Abstract
Eicosanoids are bioactive lipids derived from enzymatic metabolism of arachidonic acid via the cyclooxygenase (COX) and lipoxygenase (LOX) pathways. These lipids are newly formed and nonstorable molecules that have important roles in physiological and pathological processes. The particular interest to determine intracellular compartmentalization of eicosanoid-synthetic machinery has emerged as a key component in the regulation of eicosanoid synthesis and in delineating functional intracellular and extra-cellular actions of eicosanoids. In this chapter, we discuss the EicosaCell protocol, an assay that enables the intracellular detection and localization of eicosanoid lipid mediator-synthesizing compartments by means of a strategy to covalently cross-link and immobilize eicosanoids at their sites of synthesis followed by immunofluorescent-based localization of the targeted eicosanoid. EicosaCell assays have been successfully used to identify different intracellular compartments of synthesis of prostaglandins and leukotrienes upon cellular activation. This chapter covers basics of EicosaCell assay including its selection of reagents, immunodetection design as well as some troubleshooting recommendations.
Keywords: Eicosanoids, Prostaglandin, Leukotriene, Biosynthesis, Compartmentalization, Carbodii mide, EDAC, Lipid droplets, Phagosomes, Perinuclear
1 Introduction
Over the past decade, intracellular compartmentalization of eicosanoid-synthetic machinery has emerged as a key component of the regulation of eicosanoid synthesis [1–4]. Among bioactive lipid mediators, eicosanoids—including leukotrienes and prostaglandins—are a family of signaling lipids derived from the enzymatic oxygenation of arachidonic acid (AA) that control key processes involving cell–cell communication, including cell activation, proliferation, apoptosis, metabolism, and migration [5, 6], regulating a diverse set of homeostatic and inflammatory processes linked to numerous diseases.
In all cells, the highly regulated generation of eicosanoids is dependent on activation of specific phospholipases and specific eicosanoid-synthesizing enzymes and involves small molecules (e.g., Ca++) and activation-dependent localization of enzymes at specific compartments within cells [1–3, 7–9]. Intracellular compartmentalization of eicosanoid synthesis within leukocytes has emerged as a key feature that regulates the amount and may also regulate the eicosanoid produced. Such intracellular sites of eicosanoid formation in any cell have been inferred based on the permanent or temporary localization of specific eicosanoid forming enzymes under proper cell activation, since the direct observation of sites of eicosanoid synthesis has been hard to define as those lipid mediators are newly formed, non-storable and often rapidly released upon cell stimulation. It has been recently established that successful eicosanoid production is not merely determined by AA and eicosanoid-forming enzymes availability, but requires sequential interactions between specific biosynthetic proteins acting in cascade, and may involve very unique spatial interactions. Therefore, just by detecting eicosanoid-forming enzymes within discrete subcellular structures, one cannot assure that those sites are indeed accountable for the efficient and enhanced eicosanoid synthesis observed during inflammatory responses. The immunolocalization of eicosanoid forming proteins does not necessarily ascertain that the localized protein is functional and activated to synthesize a specific eicosanoid lipid at an intracellular site.
To develop a new strategy for in situ immunolocalization of newly formed eicosanoids to ascertain the intracellular compartmentalization of their synthesis—the EicosaCell assay—modifications of a prior technique, previously described to capture and localize the prostaglandin E2 (PGE2) released extracellularly by a nematode parasite was used [10].
The EicosaCell rationale relies on the specific features of the heterobifunctional cross-linker 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide (C8H17N3–HCl; EDAC). EDAC immobilizes newly synthesized eicosanoids intracellularly by cross-linking their eicosanoid carboxyl groups to the amines of adjacent proteins localized at eicosanoid-synthesizing sites. Such EDAC-mediated reaction forms a bond without any spacer length between the two molecules, favoring an accurate positioning of the newly synthesized eicosanoid within the cell. In addition, while other cross-linkers formed bonds that often generate foreign molecules, EDAC-driven eicosanoid-bond is homologous to native eicosanoid that allows immunoassays like EicosaCell. Besides the precise positioned coupling of an immuno-detectable eicosanoid at its sites of formation, EDAC enables: (1) the ending of cell stimulation step; (2) cell fixation; (3) cell permeabilization, allowing the penetration of both anti-eicosanoid antibody (Ab) and the secondary detecting fluorochrome-conjugated Ab into cells; and, importantly, (4) the relative preservation of lipid domains, such as membranes and lipid bodies, which dissipate with air drying or commonly used alcohol fixation.
The EicosaCell technique described herein enables one to directly pinpoint the intracellular locales of eicosanoid synthesis by detecting the newly formed lipids and has been successfully able to confirm the dynamic aspect involved in the localization of eicosanoid synthesis, providing direct evidence of compartmentalization within the perinuclear envelope [11–14], phagosomes [15] or lipid bodies in accord to cell type and stimulatory condition [11, 12, 16–18]. So far, the EicosaCell assay has been used to identify the production of leukotriene C4 (LTC4) [11, 19–21], leukotriene B4 (LTB4) [17, 22], prostaglandin E2 (PGE2) [12, 13, 16], and prostaglandin D2 (PGD2) [23, 24] in different cell types and under different stimulatory conditions. Moreover, it could in principle be adapted to intracellular detection of other lipid mediators as long as specific antibodies are available.
