamline the literature review process. This customizable information retrieval tool systematically formulates queries that describe the specific information needs. The list of retrieved documents is ranked by relevancy, and search terms are highlighted in the results for ease of interpretation. Additional publically available databases, such as ClinVar (11), dbSNP (12), The Breast Cancer Information Core (13), and internet search engines may also be manually queried.
We manually review the retrieved literature to write functional annotation of variants. Each variant is assigned a priority based on its reported frequency within the consulted databases and the existence of published functional or therapeutic data. Only alterations with clear clinical or preclinical data are published to the website.
Drug Annotation: Identifying Drug-Gene Associations
To generate a list of drugs relevant to the gene of interest, we catalog drugs using online databases and scientific literature. Such databases include the National Cancer Institute (NCI) drug dictionary (14), Selleckchem.com (15), medkoo.com (16), the Drug Gene Interaction Database (DGIdb) (17), and ClinicalTrials.gov (18). Drugs are curated and annotated within the PODS database. Although the annotator must identify some information manually (i.e., drug-gene associations and phase of drug development), the database is able to automatically retrieve much of the information (i.e., NCI definition and aliases from NCI Thesaurus (19)).
We then tag each drug with potentially relevant biomarkers. Currently for PODS, a biomarker refers to an alteration at the genomic or protein expression level. We record reported the IC50 of a given drug from the scientific literature in the context of each alteration. We then order each of the direct and indirect drug targets by specificity. A gene is considered to be a direct target of a drug if the drug is capable of inhibiting the activity of the gene-encoded protein with an IC50 < 1 μM within biochemical assays. A gene is considered to be an indirect target of a drug if the protein targeted by the drug is within the pathway of the protein encoded by the gene of interest. Additionally, there must be evidence that the drug has anti-tumor properties when an alteration within the gene of interest is present. For indirect targets, knowledge of the pathway and biologic relevance impact the decision to list a gene as applicable to that specific drug.
Drugs with FDA-approval are also tagged with each approved disease type, biomarker, and any other drugs used in combination. Only clinically available drugs (i.e., drugs that are FDA-approved or being actively investigated within clinical trials) that directly or indirectly target the gene of interest are automatically published to the “Drugs” tab of their respective gene pages.
Therapeutic Implications
We annotate therapeutic implications to describe the level of evidence supporting the effectiveness of a specific drug in a specific tumor type harboring a specific biomarker. RetriLite is utilized to retrieve scientific literature relevant to potential therapeutic implications from MEDLINE abstracts. Literature is then manually reviewed and stratified for drug, gene alteration, disease, and level of evidence.
Only literature that fulfills our previously described level of evidence criteria (2) is published to the website. The level of evidence scale is as follows: Level 1 includes prospective studies with strong evidence of therapeutic efficacy linked to a biomarker evaluation (in the context of a specific disease), usually with FDA approval of the drug in the context of a biomarker; Level 2 includes retrospective studies in a specific tumor type or clinical data from another tumor type; Level 3 includes case studies with a scientific rationale or high quality preclinical data linking a biomarker to drug efficacy (2). We also report conflicting data, as well as evidence that a biomarker may confer resistance to a particular drug or class of drugs.
Clinical Trial Annotation
We use an in-house informatics tool to identify clinical trials. Retrieval of trials leverages the direct and indirect gene target information from drugs annotated in the PODS database. Once a list of trials has been generated for the gene of interest, each trial is annotated by aggregating information from ClinicalTrials.gov and MD Anderson’s internal Clinical Trial Management System.
When a gene is entered into the system, trials using drugs that have been previously annotated in the database as being relevant to that gene are identified. The clinical trials informatics tool programmatically interacts with ClinicalTrials.gov’s web application programming interface (API). Through this, we can retrieve open trials that use one or more targeted therapies of interest to treat cancer patients. The PODS team then manually annotates these trials for drugs, diseases, and biomarkers. Trials are designated as “genotype-selected” (i.e., a trial that requires a certain genomic alteration for enrollment) or “genotype-relevant” (i.e., a trial that uses a drug relevant to a genomic alteration, but does not require that alteration for enrollment) (10).
Gene Page Development Workflow
The workflow process for developing a gene page is outlined in Figure 1. Prior to publication, scientists on the PODS team, oncologists, disease experts, and gene domain experts extensively review the content for a gene page. Level of annotator effort can be broken down as follows: approximately 30% on functional variants, 30% on therapeutic implications, 20% clinical trials, 10% on drugs, and 10% on the overviews and frequency/outcomes sections. Time to completion can vary depending on the amount of information to be reviewed and access to expert reviewers.
