Abstract
Aberrant activation of the three-amino-acid-loop extension (TALE) homeobox gene MEIS1 shortens the latency and accelerates the onset and progression of acute leukemia, yet the molecular mechanism underlying persistent activation of the MEIS1 gene in leukemia remains poorly understood. Here we used a combined comparative genomics analysis and an in vivo transgenic zebrafish assay to identify 6 regulatory DNA elements that are able to direct GFP expression in a spatiotemporal manner during zebrafish embryonic hematopoiesis. Analysis of chromatin characteristics and regulatory signatures suggest that many of these predicted elements are potential enhancers in mammalian hematopoiesis. Strikingly, one of the enhancer elements (E9) is a frequent integration site in retroviral induced mouse acute leukemia. The genomic region corresponding to enhancer E9 is differentially marked by H3K4 mono-methylation and H3K27 acetylation, hallmarks of active enhancers, in multiple leukemia cell lines. Decreased enrichment of these histone marks is associated with downregulation of MEIS1 expression during hematopoietic differentiation. Furthermore, MEIS1/HOXA9 transactivate this enhancer via a conserved binding motif in vitro, and participate in an autoregulatory loop that modulates MEIS1 expression in vivo. Our results suggest that an intronic enhancer regulates the expression of MEIS1 in hematopoiesis and contributes to its aberrant expression in acute leukemia.
Keywords: MEIS1, comparative genomic analysis, enhancer elements, hematopoiesis, leukemia
Introduction
The Myeloid Ectropic viral Integration Site 1(MEIS1) gene was first identified as a common retroviral integration site in BXH-2 leukemic mice [1]. It encodes a homeodomain-containing transcription factor belonging to the three-amino-acid loop extension (TALE) superfamily. As a cofactor of HOXA family members and PBX1, MEIS1 is critical for the development of hematopoietic cells and many other tissues including the central nervous system, vasculature, lung, and eye[2–4]. During normal hematopoiesis, the expression of the MEIS1 gene is tightly controlled. MEIS1 shows lineage and developmental stage specific expression, with high levels observed in hematopoietic stem cells and early progenitor cells. MEIS1 expression downregulates in later stages of hematopoietic development [5]. Meis1 knock out mice die by embryonic day 12.5 to 14.5 due to the lack of megakaryocytes [2].
In addition to its role in normal hematopoiesis, MEIS1 is critical to leukemogenesis. In acute leukemia patients, persistent overexpression of MEIS1 has been consistently observed[6–10]. The level of MEIS1 expression is inversely correlated with prognosis in human acute myeloid leukemia [11, 12]. Furthermore, it has been reported that overexpression of Meis1 correlated with shorter latency and accelerated progression in various leukemogenic models, such as mouse MLL-associated leukemia models [13] [14], leukemogenic cells with NUP98-HOX translocations[15], and CD34+ NPM1-mutated acute myeloid leukemia cells hoho[16]. Downregulation of MEIS1 in MLL-rearranged leukemia cell lines resulted in decreased proliferation as well as transcriptional repression of cell cycle entry related genes [8, 17, 18]. Despite the essential role of MEIS1 overexpression in acute leukemia, the molecular mechanism underlying persistent activation of the MEIS1 gene in leukemia remains poorly understood.
Cellular gene expression is critically determined by DNA regulatory elements, sequence-specific transcription factors, as well as chromatin modifications. The human MEIS1 gene is located on chromosome 2p13-2p14 and spans approximately 1300 Kb in length[19]. The strict temporal and spatial pattern of MEIS1 expression suggests that it is under the tight control of cis-regulatory sequences. The Meis1 promoter is regulated by ELF1 and CREB [20] [21]. In hematopoietic stem cells, the expression of MEIS1 gene is under the combinatorial control of multiple hematopoietic transcription factors. The binding of those factors is not limited to the promoter region and is distributed along the MEIS1 locus [22]. These data suggest that hematopoietic specific regulatory elements may exist in the MEIS1 locus. We therefore sought to determine the genetic and epigenetic mechanisms involved in the persistent expression of MEIS1 in leukemogenesis.
In this study, we report the systematic identification of distal enhancer sequences in the 1300Kb genomic region of the MEIS1 locus. Traditionally, labor-intensive and time-consuming techniques have been employed to hunt for distal regulatory elements. Comparative genomic strategies are based on the concept that sequences important for gene regulation are conserved throughout evolution [23]. When coupled with appropriate functional tests, this genomic strategy has proved to be a powerful approach for systematic discovery of enhancer sequences [24]. Using a combined comparative genomic and molecular characterization strategy, we identified 6 regulatory elements in the MEIS1 locus. These elements contributed to tissue specific gene expression of normal hematopoiesis/vasculogenesis in a zebrafish reporter assay. One of those 6 elements, designated “E9”, corresponds to a common retroviral integration site in retrovirus-induced mouse leukemia models. We demonstrate that increased levels of histone H3K4 mono-methylation (H3K4me1) and H3K27 acetylation (H3K27ac) at this intronic E9 region are associated with active MEIS1 expression in multiple human leukemic cell lines. In an inducible MLL-ENL leukemia system, the levels of those histone marks diminish when the expression of MEIS1 is downregulated during cellular differentiation. Finally, we show that HOXA9 and MEIS1 directly bind to a conserved binding motif in the E9 region. Knock-down of HOXA9 in THP1 cells results in downregulation of the MEIS1 gene. These studies suggest that expression of MEIS1 can be driven by an autoregulatory loop mediated through a distal intronic enhancer.
Material and Methods
Analysis of sequence conservation
Human, mouse, rat, fugu, zebrafish, and tetraodon sequences were downloaded from the UCSC Genome Bioinformatics (http://genome.ucsc.edu). Based on the human May 2004 assembly hg17, the coordinates for the analyzed MEIS1 human locus are: chr2:66,223,424-67,536,101. This region spans 1,312,678 bp and includes all the MEIS1 exons and introns, as well as the entire intergenic region on each side of the gene.