2 Materials
2.1 For Conventional EicosaCell
EDAC: 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide hydrochloride (C8H17N3-HCl).
Immuno reagents, including: primary rat mAb against LAMP-1, anti-eicosanoid antibodies (mouse anti-PGE2, Cayman Chemical), isotype control (IgG1 murine myeloma clone MOPC 21, Sigma-Aldrich, cat. no. M5284), DyLight488 fluorochrome-labeled goat anti-mouse and Cy3-conjugated donkey anti-rat secondary antibodies (Jackson Immunoresearch), normal serum for nonspecific blocking.
HBSS−/−: Hanks buffered salt solution without calcium chloride and magnesium chloride (Sigma-Aldrich Inc., St. Louis, MO; cat. no. H4891).
RPMI-1640: cell-culture medium containing 2 % FBS infected with Mycobacterium bovis BCG (Moreau strain) vaccine [multiplicity of infection (moi), 3:1].
Water bath or incubator at 37 °C.
Cytospin centrifuge.
Glass microscope slides and coverslips.
Mounting mediua: Aqua Poly/Mount (Polysciences Inc., Warrington, PA; cat. no. 18606) or ProLong® Gold Antifade Reagent with DAPI (Molecular Probes), or Vectashield (Vector, Inc., Burlingame, CA; cat. no. H 1000).
AlexaTM488 protein labeling kit (Molecular Probes).
PFA: paraformaldehyde fixative.
Confocal laser scanning microscope.
2.2 For Double-Labeling Purposes
To ascertain the specific intracellular compartment involved in synthesizing eicosanoids, double-labeling of at least three particular structures should be performed, such as nucleus, cytoplasmic lipid bodies, and phagolysosomes (in the case phagocytic cells). Table 1 shows the reagents used successfully for double-labeling purpose in EicosaCell preparations.
Table 1.
Double-labeling procedures for co-localization of intracellular sites known to compartmentalize eicosanoid synthesis
| Sites of eicosanoid synthesis | Materials | Double-labeling procedures |
|---|---|---|
Nucleus
|
DAPI (4,6-diamidino-2-phenylindole dihydrochloride) |
For nuclear staining, after EDAC and Ab incubation steps, EicosaCell slide preparations should be extensively washed in HBSS−/− and then incubated with DAPI (DAPI working solution 100 ng/ml or 300 nM, see Note 4) for 5 min before aqueous mounting medium application |
| TO-PRO-3 (642/661) 1 mM solution in dimethylsulfoxide (DMSO) |
To better visualize perinuclear eicosanoid synthesis by EicosaCell, a double labeling with TO-PRO-3 is recommended. TO-PRO-3 must be added together with the secondary Ab for 1 h to label nuclei | |
Lipid Body
|
BODIPY® 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene) |
To employ BODIPY 493/503 for lipid body labeling, incubate EicosaCell preparations with 1 μm BODIPY 493/503 for 45–60 min at 37 °C simultaneously with secondary Ab incubation. Then, to remove non-incorporated BODIPY 493/503, EicosaCell preparations should be washed at least twice in HBSS−/− |
| mAb anti-ADRP Mouse monoclonal antibody (mAb) to adipose differentiation-related protein (ADRP) |
For ADRP immuno-labeling, slides are incubated for 1 h with guinea pig anti-ADRP Ab together with the primary anti-eicosanoid Ab to localize lipid bodies within leukocytes. The cells are washed with HBSS for 10 min (3×) and incubated with Cy3-labeled anti-guinea pig secondary antibodies for 1 h | |
Phagolysosome
|
Abs anti-LAMP-1 | For phagolysosome double-labeling, after incubation at 37 °C with EDAC, cells should be washed with HBSS−/− and incubated with mouse anti-eicosanoid Ab and rat mAb against LAMP-1 (2.5 μg/ml) in blocking buffer (5 % normal donkey serum) for 2 h at room temperature |
3 Methods
3.1 Preparation of EDAC Solution for EicosaCell
Water soluble EDAC hydrochloride should be diluted in HBSS−/−. Refer to Note 1 (bellow) for EDAC solution handling. EDAC final concentration with cells varies according cell type and protocol used (see next subheadings).
The working solution should have twice the concentration of the final desired concentration with cells. For instance, specifically regarding purified human eosinophils stimulated as a cell suspension, EDAC final concentration with eosinophils should be 0.1 % in HBSS−/−, and therefore the EDAC working solution should be diluted to 0.2 %.