Our goal is to make major updates to therapeutic implications, frequencies, and outcomes annually using published literature and abstracts presented at major cancer meetings. Our Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy Medical Director and/or oncologists specializing in the implicated gene or diseases review any significant changes prior to publication. However, we update www.personalizedcancertherapy.org daily for alterations that are annotated for MD Anderson patient reports. This includes alterations with either deleterious or likely benign functional significance. MD Anderson clinical trials, which can capture new drugs, are curated and annotated every two weeks.
Comparison to Other Available Decision Support Resources
There are a variety of resources that contain information relevant to precision oncology (20). Given the great need for decision support for genomic medicine, several academic and commercial entities are trying to meet this need. Two other publically available websites with similar goals to our website are My Cancer Genome (www.mycancergenome.org) and OncoKB (www.oncokb.org). Similar to www.personalizedcancertherapy.org, My Cancer Genome contains information regarding alterations, actionability, and therapeutic implications. However, alteration-specific information is organized by tumor type rather than by alteration. This difference in organization may make it more difficult to extrapolate alteration information to other tumor types. Alternatively, OncoKB is organized by alteration, and also provides information regarding actionability and therapeutic implications. However, clinical trials are not included in this resource, and users must navigate to a different webpage to search for relevant clinical trials.
Case Scenarios
We present the following two case scenarios to highlight how our resource might be used in a clinical setting. We have also included a virtual tour of www.personalizedcancertherapy.org (see Video 1).
Mr. B is a patient with cholangiocarcinoma who is being seen for treatment planning. At the last visit, his physician, Dr. A, ordered a clinically-available next-generation sequencing panel test in order to identify actionable alterations. Dr. A is now reviewing the report prior to her scheduled discussion with Mr. B. The panel testing revealed an ERBB2 gene amplification. Using www.personalizedcancertherapy.org, Dr. A identifies that there are no HER2-targeted therapies that are FDA-approved for cholangiocarcinoma. However, she is able to identify several genotype-selected clinical trials that are currently available to Mr. B in his home state of California.
Mrs. C is a patient with HER2 non-overexpressing and non-amplified breast cancer who has progression of her disease. She is referred to the Phase 1 Clinic for consideration of new therapeutic options. She had previously undergone next-generation sequencing panel testing that revealed an ERBB2 single nucleotide variant S310F. The clinical research nurse in charge of enrolling on a trial for a HER2 inhibitor reviews her results in anticipation of her consult visit. Using www.personalizedcancertherapy.org, she determines that S310F is a known activating mutation, and thus is actionable. Consequently, the team is able to offer Mrs. C enrollment on their clinical trial.
Future Enhancements
One of the important aims of www.personalizedcancertherapy.org is to curate updated information for immediate clinical use. As such, the website is frequently evolving to the needs of our audience. We are currently working toward adding several additional biomarkers, including biomarkers relevant to immunotherapy: microsatellite instability status, PD1/PDL1, and tumor mutational burden. Additionally, we plan to continue to expand the filters on our clinical trial search function in order to improve generalizability to other practice settings. In the future, we hope that other institutions will be able to tailor some of the features of our website to their specific needs.
Conclusion
www.personalizedcancertherapy.org aggregates decision support information that is critical to the practice of precision oncology into a single, well-supported platform. While it is not the only resource of its kind, we believe that it is a valuable addition to the current publically-available resources. Connecting clinicians and researchers with available trials through the use of highly curated and organized complex data is a daunting task, and is becoming increasingly difficult as our knowledge base grows. The goal of our website is to provide easily-accessible and interpretable information to a broad oncology audience in order to improve the real-world practice of precision oncology.
Supplementary Material
Video 1: A virtual tour of www.personalizedcancertherapy.org.
Acknowledgments
Financial support: This work was supported in part by 1U01 CA180964 (F. Meric-Bernstam, E. Bernstam), NIH Research Training Grant T32 CA101642 (K.C. Kurnit), The Cancer Prevention and Research Institute of Texas (RP150535) (A.M. Bailey, J. Zeng, A.M. Johnson, B.C. Litzenburger, N.S. Sanchez, Y.B. Khotskaya, V. Holla, J. Mendelsohn, E. Bernstam, K. Shaw, F. Meric-Bernstam), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (A.M. Bailey, J. Zeng, A.M. Johnson, B.C. Litzenburger, N.S. Sanchez, Y.B. Khotskaya, V. Holla, J. Mendelsohn, K. Shaw, F. Meric-Bernstam), NCATS Grant UL1 TR000371 (Center for Clinical and Translational Sciences) (E. Bernstam, F. Meric-Bernstam), The Bosarge Family Foundation (G.B. Mills, F. Meric-Bernstam), and the MD Anderson Cancer Center Support Grant (P30 CA016672).