We aligned the human MEIS1 locus to its orthologs in mouse, rat, fugu, zebrafish, and tetraodon using MLAGAN [25]. Aligned sequences were scanned for statistically significant evolutionarily conserved regions using Gumby [26]. We performed human-fish (human-fugu-zebrafish-tetraodon) and human-mouse-rat comparisons, and selected top ranked elements from each analysis. Conserved non-coding sequences were also identified based on significant conservation in an alignment of 17 vertebrate genomes [27]. Selection of 14 potential regulatory sequences tested in the transgenic zebrafish assay was based on prediction by more than one analysis.
Reporter gene assay in Transgenic zebrafish
The transgenic zebrafish system were based on the Tol2 transposon [28]. Corresponding human sequences of predicted enhancer elements (Figure 1 and Supplemental Table S1) were individually cloned. Each PCR product was recombined first into the pDONR221 vector, and then into pXIG_cfos_GW, using Gateway reagents (Invitrogen).
Fig. 1. Multiple regulatory elements within the MEIS1 gene.

A). Schematic map of enhancer elements in the MEIS1 Locus. Orthologous sequences in the MEIS1 locus are compared between human and mouse (Vista), with human sequence as the base. Numbered green bars (1 to 14) correspond to predicted enhancer elements conserved in multiple vertebrate species. The elements showing positive expression in the transgenic zebrafish assay are marked in red. Hg17: chr2:66,223,424-67,536,101 (corresponding to Hg19: 66311773-66680696)
B). Identification of enhancer activity by transgenic Zebrafish assay. The GFP reporter gene is under the control of a minimal promoter and the human sequence of putative enhancers. GFP reporter constructs were injected into fertilized zebrafish eggs. Representative transgenic zebrafish embryos (24 hpf) are shown, with the PLM area framed in blue, and ALM area circled in red. The percentage of positive embryos is marked in parentheses.
Approximately 100 fertilized eggs were injected for each reporter gene construct. The transparent embryos were then examined by fluorescence microscopy for GFP expression at the hematopoiesis sites of both posterior lateral mesoderm (PLM) and anterior lateral mesoderm (ALM) over a time period of 3 days. A functional enhancer should exhibit an expression pattern consistent across more than 35% of embryos.
Cell isolation and antibody staining
HSPC cells were isolated as described [29]. In brief, bone marrow cells from femurs of 4-week-old C57/BL6 mice were incubated with the mouse Lineage Cell Depletion Kit (Miltenyi Biotec, 130-090-858) to obtain Lin− cells. To isolate HSPC cells which are Lin−, c-Kit+, and CD34+, Lin− cells were stained with an APC-conjugated anti–c-Kit antibody (clone 2B8) and a FITC-conjugated anti-CD34 antibody.
Cell culture
Leukemic cell lines were maintained in RPMI 1640 (Gibco, USA) with 10% fetal bovine serum (Hyclone, USA). Two clones of MLL-ENL inducible cell line (csh2 and csh3) were cultured in the presence of IL-3 (10 ng/mL), IL-6 (10 ng/mL), GM-CSF (10 ng/mL), SCF (100 ng/mL) [30].
Chromatin immunoprecipitation (ChIP)
The ChIP was performed as described [31]. Antibodies used were as follows, anti HOXA9 (Upstate, 07-178), anti-MEIS1 (Santa Cruz, CA sc-10599x), anti H3K4me1 (Abcam, ab8895), anti H3K4me3 (Upstate, 07-473), H3K27ac3 (Abcam, ab4729) and rabbit IgG (Santa Cruz, CA sc2027).
The amount of purified DNA was measured using the PicoGreen system (Molecular Probes, Oregon), and was subjected to PCR using the Applied Biosystems SYBR Green Master mix. The results are shown as fold enrichment of ChIP DNA over input DNA. Primers are listed in Supplemental Table S3.
Transient transfection and luciferase reporter assay
The human promoter was cloned in the proper orientation upstream of the luciferase cDNA in the pGL3 Basic construct (Promega, Madison, WI, USA). The enhancer elements were PCR cloned into polylinker sites upstream of the promoter. Site-specific point mutations and deletions were introduced into the element E9 using QuickChangeII site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The sequences were confirmed by sequencing (Supplemental Table S2).
The cells were grown in 12-well plates and transfected using Fugene (Roche Molecular Biochemicals, USA) following the manufacturer’s protocol. Activity of luciferase and β-galactosidase was measured using the Luciferase Assay System (Promega) and galacto-Light Plus (Applied Biosystems), respectively. Luciferase activity for each sample was normalized to the β-galactosidase assay control. Transfections were carried out in duplicates. All experiments are representative of at least three independent transfections.
siRNA knockdown experiments
HOXA9 siRNA (Sigma) was transfected into THP1 cells with Fugene HD transfection kit (Roche). At 24 hrs after transfection, the total RNA was extracted by TRIZOL, and treated with DNaseI Turbo kit (Ambion, AM1907). The cDNA was then obtained by reverse transcription (Promega A3500) and real-time PCR was performed. The gene expression level was normalized with the expression of the PGK1 gene.