Specifically for adherent cells, EDAC final concentration should be higher at 0.5 % in HBSS−/−, therefore the EDAC working solution should be diluted to 1.0 %. Alternatively, a mixture of EDAC 1 % plus PFA 2 % may be prepared to use with adherent macrophages, so the final concentration of these fixatives with cells are 0.5 % and 1 %, respectively.
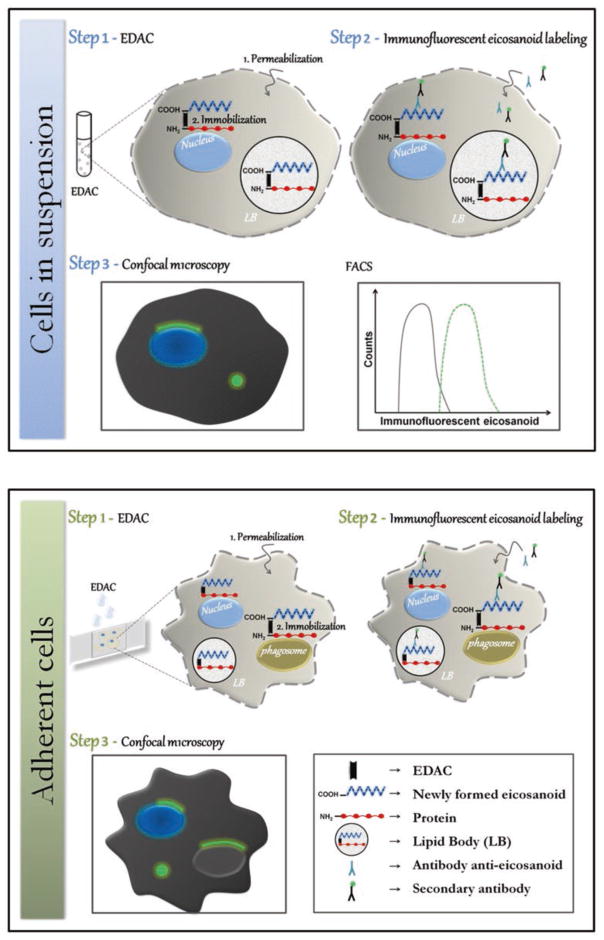
3.2 EicosaCell with Cells in Suspension
EicosaCell can be easily performed with a variety of cell types in suspension, such as purified human blood leukocytes, cell lineages, as well as peritoneal, pleural or brochoalveolar animal cells.
After in vivo or in vitro stimulation of these cell populations, incubation with EDAC should instantaneously guarantee the immobilization of eicosanoids at their synthesizing spot within the cell, just before cytospin slides are prepared to allow microscopic analysis.
As schematically illustrated in Fig. 1 (top panel), after preparing a cell suspension from, for instance, a mouse peritoneal cavity, EDAC working solution should be added to cell suspension and incubated for a period of time to ensure cell fixation and permeabilization (refer to Note 2 for details), as well as immobilization of eicosanoid.
Slide preparations can be done by cytocentrifuging the cells onto slides. Labeling of newly formed eicosanoids can be done with a variety of already tested antibodies, as already plublished elsewhere [11, 16, 17, 19].
At the end of the staining procedure, cytospun cells should be always extensively washed with HBSS−/− and then mounted in an aqueous mounting medium to be visualized with 100× objective by both phase-contrast and fluorescence microscopy (see Note 3 for details). Alternatively, for cells in suspension, instead of preparing slides to analyze by microscopy, intracellular newly synthesized eicosanoid can be detected by flow cytometry. At least two reports showed that, after incubation with EDAC and labeling with proper anti-eicosanoid antibodies, cells in EicosaCell preparations may be successfuly analyzed flow cytometry [23, 25].
The specificity of the eicosanoid immuno-labeling using EicosaCell system should always be ascertained by including some mandatory control conditions: (1) non-stimulated EDAC-treated cells labeled with the proper anti-eicosanoid antibody; (2) the incubation (1 h min before EDAC) with the eicosanoid synthesis inhibitors, such as cPLA2-α inhibitor (e.g., pyrrolidine-2; 1 mM), COX inhibitor (e.g., indomethacin; 1 mg/ml), PGD synthase inhibitor (e.g., HQL-79; 1 mg/kg or 10 μM for in vivo and in vitro treatments, respectively), FLAP inhibitor (e.g., MK886; 50 μg/animal or 10 μM for in vitro incubations), or 5-LO inhibitor (e.g., zileuton; 50 μg/animal or 10 μM for in vitro incubations) to avoid synthesis of the studied eicosanoid; and (3) the use of an irrelevant antibody control. Optionally, other suitable controls to check specificity and performance of EicosaCell are: (1) to use, instead of EDAC, paraformaldehyde which will not immobilize the newly synthesized eicosanoid within cells; (2) to, in parallel, carry out the EicosaCell in a different cell type that lacks the ability to synthesize the targeted eicosanoid (for instance, to use neutrophils to check specificity of LTC4 immunodetection by EicosaCell); or (3) to analyze mixed populations of responsive plus unresponsive cells to a specific stimulus, so you can reassure that the targeted eicosanoid is specifically detected only within stimulated cells.