Conflict of Interest: Aileron Therapeutics (FMB – Research Funding), Allostery Inc (GBM – Consulting), AstraZeneca (FMB – Research Funding; GBM – Consulting, Research Funding), Bayer (FMB – Research Funding), Catena Pharmaceuticals (GBM – Consulting, Financial), Celgene (FMB – Consulting), Clearlight Diagnostics (FMB – Consulting), Critical Outcome Technologies (GBM – Consulting, Research Funding), Curis (FMB – Research Funding), CytomX Therapeutics (FMB – Research Funding), Debiopharm (FMB – Research Funding), Effective Pharmaceuticals (FMB – Research Funding), Genetech (FMB – Honoraria, Consulting), Illumina (GBM – Research Funding), ImmunoMet (GBM – Consulting, Financial), Inflection Biosciences (FMB – Consulting), Ionis (GBM – Consulting, Research Funding), Jounce Therapeutics (FMB – Research Funding), Karus (GBM – Research Funding), Medimmune (GBM – Consulting), Merrimack Pharmaceuticals (JM – Board Member), Myriad Genetics (GBM – Licensed Technology [HRD assay]), Nanostring (GBM – Research Funding), Novartis (FMB – Consulting, Research Funding), Nuevolution (GBM – Consulting), Pieris Pharmaceuticals (FMB – Consulting), Precision Medicine (GBM – Consulting), PTV Ventures (GBM – Financial), PUMA Biotechnology (FMB – Research Funding), Roche Diagnostics (FMB – Honoraria, Consulting), Signalchem Lifesciences (GBM – Consulting), Spindletop Ventures (GBM – Financial), Symphogen (GBM – Consulting), Taiho Pharmaceutical (FMB – Research Funding), Takeda/Millenium Pharmaceuticals (GBM – Consulting, Research Funding), Tarveda (GBM – Consulting), Verastem (FMB – Research Funding); Zymeworks (FMB – Research Funding)
References
- 1.Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:2753–62. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meric-Bernstam F, Johnson A, Holla V, Bailey AM, Brusco L, Chen K, et al. A decision support framework for genomically informed investigational cancer therapy. Journal of the National Cancer Institute. 2015;107 doi: 10.1093/jnci/djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson A, Zeng J, Bailey AM, Holla V, Litzenburger B, Lara-Guerra H, et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug discovery today. 2015;20:1433–8. doi: 10.1016/j.drudis.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The University of Texas MD Anderson Cancer Center. Personalized Cancer Therapy: Knowledge Base for Precision Oncology. www.personalizedcancertherapy.org. Accessed: 14 January 2017.
- 5.Fathiamini S, Johnson AM, Zeng J, Araya A, Holla V, Bailey AM, et al. Automated identification of molecular effects of drugs (AIMED) Journal of the American Medical Informatics Association : JAMIA. 2016;23:758–65. doi: 10.1093/jamia/ocw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UniProt: the universal protein knowledgebase. Nucleic acids research. 2017;45:D158–d69. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic acids research. 2011;39:D52–7. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng J, Wu Y, Bailey A, Johnson A, Holla V, Bernstam EV, et al. Adapting a natural language processing tool to facilitate clinical trial curation for personalized cancer therapy. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science. 2014;2014:126–31. [PMC free article] [PubMed] [Google Scholar]
- 11.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic acids research. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabo C, Masiello A, Ryan JF, Brody LC. The breast cancer information core: database design, structure, and scope. Human mutation. 2000;16:123–31. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 14.National Cancer Institute. NCI Drug Dictionary. www.cancer.gov/publications/dictionaries/cancer-drug. Accessed: 17 January 2017.
- 15.Selleck Chemicals. Selleckchem.com: Inhibitor Expert. selleckchem.com. Accessed: 17 January 2017.
- 16.MedKoo Biosciences, Inc. MedKoo. www.medkoo.com. Accessed: 25 January 2017.
- 17.Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, et al. DGIdb: mining the druggable genome. Nature methods. 2013;10:1209–10. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Institutes of Health. ClinicalTrials.gov: A service of the U.S. National Institutes of Health. clinicaltrials.gov. Accessed: 14 January 2017.
- 19.National Cancer Institute. NCIthesaurus. https://ncit.nci.nih.gov/ncitbrowser. Accessed: 25 January 2017.
- 20.Bailey AM, Mao Y, Zeng J, Holla V, Johnson A, Brusco L, et al. Implementation of biomarker-driven cancer therapy: existing tools and remaining gaps. Discovery medicine. 2014;17:101–14. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1: A virtual tour of www.personalizedcancertherapy.org.