Results
Multiple hematopoietic regulatory elements in the MEIS1 gene locus identified by combined comparative-genomic analysis and functional assay
Based on the temporal and spatial expression pattern of MEIS1 during embryonic and adult hematopoiesis, we hypothesized that distal enhancer sequences are needed to control the expression of MEIS1. Comparative sequence analysis of the human MEIS1 locus showed that the evolutionary conservation of MEIS1 is not limited to exons, but also introns and adjacent downstream and upstream regions. To predict putative enhancer elements, we employed bioinformatic analytical tools including VISTA [32], GUMBY[26], PReMod [33] and ECR [34] (Supplemental Table S1), to scan the 1,312,678 bp MEIS1 locus. This scanned region includes all the MEIS1 exons and introns, as well as the entire intergenic region on each side of the gene. We identified 14 non-coding DNA elements that are conserved in vertebrate species at varying evolutionary distance, from 75 million years in rodents to 450 million years in zebrafish (See Methods and Supplemental Table S1, Figure 1A, element 1 to 14). These sequences also contain clusters of binding sites for multiple transcription factors (data not shown), a sequence characteristic of regulatory modules [33] [35]. While 5 of these DNA sequences are located in the intronic regions of the gene, the remaining 9 elements were found at the intergenic regions between MEIS1 and its neighboring genes (Figure 1A). Among these conserved regions, element E6 has nervous system associated enhancer activity in a transgenic zebrafish system [36]; and element E13 has been reported to exhibit hindbrain specific enhancer activity in a transgenic mouse model (element 831 of the VISTA enhancer browser at http://pipeline.lbl.gov/cgi-bin/gateway2).
Using a transgenic zebrafish system [28], we tested the ability of the 14 predicted regulatory sequence elements to drive GFP reporter gene expression in vivo. The corresponding human sequence of predicted enhancer elements (Supplemental Table S1) were individually cloned upstream of the minimal c-fos promoter of the reporter gene construct, and then injected into fertilized zebrafish eggs. With a minimal c-fos promoter, the GFP reporter gene showed no expression in developing embryos of transgenic zebrafish (Figure 1B). When cloned upstream of the minimal promoter, 6 out of 14 predicted regulatory elements showed hematopoietic specific gene activating activity in this in vivo transgenic zebrafish assay (Supplemental Table S1). The posterior lateral mesoderm (PLM) and anterior lateral mesoderm (ALM) are major sites of zebrafish embryonic blood development (Figure 1B). In our transgenic zebrafish system, the corresponding human sequence of E6 and E14 were able to drive GFP expression at ALM, elements E5, E7, E9 were able to drive GFP expression at PLM, and E8 element was able to drive GFP expression at both PLM and ALM at 24 hours post fertilization (Figure 1B, Supplemental Table S1). Thirty-eight out of 95 (39%) of fertilized zebrafish eggs injected with the E9 reporter gene, and 41%, 38%, 39%, 66%, 40% for E5, E6, E7, E8, and E14, respectively, consistently exhibited the same pattern of restricted GFP expression (Figure 1B). In addition, elements E6 and E9 were able to drive GFP expression in the heart. The transgenic zebrafish results suggest that these distal regulatory elements may contribute to hematopoietic expression of MEIS1 [5].
We further explored whether the enhancer elements identified in zebrafish reporter assay are active during mammalian hematopoiesis. Regulatory DNA elements such as enhancers and promoters are marked by distinct histone modification patterns, and show characteristic of open chromatin structure in the mammalian genome. H3K4me1 is commonly identified at both active and poised enhancer regions [23, 37, 38], and H3K27ac is frequently enriched at both active enhancers and promoters [39, 40]. We employed chromatin immunoprecipitation (ChIP) assay to determine the levels of acetylated H3K27 in primary mouse hematopoietic stem and progenitor cells (HSPCs) in which MEIS1 is actively expressed.
We isolated Lin- CD34+ c-kit+ hematopoietic stem and progenitor cells (HSPC), a combination of hematopoietic stem cells (HSC), common myeloid progenitors (CMP) and granulocyte/monocyte lineage progenitor (GMP) cells, from C57/BL6 mouse bone marrows by FACS sorting (Figure 2A). The sorted population had high c-kit mRNA expression, lacked expression of Gr-1, a marker for differentiated mature myeloid cells (Figure 2B). The ChIP results showed that the active histone mark H3K27ac was significantly enriched at genomic regions corresponding to enhancer elements E6 and E9, as well as the MEIS1 promoter (Figure 2C), suggesting that those regulatory sequences may play a role in regulating MEIS1 expression in primary murine HSPCs.
Fig. 2. Chromatin characteristics of MEIS1 regulatory elements in mammalian hematopoietic cells.

A). HSPC cells (Lin- CD34+ c-Kit+) were isolated from mouse bone marrow by FACS.
B). mRNA expression of c-Kit, Gr1, and Meis1 were detected in HSPC population.
C). H3K27ac enhancer marks of the Meis1 gene during hematopoiesis in mice. ChIP assay was performed with the antibody against H3K27ac in HSPC cells. The amounts of input DNA and ChIP DNA were normalized, and the data are shown as the relative enrichment ratio of precipitated DNA to input DNA. P1 is in the promoter of Meis1 gene, and D130 is located 140kb down stream of the 3′ of Meis1 gene.
The enhancer E9 region is a frequent retroviral integration site in mouse leukemia models and is specifically marked by active histone modifications in MEIS1-expressing leukemic cells
To better understand the molecular mechanism in disregulated MEIS1 expression, we examined whether any of the enhancers identified may be involved in leukemeogenesis. In mouse leukemia models, the Meis1 locus harbors frequent retrovirus intergration sites which result in overexpression of Meis1 leading to the development of AML. The genomic location of an active enhancer possesses an open chromatin structure which often renders the region susceptible to retroviral integration. Upregulation of the affected gene may result from strong regulatory element carried by the retrovirus [41]. A search in the Retrovirus Tagged Cancer Gene Database (RTCGD) (http://variation.osu.edu/rtcgd/) [42] revealed 12 mouse clones that carry proviral insertions interrupting introns or exons of Meis1. Strikingly, these viral integration events formed two distinct clusters along the MEIS1 locus. In addition to an integration hot spot at the promoter region, 3 out 12 of the viral integrations occurred at the genomic sequence corresponding to enhancer E9 region (Figure 3A). Among them, the retroviral integration sites of clone NUP53LD PCR and 403_4_9 are located within the enhancer E9, whereas clone 73_4_4 is only 52bp away from enhancer E9. In contrast, none of the remaining 13 predicted putative enhancers harbored retroviral integration sites. A survey of the UCSC genomic browser indicates a high level enrichment of H3K4me1, H3K27ac3, and presence of DNaseI hypersensitive site (HS) at the E9 region in H1-hESC, K562, and GM12878 cells (Figure 3A). Altogether, these data suggest that enhancer E9 may be functional in mammalian hematopoiesis and is likely involved in retrovirus-induced leukemogenesis. We therefore focused subsequent analysis on this regulatory element.