Fig. 1.
Schematic illustration of EicosaCell assay. Within a variety of cell types and experimental conditions, EDAC-driven cross-linking strategy of EicosaCell assay enables detection of eicosanoid synthesis within at least three known distinct intracellular compartments: the nuclear envelope, cytoplasmic lipid bodies, and phagosomes. In EicosaCell preparations with cells in suspension (top panel), which undergo EDAC-dependent cell permeabilization, as well as, capturing and immobilization of newly formed-eicosanoids at their sites of synthesis, cells can be cytospun and analyzed by fluorescent microscopy or flow cytometry. With adherent cells (bottom panel), successful EicosaCell analysis by microscopy depends on conservation of good cell morphology, therefore, besides EDAC-driven cell permeabilization and eicosanoid fixation, cell fixation by paraformaldehyde in association to EDAC is advisable
3.3 EicosaCell with Adherent Cells
To study the intracellular compartmentalization of eicosanoid synthesis by EicosaCell in adherent cells, extra care should be taken to ensure the conservation of cell adherence and morphology during EDAC step. As published, EicosaCell assays have succeeded to immunolocalize PGD2, detected within in hepatic stellate cells obtained from S. mansoni infected mice [24]. In addition, PGE2 has been also immuno-detected within at least five distinct adherent cell types: plated murine peritoneal macrophages [26], two lineages of intestinal cells, CACO-2 (a human colon adenocarcinoma cell line; [13]) and IEC-6 (a rat epithelial cell line; [12]), human amniotic WISH epithelial cell line [21], and bone-marrow derived murine macrophages (Fig. 2).
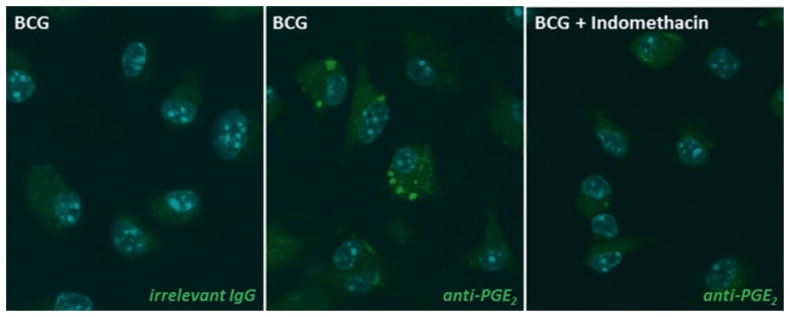
Fig. 2.
EicosaCell for PGE2 immunolocalization within adherent bone-marrow differentiated murine macrophages (BMDMs). The middle panel shows fluorescence microscopy of BMDMs labeled for newly formed PGE2 (green staining). Right and left panels show lack of PGE2 immunolabeling within BMDCs which were, respectively, incubated with isotyping matching antibodies as control, or treated with indomethacin (1 mM) for 20 h before EDAC/PF0. Briefly, BMDMs were fixed and permeabilized during 20 min at 37 °C with a mixture of EDAC and PFA (0.5 % and 1 % in HBSS−/−, respectively). Then, cells were washed/blocked with HBSS−/− containing bovine serum albumin (BSA) for 30 min before overnight incubation (4 °C) with anti-PGE2 monoclonal antibody (Cayman Chemicals). Cells were washed with HBSS−/− and incubated with DyLight488-labeled goat anti-mouse IgG antibody (Jackson) for 1 h. Nucleus are blue, since mounting medium containing DAPI was used for double-labeling purpose
While adherent CACO-2 cells (on glass coverslips), for instance, can be incubated during 1 h at 37 °C with EDAC at 0.5 % in HBSS−/− to cross-link PGE2 carboxyl groups to amines in adjacent proteins without affecting cell morphology, IEC-6 cells can be incubated for at most 30 min (at same concentration; 0.5 % in HBSS−/−) to retain reasonable cell morphology and PGE2 immuno-detection at synthesizing compartments (refer to the original articles [12, 13] for details of blocking and staining conditions with anti-PGE2 monoclonal antibody (Cayman Chemicals) and proper secondary antibodies).
Different from the intestinal cells, adherent macrophages appear to be more sensitive to EDAC treatment and simple adjustments of EDAC concentration or time of incubation were not enough to overcome problems with maintenance of proper cell morphology (Fig. 1; bottom panel). Therefore, to solve this issue we included a paraformaldehyde co-fixation in parallel to EDAC incubation (see Fig. 2 and Note 3 for details) to improve macrophage morphology in EicosaCell preparations.
At the end of the staining procedure, coverslips-adhered cells should be always extensively washed with HBSS and then mounted in an aqueous mounting medium to be visualized with 100× objective by both phase-contrast and fluorescence microscopy. As for cytospun cells, the specificity of the eicosanoid immuno-labeling using EicosaCell system should be ascertained by including mandatory controls listed in Subheading 3.2, step 6.