Fig. 3. Retroviral integration sites and enriched histone H3K4me1 at enhancer E9; correlation with active MEIS1 expression in acute myeloid leukemia cells.
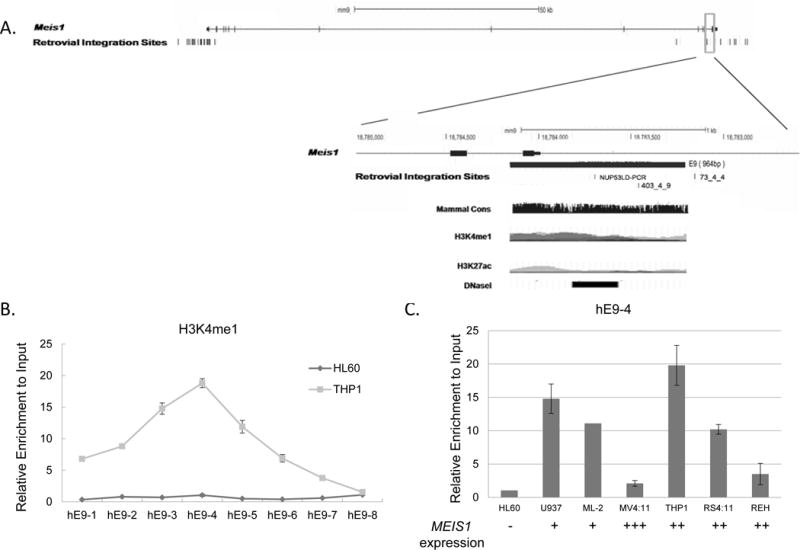
A). Retroviral integration sites in the mouse Meis1 gene. The 3′ region of the mouse Meis1 gene (UCSC genomic browser, NCBI36/mm9, chr11:18,780,325-18,786,183) is enlarged. The genomic location of the predicted enhancer E9 is marked in the gene structure as a black solid box. The open boxes below the gene structure represent the four genomic sequences recovered from the integration sites in individual leukemia mouse clones (data obtained from Retrovirus Tagged Cancer Gene Database). Retroviral integration site of clone NUP53LD PCR and 403_4_9 are located within the enhancer E9, whereas clone 73_4_4 is only 52bp away from enhancer E9. The DNA sequence of the enhancer E9 is aligned with the human MEIS1 genomic region in UCSC genomic browser. Histone modifications of H3K4me1, H3K4me3 and H3K27ac are the ChIP seq results from the H1-hESC, K562, and GM12878 cells in the UCSC genomic browser, and the DNase I HS (ENCODE project) and the mammalian conservation is presented for the enhancer element E9 region.
B, C). Histone H3K4me1 at enhancer E9 is associated with MEIS1 activation. Crosslinked chromatin from THP-1(rectangle) and HL-60 (open circle) were immunoprecipitated with antibodies against H3K4me1, a marker for potential enhancers. The precipitated DNA was amplified using 8 primer pairs (hE9-1 to hE9-8) spanning the 2.1 Kb enhancer E9. Open bars are HL60 cells, and grey bars are THP1 cells. Data are shown as fold change versus input DNA. C) Crosslinked chromatin was immunoprecipitated with antibodies against H3K4me1 in multiple leukemic cell lines. The precipitated DNA was amplified using primer hE9-4. Data are shown as fold change versus input DNA. The mRNA expression level of each cell line is marked under the each column, and were shown as the “-, +, ++, +++”, representing negative, or a graded relative expression level.
To explore whether enhancer element E9 is active in human leukemias, we determined the level of mono- methylated H3K4, a hallmark of distal enhancers [39], in patient-derived leukemia cell lines. In MEIS1-expressing THP-1 cells, the genomic region of enhancer E9 was marked by extensive mono-methylation of H3K4 (Fig. 3B). In contrast, no enrichment of H3K4me1 was observed in HL-60 cells, which have a low level of MEIS1 mRNA. We did not detect significant level of H3K4me1 in the genomic regions corresponding to the other 5 identified enhancers (data not shown). MEIS1 is expressed at high levels in certain subtypes of AML patients. We further selected a panel of leukemic cell lines that exhibit varied levels of MEIS1 expression and examined the enhancer E9 region for the presence of H3K4me1. We found that the presence of H3K4 monomethylation is associated with the active expression of MEIS1 (Figure 3C). Comparing to leukemia cell line HL60, the level of H3K4me1 enrichment is significantly high in cells harboring MLL fusion proteins (THP1, RS4;11, and ML-2). U937 cells, which carry the CALM-AF10 fusion gene, also exhibit a high level of H3K4me1 (Figure 3C).
Considering the developmental stage-specific expression of MEIS1, we sought to determine the chromatin characteristics of enhancer E9 during induced differentiation of leukemic cells. We employed a murine Meis1-expressing myeloblast cell line, which harbors an oncogenic MLL-ENL gene that can be induced to terminal differentiation upon inactivation of the MLL-ENL fusion protein [43][44]. Consistent with reports that the level of Meis1 is down-regulated when myeloid progenitor cells undergo terminal differentiation to neutrophils [5], Meis1 expression diminishes with induced differentiation in this MLL-ENL cell line (Figure 4A). Using this inducible MLL-ENL cell line, we determined the levels of H3K4me1, H3K4me3, and H3K27ac3 enrichment at both the promoter and enhancer E9 regions. When endogenous Meis1 expression was downregulated after inactivation of MLL-ENL, active histone marks H3K4me1 and H3K27ac were significantly decreased across the E9 region (Figure 4B, 4C). In contrast, a high level of histone H3K4me3 modification, a marker at the promoter of actively expressed genes, remained largely unchanged at the promoter region upon inactivation of the MLL-ENL fusion protein (Figure 4D). As the presence of both H3K4me1 and H3K27ac have been associated with active enhancers, these data suggest that enhancer E9 is a functional regulatory sequence in controlling expression of Meis1 in MLL-rearranged leukemic cells.
Fig. 4. Regulatory element E9 is active in MLL-ENL leukemic cells.

A). MLL-ENL-ER cells were cultured with 4-HT (MLL-ENL induced), or without 4-HT for 72 hours (no MLL-ENL). The expression of MEIS1 was detected by real-time PCR.
(B–D). Enriched histone marks at E9 correlated with active Meis1 expression in acute myeloid leukemic cells. Crosslinked chromatin was immunoprecipitated with antibodies against H3K4me1 (B), H3K27ac (C) and H3K4me3 (D) in the MLL-ENL fusion cell line before and after adding 4HT. The precipitated DNA was amplified using primers spanning the Meis1 promoter (mP1, mP2) and the enhancer E9 region (mE9-1, mE9-2, mE9-3, mE9-4) using qPCR. Data are shown as fold change of precipitated DNA versus input DNA. Each experiment was repeated at least three times.
MEIS1/HOXA9 regulates MEIS1 expression through enhancer E9
In an effort to explore the molecular basis underlying the observed regulatory function of enhancer E9, we discovered several evolutionary conserved MEIS1/HOXA9 binding sites at the enhancer E9 region using the Genomatix tool (http://www.genomatix.de, Germany) (Figure 5A and 5B). To test possible recruitment of these two proteins to enhancer E9 in vivo, ChIP-qPCR primers were designed to amplify 8 locations across the 2.1 kb enhancer, with an average spacing of 200-300bp. The binding of MEIS1 and HOXA9 proteins was enriched both at the MEIS1 promoter (data not shown) and the primer hE5 region corresponding to two overlapping conserved sites (M2/M3 in Fig. 5A and 5B) in MEIS1-expressing THP-1 cells. The MEIS1 and HOXA9 binding signals were diminished at flanking regions approximately 300bp in both directions (Figure 5C).
Fig. 5. MEIS1/HOXA9 binds to a predicted consensus site in the enhancer E9.
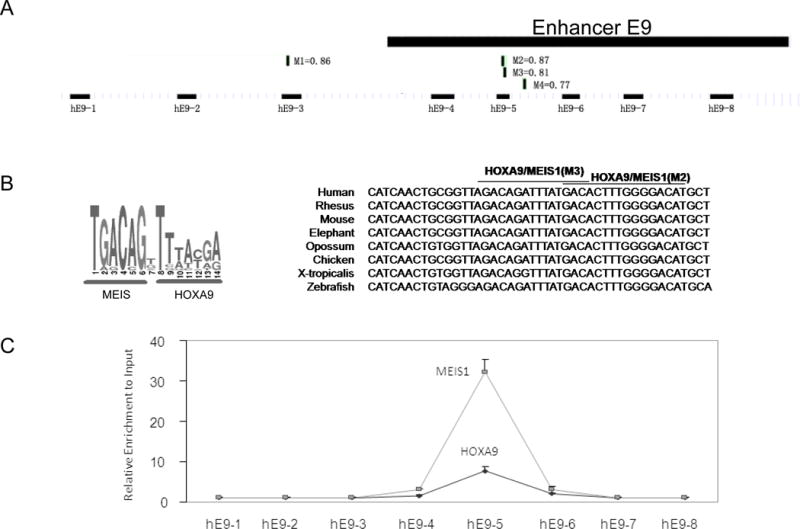
A). Schematic map of the enhancer E9, predicted MEIS1/HOXA9 binding sites and the sequence alignment of the detected PCR replicon in ChIP assay. Four HOXA9/MEIS1binding sites (M1 to M4) were predicted by bioinformatics analysis (http://www.genomatix.de) with the simultaneous matrix shown (Also see Supplementary Table S2).
B). HOXA9/MEIS1 binding motif by Genomatix and the conserved sequence of MEIS1/HOXA9 binding sites from various vertebrate species.
C). Crosslinked chromatin from THP-1 was immunoprecipitated with antibodies to MEIS1 and HOXA9. The precipitated DNA was amplified using the primer pairs spanning the enhancer E9 region. A higher binding signal was detected with the primer pair hE9-5.
To examine whether the predicted MEIS1/HOXA9 consensus sites are critical for the function of enhancer E9, we mutated the MEIS1/HOXA9 binding sites and determined their contributions to the enhancer activity in REH cells (Fig 6A). A 14-bp deletion/mutations were introduced into the E9 element to destroy the two overlapping MEIS1/HOXA9 binding sites (M2/M3). Compared to the construct with a CMV promoter only, addition of enhancer E9 upstream of the promoter increased the luciferase activity in a transient transfection assay. Mutation of the MEIS1/HOXA9 binding sites diminished the enhancer activation activity (Figure 6A and Supplemental Table S2). These results suggest that the binding of MEIS1/HOXA9 to enhancer E9 might regulate the expression level of MEIS1 itself. Furthermore, siRNA knock down of HOXA9 resulted in a down-regulation of MEIS1 expression in the THP1 cell line (Figure 6B). Altogether, these results demonstrated that MEIS1/HOXA9 may transactivate the MEIS1 gene through a conserved binding site in the enhancer E9.