As shown in Fig. 2, EicosaCell system was successfully employed on adherent macrophages in vitro differentiated from murine bone marrow cells (BMDMs) to detect intracellular newly formed PGE2 at its intracellular sites of synthesis. Briefly, cells recovered from flushed bone marrows of naïve C57BL/6 mice were grown and differentiated in the presence of 30 % of the supernatant obtained from L929 cells, as a source of M-CSF. After 7 days, BMDMs (1 × 105 cells/ml) were adhered in coverslips within culture plates (24 wells; 500 μl/well) overnight with RPMI-1640 cell-culture medium containing 2 % FBS and then infected with Mycobacterium bovis BCG vaccine. At the end of 20 h infection, adherent macrophages were immediately incubated with 500 μl of a mixture of EDAC (0.5 % in HBSS−/−) and PFA (1 % in HBSS−/−), to fix and cross-link the carboxyl groups of newly synthesized PGE2 to amine groups in adjacent proteins without affecting macrophage morphology. Alternatively, cells adhered in Lab-Tek chambers can be used. After 20 min incubation at 37 °C with EDAC–PFA mixture, adherent macrophages were then washed with HBSS−/− containing 1 % bovine serum albumin (BSA) in HBSS−/− for 30 min at room temperature. Adherent macrophages were then incubated with the primary antibody mouse anti-PGE2 in 0.1 % normal goat serum overnight at 4 °C (some antibodies may be alternatively incubated for 1 h at room temperature). Isotyping matching antibodies IgG1 were used as controls, to check for unspecific labeling with fluorochrome-labeled secondary antibody. In addition, although its use was not necessary for this specific assay, the non-immune serum from the animal where the secondary antibody was produced may be added to the primary antibody so as to decrease unspecific labeling. Cells were washed twice with HBSS−/− for 5 min and incubated for 1 h with fluorochrome-labeled secondary antibody. The slides were washed (twice, 10 min each) and mounted with anti-fade mounting medium containing DAPI. Cells were analyzed by both phase-contrast (to ascertain cell morphology) and confocal laser fluorescence microscopy. The specificity of PGE2 immuno-labeling using EicosaCell system was ascertained by including the mandatory control with PGE2 synthesis inhibitor (Fig. 2, left panel), indomethacin (1 μM) added together with BCG and incubated for 20 h at 37 °C.
3.4 EicosaCell with Cells Embedded in a Gel Matrix
In contrast to analyzing cytospun cells which do not preserve in situ morphology, cells embedded in an agarose matrix, that are kept in a hydrated system with a substrate where they can crawl, display tissue-like cell morphology exhibiting polarization and other characteristics of activated leukocyte, for instance. Therefore, by immunolocalizing eicosanoids at its formation sites within agarose-embedded cells (as schematically illustrated in Fig. 1b), generated products may be microscopically localized at cell structures assembled during stimulation and preserved in cells that are not cytospun into slides [11, 19].
To prepare the agarose matrix, 2.5 % agarose (24 °C gelling point) (Promega, Madison, WI) in sterile distilled H2O is melted at 70 °C; and while liquid at 37 °C, 9 volumes of agarose are mixed with 1 volume of 10× concentrated RPMI 1640 medium.
One volume of this medium-supplemented agarose is mixed with one volume of RPMI 1640 medium containing 2 % fatty acid free-albumin at 37 °C and with three volumes of the studied cell, exemplified here as human eosinophils (Fig. 1b), which should be at 15 × 106 cells/ml in RPMI 1640 medium containing 1 % fatty acid free-human albumin.
Stimuli are then added in 0.1 volumes to agarose/eosinophil mixtures. As schematically illustrated in Fig. 1b, immediately thereafter, 20 μl samples are gently spread onto microscope slides and covered with CoverWellTM chambers (Grace Bio-Labs, Bend, OR).
Each slide is overlaid with RPMI 1640 medium containing 1 % albumin and an identical concentration of the stimulus present in the agarose/eosinophil mixture.
Slides can be incubated (37 °C, humidified 5 % CO2) for varying periods of time.
Overlying medium should be removed and replaced with RPMI 1640, 1 % albumin medium, that may or may not contain 0.1 μM A23187, and slides can be incubated for extra 15 min (37 °C; 5 % CO2).
Stimulations are stopped by removing the chambers and adding EDAC. Fixation and permeabilization of cells with proper immobilization of newly formed eicosanoids at its intracellular sites of synthesis are achieved by immersing the slides containing stimulated cells in 0.5 % EDAC (in HBSS−/−) for 30 min.
After three washes (5 min each) with HBSS, the fluorochrome-labeled anti-eicosanoid antibody (for instance 400 μl of 10 μg/ml Alexa488-labeled rat anti-cysteinyl LT detection mAb) should be added for 1 h.
Slides need to be extensively washed with HBSS, and an aqueous mounting medium should be applied to each slide before coverslip attachment. Slides can be viewed by both phase-contrast and fluorescence microscopy.