Fig. 6. MEIS1/HOXA9 transactivates the E9 enhancer.
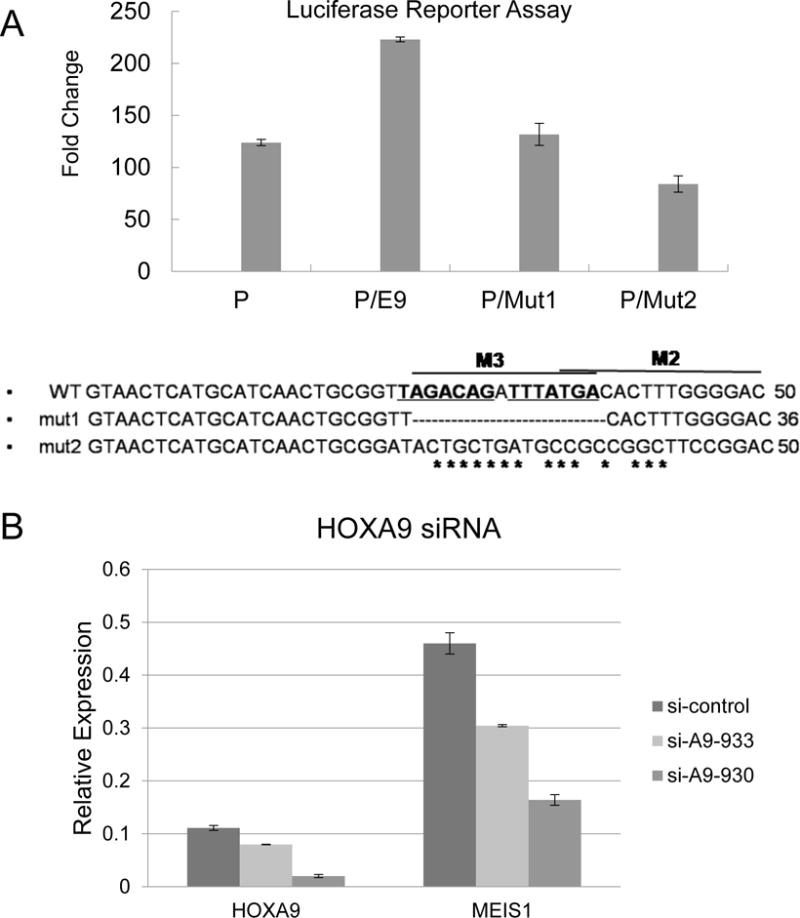
A). REH cells were transfected with a luciferase reporter gene driven by the Meis1 promoter only (P), Meis1 promoter together with enhancer E9 (P/E9), or the enhancer E9 with the designated mutations (P/Mut1, P/Mut2). The mutation sequences of Mut 1 and Mut 2 are shown in the supplementary data S2.
B). HOXA9 siRNA in THP1 cells. Two HOXA9 siRNA sequences were applied to knockdown the HOXA9, and the expression of HOXA9 and MEIS1 detected by RT-PCR.
Discussion
As its level of expression is inversely correlated with prognosis, MEIS1 expression is an important predictive marker in human acute leukemia. Furthermore, knockdown of Meis1 delays development of overt leukemia in mouse models [8]. Despite the essential role of MEIS1 overexpression in acute leukemia, the underlying genetic and epigenetic mechanisms regulating its expression are largely unknown. Using combined comparative genomics analysis and functional assays, we identified multiple intronic regulatory DNA elements which may play a role in regulating MEIS1 expression in zebrafish and mammalian hematopoiesis. One of the identified enhancer elements, designated E9, may mediate the auto-regulation of MEIS1 through an evolutionarily conserved binding motif.
Considering the tightly controlled expression pattern of MEIS1 during embryonic and adult hematopoiesis, we explored the hypothesis that tissue specific distal enhancer elements exist in the 1300Kb genomic region of MEIS1 locus, which may participate in overexpression of MEIS1 during leukemogenesis. The identification of regulatory sequences embedded in the ~98% non-coding fraction of the genome remains a challenge [35]. Regulatory elements such as enhancers are difficult to identify due to their degenerate nature and lack of a well-defined signature. In this study, we employed a combined genomic sequence analysis and in vivo transgenic zebrafish assay to systematically discover enhancer sequences in the 1300 Kb of MEIS1 locus. Zebrafish uses shared genetic pathways similar to those found in humans to regulate the complex process of blood cell development [45, 46]. The spatio-temporal expression patterns of many key hematopoietic genes are highly conserved between zebrafish and human, supporting the usage of zebrafish as an informative model to study transcriptional regulation of blood cell development [45, 46] .
Enhancers identified in this study were able to drive GFP expression at either ALM or PLM (Figure 1B and Supplemental Table S1), where primitive hematopoiesis occurs [24] [47]. ALM and PLM also produce the vascular progenitor cells, supporting these sites as a common location for both hematopoiesis and vasculogenesis [48] [49]. Consistent with the observation that MEIS1 is expressed in both the hematopoietic and vascular systems [2], enhancers E6 and E9 also drove GFP expression in the developing heart of the transgenic zebrafish (Supplemental Table S1), suggesting that some of these newly identified elements may be involved in vascular development. Two of the total 14 predicted DNA elements showed active chromatin modifications in murine HSPC cells (Figure 2C). However, it is possible that some of the remaining 12 elements may control MEIS1 expression in distinct hematopoietic cell populations that were not examined in this study. It is worthy to note that the prediction of enhancer DNA elements in this study was purely based on genomic sequence analysis. As a proportion of regulatory elements are not sufficiently conserved to be detected by comparative genomic methods [50–52], it is likely that additional blood specific enhancers exist in the region.