Mandatory control conditions, as listed in Subheading 3.2, step 6, should be always included as for cytospun and adherent cells to ascertain the specificity of the eicosanoid immuno-labeling using EicosaCell system.
3.5 Identification of Eicosanoid-Synthesizing Intracellular Sites by Double-Labeling Procedures
To successfully identify intracellular compartments of eicosanoid-synthesis, double labeling of the potential eicosanoid-synthesizing sites is required. Table 1 lists the three intracellular compartments known to localize eicosanoid synthesis and the double-labeling procedures used to their identification.
3.5.1 Nuclear Localization
To better visualize perinuclear eicosanoid synthesis by EicosaCell, a double labeling with DAPI is advised.
After EDAC and antibody incubation steps, EicosaCell slides preparations should be extensively washed in HBSS−/− and then incubated with DAPI (DAPI working solution 100 ng/ml or 300 nM, see Notes 4 and 5) for 5 min before aqueous mounting medium application.
The morphology of the cells’ nuclei is observed using a fluorescence microscope at excitation wavelength 350 nm.
3.5.2 Phagosomal Localization
As performed by Balestrieri and coworkers [15], phagosome involvement in eicosanoid synthesis can be ascertained by co-localizing the phagosomal protein marker LAMP-1 in EicosaCell preparations.
After incubation at 37 °C with EDAC, cells should be washed with HBSS−/−, cytospun onto glass slides, and incubated with mouse anti-eicosanoid antibody and rat mAb against LAMP-1 (2.5 μg/ml) in blocking buffer (5 % normal donkey serum) for 2 h at room temperature (see Note 6).
Negative control cells are instead incubated for 2 h with rat IgG.
After 2 h, the cells are washed extensively with HBSS−/− and incubated for 1 h at room temperature with fluorescent-labeled secondary antibody to detect the anti-eicosanoid antibody and with Cy3-conjugated donkey anti-rat IgG (1:200).
The cells should be washed five times with HBSS−/− and then mounted in aqueous mounting medium.
3.5.3 Lipid Body Localization
To investigate roles of lipid bodies in eicosanoid synthesis by EicosaCell assay, two double-labeling strategies can be employed: BODIPY or anti-ADRP immunostaining (for further information on lipid body labeling refer to [27]). Both approaches can be used for adherent, suspension, or agarose-embedded cells.
To employ BODIPY strategy, incubate EicosaCell preparations (coverslips or slides) with 1 μm BODIPY (working solution) for 45–60 min at 37 °C (water bath) simultaneously to secondary antibody incubation.
To remove free BODIPY after incubation, EicosaCell preparations should be washed at least twice in HBSS−/− before aqueous mounting medium application and coverslip attachment to slides.
Alternatively, to visualize lipid bodies anti-ADRP immuno-labeling may be performed, as an example, in Subheading 3.2.
Acknowledgments
The work of the authors is supported by PRONEX-MCT, Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq, Brazil), PAPES-FIOCRUZ, Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ, Brazil), and NIH grants (AI022571, AI020241, AI051645).
Footnotes
EDAC working solution should be prepared fresh, keep protected from light and discarded after each experiment.
Incubation of cells with EDAC can be carried out on either cell in suspension or with the cells already cystopun onto slides by dropping EDAC on top of cells. Even though the latter method is less costly, some differences in preservation of cell morphology, cell permeabilization, and eicosanoid detection may occur and should be analyzed with care.
Concentrated solutions of the EDAC–PFA mixture (twice of final working concentration) should also be prepared fresh, in HBSS−/−, preferably from PFA methanol free aqueous concentrated solutions (Electron Microscopy Sciences).
Analysis of EicosaCell preparations should be performed as soon as the slides are mounted, inasmuch as immunofluorescent labeling is usually not stable for a long period bleaching after a certain time. Even though freezing may preserve fluorescence overnight, EDAC-treated cells may display altered cell appearance after freezing-thawing cycle.
Prepare DAPI stock solution by dissolving 1 mg/ml of powder in distilled water. Aliquots should be stored at −20 °C protected from light.
The following are the common problems and non-obvious features found in immunofluorescent-detection of eicosanoids in EDAC preparations by EicosaCell with their possible explanations and potential solutions:
- Lack of eicosanoid detection: When few or no eicosanoid specific immunostaining is observed (but expected), the problem usually lies in the improper fixation (e.g., EDAC-driven cross-linking) of targeted eicosanoid at its sites of synthesis. Thus, the newly formed eicosanoid would be washed out from the EicosaCell preparation rendering detection impossible. Resolution of this problem is normally achieved by adjusting (slight increase) concentration and/or time of incubation of EDAC. Alternatively, the lack of immuno-detection of newly formed eicosanoids can be due to inefficient stimulation; a positive control with a known agonist should be always included in the experiments.