Genome wide studies have shown that genomic regions of functional regulatory sequences are marked by specific histone modifications [38]. The presence of such marks can be a strong indication of an active chromatin domain involving in regulating gene expression. In the genome, active enhancers are often associated with monomethylation of H3K4, while promoter regions are often characterized by trimethylated H3K4 [23, 37]. As a characteristic of active enhancers, H3K4me1 has also been shown to play a causal role in defining tissue-specific binding patterns of hematopoietic specific transcription factor PU.1 [53]. In this study, the genomic region of this enhancer E9 is marked by monomethylated H3K4 in a series of human acute leukemic cells, and the active status of the enhancer is strongly associated with high level of expression of MEIS1 (Figure 3C). In addition, down-regulation of MEIS1 in MLL-ENL cells (Figure 4) was associated with loss of the active histone mark H3K4me1 at the enhancer region, but not with H3K4me3 at the promoter region, suggesting a more prominent role of enhancer E9 in the regulation of the gene. Taken together with the high frequency of retroviral integration events at this site (Figure 3A), the enhancer E9 may be an important regulatory element that mediates MEIS1 expression in leukemogenesis.
In this study, we found that MEIS1 and HOXA9 are bound to the E9 enhancer at a functional consensus binding site in vivo (Fig. 5C). Binding of MEIS1 protein was also identified at the promoter region of MEIS1 gene (data not shown). In addition, knockdown of HOXA9 resulted in decreased expression of MEIS1. Our data suggest that MEIS1, with its heterodimeric partner HOXA9, can regulate its own expression. This result is consistent with several recent studies investigating the functional role of MEIS1 and HOXA9 in normal hematopoiesis and leukemia development. Faber et al showed that loss of Hoxa9 causes downregulation of the Meis1 mRNA, while overexpression of HOXA9 upregulates Meis1 [54]. Suppression of HOXA9 by shRNA induced immediate downregulation of MEIS1 in multiple human leukemia cell lines examined [55]. Furthermore, MEIS1 binding was detected in the promoter and several intronic regions of the Meis1 gene in multipotent hematopoietic progenitor cell line 7 (HPC-7) in a recent genome wide study [22]. We note that MEIS1 and/or HOXA9 binding were not detected in the proximity of the MEIS1 locus in two ChIP sequencing analyses both of which employed HA-tagged MEIS1 and/or HOXA9 expression systems in mouse bone marrow cells [14, 54, 56]. The apparent discrepancy between these results and findings from our study may be due to differences in the stage of differentiation of the analyzed cells, levels of sensitivity of the assay, and the antibodies and biological systems used. The functional assays we performed validate the potential for the E9 enhancer to regulate the expression of MEIS1. Some HOXA genes are known to be autoregulated during development [57]. Regulation of MEIS1 by its own product and HOXA9 suggests that coexpression of these two genes in leukemia is reinforced, at least in part, by a common genetic pathway through a newly identified enhancer. Our data provide a mechanism for the direct regulation of MEIS1 by the MEIS1/HOXA complex.
Taken together, our study identifies potential enhancer elements controlling the expression of MEIS1 in mammalian hematopoiesis. Furthermore, one of these regulatory elements located in an intronic region, may mediate an auto-regulatory loop that contributes to the sustained expression of MEIS1 in acute leukemia. This study provides important insights into the molecular mechanisms that underlie the regulation of MEIS1 in normal hematopoiesis and its aberrant activation in acute leukemia. Understanding the regulation of oncogenic gene expression should facilitate the development of targeted therapies with the potential to correct the inappropriate transcriptional regulation of MEIS1.
Supplementary Material
Acknowledgments
We thank Dr Robert K. Slany (University Erlangen) for providing the MLL-ENL–inducible cell line and Dr. Andrew S. McCallion for providing the pXIG_cfos_GW plasmid. We thank Qianben Wang (Ohio State University) for his help with the ChIP assay, and advice on the experimental design.
This research was supported by National Institutes of Health grant CA105049 (M.J.T.), a Leukemia & Lymphoma Society Translational Research Program grant (M.J.T.), the family of Jerome Thrall, the National Natural Science Foundation of China (Grant No.81070442 [Q.-f.W.], and by the Young Investigator Award from the Cancer Research Foundation (Q.-f.W.). This work was also supported in part by the National Natural Science Foundation of China (Grant No. 81100380 [Y.-j.L.]), 100 Talents Program from Chinese Academy of Sciences (to Q.-f.W.), and the Knowledge Innovation Program of the Chinese Academy of Science (to Y.-j.L.).