- Eicosanoid detection within non-stimulated cells: Eicosanoids are lipid mediators not stored in the cell and newly formed upon stimulation, therefore non-stimulated cells should not show any immunostaining for the targeted eicosanoid. Thus, non-stimulated cells should always be included as an important negative control. However, cell activation during the procedures including cell incubation at 37°C or cell fixation/permeabilization with EDAC can lead to spontaneous, stimulus-“independent” eicosanoid synthesis. Throughout cell preparation, care is needed to ensure that cells are not mechanically, chemically or immunologically stimulated. Unexpected eicosanoid detection within EicosaCell preparantions can also result from nonspecific detection (discussed below).
- Nonspecific detection: Fluorescent detection antibodies may non-specifically bind to other lipids found within cells or bind to other cellular structures. The cross-linking properties of EDAC may favor the tendency for cells to be sticky; therefore antibodies could interact through low affinity non-antigen binding site. To investigate nonspecific binding in EicosaCell preparations, a proper control using host/isotype-matched irrelevant antibodies must be always included. An additional mandatory control that needs to be always included in the experimental design to rule out nonspecific immuno-staining is the condition with a synthesis inhibitor of the targeted eicosanoid. Synthesis inhibitor-treated controls should show no immune-labeling confirming specific detection of targeted eicosanoid. If nonspecific staining is too high (>10 % positive), there are several possible remedies. The detecting antibody may be diluted further, or a different one from a different host may be tried. Also, it is possible to try an adsorbing reagent that effectively blocks out non-specific sites, such as a normal serum (same host of the detecting antibody). Nonspecific fluorescence can also be detected when the solution of detecting antibody contains a high degree of aggregated antibody; therefore it is important to centrifuge the detecting antibody before adding to cell preparations.
- Poor preservation of cell morphology: During EDAC incubation step of EicosaCell assays, cell appearance may change from unimportant to severe modification of typical cell morphology. This undesirable effect of EDAC on cells can be avoided by adjusting both EDAC concentration and incubation time.
- Losing cell adherence with EDAC: Similar to unwanted EDAC effect on cell morphology, the ability of cells to stay adhered to coverslips or other substrates can be affected by EDAC incubation. Again, previous careful setting of EDAC incubation step is obligatory and should be adjusted for each cell type.
- Loss of cell integrity: Eicosanoid localization within cells by EicosaAssay may be tricky sometimes since some cell types are destroyed during EDAC-driven cross-linking/permeabilization step. For instance, even though lipid bodies of human neutrophils and basophils are sites of 5-LO localization [4, 11], EicosaCell assays with agarose-embedded neutrophils and basophils were not feasible since these cells did not endure to EDAC-driven fixation/permeabilization process which precedes eicosanoid immuno-detection by EicosaCell, indicating that the combination of gel matrix with EDAC step may be useful to study only a small group of tough cells, like eosinophils. Compartmentalization studies of eicosanoid synthesis within more fragile cells like neutrophils and basophils, however, can be carried out with EicosaCell system in non-gel solutions.
References
- 1.Bozza PT, Magalhaes KG, Weller PF. Leukocyte lipid bodies—biogenesis and functions in inflammation. Biochim Biophys Acta. 2009;1791(6):540–551. doi: 10.1016/j.bbalip.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandal AK, Skoch J, Bacskai BJ, et al. The membrane organization of leukotriene synthesis. Proc Natl Acad Sci U S A. 2004;101(17):6587–6592. doi: 10.1073/pnas.0308523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters-Golden M, Brock TG. Intracellular compartmentalization of leukotriene synthesis: unexpected nuclear secrets. FEBS Lett. 2001;487(3):323–326. doi: 10.1016/s0014-5793(00)02374-7. [DOI] [PubMed] [Google Scholar]
- 4.Bandeira-Melo C, Bozza PT, Weller PF. The cellular biology of eosinophil eicosanoid formation and function. J Allergy Clin Immunol. 2002;109(3):393–400. doi: 10.1067/mai.2002.121529. [DOI] [PubMed] [Google Scholar]
- 5.Yaqoob P. Fatty acids as gatekeepers of immune cell regulation. Trends Immunol. 2003;24(12):639–645. doi: 10.1016/j.it.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9(2):162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 7.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 8.Diaz BL, Arm JP. Phospholipase A(2) Prostaglandins Leukot Essent Fatty Acids. 2003;69(2–3):87–97. doi: 10.1016/s0952-3278(03)00069-3. [DOI] [PubMed] [Google Scholar]