Footnotes
Contributions: Q.-f.W. designed research, performed the experiments, contributed to data analysis, and wrote the paper; M.J.T. supervised the project, design the research and wrote the paper; Y.-j. L. performed the experiments, analyzed the data, prepared the figures and wrote the paper; B.L., L. Zh., J.-f. D., J. Zh., performed experiments, F.E.A., M.A.N. performed the zebrafish assay; S. P., F.-h. H. and J. W. performed genomic analysis; and J. N, R. M, J. K, R. T. L and C. W performed PCR assay, construct cloning and mutagenesis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Supplementary information is available at Leukemia’s website
References
- 1.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM, et al. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minehata K, Kawahara A, Suzuki T, et al. meis1 regulates the development of endothelial cells in zebrafish. Biochem Biophys Res Commun. 2008;374:647–652. doi: 10.1016/j.bbrc.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 4.Stankunas K, Shang C, Twu KY, Kao SC, Jenkins NA, Copeland NG, et al. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pineault N, Helgason CD, Lawrence HJ, Humphries RK, et al. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 6.Imamura T, Morimoto A, Takanashi M, Hibi S, Sugimoto T, Ishii E, et al. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br J Haematol. 2002;119:119–121. doi: 10.1046/j.1365-2141.2002.03803.x. [DOI] [PubMed] [Google Scholar]
- 7.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE, et al. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 8.Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q, et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 10.Kohlmann A, Schoch C, Dugas M, Schnittger S, Hiddemann W, Kern W, et al. New insights into MLL gene rearranged acute leukemias using gene expression profiling: shared pathways, lineage commitment, and partner genes. Leukemia. 2005;19:953–964. doi: 10.1038/sj.leu.2403746. [DOI] [PubMed] [Google Scholar]
- 11.Serrano E, Lasa A, Perea G, Carnicer MJ, Brunet S, Aventin A, et al. Acute myeloid leukemia subgroups identified by pathway-restricted gene expression signatures. Acta Haematol. 2006;116:77–89. doi: 10.1159/000093636. [DOI] [PubMed] [Google Scholar]
- 12.Rozovskaia T, Feinstein E, Mor O, Foa R, Blechman J, Nakamura T, et al. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4 : 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 13.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML, et al. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuser M, Yun H, Berg T, Yung E, Argiropoulos B, Kuchenbauer F, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell. 2011;20:39–52. doi: 10.1016/j.ccr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pineault N, Abramovich C, Humphries RK, et al. Transplantable cell lines generated with NUP98-Hox fusion genes undergo leukemic progression by Meis1 independent of its binding to DNA. Leukemia. 2005;19:636–643. doi: 10.1038/sj.leu.2403696. [DOI] [PubMed] [Google Scholar]
- 16.Woolthuis CM, Han L, Verkaik-Schakel RN, Van Gosliga D, Kluin PM, Vellenga E, et al. Downregulation of MEIS1 impairs long-term expansion of CD34+ NPM1-mutated acute myeloid leukemia cells. Leukemia. 2012;26:848–853. doi: 10.1038/leu.2011.277. [DOI] [PubMed] [Google Scholar]
- 17.Kumar AR, Sarver AL, Wu B, Kersey JH, et al. Meis1 maintains stemness signature in MLL-AF9 leukemia. Blood. 2010;115:3642–3643. doi: 10.1182/blood-2010-01-264564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, et al. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc Natl Acad Sci U S A. 2011;108:7956–7961. doi: 10.1073/pnas.1103154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JE, Jr, Bollekens JA, Inghirami G, Takeshita K, et al. Cloning and mapping of the MEIS1 gene, the human homolog of a murine leukemogenic gene. Genomics. 1997;43:99–103. doi: 10.1006/geno.1997.4766. [DOI] [PubMed] [Google Scholar]
- 20.Xiang P, Lo C, Argiropoulos B, Lai CB, Rouhi A, Imren S, et al. Identification of E74-like factor 1 (ELF1) as a transcriptional regulator of the Hox cofactor MEIS1. Exp Hematol. 2010;38:798–798. 808 e791–792. doi: 10.1016/j.exphem.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esparza SD, Chang J, Shankar DB, Zhang B, Nelson SF, Sakamoto KM, et al. CREB regulates Meis1 expression in normal and malignant hematopoietic cells. Leukemia. 2008;22:665–667. doi: 10.1038/sj.leu.2404933. [DOI] [PubMed] [Google Scholar]
- 22.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009 doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsia N, Zon LI, et al. Transcriptional regulation of hematopoietic stem cell development in zebrafish. Exp Hematol. 2005;33:1007–1014. doi: 10.1016/j.exphem.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, et al. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Yan X, Sashida G, Zhao X, Rao Y, Goyama S, et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood. 2012;120:1118–1129. doi: 10.1182/blood-2012-02-412379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiner SA, Garcia-Cuellar MP, Fey GH, Slany RK, et al. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia. 1999;13:1525–1533. doi: 10.1038/sj.leu.2401534. [DOI] [PubMed] [Google Scholar]
- 31.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L, et al. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boffelli D, Nobrega MA, Rubin EM, et al. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 36.Royo JL, Bessa J, Hidalgo C, Fernandez-Minan A, Tena JJ, Roncero Y, et al. Identification and analysis of conserved cis-regulatory regions of the MEIS1 gene. PLoS One. 2012;7:e33617. doi: 10.1371/journal.pone.0033617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Schones DE, Zhao K, et al. Characterization of human epigenomes. Curr Opin Genet Dev. 2009 doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trono D, et al. Virology. Picking the right spot. Science. 2003;300:1670–1671. doi: 10.1126/science.1086238. [DOI] [PubMed] [Google Scholar]
- 42.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG, et al. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Boer J, Walf-Vorderwulbecke V, Williams O, et al. In focus: MLL-rearranged leukemia. Leukemia. 2013;27:1224–1228. doi: 10.1038/leu.2013.78. [DOI] [PubMed] [Google Scholar]
- 45.Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 46.Berman JN, Kanki JP, Look AT, et al. Zebrafish as a model for myelopoiesis during embryogenesis. Exp Hematol. 2005;33:997–1006. doi: 10.1016/j.exphem.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 47.Patterson LJ, Gering M, Patient R, et al. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–3511. doi: 10.1182/blood-2004-09-3547. [DOI] [PubMed] [Google Scholar]
- 48.Zhong TP, Childs S, Leu JP, Fishman MC, et al. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414:216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]
- 49.Roman BL, Weinstein BM, et al. Building the vertebrate vasculature: research is going swimmingly. Bioessays. 2000;22:882–893. doi: 10.1002/1521-1878(200010)22:10<882::AID-BIES3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 50.Cooper GM, Brown CD, et al. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 2008;18:201–205. doi: 10.1101/gr.7205808. [DOI] [PubMed] [Google Scholar]
- 51.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mcgaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, Mccallion AS, et al. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Hu YL, Fong S, Ferrell C, Largman C, Shen WF, et al. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol. 2009;29:5181–5192. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, Van Den Heuvel-Eibrink M, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelly M, Daftary G, Taylor HS, et al. An autoregulatory element maintains HOXA10 expression in endometrial epithelial cells. Am J Obstet Gynecol. 2006;194:1100–1107. doi: 10.1016/j.ajog.2005.12.025. discussion 1107–1109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