- 9.Bandeira-Melo C, Weller PF. Eosinophils and cysteinyl leukotrienes. Prostaglandins Leukot Essent Fatty Acids. 2003;69(2–3):135–143. doi: 10.1016/s0952-3278(03)00074-7. [DOI] [PubMed] [Google Scholar]
- 10.Liu LX, Buhlmann JE, Weller PF. Release of prostaglandin E2 by microfilariae of Wuchereria bancrofti and Brugia malayi. Am J Trop Med Hyg. 1992;46(5):520–523. doi: 10.4269/ajtmh.1992.46.520. [DOI] [PubMed] [Google Scholar]
- 11.Bandeira-Melo C, Phoofolo M, Weller PF. Extranuclear lipid bodies, elicited by CCR3-mediated signaling pathways, are the sites of chemokine-enhanced leukotriene C4 production in eosinophils and basophils. J Biol Chem. 2001;276(25):22779–22787. doi: 10.1074/jbc.M101436200. [DOI] [PubMed] [Google Scholar]
- 12.Moreira LS, Piva B, Gentile LB, et al. Cytosolic phospholipase A2-driven PGE2 synthesis within unsaturated fatty acids-induced lipid bodies of epithelial cells. Biochim Biophys Acta. 2009;1791(3):156–165. doi: 10.1016/j.bbalip.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Accioly MT, Pacheco P, Maya-Monteiro CM, et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 2008;68(6):1732–1740. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 14.Tedla N, Bandeira-Melo C, Tassinari P, et al. Activation of human eosinophils through leukocyte immunoglobulin-like receptor 7. Proc Natl Acad Sci U S A. 2003;100(3):1174–1179. doi: 10.1073/pnas.0337567100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balestrieri B, Hsu VW, Gilbert H, et al. Group V secretory phospholipase A2 translocates to the phagosome after zymosan stimulation of mouse peritoneal macrophages and regulates phagocytosis. J Biol Chem. 2006;281(10):6691–6698. doi: 10.1074/jbc.M508314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Avila H, Melo RC, Parreira GG, et al. Mycobacterium bovis bacillus Calmette-Guerin induces TLR2-mediated formation of lipid bodies: intracellular domains for eicosanoid synthesis in vivo. J Immunol. 2006;176(5):3087–3097. doi: 10.4049/jimmunol.176.5.3087. [DOI] [PubMed] [Google Scholar]
- 17.Pacheco P, Vieira-de-Abreu A, Gomes RN, et al. Monocyte chemoattractant protein-1/CC chemokine ligand 2 controls microtubule-driven biogenesis and leukotriene B4-synthesizing function of macrophage lipid bodies elicited by innate immune response. J Immunol. 2007;179(12):8500–8508. doi: 10.4049/jimmunol.179.12.8500. [DOI] [PubMed] [Google Scholar]
- 18.Mesquita-Santos FP, Vieira-de-Abreu A, Calheiros AS, et al. Cutting edge: prostaglandin D2 enhances leukotriene C4 synthesis by eosinophils during allergic inflammation: synergistic in vivo role of endogenous eotaxin. J Immunol. 2006;176(3):1326–1330. doi: 10.4049/jimmunol.176.3.1326. [DOI] [PubMed] [Google Scholar]
- 19.Vieira-de-Abreu A, Assis EF, Gomes GS, et al. Allergic challenge-elicited lipid bodies compartmentalize in vivo leukotriene C4 synthesis within eosinophils. Am J Respir Cell Mol Biol. 2005;33(3):254–261. doi: 10.1165/rcmb.2005-0145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devchand PR, Keller H, Peters JM, et al. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384(6604):39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 21.Dvash E, Har-Tal M, Barak S, et al. Leukotriene C4 is the major trigger of stress-induced oxidative DNA damage. Nat Commun. 2015;6:10112. doi: 10.1038/ncomms10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva AR, Pacheco P, Vieira-de-Abreu A, et al. Lipid bodies in oxidized LDL-induced foam cells are leukotriene-synthesizing organelles: a MCP-1/CCL2 regulated phenomenon. Biochim Biophys Acta. 2009;1791(11):1066–1075. doi: 10.1016/j.bbalip.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Luna-Gomes T, Magalhães KG, Mesquita-Santos FP, et al. Eosinophils as a novel cell source of prostaglandin D2: autocrine role in allergic inflammation. J Immunol. 2011;187(12):6518–6526. doi: 10.4049/jimmunol.1101806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paiva LA, Coelho KA, Luna-Gomes T, et al. Schistosome infection-derived Hepatic Stellate Cells are cellular source of prostaglandin D2: role in TGF-β-stimulated VEGF production. Prostaglandins Leukot Essent Fatty Acids. 2015;95:57–62. doi: 10.1016/j.plefa.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Plotkowski MC, Brandão BA, de Assis MC, et al. Lipid body mobilization in the ExoU-induced release of inflammatory mediators by airway epithelial cells. Microb Pathog. 2008;45(1):30–37. doi: 10.1016/j.micpath.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 26.D’Avila H, Freire-de-Lima CG, Roque NR, et al. Host cell lipid bodies triggered by Trypanosoma cruzi infection and enhanced by the uptake of apoptotic cells are associated with prostaglandin E2 generation and increased parasite growth. J Infect Dis. 2011;204(6):951–961. doi: 10.1093/infdis/jir432. [DOI] [PubMed] [Google Scholar]
- 27.Melo RC, D’Ávila H, Bozza PT, et al. Imaging lipid bodies within leukocytes with different light microscopy techniques. Methods Mol Biol. 2011;689:149–161. doi: 10.1007/978-1-60761-950-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]